Abstract
Context:
A previous genome-wide association study in Chinese women with polycystic ovary syndrome (PCOS) identified a region on chromosome 2p16.3 encoding the LH/choriogonadotropin receptor (LHCGR) and FSH receptor (FSHR) genes as a reproducible PCOS susceptibility locus.
Objective:
The objective of the study was to determine the role of the LHCGR and/or FSHR gene in the etiology of PCOS in women of European ancestry.
Design:
This was a genetic association study in a European ancestry cohort of women with PCOS.
Setting:
The study was conducted at an academic medical center.
Participants:
Participants in the study included 905 women with PCOS diagnosed by National Institutes of Health criteria and 956 control women.
Intervention:
We genotyped 94 haplotype-tagging single-nucleotide polymorphisms and two coding single-nucleotide polymorphisms mapping to the coding region of LHCGR and FSHR plus 20 kb upstream and downstream of the genes and test for association in the case control cohort and for association with nine quantitative traits in the women with PCOS.
Results:
We found strong evidence for an association of PCOS with rs7562215 (P = 0.0037) and rs10495960 (P = 0.0046). Although the marker with the strongest association in the Chinese PCOS genome-wide association study (rs13405728) was not informative in the European populations, we identified and genotyped three markers (rs35960650, rs2956355, and rs7562879) within 5 kb of rs13405728. Of these, rs7562879 was nominally associated with PCOS (P = 0.020). The strongest evidence for association mapping to FSHR was observed with rs1922476 (P = 0.0053). Furthermore, markers with the FSHR gene region were associated with FSH levels in women with PCOS.
Conclusions:
Fine mapping of the chromosome 2p16.3 Chinese PCOS susceptibility locus in a European ancestry cohort provides evidence for association with two independent loci and PCOS. The gene products LHCGR and FSHR therefore are likely to be important in the etiology of PCOS, regardless of ethnicity.
Polycystic ovary syndrome (PCOS) is a complex trait characterized by hyperandrogenemia and anovulation/oligoovulation and is associated with obesity, insulin resistance, and a 7-fold increased risk of developing type 2 diabetes mellitus (1). A recent genome-wide association study (GWAS) identified three PCOS susceptibility loci (2p16.3, 2p21, and 9q33.3) in the Han Chinese population (2). The genes encoding LH/choriogonadotropin receptor (LHCGR), FSH receptor (FSHR), and general transcription factor IIA, 1-like isoform (GTF2A1L) map to the 2p16.3 locus.
LHCGR is the receptor for two structurally homologous glycoproteins: LH and human chorionic gonadotropin (hCG). LH triggers ovulation of the mature follicle and formation of the corpus luteum, and hCG is required for the maintenance of pregnancy. FSHR is the receptor for FSH. Early in the menstrual cycle, FSH acts to induce expression of the LH/hCG receptor in small- and medium-sized follicles. LH binds to its receptor on early-stage follicles supporting further follicular development and steroidogenesis. In the middle of the cycle, the LH surge is necessary to trigger ovulation (3). GTF2A1L is a poorly characterized gene expressed at high levels in the testis (4).
It is unknown whether the 2p16.3 region is associated with PCOS in non-Chinese populations. The variant with the strongest evidence for association with PCOS in the Chinese population (rs13405728) is not informative in European-derived populations [minor allele frequency (MAF) 0.049] and does not show evidence for association with PCOS in preliminary studies (5–8). We therefore tested for association between PCOS and 96 single-nucleotide polymorphisms (SNPs) mapping to the genomic region encompassing LHCGR and FSHR in a US Caucasian cohort.
Materials and Methods
Subjects
This study was approved by the Institutional Review Boards of the Brigham and Women's Hospital, Northwestern University Feinberg School of Medicine, and Pennsylvania State University College of Medicine. Written informed consent was obtained from all participants. We studied 905 index cases (probands) with PCOS and 956 control women (108 intensively phenotyped subjects and 848 minimally phenotyped subjects from a DNA repository).
PCOS cases
PCOS was defined according to the Eunice Kennedy Shriver National Institute of Child Health and Human Development criteria as previously described (9) and fulfilled the Eunice Kennedy Shriver National Institute of Child Health and Human Development, Rotterdam, and Androgen Excess Society criteria for the diagnosis of PCOS (10–12). PCOS was diagnosed by a history of six or fewer menses per year and elevated levels of total (T) or non-SHBG bound testosterone (uT) with the exclusion of specific ovarian, adrenal, or pituitary disorders and in the absence of confounding medication.
Controls
Intensively phenotyped control women (n = 108) had normal androgen levels, regular menses, were of age and ethnicity similar to the PCOS cases, and had not received confounding medications (9). Minimally phenotyped women (n = 848) were selected from NUgene (http://www.nugene.org), a large-scale GenBank with centralized genomic DNA sample collection (13). The majority of these subjects (n = 703) were screened using a questionnaire, and 3% of the subjects were excluded for oligomenorrhea or known diagnosis of PCOS. We therefore expect that our control cohort includes fewer women with PCOS than the 5–10% population prevalence of PCOS (14).
Clinical measurements
Anthropometric measurements (blood pressure, waist circumference, weight, and height) were taken (15). A 75-g oral glucose tolerance test was carried out after a 300-g carbohydrate diet and overnight fast (16). Morning fasting blood samples were obtained for baseline insulin and glucose levels and 2 h after oral glucose tolerance test. A morning fasting blood sample was obtained for fasting reproductive and metabolic (9) hormones.
Biochemical assays
Circulating levels of glucose, insulin, proinsulin, T, uT, LH, FSH, and triglycerides (TG) were determined (9).
Genotyping
We genotyped 102 SNPs mapping to LHCGR, FSHR, and GTF2A1L, plus 20 kb of genomic sequence upstream and downstream of each gene using an Illumina iSelect custom array (Illumina, San Diego, CA). SNPs included haplotype-tagging SNPs, (r2 ≥ 0.80 and MAF of 0.05 or greater in the HapMap CEU, HapMap Tagger, http://www.hapmap.org), and known nonsynomymous coding SNPs.
Genetic analysis
Genotypes were called using the Illumina GenomeStudio Genotyping Module (http://www.illumina.com/software/genomestudio_software.ilmn), with a no-call threshold of 0.15. Nineteen DNA samples with P10 quality score of less than 0.40 or P50 quality score of less than 0.60 were excluded.
Principal component analysis (PCA)
We used EIGENSTRAT/SmartPCA in the EIGENSOFT software package (http://genepath.med.harvard.edu/∼reich/Software.htm), implemented in the BC/SNPmax platform (http://www.bcplatforms.com/solutions/solutions_1_1.html) to calculate principal components (PCs) from 253 ancestry informative markers (17). SmartPCA identified five subjects who were genetic outliers, using five maximum outlier removal iterations, 10 PCs along which to remove outliers during each iteration, and a sd threshold of 6.0 for outlier removal. These subjects were removed from subsequent analyses. The first two PCs derived from the PCA were used as covariates to adjust for population stratification.
Association testing
We tested for association with PCOS using four models. Model 1 adjusted for population stratification. Model 2 adjusted for body mass index (BMI) and population stratification. Model 3 adjusted for age, BMI, and population stratification, and model 4 adjusted for age and population stratification. Linkage disequilibrium (LD) plots were generated using Haploview 4.2 implemented in the BC/SNPmax platform (18). We assessed the impact of genetic variation on the distribution of nine quantitative traits [BMI (n = 905), T (n = 885), uT (n = 868), fasting insulin (n = 805), fasting glucose (n = 860), 2 h glucose (n = 425), LH (n = 817), FSH (n = 816), and TG (n = 778)] using the four models described above. For BMI the second and third models were not evaluated because BMI was used as a covariate. All quantitative trait analyses were carried out only in the PCOS subjects. These analyses were implemented in PLINK, version 1.06, within the BC/SNPmax (19).
Results
Study participant characteristics
All participants were women of European ancestry and fulfilled National Institutes of Health and Rotterdam criteria for PCOS (10–12). PCOS cases were significantly younger than minimally phenotyped controls and had significantly higher BMI than the intensively and minimally phenotyped controls (Supplemental Table 1, published on The Endocrine Society's Journals Online web site at http://jcem.endojournals.org). T, uT, fasting insulin, 2-h glucose, LH, and FSH levels were significantly elevated in the women with PCOS compared with the intensively phenotyped control women. Fasting glucose levels did not differ between women with PCOS and intensively phenotyped control women.
Genotyping
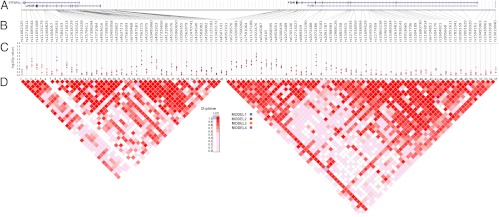
Six SNPs failed genotyping, whereas 94 haplotype-tagging SNPs and two nonsynomymous coding SNPs were successfully genotyped (Fig. 1). The average genotype call rate was greater than 99.9%, MAF was 0.26, and genotype concordance in DNA replicates was greater than 99.9%.
Fig. 1.
Schematic of the 2p16.3 region genetic analysis. A, The LHCGR and FSHR genomic region as derived from the University of California, Santa Cruz, genome browser National Center for Biotechnology Information Build 36.1 (http://genome.ucsc.edu). B, The relative locations of the SNPs studied in this report are indicated. C, P values plotted as −log10 values, generated using the Synthesis-View application (https://chgr.mc.vanderbilt.edu/synthesisview/). D, The pair-wise LD (D′) plot was generated from the PCOS case-control cohort using Haploview. The PCOS case-control cohort is of Caucasian European ancestry. Dark red indicates strong LD and white indicates no LD.
Association testing
The strongest PCOS association was with rs10495960 and rs7562215. rs1049560 maps within the first intron of LHCGR and the last exon of GTF2A1L, encoding an alanine to threonine missense mutation in GTF2A1L and 1345 bp downstream of rs7562215 (http://genome.ucsc.edu, Fig. 1A). The model 1 P value for rs10495960 was 0.0046, with an odds ratio (OR) of 1.31. The model 2 had a P = 0.0050, with an OR of 1.36. The model 3 had a P = 0.0244, with an OR of 1.36. rs7562215 showed significant evidence for association with PCOS after adjusting for ethnicity, age, and BMI (P = 0.0036; OR 0.56). Several markers mapping to the vicinity of rs10495960 also were nominally associated with PCOS (Fig. 1B, Table 1).
Table 1.
Results of genetic analyses
| Trait | SNP | Allele | MAFa | Model 1 |
Model 2 |
Model 3 |
Model 4 |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | P value | OR | P value | OR | P value | OR | P value | ||||
| Qualitative analysis | |||||||||||
| LHCGR variants | |||||||||||
| Case/control | rs7562215 | T | 0.069 | 0.71 | 0.013 | 0.72 | 0.044 | 0.56 | 0.0036 | 0.58 | 8.53 × 10−4 |
| Case/control | rs4319975 | A | 0.268 | 1.1 | 0.192 | 1.13 | 0.178 | 1.11 | 0.353 | 1.07 | 0.493 |
| Case/control | rs10495960 | A | 0.158 | 1.31 | 0.0046 | 1.36 | 0.005 | 1.36 | 0.024 | 1.29 | 0.025 |
| Case/control | rs4952923 | G | 0.193 | 1.22 | 0.022 | 1.29 | 0.012 | 1.26 | 0.07 | 1.17 | 0.125 |
| Case/control | rs17326656 | T | 0.227 | 0.9 | 0.203 | 0.9 | 0.279 | 0.94 | 0.576 | 0.98 | 0.83 |
| Case/control | rs4131886 | G | 0.264 | 0.93 | 0.336 | 0.94 | 0.501 | 0.97 | 0.811 | 1 | 0.996 |
| Case/control | rs4519576 | C | 0.491 | 0.89 | 0.085 | 0.85 | 0.039 | 0.94 | 0.524 | 0.99 | 0.903 |
| Case/control | rs35960650 | T | 0.349 | 1.11 | 0.127 | 1.15 | 0.103 | 1.2 | 0.078 | 1.11 | 0.218 |
| Case/control | rs13405728b | NA | 0.049 | NA | NA | NA | NA | NA | NA | NA | NA |
| Case/control | rs2956355 | T | 0.093 | 1.08 | 0.492 | 0.96 | 0.754 | 0.88 | 0.45 | 1.01 | 0.916 |
| Case/control | rs7562879 | A | 0.108 | 0.82 | 0.068 | 0.74 | 0.022 | 0.7 | 0.021 | 0.783 | 0.065 |
| FSHR variants | |||||||||||
| Case/control | rs6739570 | T | 0.066 | 1.37 | 0.038 | 1.5 | 0.019 | 1.36 | 0.162 | 1.26 | 0.202 |
| Case/control | rs10205982 | A | 0.066 | 0.73 | 0.023 | 0.68 | 0.021 | 0.71 | 0.093 | 0.74 | 0.075 |
| Case/control | rs17500266 | G | 0.46 | 0.88 | 0.061 | 0.88 | 0.11 | 0.83 | 0.07 | 0.85 | 0.061 |
| Case/control | rs7591064 | C | 0.168 | 1.21 | 0.021 | 1.28 | 0.013 | 1.3 | 0.033 | 1.2 | 0.071 |
| Case/control | rs11125188 | T | 0.487 | 0.87 | 0.048 | 0.85 | 0.041 | 0.77 | 0.012 | 0.82 | 0.019 |
| Case/control | rs1922476 | C | 0.359 | 1.23 | 0.004 | 1.23 | 0.013 | 1.34 | 0.005 | 1.31 | 0.002 |
| Beta | P value | Beta | P value | Beta | P value | Beta | P value | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Quantitative analysis | |||||||||||
| FSH (mlU/ml) | rs17038320 | T | 0.164 | −0.83 | 1.0 × 10−3 | −0.81 | 1.4 × 10−3 | −0.81 | 1.3 × 10−3 | −0.84 | 9.3 × 10−4 |
| FSH (mlU/ml) | rs17038059 | T | 0.195 | −0.7 | 3.1 × 10−3 | −0.69 | 3.2 × 10−3 | −0.69 | 3.1 × 10−3 | −0.7 | 3.1 × 10−3 |
| FSH (mlU/ml) | rs17038141 | A | 0.159 | −0.76 | 3.6 × 10−3 | −0.74 | 4.4 × 10−3 | −0.77 | 3.2 × 10−3 | −0.79 | 2.7 × 10−3 |
| TG (mg/dl) | rs7591064 | C | 0.168 | −24 | 6.9 × 10−4 | −23 | 1.1 × 10−3 | −23 | 1.1 × 10−3 | −24 | 7.1 × 10−3 |
| TG (mg/dl) | rs4952931 | G | 0.42 | −18 | 1.8 × 10−3 | −18 | 1.7 × 10−3 | −18 | 1.7 × 10−3 | −18 | 1.8 × 10−3 |
| TG (mg/dl) | rs17500266 | G | 0.46 | 18 | 1.9 × 10−3 | 17 | 3.0 × 10−3 | 17 | 3.2 × 10−3 | 18 | 2.2 × 10−3 |
Model 1, PCA adjusted; model 2, BMI and PCA adjusted; model 3, age, BMI, and PCA adjusted; model 4, age, and PCA adjusted. NA, Not available, did not genotype.
Minor allele frequency in CEU.
Chinese PCOS locus.
rs13405728, the marker with strongest evidence for association in the Han Chinese population (2) was not informative in our cohort (5). The Han Chinese HapMap population has more extensive LD than the European HapMap trios (CEU). In the Chinese HapMap population, a single 22-kb LD block encompasses rs10495960 and rs13405728, whereas in the CEU, this region contains three LD blocks, with rs10495960 and rs13405728 mapping to distinct blocks (Supplemental Fig. 1).
The strongest evidence for association within the FSHR gene region was with rs1922476, (∼5.3 kb upstream of FSHR; Fig. 1B; PCA adjusted P = 0.004; OR 1.23, BMI and PCA adjusted P = 0.013; OR 1.23; and age, BMI, and PCA adjusted P = 0.005; OR 1.34). The coding SNP rs6165, which encodes an alanine to threonine missense mutation in FSHR, was nominally associated with PCOS (PCA adjusted P = 0.0401, OR 1.15, BMI and PCA adjusted P = 0.069; OR 1.16; and age, BMI, and PCA adjusted P = 0.0258; OR 1.25; Supplemental Table 2).
Quantitative trait analysis
Although common variants (MAF > 0.1) within LHCGR did not show evidence for association with quantitative traits, multiple variants within FSHR showed evidence for association with PCOS quantitative traits (Table 1 and Supplemental Table 3). The strongest evidence for association was observed with TG levels and rs4952931, rs7591064, and rs17500266 and FSH levels and rs17038320, rs17038320, and rs17038141 (Table 1 and Supplemental Table 3).
All given P values are nominal. Given the number of SNPs tested, an adjustment for multiple testing needs to be applied. However, the high degree of correlation between the SNPs genotyped for this study (see Fig. 1) makes Bonferroni correction for the number of SNPs too conservative. An alternative approach is to correct for the number of independent haplotype blocks and SNPs (SNPs not contained within haplotype blocks) covered by the SNPs in the study. The number of haplotype blocks plus independent SNPs was 21; therefore, results with P < 2.38 × 10−3 can be considered significant after correction for multiple testing.
Discussion
Although the Chinese chromosome 2p16.3 PCOS susceptibility locus SNPs (rs13405728 and rs6732721) did not show evidence for association with PCOS in European-derived populations (5, 6, 8), these SNPs are not informative in individuals of European ancestry. Here we fine-mapped this locus by testing for an association between PCOS and 96 SNPs mapping to the region encompassing LHCGR and FSHR in a U.S. cohort of European ancestry. This analysis identified two independent PCOS susceptibility loci, thereby replicating the findings of the Chinese study and demonstrating that this region does contribute to the PCOS phenotype in women of multiple ethnicities.
The chromosome 2p16.3 region contains three genes with known reproductive functions: LHCGR, FSHR, and GTF2A1L. The strongest evidence for association in our cohort is for rs10495960 and rs1922476. In the Chinese population, rs10495960, the marker with the strongest evidence for an association with PCOS in our cohort, and rs13405728, the marker with the strongest evidence for an association with PCOS in the Chinese cohorts, map to the same LD block; therefore, both SNPs could tag the same susceptibility locus. By contrast, reduced LD in Europeans separates the two SNP into distinct LD blocks, and therefore they can no longer tag the same locus. The results from the Chinese GWAS and our candidate gene analysis are consistent with a chromosome 2p13.6 PCOS susceptibility locus mapping near rs10495960 that is shared by both Chinese and European-derived populations.
The second SNP, rs1922476, maps approximately 5.3 kb upstream of the FSHR transcription start site. Chen et al. (2) also identified 13 SNPs mapping to FSHR with nominal evidence for association with PCOS. The FSHR (rs10495960) and LHCGR (rs1922476) loci are separated by 124 kb and are not in linkage disequilibrium in either the Chinese or Caucasian cohorts. The two SNPs therefore map to genetically distinct loci.
Fine-mapping the 2p16.3 PCOS susceptibility locus in a Caucasian cohort demonstrates that the Chinese locus is also associated with PCOS in European-derived populations, a finding that was completely missed in studies attempting to replicate only the Chinese PCOS susceptibility SNP in Caucasians. This underscores the importance of accounting for differences in LD pattern when comparing association study results across different ethnicities. Fine-mapping further resolved the association signal into two independent loci mapping to LHCGR and FSHR.
Acknowledgments
We thank all the women for participating in this study. Editorial assistance was provided by Stacey C. Tobin, Ph.D.
This work was supported by National Institutes of Health Grants U54HD041857 (to E.G., B.P.B., L.D.S., T.K.W., A.D., and M.U.), U54 HD34449 (to A.D.), R01HD057450 (O.G. and M.U.), P50HD044405 (to A.D. and M.U.), and M01 RR10732 and C06 RR016499 (to Pennsylvania State University General Clinical Research Center); and American Diabetes Association Career Development Award 7-09-CD-13 (to MU). This project was funded, in part, under a grant with the Pennsylvania Department of Health using Tobacco Settlement Funds. The Department of Health specifically disclaims responsibility for any analyses, interpretations, or conclusions. Some of the hormone assays were performed at the University of Virginia, Center for Research in Reproduction, Ligand Assay and Analysis Core, which is supported by Eunice Kennedy Shriver National Institute of Child Health and Human Development Grant U54-HD28934.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- BMI
- Body mass index
- CEU
- Han Chinese European HapMap trios
- FSHR
- FSH receptor
- GTF2A1L
- general transcription factor IIA, 1-like isoform
- GWAS
- genome-wide association study
- hCG
- human chorionic gonadotropin
- LD
- linkage disequilibrium
- LHCGR
- LH/choriogonadotropin receptor
- MAF
- minor allele frequency
- OR
- odds ratio
- PC
- principal component
- PCA
- principal component analysis
- PCOS
- polycystic ovary syndrome
- SNP
- single-nucleotide polymorphism
- T
- total testosterone
- TG
- triglycerides
- uT
- non-SHBG bound testosterone.
References
- 1. Sam S, Dunaif A. 2003. Polycystic ovary syndrome: syndrome XX? Trends Endocrinol Metab 14:365–370 [DOI] [PubMed] [Google Scholar]
- 2. Chen ZJ, Zhao H, He L, Shi Y, Qin Y, Li Z, You L, Zhao J, Liu J, Liang X, Zhao X, Zhao J, Sun Y, Zhang B, Jiang H, Zhao D, Bian Y, Gao X, Geng L, Li Y, Zhu D, Sun X, Xu JE, Hao C, Ren CE, Zhang Y, Chen S, Zhang W, Yang A, Yan J, Li Y, Ma J, Zhao Y. 2011. Genome-wide association study identifies susceptibility loci for polycystic ovary syndrome on chromosome 2p16.3, 2p21 and 9q33.3. Nat Genet 43:55–59 [DOI] [PubMed] [Google Scholar]
- 3. Dufau ML. 1998. The luteinizing hormone receptor. Annu Rev Physiol 60:461–496 [DOI] [PubMed] [Google Scholar]
- 4. Upadhyaya AB, Lee SH, DeJong J. 1999. Identification of a general transcription factor TFIIAα/β homolog selectively expressed in testis. J Biol Chem 274:18040–18048 [DOI] [PubMed] [Google Scholar]
- 5. Goodarzi MO, Jones MR, Li X, Chua AK, Garcia OA, Chen YD, Krauss RM, Rotter JI, Ankener W, Legro RS, Azziz R, Strauss JF, 3rd, Dunaif A, Urbanek M. 2012. Replication of association of DENND1A and THADA variants with polycystic ovary syndrome in European cohorts. J Med Genet 49:90–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lerchbaum E, Trummer O, Giuliani A, Gruber HJ, Pieber TR, Obermayer-Pietsch B. 2011. Susceptibility loci for polycystic ovary syndrome on chromosome 2p16.3, 2p21, and 9q33.3 in a cohort of Caucasian women. Horm Metab Res 43:743–747 [DOI] [PubMed] [Google Scholar]
- 7. Eriksen MB, Brusgaard K, Andersen M, Tan Q, Altinok ML, Gaster M, Glintborg D. 2012. Association of polycystic ovary syndrome susceptibility single nucleotide polymorphism rs2479106 and PCOS in Caucasian patients with PCOS or hirsutism as referral diagnosis. Eur J Obstet Gynecol Reprod Biol 163:39–42 [DOI] [PubMed] [Google Scholar]
- 8. Welt CK, Styrkarsdottir U, Ehrmann DA, Thorleifsson G, Arason G, Gudmundsson JA, Ober C, Rosenfield RL, Saxena R, Thorsteinsdottir U, Crowley WF, Stefansson K. 2012. Variants in DENND1A are associated with polycystic ovary syndrome in women of European ancestry. J Clin Endocrinol Metab 97:E1342–E1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Legro RS, Driscoll D, Strauss JF, 3rd, Fox J, Dunaif A. 1998. Evidence for a genetic basis for hyperandrogenemia in polycystic ovary syndrome. Proc Natl Acad Sci USA 95:14956–14960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zawadzki JK, Dunaif A. 1992. Diagnostic criteria for polycystic ovary syndrome: towards a rational approach. In: Dunaif A, Givens JR, Haseltine FP, Merriam GR, eds. Polycystic ovary syndrome. Boston: Blackwell Scientific; 377–384 [Google Scholar]
- 11. Rotterdam ESHRE/ASRM-Sponsored PCOS Consensus Workshop Group 2004. Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome. Fertil Steril 81:19–25 [DOI] [PubMed] [Google Scholar]
- 12. Azziz R, Carmina E, Dewailly D, Diamanti-Kandarakis E, Escobar-Morreale HF, Futterweit W, Janssen OE, Legro RS, Norman RJ, Taylor AE, Witchel SF, Androgen Excess S. 2006. Positions statement: criteria for defining polycystic ovary syndrome as a predominantly hyperandrogenic syndrome: an Androgen Excess Society guideline. J Clin Endocrinol Metab 91:4237–4245 [DOI] [PubMed] [Google Scholar]
- 13. Lemke AA, Wolf WA, Hebert-Beirne J, Smith ME. 2010. Public and biobank participant attitudes toward genetic research participation and data sharing. Public Health Genomics 13:368–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kahsar-Miller MD, Nixon C, Boots LR, Go RC, Azziz R. 2001. Prevalence of polycystic ovary syndrome (PCOS) in first degree relatives of patients with PCOS. Fertil Steril 75:53–58 [DOI] [PubMed] [Google Scholar]
- 15. Legro RS, Bentley-Lewis R, Driscoll D, Wang SC, Dunaif A. 2002. Insulin resistance in the sisters of women with polycystic ovary syndrome: association with hyperandrogenemia rather than menstrual irregularity. J Clin Endocrinol Metab 87:2128–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sam S, Sung YA, Legro RS, Dunaif A. 2008. Evidence for pancreatic β-cell dysfunction in brothers of women with polycystic ovary syndrome. Metab Clin Exp 57:84–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Price AL, Butler J, Patterson N, Capelli C, Pascali VL, Scarnicci F, Ruiz-Linares A, Groop L, Saetta AA, Korkolopoulou P, Seligsohn U, Waliszewska A, Schirmer C, Ardlie K, Ramos A, Nemesh J, Arbeitman L, Goldstein DB, Reich D, Hirschhorn JN. 2008. Discerning the ancestry of European Americans in genetic association studies. PLoS Genet 4:e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Barrett JC, Fry B, Maller J, Daly MJ. 2005. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265 [DOI] [PubMed] [Google Scholar]
- 19. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]