Abstract
Embryonic stem cells repress retroviral infection through transcriptional silencing of proviral DNAs. We characterized two distinct mechanisms of silencing in embryonic mouse cells infected by Moloney murine leukaemia virus (MLV): a highly efficient one targeting the proline transfer RNA primer-binding site (PBSpro), and a less efficient one operating independently of the PBS. Rare virus-expressing populations were isolated, and the timing and efficiency of establishment of silencing were determined. Superinfection of the selected virus-expressing cells with a second virus carrying a distinguishable reporter revealed that the PBSpro-directed silencing was still largely intact, whereas the PBS-independent silencing was partially reduced. The timing and stability of silencing, and the associated chromatin modifications on newly established and endogenous proviruses were determined. The results indicate that epigenetic mechanisms with different specificity and efficiency are used to silence the exogenous retroviral sequences in embryonic cells.
Keywords: chromatin modifications, embryonic stem cells, endogenous retroviruses, epigenetic silencing, primer-binding site
Introduction
Embryonic stem (ES) and embryonic carcinoma (EC) cell lines typically repress transcription of newly introduced retroviral DNAs [1, 2]. This silencing mechanism in murine ES cells blocks replication of murine leukaemia viruses (MLV), and limits the expression of genes delivered by retroviral vectors [3]. A chief component of the silencing machinery in murine ES cells is targeted to a repressor-binding site that overlaps with the primer-binding site (PBS) complementary to the 3′ end of proline transfer RNA, the most common transfer RNA primer used for initiation of reverse transcription by MLVs [4, 5]. This silencing is mediated by the zinc-finger DNA -binding protein, ZFP809, which binds to the proline PBS sequence, and a universal silencing protein, TRIM28/Kap-1/Tif1β, which interacts with ZFP809 [6, 7]. Retroviral genomes that use a distinct PBS, such as one complementary to the 3′ end of glutamine tRNA, are not recognized by the silencing machinery and partially escape the transcriptional repression [5, 8, 9]. Retroviral vectors utilizing these alternate PBS sequences are still subject to some transcriptional repression by other mechanisms [10, 11, 12].
The silencing machinery in ES cells might protect them from spreading virus infections, and might also have a more important role in repressing expression of endogenous retroviruses (ERVs) and retrotransposons in the genome. Germline mutation of the TRIM28 gene results in activation of endogenous proviruses and early embryonic lethality [13, 14]. This silencing was initially attributed to DNA methylation [15] but other epigenetic mechanisms were shown to be responsible [12, 16]. The chromatin of endogenous retroviral elements in ES cells is marked by repressive H3K9 lysine methylation and by H3K4 demethylation, and these modifications are required to maintain silencing of endogenous retroelements [14, 16, 17, 18]. Recent work suggests that silencing of exogenous proviral DNAs in ES cells is also coupled with histone modifications in chromatin laid down onto the new proviruses [19]. It is unknown whether these modifications are linked to the PBSpro-specific silencing machinery or to the other mechanisms involved in more general PBSpro-independent silencing [10].
To further characterize the ability of ES cells to silence retroviral DNAs, we monitored expression of reporter genes delivered to ES and EC cells by various retroviral vectors. We found that viruses utilizing the proline PBS were rapidly and completely silenced, whereas other viruses were more slowly and incompletely silenced. Cells transiently expressing PBSpro virus DNAs could be selected, but these rapidly silenced the viruses; cells expressing other virus DNAs only slowly imposed silencing. Although sorted cell populations were not amenable to analysis of chromatin modifications, we were able to examine those populations that exhibited stable expression phenotypes for histone modifications. We show that strong epigenetic silencing mechanisms are applied to all incoming viruses as well as to ERVs.
Results and discussion
Infection with viruses utilizing PBSpro or PBSproB2
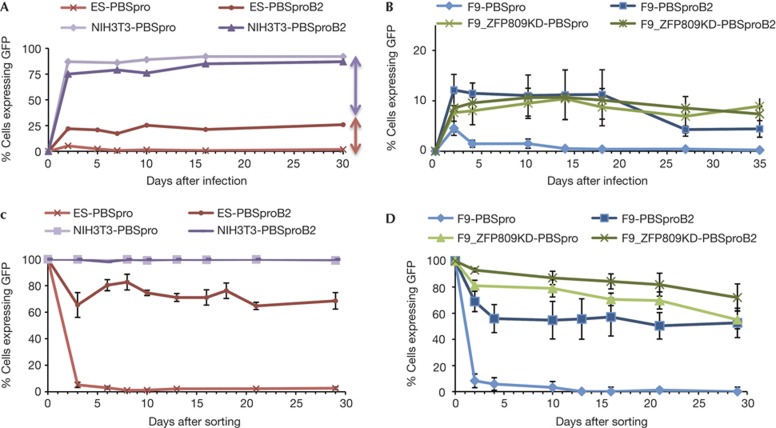
To monitor the course of retroviral restriction in ES cells, MLV genomes expressing a green fluorescent protein (GFP) reporter and utilizing either the wild-type (wt) PBSpro sequence or a variant PBSproB2 were packaged into virus particles and used to infect various cell populations. Cells were analysed by flow cytometry at various times after infection to determine the per cent GFP-positive cells. The PBSpro and PBSproB2 viruses were both efficiently expressed in differentiated NIH/3T3 (Fig 1A), Rat2, L and 293A cells (data not shown). In contrast, the PBSpro virus was profoundly and rapidly silenced in ES (Fig 1A; supplementary Fig S1A online) and F9 EC cells (Fig 1B; supplementary Fig S1B online). The PBSproB2 virus was also partially silenced in these cells, typically giving rise to 10–25% GFP-positive cells. Similar results were observed with a virus vector containing PBSgln (supplementary Fig S1B online). In F9 cells in which expression of ZFP809 was suppressed by RNA-mediated interference (RNAi)-mediated knockdown ([7], see supplementary Fig S1C online), the high-level silencing of PBSpro virus genomes was lost (Fig 1B). These differences in reporter gene expression were not attributable to differences in proviral DNA copy number; infections were performed at multiplicities <1, and DNA copy numbers of expressing and nonexpressing cell populations were comparable (supplementary Fig S2A online).
Figure 1.
Kinetics of silencing. Kinetics of viral silencing in ES cells using a GFP reporter virus. (A) Flow analysis of GFP-positive cells at different time points after infection (day 0) by wt (pro) and mutant (proB2) PBS virus in ES cells or differentiated cells. Representative experiment is shown (see Supplementary Fig S1A online for mean±s.d.). Arrows denote extent of silencing due to PBSpro-dependent (red) and PBSpro-independent mechanisms (purple). (B) Same assay on F9 scrambled pool and on ZFP809 KD F9 pool 9 ([7], see also Supplementary Fig S1C online). Note change of y axis from A. Averages±s.e.m. from three independent experiments are shown. For F9 EC cell line results see Supplementary Fig S1B online. (C) Flow analysis after FACS sorting of cells that escape restriction and express GFP. NIH/3T3 cells are shown as control. (D) Same analysis in F9 cells and F9 ZFP809 KD clone. Error bars show standard error of the mean (s.e.m.) for n=3 (C) or n=4 (D) biological replicates. See also Supplementary Fig S2 online for analyses of proviral DNA copy number and Supplementary Fig S3 online for ES-specific markers analysis. ES, embryonic stem; FACS, fluorescence-activated cell sorting; GFP, green fluorescent protein; PBSpro, proline tRNA primer-binding site; wt, wild-type.
To assess the level of expression per cell, the mean GFP fluorescence of the positive cells in each population was determined by flow cytometry (supplementary Fig S3 online). Although both viruses induced high-level expression in NIH/3T3 cells, levels of expression in GFP-positive ES and EC cells were several 100-fold lower. This might reflect weak activity of the viral long terminal repeat (LTR) promoter in ES and EC lines.
Time course of silencing in embryonic cells
Rare EC cells that stably express the PBSpro genome (∼10−6 of infected cells) have previously been isolated and found to have mutations of the PBSpro or to have integrated the virus into rare expressing sites [2]. However, cells that transiently express the genomes were more abundant and readily isolated. We infected ES cells with GFP viral vectors utilizing PBSpro or PBSproB2, used fluorescence-activated cell sorting (FACS) to isolate GFP-positive cells after 6 days, cultured the sorted cells and analysed them for continued GFP expression by cytometry (Fig 1C). ES cells sorted for initial expression of the PBSpro viruses rapidly and efficiently silenced their GFP expression. In contrast, ES cells sorted for expression of PBSproB2 viruses only partially silenced their expression, with ∼70% of cells remaining GFP positive after 30 days in culture (Fig 1C). Differentiated NIH/3T3 cells infected with either PBSpro or PBSproB2 viruses, and sorted for GFP expression, remained fully GFP positive for >50 days.
Similar tests for stability of expression were also performed in F9 cells expressing shRNAs targeting ZFP809 or scrambled control (Fig 1D). In the control line, we observed kinetics similar to that in ES-sorted cells. In F9 ZFP809KD cells, both PBSpro and PBSproB2 viruses were only partially silenced, with 70–80% of the cells remaining GFP positive, and the silencing was slow (Fig 1D). These experiments indicate that there is a rapid, efficient silencing of the PBSpro virus in ES and F9 cells that depended on ZFP809, and a slow, partial silencing of virus expression by a PBS-independent mechanism. Differences in reporter gene expression were not attributable to different proviral DNA copy number (supplementary Fig S2B online) or to changes in the differentiation state of the cells, as judged by Oct4 expression levels and flow analysis of standard ES cell markers (supplementary Fig S4A,B online). GFP expression of sorted cells was verified by reverse transcription quantitative PCR (RT–qPCR; supplementary Fig S4C online).
These results show that the expression in rare GFP-positive populations (escapees) is not stable. The silencing machinery is readily reactivated, and expression is extinguished soon after the isolation of these cells. The temporary escape from silencing is thus not due to a permanent loss of the silencing factors. Rather, these cells seem to have stochastically and very transiently escaped silencing and in days are able to reestablish it. PBSpro viruses were silenced nearly completely in 3 days, suggesting that the rapid silencing mechanism is still present in a latent form. It is unclear whether new protein synthesis is required to establish this quick silencing.
It is unclear whether the expressing ‘escapees’ have acquired proviruses integrated into special regions of the genome. The overall several proviruses per cell in this population (1.1) is only slightly higher than in the unselected cells (0.5), in accordance with the requirement that they have at least one provirus, so increased copy number alone is not a likely explanation for their expression (see supplementary Fig S2 online). Integration by MLV and MLV-based vectors is thought to be nearly random, with some bias towards integration into the 5′ ends of genes [20]. The transiently PBSproB2-expressing cells represent a sufficiently large proportion of the total cells (∼20%) that even if they acquired proviral integrants in ‘special’ sites, many such sites must be present in the genome. Therefore, we favour the model that integrated proviruses at most sites, and, perhaps at any site, can be activated at low frequency and for short periods of time before being silenced again, as is the case for other exogenous and endogenous retroviruses [21, 22].
Superinfection of ES cells selected for expression
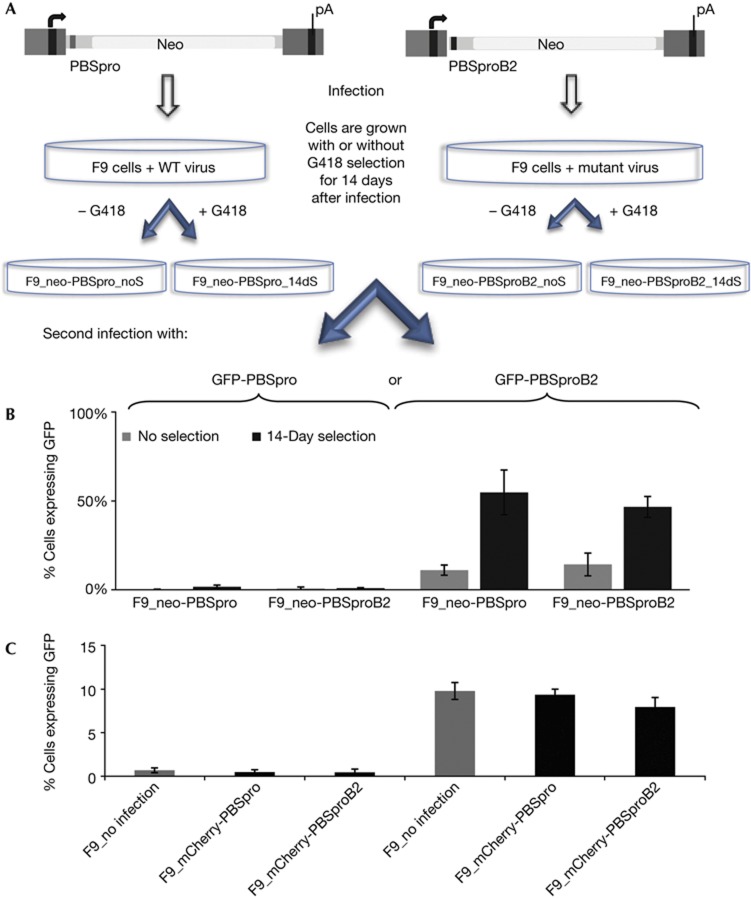
To examine the stability and longevity of the silencing machinery in ES cells, we selected for cells that escaped silencing of virus carrying a drug-resistance marker, and rechallenged both selected and unselected populations with a second viral genome. F9 EC cells were first infected with retroviral genomes expressing the neor marker, encoding resistance to G418, and utilizing either PBSpro or PBSproB2. The cells were selected for expression by growth in medium containing G418 for 2 weeks or grown without selection. No main differences were seen in viral DNA copy number in the populations (supplementary Fig S5 online). Cells were then superinfected with a viral genome expressing the GFP reporter, again using PBSpro or PBSproB2, and scored for expression by cytometry 48 h after infection; the PBSpro-targeted silencing machinery, although slightly reduced after the initial round of selection, was still largely intact (Fig 2). The rare cells selected for neor after infection with the first PBSpro virus were still able to significantly silence the incoming second PBSpro virus (3% of cells GFP positive versus 1% without previous selection). The more abundant neor cells selected after infection with the PBSproB2 neor virus also efficiently silenced an incoming PBSpro virus essentially like an unselected cell population (1% remaining GFP positive; Fig 2B). These results indicate that the escape from silencing of the initial PBSpro neor virus was local to the initial proviruses, and not due to a global loss of the PBS-directed silencing system.
Figure 2.
GFP reporter gene expression in cell populations after long-term selections or cell sorting. (A) F9 cells were infected first with VSV-G pseudotyped MLV containing neor reporter with PBSpro (WT) or PBSproB2 (mutant) constructs. Half the population was selected for 2 week using G418 (14dS) and half was grown without selection (noS). Both populations were then infected with GFP virus with PBSpro or PBSproB2 and analysed by flow cytometry. (B) Percent of GFP expressing cells after second infection with PBSpro or PBSproB2. Infection efficiency in each cell line normalized with NIH/3T3=100%. (C) Cells initially infected with mCherry viruses were sorted and then subjected to a second round of infections with the GFP viruses, and analysed by flow cytometry. Percent of GFP expressing cells after second infection with wt PBSpro or mutant PBSproB2 is shown. Infection efficiency in each cell line normalized with NIH/3T3=100%. Averages±s.e.m. for n=3 (B) or n=5 (C) biological replicates are shown. See Supplementary Fig S5 online for analyses of proviral DNA copy number and ES-specific marker. ES, embryonic stem; GFP, green fluorescent protein; MLV, murine leukaemia virus; PBSpro, proline tRNA primer-binding site; wt, wild-type.
The long-term selection of expressing cells after infection with neor viruses did result in a modest reduction in the ability of the cells to silence a second infection with PBSproB2 virus. Cells selected after infection with the PBSpro or PBSproB2 neor virus could only partially restrict the incoming GFP PBSproB2 virus (45% or 55% scored GFP positive, respectively; Fig 2B). Thus, the non-PBS-directed silencing suffered some global loss of function in response to the initial selection. This effect seemed to require a prolonged period of selection for virus reporter gene expression.
To test whether these changes required long-term selection for expression, we isolated mCherry-positive cells by sorting only 6 days after infection with mCherry-expressing PBSpro or PBSproB2 virus, and then challenged with GFP PBSpro or PBSproB2 virus. No significant loss of GFP silencing was observed (Fig 2C). Thus, either the different length of time in selection or the different levels of expression demanded in these two isolation methods could account for this difference. Neither DNA copy number nor differentiation state of the cells accounted for the changes in reporter gene expression (supplementary Fig S5 online).
Histone modifications at proviral DNAs
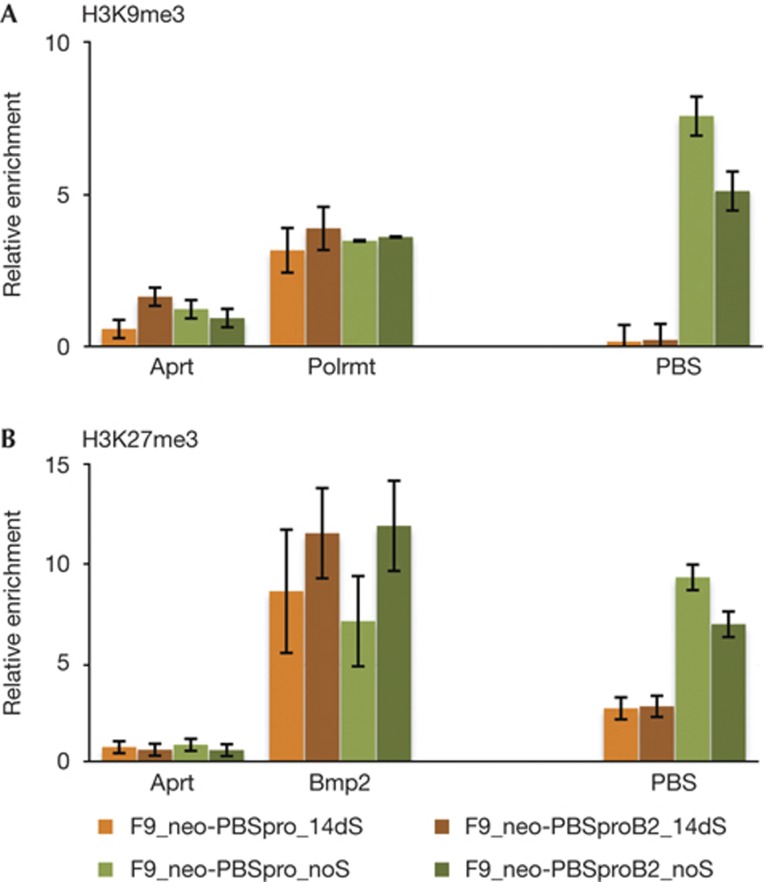
To determine whether changes in viral expression were correlated with a change in the chromatin state of the newly integrated proviral DNAs, the selected and unselected cell populations were analysed by chromatin immunoprecipitation (ChIP). In F9 cells grown without selection, in which expression was heavily silenced, the PBSpro and PBSproB2 virus sequences were both highly enriched in the H3K9me3 fraction (green bars), whereas virus sequences in cells grown with selection for neor expression (orange bars) were much less enriched (Fig 3A). Analysis with anti-H3K27me3 gave similar results (Fig 3B). Thus, these marks did not require the rapid PBS-mediated silencing. It is not clear whether both marks are present on all silenced proviruses or whether individual proviruses are marked only by one or the other modification. Recent findings in human [23] and mouse ES cells [24] indicate that although rare, the mutual occupancy of these marks is possible. The data indicate that at least two distinct histone methyltransferases are simultaneously active in marking the viral chromatin of the newly incoming MLVs, and the two marks are consistent with the existence of redundant pathways mediating the virus silencing.
Figure 3.
ChIP analysis of infected F9 cells subjected to long-term selection. ChIP-based measurement of (A) H3K9me3 and (B) H3K27me3 at the viral PBS sequence of cells infected with MLV containing neor gene, with PBSpro or PBSproB2 and selected for 14 days or infected without selection. Averages±s.e.m. from three independent experiments are shown. For each experiment, relative enrichment values (% of input) are shown, normalized to the signal of negative control (Gapdh). Results for negative (Aprt) and positive (Polmrt, BMP2) control genes are shown. ChIP, chromatin immunoprecipitation; MLV, murine leukaemia virus; PBSpro, proline tRNA primer-binding site.
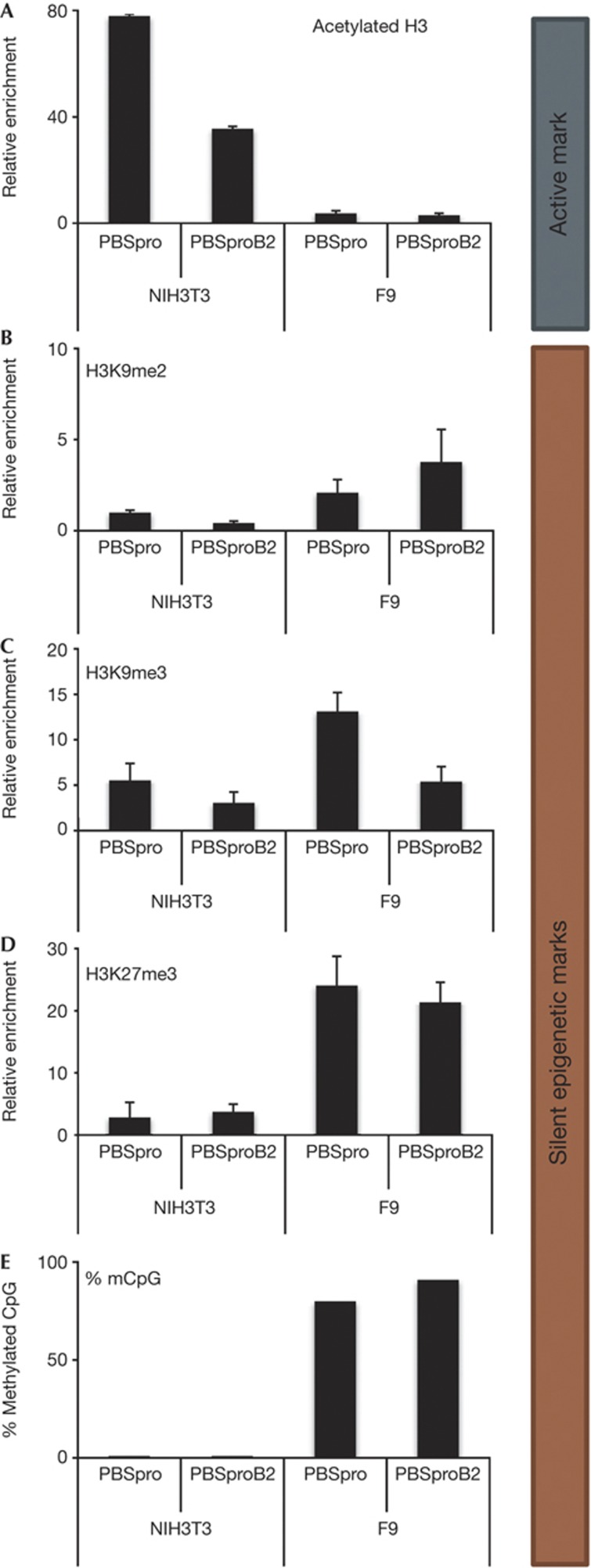
Similar experiments were performed on infected F9 and NIH/3T3 cells to examine histone H3 acetylation, a mark of active chromatin. Infection of NIH/3T3 cells with PBSpro GFP or PBSproB2 GFP viruses for 14 days resulted in the establishment of proviruses with heavily acetylated chromatin (Fig 4A). In contrast, infection of F9 cells resulted in low levels of H3 acetylation. The mirror image was seen for H3K9me2, H3K9me3 and H3K27me3 marks (Fig 4B–D, respectively). The absence of H3 acetylation is thus correlated with heavy H3K9 and 27 trimethylation, and with provirus silencing in F9 cells. We also tested for DNA methylation by bisulfite sequencing (Fig 5E). NIH/3T3 cells showed no DNA methylation in the provirus 14 days after infection, whereas 80–90% of CpGs in proviruses in F9 cells were methylated, in accordance with published data, as well as with our own, showing that in embryonic cells the provirus is silenced in an epigenetic manner in a PBS-independent way [25].
Figure 4.
Differences between 3T3 and F9 cells in chromatin marks and DNA methylation levels. ChIP-based measurement of (A) H3Ac, (B) H3K9me2, (C) H3K9me3 and (D) H3K27me3 at the viral PBS sequence of cells infected with GFP reporter virus with PBSpro or PBSproB2. As the two cell types gave different background binding, the signal was first normalized to IgG control ChIP, and normalized signal of negative control α-crystalline (H3Ac) or the Gapdh promoters (H3K9me2/3, H3K27me3) was set to 1. Averages±s.e.m. from three independent experiments are shown. Negative and positive control genes gave the expected enrichment values (not shown). (E) Bisulfite sequencing analysis of the 5′LTR of the infecting virus was performed on NIH/3T3 and F9 cells; Oct4 was used as control. Percentages of methylated CpGs are shown from 10 to 15 cloned DNA molecules per cell and infection type (Supplementary Fig S6 online). ChIP, chromatin immunoprecipitation; GFP, green fluorescent protein; 5′LTR, 5′ long terminal repeat; PBSpro, proline tRNA primer-binding site.
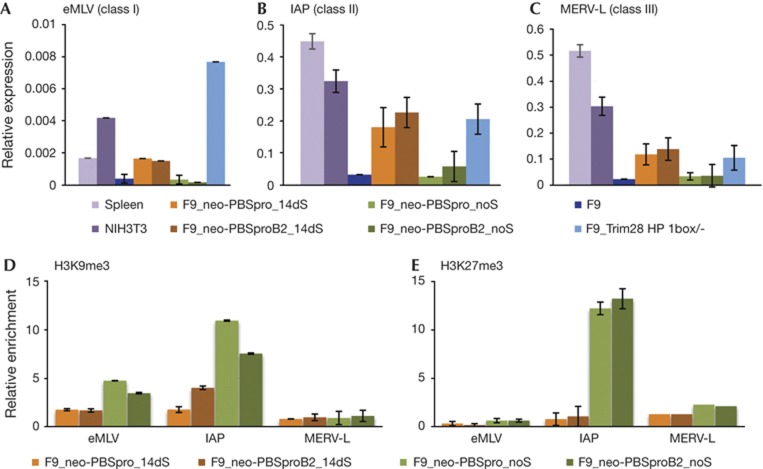
Figure 5.
Expression pattern and chromatin modifications of classes I, II and III ERVs. Quantitative RT–PCR analysis of (A) eMLVs (class I), (B) IAP (class II) and (C) MERV-L (class III) messenger RNA expression in embryonic and differentiated cells. Bars represent the mean±s.e.m. of three independently prepared samples, relative to three control genes (UBC, CYCA and Gapdh). ChIP-based measurement of (D) H3K9me3 and (E) H3K27me3 at the viral sequence of cells infected with MLV containing neor with PBSpro or PBSproB2 and selected for 14 days or infected without selection. Relative enrichment values (% of input) in all ChIP experiments were normalized to the relative enrichment at the Gapdh promoter. Negative and positive control genes gave the expected enrichment (not shown). ChIP, chromatin immunoprecipitation; MLV, murine leukaemia virus; ERV, endogenous retrovirus; PBSpro, proline tRNA primer-binding site; RT–PCR, reverse transcription PCR.
Analysis of ERV expression and chromatin modification
To test whether cells selected for long-term expression of PBSpro or PBSproB2 incoming viruses become less efficient at silencing ERVs, we used quantitative RT–PCR to measure the RNA levels of the three classes of ERVs in the mouse genome. In all cases, expression levels were elevated in the selected cells relative to wt F9 cells and unselected infected cells (Fig 5A–C). In positive control cells with a mutant TRIM28 gene lacking the HP1 box, and therefore unable to bind HP1 [26], expression levels of all ERV classes were constitutively higher than in wt cells. ChIP experiments on the different ERV sequences showed that the change in expression correlated with loss of both H3K9 and K27 trimethylation (Fig 5D,E) for ERV classes I and II, but not III, in accordance with earlier reports [17, 18, 27].
These data strongly suggest that there is considerable overlap in the machinery for the silencing of exogenous retroviruses and endogenous proviruses and in the chromatin marks placed on the proviral DNAs. When we select for loss of silencing of exogenous viral DNAs, we observe loss of silencing of the endogenous DNAs as well. When we see changes in histone marks on exogenous viruses, we see similar changes in the chromatin marks on classes I and II endogenous viruses. Furthermore, these and previous data [14, 18] show that TRIM28 has a major role in both silencing mechanisms.
The existence of the two silencing systems examined in this study suggests that the suppression of virus gene expression is an important aspect of the ES cell state.
The two silencing systems might be important in suppressing viral infections and endogenous proviruses, and might also have a might role in regulation of host gene expression in general and especially in ES cell development. There is extensive overlap in the patterns of regulation of endogenous retroviruses and of host genes, and in the silencing machinery that establishes these patterns. TRIM28 seems to be a master gene in both silencing systems [28, 29]. Recently, the phosphorylated form of TRIM28 (as TIF1b) was shown to cooperate with Oct4 in the maintenance of pluripotency in ES cells, helping to regulate the expression of Oct4 target genes [30, 31]. Phosphorylated TRIM28 can also bind Suz12, a member of the PRC2 family of histone methyltransferases, and might directly lead to histone methylation. The overlap of viral and host gene regulation might not be coincidental but might reflect the acquired use of viral LTRs and promoters during evolution to control critical embryonic gene expression [17]. The endogenous retroviruses themselves might even have important roles in development [32].
Methods
Cell culture and stable RNAi cell line production and transduction. Cells were cultured as described [11]. RNAi knockdown was performed as in Wolf and Goff [7]. For primers and detailed description see supplementary Data online. For viral transduction assays, viruses were prepared as before using LJ-PAdMLPEnh or LJB2-ADMLPEnh vectors [9] or pNCA-GFP/mCherry vectors [33]. Each experiment was repeated three times or more.
Flow cytometry and sorting. GFP-positive cells were isolated on a cell sorter (FACSAria Cell Sorter; BD Biosciences). Data were acquired on an automated cell analyser (LSR II; BD Biosciences) and analysed with FlowJo software (Treestar)
Chromatin immunoprecipitations (ChIP) was performed with Magna ChIP kit (Millipore), and DNA was purified using QIAquick PCR purification kit (Qiagen). Antibodies and primers are listed in supplementary Methods online and supplementary Table S1 online
Bisulfite conversion of genomic DNA was carried out using the Zymo EZ DNA Methylation-Gold Kit. PCR primers were designed using Methyl Primer Express software version 1.0 (https://www2.appliedbiosystems.com; see supplementary Table S1 online)
RNA Extraction and RT–PCR protocols are presented in supplementary Methods online.
Statistical analysis. Statistical analysis of the data was performed with a two-tailed Student’s t-test or one-way analysis of variance. Data presented as the mean±s.e.m. of three or more independent biological replicates.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
In memory of Daniel Wolf. This work was supported by NCI grant R37 CA 30488 and NYSTEM grant N08G-152/contract #C024329 from the New York State Department of Health. S.P.G. is an Investigator of the Howard Hughes Medical Institute.
Author contributions: S.S. performed the experiments, and S.S. and S.P.G. wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Teich NM, Weiss RA, Martin GR, Lowy DR (1977) Virus infection of murine teratocarcinoma stem cell lines. Cell 12: 973–982 [DOI] [PubMed] [Google Scholar]
- Barklis E, Mulligan RC, Jaenisch R (1986) Chromosomal position or virus mutation permits retrovirus expression in embryonal carcinoma cells. Cell 47: 391–399 [DOI] [PubMed] [Google Scholar]
- Ellis J (2005) Silencing and variegation of gammaretrovirus and lentivirus vectors. Hum Gene Ther 16: 1241–1246 [DOI] [PubMed] [Google Scholar]
- Harada F, Peters GG, Dahlberg JE (1979) The primer tRNA for Moloney murine leukemia virus DNA synthesis. Nucleotide sequence and aminoacylation of tRNAPro. J Biol Chem 254: 10979–10985 [PubMed] [Google Scholar]
- Petersen R, Kempler G, Barklis E (1991) A stem cell-specific silencer in the primer-binding site of a retrovirus. Mol Cell Biol 11: 1214–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Goff SP (2007) TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 131: 46–57 [DOI] [PubMed] [Google Scholar]
- Wolf D, Goff SP (2009) Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458: 1201–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grez M, Akgun E, Hilberg F, Ostertag W (1990) Embryonic stem cell virus, a recombinant murine retrovirus with expression in embryonic stem cells. Proc Natl Acad Sci USA 87: 9202–9206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modin C, Lund AH, Schmitz A, Duch M, Pedersen FS (2000) Alleviation of murine leukemia virus repression in embryonic carcinoma cells by genetically engineered primer binding sites and artificial tRNA primers. Virology 278: 368–379 [DOI] [PubMed] [Google Scholar]
- Pannell D, Ellis J (2001) Silencing of gene expression: implications for design of retrovirus vectors. Rev Med Virol 11: 205–217 [DOI] [PubMed] [Google Scholar]
- Cherry SR, Biniszkiewicz D, van Parijs L, Baltimore D, Jaenisch R (2000) Retroviral expression in embryonic stem cells and hematopoietic stem cells. Mol Cell Biol 20: 7419–7426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannell D, Osborne CS, Yao S, Sukonnik T, Pasceri P, Karaiskakis A, Okano M, Li E, Lipshitz HD, Ellis J (2000) Retrovirus vector silencing is de novo methylase independent and marked by a repressive histone code. EMBO J 19: 5884–5894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammas F, Mark M, Dolle P, Dierich A, Chambon P, Losson R (2000) Mice lacking the transcriptional corepressor TIF1beta are defective in early postimplantation development. Development 127: 2955–2963 [DOI] [PubMed] [Google Scholar]
- Rowe HM et al. (2010) KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463: 237–240 [DOI] [PubMed] [Google Scholar]
- Jahner D, Stuhlmann H, Stewart CL, Harbers K, Lohler J, Simon I, Jaenisch R (1982) De novo methylation and expression of retroviral genomes during mouse embryogenesis. Nature 298: 623–628 [DOI] [PubMed] [Google Scholar]
- Karimi MM et al. (2011) DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell 8: 676–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan TS et al. (2011) Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes Dev 25: 594–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y (2010) Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464: 927–931 [DOI] [PubMed] [Google Scholar]
- Leung DC et al. (2011) Lysine methyltransferase G9a is required for de novo DNA methylation and the establishment, but not the maintenance, of proviral silencing. Proc Natl Acad Sci USA 108: 5718–5723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman FD, Craigie R (1990) Sequence requirements for integration of Moloney murine leukemia virus DNA in vitro. J Virol 64: 5645–5648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz RA, Jack-Scott E, Narezkina A, Palagin I, Boimel P, Kulkosky J, Nicolas E, Greger JG, Skalka AM (2007) High-frequency epigenetic repression and silencing of retroviruses can be antagonized by histone deacetylase inhibitors and transcriptional activators, but uniform reactivation in cell clones is restricted by additional mechanisms. J Virol 81: 2592–2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL (2012) Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Geen H, Squazzo SL, Iyengar S, Blahnik K, Rinn JL, Chang HY, Green R, Farnham PJ (2007) Genome-wide analysis of KAP1 binding suggests autoregulation of KRAB-ZNFs. PLoS Genet 3: e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram O et al. (2011) Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. Cell 147: 1628–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung DC, Lorincz MC (2012) Silencing of endogenous retroviruses: when and why do histone marks predominate? Trends Biochem Sci 37: 127–133 [DOI] [PubMed] [Google Scholar]
- Cammas F, Herzog M, Lerouge T, Chambon P, Losson R (2004) Association of the transcriptional corepressor TIF1beta with heterochromatin protein 1 (HP1): an essential role for progression through differentiation. Genes Dev 18: 2147–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeb M, Pasini D, Novatchkova M, Jaritz M, Helin K, Wutz A (2010) Polycomb complexes act redundantly to repress genomic repeats and genes. Genes Dev 24: 265–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar S, Farnham PJ (2011) KAP1 protein: an enigmatic master regulator of the genome. J Biol Chem 286: 26267–26276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Trono D (2011) Dynamic control of endogenous retroviruses during development. Virology 411: 273–287 [DOI] [PubMed] [Google Scholar]
- Seki Y, Kurisaki A, Watanabe-Susaki K, Nakajima Y, Nakanishi M, Arai Y, Shiota K, Sugino H, Asashima M (2010) TIF1beta regulates the pluripotency of embryonic stem cells in a phosphorylation-dependent manner. Proc Natl Acad Sci USA 107: 10926–10931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Kim J, Xu Q, Leng Y, Orkin SH, Elledge SJ (2009) A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev 23: 837–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoye JP (2012) Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat Rev Microbiol 10: 395–406 [DOI] [PubMed] [Google Scholar]
- Ooi SK, Wolf D, Hartung O, Agarwal S, Daley GQ, Goff SP, Bestor TH (2010) Dynamic instability of genomic methylation patterns in pluripotent stem cells. Epigenetics Chromatin 3: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.