Abstract
Active zones are specialized presynaptic structures critical for neurotransmission. We show that a neuronal maintenance factor, nicotinamide mononucleotide adenylyltransferase (NMNAT), is required for maintaining active zone structural integrity in Drosophila by interacting with the active zone protein, Bruchpilot (BRP), and shielding it from activity-induced ubiquitin–proteasome-mediated degradation. NMNAT localizes to the peri-active zone and interacts biochemically with BRP in an activity-dependent manner. Loss of NMNAT results in ubiquitination, mislocalization and aggregation of BRP, and subsequent active zone degeneration. We propose that, as a neuronal maintenance factor, NMNAT specifically maintains active zone structure by direct protein–protein interaction.
Keywords: NMNAT, active zone, BRP, chaperone, protein homeostasis
Introduction
Active zones are highly specialized presynaptic sites for synaptic vesicle docking and fusion, and fast, efficient and precise neurotransmission relies on their structural integrity. The protein composition of active zones was identified recently [1, 2, 3], but the mechanism for maintaining their structural integrity is largely unknown. Chaperones have been implicated in synaptic function; for example, cysteine-string protein (CSP), a main synaptic vesicle and secretory granule protein, is known to act as a chaperone maintaining synaptic integrity [4, 5, 6, 7]. It is likely that molecular chaperones, the main mediators in maintenance of protein homeostasis, facilitate the localization of synaptic proteins and maintain synaptic structural integrity during neuronal activity. Here, we examine the role of a newly identified chaperone, nicotinamide mononucleotide adenylyltransferase (NMNAT), in active zone maintenance. We showed previously that NMNAT is a protective factor required for maintaining neuronal integrity [8, 9]. Importantly, the neuroprotective ability of NMNAT was attributed partly to its chaperone function, where NMNAT was able to interact with misfolded protein oligomers and prevent the formation of large aggregates and also reduce the cellular load of aggregates, partly through the ubiquitin–proteasome pathway [8, 9]. In Drosophila, NMNAT is ubiquitously expressed and is localized at the synapse as well as in the cell body [8]. Here, we directly examine the specific role of NMNAT at the synapse, and report a new mechanism of active zone maintenance by NMNAT, in which it stabilizes the primary active zone structure protein Bruchpilot (BRP). BRP was recently identified as an integral component of T-bar, the dense projection of the active zone [10], and was found to be essential for structural and functional integrity of the active zone [11]. We found that loss of NMNAT was in parallel with loss of synaptic BRP levels, leading to ubiquitination and redistribution of BRP from the synapse to the cell body, resulting in subsequent active zone degeneration. Moreover, we show that NMNAT interacts with BRP in an activity-dependent manner. Our findings suggest that NMNAT stabilizes BRP and shields it from activity-induced ubiquitin–proteasome-mediated protein degradation, thereby maintaining active zone structural integrity during neuronal activity. Our work describes NMNAT as an essential active zone maintenance factor and provides a new mechanism by which it sustains the proper structural integrity of active zones.
Results and Discussion
BRP aggregation and ubiquitination by loss of NMNAT
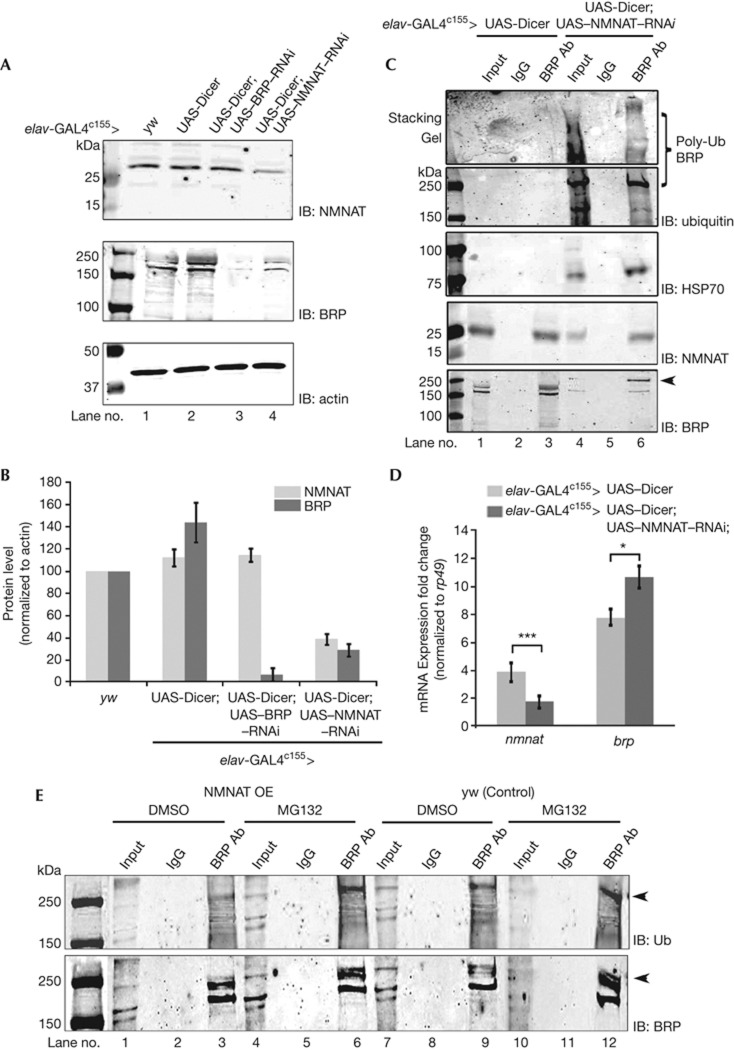
To investigate the specific role of NMNAT in synapses, we first examined the changes in synaptic proteins in NMNAT-deficient neurons. By means of mosaic analysis with a repressible cell marker (MARCM) with eyFLP [12], we generated mosaic nmnat-null and heterozygous synapses in the lamina and observed that synaptic proteins, including synaptobrevin, synaptotagmin and BRP, were reduced in nmnat mutant patches (Fig 1A,B). We then analysed central brain synapses where NMNAT expression was reduced using an RNA interference (RNAi)-mediated knockdown approach with a pan-neural c155-GAL4 driver, which achieved 76% knockdown of the NMNAT protein level (supplementary Fig S1A,B online). Consistently, we observed reduced staining of synaptic proteins in central brain synapses, including DLAR, synaptotagmin, CSP and BRP (Fig 1C,D; supplementary Fig S2 online), and reduced protein levels as quantified by western analysis (supplementary Fig S1A,B online). These results indicate a loss of synapses induced by loss of NMNAT. Interestingly, BRP displayed a unique mislocalization phenotype. In contrast to the exclusively synaptic localization in wild-type synapses (Fig 1C), in NMNAT RNAi-knockdown synapses BRP was found clustered in cell bodies, colocalizing with remnant NMNAT protein (Fig 1D,D′). The mislocalization was specific to BRP, as the distribution of other synaptic proteins including DLAR, synaptotagmin and CSP was not altered in NMNAT-knockdown brains (supplementary Fig S2 online), despite similar reduction in protein levels (supplementary Fig S1A,B online). These results indicate that NMNAT is required to maintain the level of synaptic proteins, and also specifically the synaptic localization of BRP. The specificity of NMNAT RNAi-mediated knockdown was confirmed by a rescue experiment in which overexpressing wild-type human NMNAT3 not targeted by Drosophila NMNAT RNAi suppressed the pupal lethality phenotype induced by NMNAT knockdown in motor neurons, and rescued the eclosion rate (supplementary Fig S3 online). Furthermore, the maintenance role of NMNAT on BRP is likely unidirectional, as knockdown of NMNAT reduced the BRP level; however, knockdown of BRP in the brains did not change the protein level of NMNAT (Fig 2A,B).
Figure 1.
Loss of NMNAT causes loss of synaptic proteins and mislocalization of BRP. (A,B) MARCM analysis of adult brain lamina showing that synaptobrevin (A1), synaptotagmin (A3) and BRP (B3) levels are reduced in nmnat-null (GFP positive) lamina synaptic terminals. Dashed lines indicate nmnat-null (GFP positive) clones. Scale bars indicate 5 μm. (C) Midbrain structure of 2 days after eclosion control flies overexpressing UAS-Dicer with elav-GAL4C155 showing NMNAT in both cell bodies and synapses (C1) and BRP enriched at the synapses (C2). (D) In NMNAT–RNAi flies, BRP protein is drastically reduced in neuropil and forms clusters in the cell bodies. (D1′–D4′) Higher magnification of boxed areas in C reveals a high degree of colocalization of the remaining NMNAT with BRP clusters. Scale bars indicate 20 μm. BRP, Bruchpilot; DAPI, 4′6-diamidino-2-phenylindole; GFP, green fluorescent protein; MARCM, mosaic analysis with a repressible cell marker; NMNAT, nicotinamide mononucleotide adenylyltransferase; RNAi, RNA interference.
Figure 2.
Reduction in NMNAT level causes BRP ubiquitination and aggregation. (A) Western analysis of brain lysates from flies overexpressing UAS-Dicer or UAS-Dicer; UAS–NMNAT–RNAi reveals that BRP level is down regulated with a reduced level of NMNAT in NMNAT RNAi brains; however, the NMNAT level is unchanged in BRP RNAi brains. (B) Quantification of the protein level of NMNAT and BRP in A. n=3. All data were presented as mean±s.e.m. (C) Immunoprecipitation of brain lysates from flies overexpressing UAS-Dicer or UAS-Dicer; UAS–NMNAT–RNAi with BRP antibody reveals significant ubiquitination of BRP in NMNAT–RNAi brains, including poly-ubiquitinated BRP (marked by square bracket). Ubiquitinated BRP also recruits HSP70 and remaining NMNAT. BRP ubiquitination is further shown by an upshifted band detected with anti-BRP (marked by arrowhead) in NMNAT–RNAi brain. (D) Real-time PCR shows that nmnat transcript is reduced, while brp transcript is slightly increased in NMNAT-knockdown flies, compared with flies overexpressing UAS-Dicer. All data were presented as mean±s.e.m. Significance level was established by t-test. *P<0.05, ***P<0.001, n=3. (E) MG132 treatment induces ubiquitination of BRP. One day after eclosion flies were fed with 50 μM MG132 or DMSO for 24 h and brain lysate was prepared immediately after drug feeding. Multiplex western analysis was carried out with anti-BRP and anti-ubiquitin antibodies and respective IRDye 700DX- and IRDye 800DX-conjugated secondary antibodies. Ubiquitinated BRP is marked by arrowheads. BRP, Bruchpilot; DMSO, dimethyl sulphoxide; IB, immunoblotting; NMNAT, nicotinamide mononucleotide adenylyltransferase; RNAi, RNA interference; Ub, ubiquitin.
The clustering and mislocalization of BRP is intriguing and cannot be explained by loss of synapses; therefore, we investigated the underlying mechanism. Previous work indicated the role of the ubiquitin–proteasome pathway in regulating synaptic protein levels, such as dUNC-13 and Liprin α [13]. To determine the biochemical nature of the mislocalized BRP protein clusters, we immunoprecipitated BRP protein with nc82 antibody from whole brain lysates of 2 DAE (days after eclosion) NMNAT RNAi flies (genotype: elav-GAL4C155>UAS-Dicer,UAS-NMNAT-RNAi), and probed for ubiquitin and HSP70, a molecular chaperone serving as a protein marker for aggregation [14]. As shown in Fig 2C, BRP protein in NMNAT RNAi flies was significantly ubiquitinated with several bands of different molecular weight, indicating the presence of poly-ubiquitinated BRP (Fig 2C, lane 6), while no ubiquitination of BRP was observed in control flies (Fig 2C, lane 3). Interestingly, HSP70 was present in the NMNAT RNAi brains but not in the control brains and, more importantly, interacted with BRP (Fig 2C, lanes 4 and 6), indicating the expression of stress response protein HSP70 and the formation of BRP aggregates. As expected, the level of NMNAT pulled down by BRP antibody was reduced as a result of NMNAT RNAi knockdown and reduced BRP level (Fig 2C). Consistent with the results described above (Figs 1, 2A,B), the level of BRP in NMNAT RNAi brains was reduced, but a higher molecular weight modification of BRP was detected (Fig 2C, arrow head), likely ubiquitinated BRP. In contrast, ubiquitination of other synaptic proteins including DLAR, synaptotagmin and CSP was not observed in NMNAT-knockdown neurons (supplementary Fig S4 online), suggesting a specific effect on BRP.
To further dissect the mechanism underlying the reduction and ubiquitination of BRP, we first investigated the transcription of BRP utilizing real-time PCR analysis, and observed that BRP transcripts were slightly increased on NMNAT knockdown (Fig 2D). Therefore, the reduction of BRP protein is not owing to reduced transcription. Next, we investigated whether the ubiquitin proteasome pathway was involved in regulating BRP protein degradation. We blocked proteasome function by feeding flies a specific inhibitor MG-132 [15], and examined the level of BRP ubiquitination in wild-type or NMNAT overexpression flies. To specifically analyse the ubiquitinated pool of BRP protein, we immunoprecipated BRP, probed with both anti-BRP and anti-ubiquitin antibody, and determined the percentage of ubiquitinated BRP using multiplex western analysis, where only the protein bands detected by both BRP and ubiquitin antibodies were considered as ubiquitinated BRP (supplementary Fig S5A online, arrows indicate ubiquitinated BRP bands, shown by yellow colour in the merged image). In wild-type flies, the percentage of ubiquitinated BRP increased when proteasome function was inhibited with MG132 (Fig 2E; supplementary Fig S5B online), indicating an accumulation of ubiquitinated BRP on proteasome inhibition. In NMNAT-overexpressing flies with three copies of the nmnat gene, the percentage of ubiquitinated BRP was lower than that in wild-type with either dimethyl sulphoxide or MG132 treatment (Fig 2E; supplementary Fig S5 online). This suggests that the ubiquitin–proteasome pathway is involved in regulating BRP protein degradation and that a higher level of NMNAT reduces the ubiquitination of BRP. Therefore, loss of NMNAT causes specifically the ubiquitination and aggregation of BRP and subsequent reduction in synaptic BRP protein level, likely through the proteasome pathway.
NMNAT's synaptic localization and interaction with BRP
Our loss-of-NMNAT studies indicate that under normal conditions, NMNAT functions to maintain synaptic BRP protein levels by preventing ubiquitination and aggregation of BRP. The dual function of NMNAT, as an nicotinamide adenine dinucleotide synthase and a chaperone [9], suggests two possible mechanisms: an indirect mechanism expressed through NMNAT-mediated synthesis of small molecules including nicotinamide adenine dinucleotide and other adaptor proteins; and a direct mechanism through protein–protein interactions, consistent with its chaperone function. To distinguish these, we first examined the localization of NMNAT. We have shown that BRP and NMNAT localize in photoreceptor and central brain synapses ([8] and Fig 1B,C); however, the compact size of these synapses precludes a high-resolution analysis. Inasmuch as close proximity is a prerequisite for a ‘direct’ mechanism, the Drosophila larval neuromuscular junction (NMJ) is ideal for analysing synaptic localization, given its suitable spatial resolution. By confocal microscopy, we observed that NMNAT is present at the NMJ and colocalizes strongly with BRP (Fig 3A). With 3D-SIM Super-Resolution imaging (resolution 120 nm on XY axis), we observed NMNAT localization adjacent to synaptic membranes labelled with horseradish peroxidase (HRP) staining (Fig 3B) and some NMNAT puncta colocalized strongly with BRP puncta (Fig 3C,C′), with 48±11% of BRP colocalizing with NMNAT and 52±12% of NMNAT colocalizing with BRP (Fig 3D), suggesting that NMNAT is localized within close proximity to the active zone and BRP. We next observed that NMNAT co-immunoprecipitated specifically and reciprocally with BRP (Fig 3E), but not with DLAR, synaptotagmin, CSP, syntaxin or DLG (Fig 3F). These results indicate that NMNAT is localized to synaptic active zones and specifically interacts with the active zone protein BRP.
Figure 3.
NMNAT localizes to the active zone and interacts with BRP. (A–C) Immunostaining of the NMJ at muscle 6/7 in L3 larvae reveals colocalization of NMNAT with BRP. (A1–A4) Conventional confocal imaging of synaptic structure reveals a punctate distribution of NMNAT at the synapse, similar to that for BRP staining (marked by arrowheads). (B,C) 3D-SIM super-resolution imaging of single NMJ boutons reveals NMNAT localization at the synapse (B1, B3), close to the plasma membrane marked by HRP (B2, B3) and NMNAT localization to the active zone (C1, C3), colocalizing with BRP puncta (C2, C3, marked by arrowheads). (C′) Higher magnification of boxed area in C. Arrowheads indicate the colocalized BRP and NMNAT labelling. (D) Quantification of colocalization between BRP (in green) and NMNAT (in red). n=4. Scale bars indicate 10 μm in A, 1 μm in B–C and 500 nm in C′. (E–F) NMNAT interacts with BRP. (E) Reciprocal immunoprecipitation from Drosophila brain lysate with NMNAT antibody or BRP antibody reveals biochemical interaction between NMNAT and BRP. (F) NMNAT did not interact with synaptic proteins, DLAR, synaptotagmin, CSP, syntaxin or DLG. BRP, Bruchpilot; HRP, horseradish peroxidase; IB, immunoblotting; NMNAT, nicotinamide mononucleotide adenylyltransferase; NMJ, neuromuscular junction.
Activity-dependent interaction of NMNAT and BRP
Recent studies have revealed the dynamic changes of active zones and active zone proteins responding to neuronal activity [16, 17, 18, 19, 20]. To examine the functional relevance of NMNAT–BRP interaction during activity, we utilized the Drosophila visual system as synaptic activity can be easily manipulated by altering light exposure. We examined the synaptic and active zone structure of wild-type and nmnat-null synapses under normal (12 h dark/light condition, DL) or reduced activity (12 h dark/dark, DD). As shown in Fig 4B,B′, in nmnat-null neurons the photoreceptor terminal and the active zone structure were compromised in 1 DAE flies under normal conditions, consistent with our previous report [8]. However, when photoreceptor activity was diminished in complete darkness, active zone structure, as measured by the width of the T-bar, was maintained in nmnat-null synaptic terminals for as long as 10 days (Fig 4C′). To further strengthen this finding, we took an alternative approach to block neuronal activity utilizing a genetic mutation in NorpA (no receptor potential A) [21], which attenuates the phototransduction cascade and synaptic transmission in photoreceptors. We introduced the NorpA mutation in the background of nmnat null and analysed the synaptic structures in NorpA and nmnat double-mutant photoreceptor terminals. In nmnat-null terminals, the active zone (T-bar) size declined with age under the normal (dark/light) condition (Fig 4H), indicating age- and activity-dependent synaptic degeneration. Interestingly, in NorpA/nmnat double-mutant photoreceptor terminals, we observed larger T-bars compared with those in nmnat-null terminals at the same age (Fig 4E′), indicating reduced active zone degeneration when synaptic activity was attenuated. Our results also indicate that NMNAT is not required for the assembly of T-bar, as normal active zone structure is observed in nmnat-null neurons in dark-reared flies (Fig 4C′) and in pharate adults (Fig 4H, [8]), suggesting that during development the transport of BRP and synapse assembly was intact without NMNAT. Collectively, these results indicate that NMNAT protein is required for the maintenance of active zone only when synaptic activity is present.
Figure 4.
NMNAT maintains active zone structure and interacts with BRP in an activity-dependent manner. Flies with wild-type or nmnat-null photoreceptors were maintained in 12 h DL or 12 h DD conditions. (A–F) Transmission electron microscopy micrographs of lamina cartridges in control (A), nmnat-null (B–E), and NorpA and nmnat double-mutant (F) photoreceptors. Demarcating glia are coloured blue and photoreceptor terminals green to define the structures. (A′–F′) Individual synapses boxed in (A–F) reveals the disintegrated active zone structure in nmnat-null photoreceptors (B′) compared with control (A′). Blocking light activity delays the degeneration of T-bar structure in nmnat-null photoreceptors (C′, D′), measured by T-bar size (marked by arrowheads). (E–F) Under DL condition, T-bar in NorpA, nmnat double-mutant photoreceptors (F′) is better maintained than in nmnat-null photoreceptors (E′). Scale bar for (A–F) indicates 1 μm. Scale bar for (A′–F′) indicates 200 nm. (G) Quantification of T-bar size in control and nmnat-null photoreceptors in DL and DD conditions. All data were presented as mean±s.e.m. Significance level was established by t-test. *P<0.05. n=80, 73 and 92, respectively. (H) Quantification of T-bar size in nmnat-null photoreceptors at 100 h after puparium formation, 1 DAE and 2DAE and NorpA, nmnat double-mutant photoreceptor terminals. All data were presented as mean±s.e.m. Significance level was established by t-test.*P<0.05. n=28, 73, 70 and 30, respectively. (I–K) Immunoprecipitation of BRP with HA or NMNAT antibody in wild-type or HA–NMNAT-overexpressing photoreceptors from flies reared under DD or DL conditions. (I) The amount of BRP pulled down by HA–NMNAT quantified in DL and DD flies. All data were presented as mean±s.e.m. Significance level was established by t-test. *P<0.05. n=5. BRP, Bruchpilot; DAE, day after eclosion; DD, dark/dark; DL, dark/light; HA, haemagglutinin; NMNAT, nicotinamide mononucleotide adenylyltransferase.
To examine whether the interaction between NMNAT and BRP is also affected by neuronal activity, we expressed functional haemagglutinin (HA)-tagged NMNAT [8] in the photoreceptors (using GMR-GAL4) and manipulated photoreceptor activity level by light exposure. We used HA antibody to isolate the pool of NMNAT protein from the neurons (photoreceptors) subjected to activity alteration. At 10 DAE, we immunoprecipitated whole brain lysates with an HA antibody and probed for BRP. As shown in Fig 4, in brains where HA–NMNAT is not expressed (GMR-GAL4 control flies), no BRP was immunoprecipitated by HA antibody (Fig 4J, lane 3). When HA–NMNAT was expressed in the photoreceptors, HA antibody immunoprecipitated significantly more BRP from flies raised under normal activity conditions (DL) than from flies reared in darkness (DD) (Fig 4J, lanes 6 and 9, and quantified in Fig 4I). This difference is specific for HA–NMNAT expressed in the photoreceptors, as a similar level of BRP was immunoprecipitated by the anti-NMNAT antibody, indicating that the interaction between BRP and endogenous NMNAT in the entire brain was not significantly affected by the light exposure (Fig 4J, bottom blot). We also note that the reduction of BRP immunoprecipitated by HA antibody was not owing to reduced HA–NMNAT expression in flies reared in the dark, as we observed a similar amount of expressed HA–NMNAT (input lanes in Fig 4K) and similar immunoprecipitation efficiency (unbound and +lanes in Fig 4K) between normal (DL) and dark (DD) conditions (Fig 4K). These results indicate that the NMNAT–BRP interaction is affected by neuronal activity, and that increased activity leads to increased NMNAT–BRP interaction.
Our findings of ubiquitinated, clustered and mislocalized BRP in loss-of-NMNAT neurons, and that increased activity leads to increased NMNAT–BRP interaction, together with the observation that active zone structure is maintained in nmnat-null neurons when neuronal activity is reduced, suggest the following model of the activity-dependent role of NMNAT in active zone maintenance. Under normal activity conditions, NMNAT is required to maintain active zone structure by interacting with BRP and to prevent the ubiquitination of BRP, inasmuch as loss of NMNAT results in BRP ubiquitination, mislocalization, aggregation and reduced active zone size. When neuronal activity is minimized (for example, by blocking light stimulation (dark rearing), or by blocking phototransduction (NorpA)), the demand on maintenance by NMNAT is reduced. Our studies here have revealed a specific role of NMNAT in regulating homeostasis of the active zone protein BRP. The role of NMNAT in protein–protein interaction is consistent with the chaperone function of NMNAT. Chaperones, such as CSP, have been implicated in maintaining synaptic integrity [4, 5, 6, 7]. Moreover, recent studies have shown that an elevated activity level poses stress to synaptic proteins by highlighting the effect of CSP in maintaining synaptic function [22]. It is expected that increased neuronal and/or synaptic activity will lead to increased protein misfolding and turnover, and therefore to an increase in the load/demand of maintenance of synaptic protein homeostasis. This notion is supported by a study showing that the level of ubiquitin conjugation of synaptic proteins is altered by the level of synaptic activity [23]. Our studies describe NMNAT as a synapse maintenance factor under normal activity conditions post assembly, when most of the BRP protein is present at the active zone and NMNAT protein is localized to the active zone area to carry out its maintenance function. The interesting observation of clustered BRP protein in the cell body away from the synapse in loss-of-NMNAT neurons indicates a possible defect in the transport of BRP. Two possibilities might explain this phenotype. One, NMNAT facilitates the anterograde transport of BRP during activity. Reduced NMNAT level leads to inefficient transport and subsequent clustering of BRP in the cell body. Two, these BRP clusters are retrogradely transported from the active zone en route to degradation in the cell body. Further work will be required to determine the direction of transport. In summary, our work has identified NMNAT as a chaperone for maintaining active zones, and for facilitating their maintenance during neuronal activity by binding to active zone structural protein BRP, adding NMNAT to the list of synaptic chaperones that are required to maintain functional and structural integrity in neurons.
Methods
MG-132 feeding was carried out as described [15]. MARCM analysis, electron microscopy, and immunocytochemistry and fluorescence microscopy were carried out as described [8]. DSIM super-resolution imaging was performed on a DeltaVison OMX version 3 (Applied Precision, Inc.). Confocal microscopy was performed with an Olympus IX81 confocal microscope. Fluorescence analysis was performed with MetaMorph 5.07 (Universal Imaging Corp.), Amira 3.0 (TGS, Inc.) and Adobe Photoshop CS4. Immunoprecipitation and western blot analysis were carried out using infrared dye-conjugated secondary antibodies and imaged on an Odyssey system (LI-COR Biosciences). Real-time PCR (Bio-Rad) was carried out using nmnat- and brp-specific probes (PE Applied Biosystems), with rp49 as an internal control.
For detailed information of methods see supplementary information online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank Janine Babulski, Adrian Quintanilla and Leanna Ferrand from Applied Precision Inc. for assistance with super-resolution imaging; George McNamara, Wilfredo Escala and Zoriada Diaz-Perez for technical support; Dr Brant Watson for critically reviewing the manuscript; and Peggy Bates and the Electron Microscopy Core for EM assistance. Y.O.A. was supported by the American Heart Association. R.G.Z. is supported by the Neuroscience Centre at the University of Miami, the Pew Charitable Trust and NIH/NINDS (1R01NS064269).
Author contribution: S.Z., Y.O.A. and R.G.Z. conceived and designed the experiments and initiated the project. S.Z., Y.O.A., K.R. and R.G.Z. performed the experiments. S.Z., Y.O.A. and R.G.Z. analysed the data. S.Z., Y.O.A., K.R. and R.G.Z. contributed reagents, materials and analysis tools. S.Z., Y.O.A. and R.G.Z. wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Kittel RJ et al. (2006) Bruchpilot promotes active zone assembly, Ca2+ channel clustering, and vesicle release. Science 312: 1051–1054 [DOI] [PubMed] [Google Scholar]
- Schoch S, Gundelfinger ED (2006) Molecular organization of the presynaptic active zone. Cell Tissue Res 326: 379–391 [DOI] [PubMed] [Google Scholar]
- Zhai RG, Bellen HJ (2004) The architecture of the active zone in the presynaptic nerve terminal. Physiology 19: 262–270 [DOI] [PubMed] [Google Scholar]
- Braun JE, Scheller RH (1995) Cysteine string protein, a DnaJ family member, is present on diverse secretory vesicles. Neuropharmacology 34: 1361–1369 [DOI] [PubMed] [Google Scholar]
- Buchner E, Gundersen CB (1997) The DnaJ-like cysteine string protein and exocytotic neurotransmitter release. Trends Neurosci 20: 223–227 [DOI] [PubMed] [Google Scholar]
- Graham ME, Burgoyne RD (2000) Comparison of cysteine string protein (Csp) and mutant alpha-SNAP overexpression reveals a role for csp in late steps of membrane fusion in dense-core granule exocytosis in adrenal chromaffin cells. J Neurosci 20: 1281–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves J, Salaun C, Fukata Y, Fukata M, Chamberlain LH (2008) Palmitoylation and membrane interactions of the neuroprotective chaperone cysteine-string protein. J Biol Chem 283: 25014–25026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai RG, Cao Y, Hiesinger PR, Zhou Y, Mehta SQ, Schulze KL, Verstreken P, Bellen HJ (2006) Drosophila NMNAT maintains neural integrity independent of its NAD synthesis activity. PLoS Biol 4: e416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai RG, Zhang F, Hiesinger PR, Cao Y, Haueter CM, Bellen HJ (2008) NAD synthase NMNAT acts as a chaperone to protect against neurodegeneration. Nature 452: 887–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouquet W, Owald D, Wichmann C, Mertel S, Depner H, Dyba M, Hallermann S, Kittel RJ, Eimer S, Sigrist SJ (2009) Maturation of active zone assembly by Drosophila Bruchpilot. J Cell Biol 186: 129–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagh DA et al. (2006) Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila. Neuron 49: 833–844 [DOI] [PubMed] [Google Scholar]
- Wu JS, Luo L (2006) A protocol for mosaic analysis with a repressible cell marker (MARCM) in Drosophila. Nat Protoc 1: 2583–2589 [DOI] [PubMed] [Google Scholar]
- Haas KF, Broadie K (2008) Roles of ubiquitination at the synapse. Biochim Biophys Acta 1779: 495–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings CJ, Mancini MA, Antalffy B, DeFranco DB, Orr HT, Zoghbi HY (1998) Chaperone suppression of aggregation and altered subcellular proteasome localization imply protein misfolding in SCA1. Nat Genet 19: 148–154 [DOI] [PubMed] [Google Scholar]
- Hrizo SL, Palladino MJ (2010) Hsp70- and Hsp90-mediated proteasomal degradation underlies TPI sugarkill pathogenesis in Drosophila. Neurobiol Dis 40: 676–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilestro GF, Tononi G, Cirelli C (2009) Widespread changes in synaptic markers as a function of sleep and wakefulness in Drosophila. Science 324: 109–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallermann S, Fejtova A, Schmidt H, Weyhersmuller A, Silver RA, Gundelfinger ED, Eilers J (2010) Bassoon speeds vesicle reloading at a central excitatory synapse. Neuron 68: 710–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz J, Gilyan A, Kolar A, McCarvill T, Krueger SR (2010) Rapid structural alterations of the active zone lead to sustained changes in neurotransmitter release. Proc Natl Acad Sci USA 107: 8836–8841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao-Cheng JH (2006) Activity-related redistribution of presynaptic proteins at the active zone. Neuroscience 141: 1217–1224 [DOI] [PubMed] [Google Scholar]
- Weyhersmuller A, Hallermann S, Wagner N, Eilers J (2011) Rapid active zone remodeling during synaptic plasticity. J Neurosci 31: 6041–6052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomquist BT, Shortridge RD, Schneuwly S, Perdew M, Montell C, Steller H, Rubin G, Pak WL (1988) Isolation of a putative phospholipase C gene of Drosophila, norpA, and its role in phototransduction. Cell 54: 723–733 [DOI] [PubMed] [Google Scholar]
- Garcia-Junco-Clemente P, Cantero G, Gomez-Sanchez L, Linares-Clemente P, Martinez-Lopez JA, Lujan R, Fernandez-Chacon R (2010) Cysteine string protein-alpha prevents activity-dependent degeneration in GABAergic synapses. J Neurosci 30: 7377–7391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers MD (2003) Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat Neurosci 6: 231–242 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.