Abstract
Although phosphatidylinositol 5-phosphate (PtdIns5P) is present in many cell types and its biogenesis is increased by diverse stimuli, its precise cellular function remains elusive. Here we show that PtdIns5P levels increase when cells are stimulated to move and we find PtdIns5P to promote cell migration in tissue culture and in a Drosophila in vivo model. First, class III phosphatidylinositol 3-kinase, which produces PtdIns3P, was shown to be involved in migration of fibroblasts. In a cell migration screen for proteins containing PtdIns3P-binding motifs, we identified the phosphoinositide 5-kinase PIKfyve and the phosphoinositide 3-phosphatase MTMR3, which together constitute a phosphoinositide loop that produces PtdIns5P via PtdIns(3,5)P2. The ability of PtdIns5P to stimulate cell migration was demonstrated directly with exogenous PtdIns5P and a PtdIns5P-producing bacterial enzyme. Thus, the identified phosphoinositide loop defines a new role for PtdIns5P in cell migration.
Keywords: PtdIns5P, PI3KIII, MTMR3, PIKfyve, cell migration
Introduction
Phosphoinositides are an important group of membrane lipids that reversibly anchor cytosolic proteins to cellular membranes, thereby modulating their localization and activity. They are crucial in cell migration by regulating signal transduction pathways, cytoskeletal dynamics and membrane transport [1]. Phosphoinositide metabolism has also been shown to be important for focal adhesion dynamics during cell migration [2, 3]. The last identified member of the seven known phosphoinositides is PtdIns5P [4]. PtdIns5P is relatively low abundant (1–2% of PtdIns4P levels) [5] but, interestingly, levels increase on different stimuli [6, 7, 8, 9, 10, 11, 12]. Little is known about its function, although it is present in many cell types [5]. A nuclear role for PtdIns5P in regulating apoptosis by binding to ING2 has been proposed [13]. Furthermore, in cells expressing an oncogenic tyrosine kinase, high levels of PtdIns5P were observed, linking this phosphoinositide potentially to cancer [14]. During infection with Shigella flexneri [15, 16] or Salmonella typhimurium [17], the injection of bacterial phosphoinositide phosphatases (IpgD and SopB), which produce PtdIns5P from PtdIns(4,5)P2, facilitates infection [18].
Here, we show that a phosphoinositide loop producing PtdIns5P from PtdIns3P via PtdIns(3,5)P2 is crucial in cell migration.
Results and Discussion
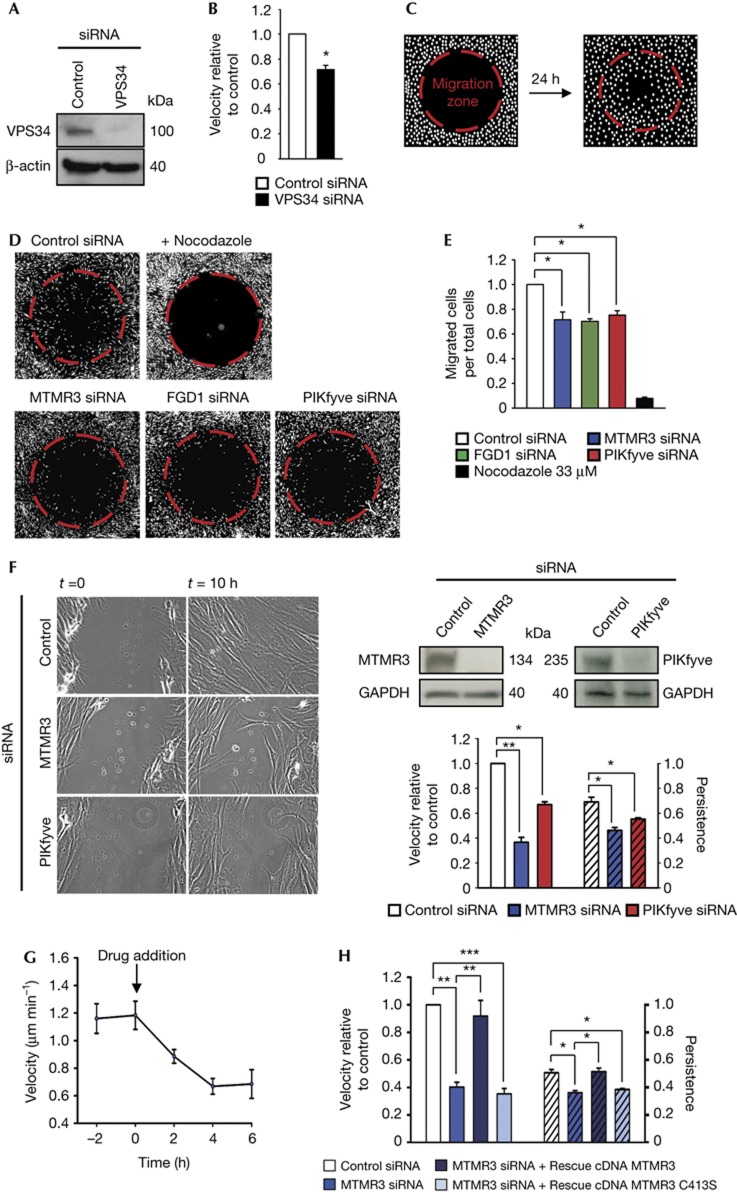
To test whether class III PI3K (PI3KIII), which produces PtdIns3P, is involved in cell migration in human BJ fibroblasts, we depleted cells for the catalytic PI3KIII subunit VPS34 and performed time-lapse live-cell imaging to monitor cell motility. Confluent BJ cell monolayers were scratch-wounded, stimulated with FGF1 to induce cell motility and observed using a live-cell microscope. After tracking cell movement, we observed a significant decrease in cell velocity on depletion of VPS34 compared with control short interfering RNA-transfected cells (Fig 1A,B).
Figure 1.
siRNA screening for proteins containing PtdIns3P-binding motifs: MTMR3 and PIKfyve control cell migration. (A) Western blot analysis demonstrating efficient decrease in protein levels of VPS34 after siRNA treatment. (B) Quantification of BJ cell velocity in control siRNA and VPS34 siRNA-transfected cells in a wound healing assay. Cells analysed in total: 130 (control siRNA); 110 (VPS34 siRNA). (C) Schematic drawing of the observed area of a well with the migration zone (red dotted lines) before and after migration in the siRNA screen assay. White dots illustrate Hoechst 33342-stained cells. (D) Representative images of the observed area with the migration zone (red) after 24 h of migration on FGF1 stimulation for control siRNA, nocodazole and siRNA treatments of selected hits from the siRNA screen. (E) Values representing fraction of cells that have migrated into the migration zone for selected hits from (D). (F) Left: wound healing assay of siRNA-treated and FGF1-stimulated BJ cells at time point 0 and after 10 h. Right, upper panel: western blot analysis demonstrating efficient decrease in protein levels of MTMR3 and PIKfyve after siRNA treatment. Right, lower panel: quantification of velocity and persistence. Cells analysed in total: 190 (control siRNA); 200 (MTMR3 siRNA); 180 (PIKfyve siRNA). (G) Perfusion assay with the inhibitor of PIKfyve, YM201636, which is added at time point 0. Each point represents the velocity in 2 h. Cells analysed in total: 195 (time point −2); 175 (time point 0); 200 (time point 2); 210 (time point 4); 210 (time point 6). The difference in velocity before and after drug addition is statistically significant. (H) Quantification of rescue of MTMR3 knockdown with siRNA-resistant cDNA of MTMR3 wild type and an inactive mutant, MTMR3 C413S; both fused to EGFP to identify transfected cells. Cells analysed in total: 130 (control siRNA); 135 (MTMR3 siRNA); 25 (rescue MTMR3 WT) and 40 (rescue MTMR3 C413S). For all panels, error bars represent the mean±s.e.m. of three independent experiments. *P<0.05; **P<0.01; ***P<0.001; Student’s t-test.
As depletion of VPS34 caused decreased cell motility, we performed an siRNA screen to identify effectors of PtdIns3P. We used an siRNA library targeting proteins with known PtdIns3P-binding domains, the PX and FYVE domains [19], combined with a cell migration assay, that enables to screen in a 96-well format. Each well contained a cell-seeding stopper to create a migration zone in the centre of each well (Fig 1C). Control siRNA-treated cells moved into the migration zone, whereas cells where microtubule polymerization was inhibited by nocodazole, as expected, did not (Fig 1D). Results of the screen were summarized as average ratio of migrated cells per total cells in the imaged area (supplementary Fig S1A, B online). The fact that Hrs was identified as a strong hit was taken as a validation of the screen, as this protein was recently shown to promote cell migration through mediating degradation of fibronectin–integrin complexes [20]. Among the best hits for decreased cell migration were two guanine nucleotide exchange factors for the small GTPase Cdc42, FGD1 and FGD2, whose overexpression has been shown to promote invadopodia formation and cell migration [21, 22]. The atypical FYVE domain of FGD1 not only binds PtdIns3P but also the structurally related PtdIns5P [23]. Intriguingly, two other major hits in the migration screen were enzymes proposed to be involved in PtdIns5P biogenesis, MTMR3, which is a phosphoinositide 3-phosphatase specific for PtdIns3P and PtdIns(3,5)P2 and PIKfyve, which produces PtdIns(3,5)P2 from PtdIns3P. PtdIns5P is thought to be formed by dephosphorylation of PtdIns(3,5)P2 by MTMR3 [7, 24, 25] (Fig 4F), and our migration screen thus identified a possible role for PtdIns5P in cell migration.
As the siRNA screen was carried out with pools consisting of four siRNAs, we verified results in the cell migration assay using the deconvoluted siRNAs for MTMR3, PIKfyve and FGD1 and compared them with control siRNA-treated cells (Fig 1D,E). All four different MTMR3 and FGD1 siRNAs caused a decrease in number of migrated cells, concordant with the knockdown on mRNA and protein level (supplementary Fig S1C–E online). For PIKfyve, individual siRNAs gave a relatively poor knockdown as reported previously [26]. To deplete PIKfyve efficiently, we therefore performed double knockdown using two siRNA oligonucleotides from the siRNA pool and could in this way observe a significant decrease in cell migration (Fig 1D,E).
To verify the screening results further, we studied the role of PIKfyve and MTMR3 by live-cell imaging of FGF1-stimulated BJ fibroblasts in a wound healing assay. Knockdown of PIKfyve (Fig 1F) resulted in a significant decrease in cell migration velocity and persistence (Fig 1F). In addition, inhibiting PIKfyve with its specific inhibitor, YM201636, reduced cell velocity by 50% (Fig 1G). On depletion of MTMR3 (Fig 1F), BJ cells were unable to migrate into the wound and showed a significant decrease in velocity of ∼60% and a decrease in persistence (Fig 1F; supplementary Movie 1 online). Manipulating PIKfyve or MTMR3 in BJ cells did not cause any overt toxicity, which could potentially interfere with cell migration (supplementary Fig S2 online). Retransfection with siRNA-resistant EGFP-MTMR3 complementary DNA restored the control phenotype (Fig 1H; supplementary Movie 2 online). Interestingly, a point mutant of MTMR3 (C413S) without catalytic activity [24] was unable to rescue the knockdown phenotype (Fig 1H), indicating that MTMR3 acts enzymatically on phosphoinositides during cell migration. Depletion of PIKfyve or MTMR3 also inhibited the random migration of unstimulated cells (supplementary Fig S3 online). Together, these data indicate that both PIKfyve and MTMR3 are involved in cell migration.
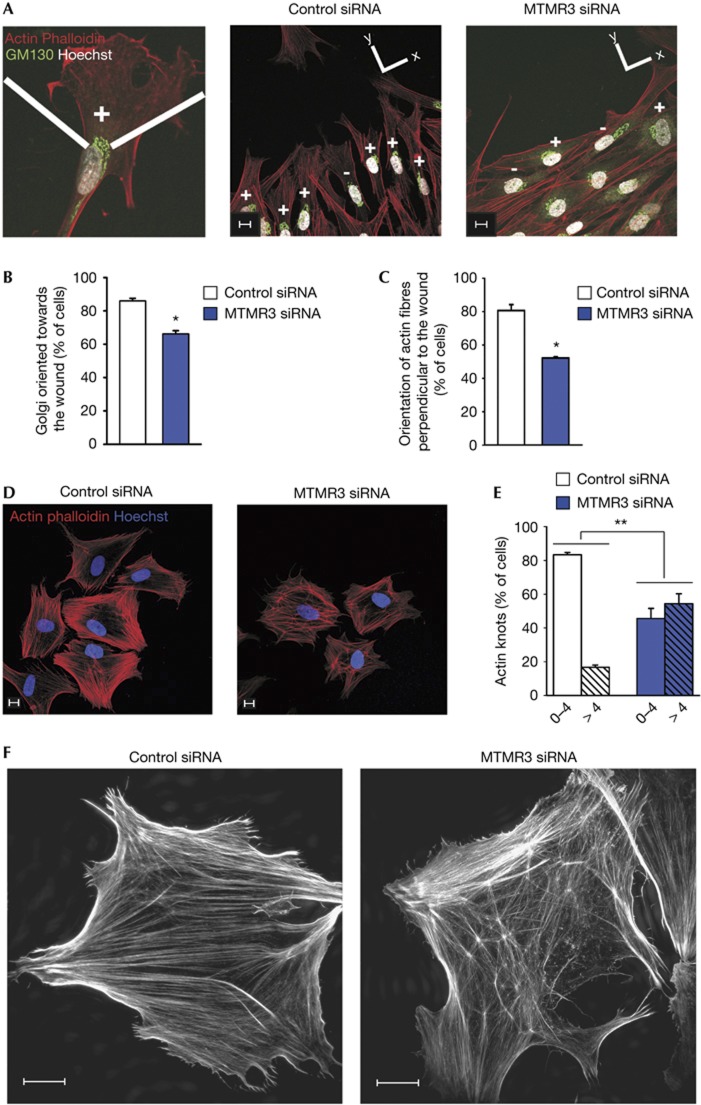
To explore the role of MTMR3 in directed cell migration, we monitored the ability of BJ cells to polarize and orientate towards a wound. The position of the Golgi can be used as a marker of cell polarization, as it reorients rapidly between the nucleus and the leading edge during directed cell migration [27]. On FGF1 stimulation, 86% of control cells oriented themselves towards the wound, whereas only 66% of MTMR3-depleted cells oriented successfully (Fig 2A,B). Consistent with this observation, we also observed that MTMR3-depleted cells were less perpendicular to the wound (Fig 2A). Quantification showed that 80% of control cells presented actin fibres perpendicular to the wound, whereas only 52% of the cells showed this on MTMR3 depletion (Fig 2C). This fits well with the observation that MTMR3-depleted cells showed decreased persistence, as cells unable to orientate their Golgi and cytoskeleton towards a signal are presumably incapable of undergoing persistent cell migration.
Figure 2.
Implication of PtdIns5P in remodelling of the actin cytoskeleton. (A) For polarization studies, control or MTMR3 siRNA-treated cells were stained as indicated. The wound is located at the top of the images. (B) Quantification of cells oriented towards the wound. Golgi apparatus staining located in a 120° angle facing the wound was scored as being oriented (+); or not oriented (−) if the majority lay outside this angle. Cells analysed in total: 100 (control siRNA); 100 (MTMR3 siRNA). (C) Quantification of actin fibre orientation. Cells oriented in 90° of the y-axis were scored as being perpendicular to the wound. Cells analysed in total: 130 (control siRNA); 130 (MTMR3 siRNA). (D) Actin cytoskeleton organization in cell-spreading assays. Quantification in (E), Cells analysed in total: 197 (control siRNA); 164 (MTMR3 siRNA). (F) Structured illumination super-resolution microscopy of actin fibres. Scale bars, 10 μm. For all panels, error bars represent the mean±s.e.m. of three independent experiments. *P<0.05; **P<0.01; ***P<0.001; Student’s t-test.
In an attempt to further investigate the role of MTMR3 and PIKfyve in cell migration, we tested whether their depletion influences the trafficking of integrins. We were unable to detect any differences in α5β1 integrin recycling (supplementary Fig S4A online). Total and cell-surface levels of α5, β1 and β5 integrins were also unchanged (supplementary Fig S4B online). In addition, α5 integrin localization remained unchanged (supplementary Fig S4C online). The integrity of focal adhesions as measured by immunostaining with antibodies against vinculin, talin and FAK, was also unchanged on depletion of MTMR3 (supplementary Fig S4D–F online).
Furthermore, to investigate the dynamics of focal adhesions, we tested the ability of depleted cells to spread on a fibronectin-coated surface, a process mediated by focal complexes. As shown in supplementary Fig S5G online, the knockdown cells spread similar to control cells. Interestingly, however, we observed a clear difference in the organization of the actin skeleton (Fig 2D). In MTMR3-depleted cells, there was a high proportion of actin ‘knots’ compared with control cells (Fig 2E). Structured illumination super-resolution microscopy revealed that these ‘knots’ could be nucleation centres from where actin fibres sprout out (Fig 2F). Although the exact nature of these structures remains to be elucidated, this observation clearly supports that PtdIns5P is implicated in the remodelling of the actin cytoskeleton. This is corroborated by the data in Fig 2A, showing that the actin fibres in knockdown cells are unable to remodel to orientate towards the wound.
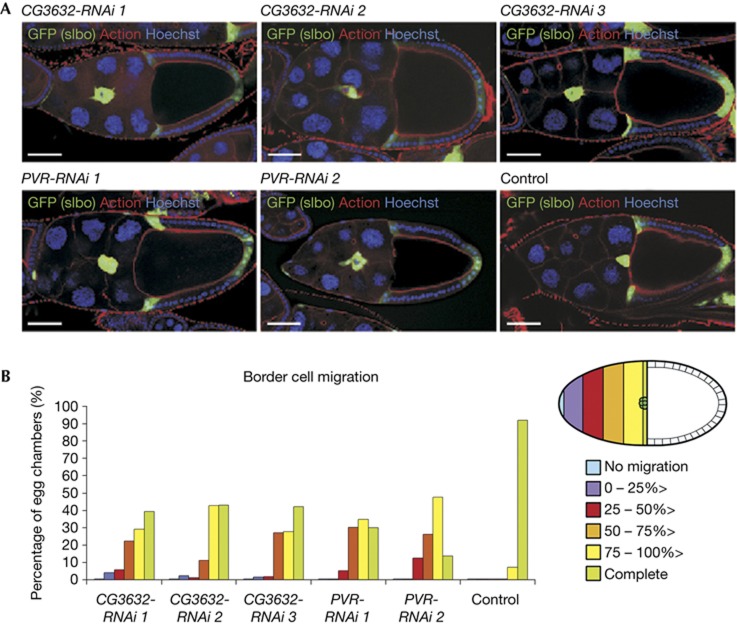
We next addressed whether MTMR3 might have a role in cell migration in vivo by studying the effect of RNAi-mediated gene silencing of the putative Drosophila melanogaster homologue of human MTMR3, Myotubularin-like (CG3632; supplementary Table S1 online), on border cell migration during Drosophila oogenesis. We expressed RNAi in border cells using the GAL4-UAS system and the driver line slbo-GAL4, UAS-GFP. RNAi against the Drosophila platelet-derived growth factor (PDGF)/vascular endothelial growth factor (VEGF)-receptor related (PVR) served as a positive control, as lack of PVR signalling is known to delay border cell migration [28]. PVR-RNAi expression in border cells using two different RNAi lines (PVR-RNAi 1-2) indeed caused a delay of their migration in more than half of the egg chambers as compared with border cells in negative control egg chambers (control), of which more than 90% had reached the oocyte at stage 10 (Fig 3A,B). Knockdown of Myotubularin-like using each of three different RNAi lines (CG3632-RNAi 1-3) also resulted in delayed border cell migration in about half of the egg chambers (Fig 3A,B). These data indicate that Myotubularin-like has a role in border cell migration in D. melanogaster and implicate an evolutionarily conserved role of this family of proteins in cell migration.
Figure 3.
The Drosophila MTMR3 homologue is required for proper border cell migration in vivo. (A) Representative early-stage 10 egg chambers expressing RNAi in border cells as indicated (CG3632, Myotubularin-like; PVR, PDGF/VEGF-receptor related) with the indicated staining. Border cell clusters express GFP (green). Scale bars, 50 μm. (B) Left: quantification of border cell migration averaged from three independent experiments, using the scale described on the right. Total egg chambers: 43 (CG3632-RNAi 1); 59 (CG3632-RNAi 2); 55 (CG3632-RNAi 3); 80 (PVR-RNAi 1); 15 (two independent experiments (PVR-RNAi 2); 70 (control). Differences are significant using analysis of variance with Tukey’s HSD correction. Right: schematic of a stage 10 egg chamber and scale used to categorize border cell migration defects. Anterior is to the left. PDGF, platelet-derived growth factor; PVR, PDGF/VEGF-receptor related; VEGF, vascular endothelial growth factor.
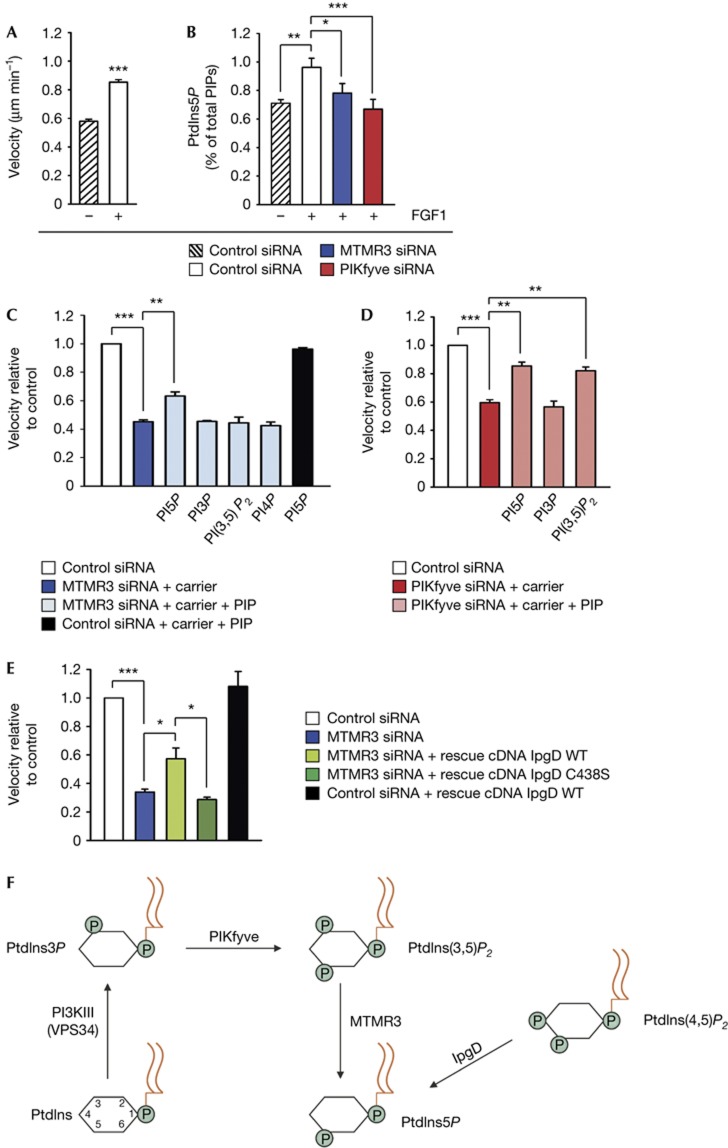
We next asked if the level of PtdIns5P was changed after depletion of either MTMR3 or PIKfyve in BJ cells. For this purpose, we extracted total cellular phosphoinositides from 3H-inositol-labelled cells, and performed HPLC analyses to measure PtdIns5P levels [5]. Interestingly, FGF1 treatment markedly increased PtdIns5P levels (Fig 4B), correlating with the higher motility of FGF1-treated BJ cells (Fig 4A). Moreover, on depletion of MTMR3 or PIKfyve, we observed a significant decrease in PtdIns5P levels consistent with the possibility that these enzymes regulate PtdIns5P production in BJ cells. A decrease in PtdIns5P levels was also observed in unstimulated cells on knockdown of MTMR3 or PIKfyve, although this was not significant (supplementary Fig S5C online). Levels of PtdIns3P and PtdIns4P were not significantly changed, neither by FGF1 treatment nor after knockdown (supplementary Fig S5A, B online). Unfortunately, PtdIns(3,5)P2 was undetectable [5].
Figure 4.
PtdIns5P promotes cell migration. (A) Stimulation of cell migration by FGF1. Quantification of BJ cell velocity in a wound healing assay without or after stimulation with FGF1 (200 ng ml−1). Cells analysed in total: 90 (without FGF1); 90 (with FGF1). (B) PtdIns5P levels (percentage of total phosphoinositides) in BJ cells stimulated with FGF1 and different siRNA treatments as indicated. Values are means±s.e.m. of five independent experiments. (C) Quantification of cell velocity on MTMR3 knockdown after treatment with exogenous phosphoinositides as indicated. Cells analysed in total: 225 (control siRNA); 225 (MTMR3 siRNA); 230 (rescue PtdIns5P); 150 (rescue PtdIns3P); 200 (rescue PtdIns(3,5)P2); 165 (rescue PtdIns4P); 145 (rescue PtdIns5P on control siRNA). (D) Quantification of cell velocity on PIKfyve knockdown after treatment with exogenous phosphoinositides as indicated. Cells analysed in total: 150 (control siRNA); 200 (PIKfyve siRNA); 170 (rescue PtdIns5P); 175 (rescue PtdIns3P); 205 (rescue PtdIns(3,5)P2). (E) Quantification of cell velocity after rescue with cDNA of the bacterial enzyme IpgD. Cells analysed in total: 65 (control siRNA); 40 (MTMR3 siRNA); 10 (rescue IpgD); 16 (rescue IpgD C438S); 23 (rescue IpgD on control siRNA). (F) Schematic representation of pathways for PtdIns5P synthesis. The production of PtdIns5P by the bacterial enzyme IpgD is also indicated. For all panels, error bars represent the mean±s.e.m. of three independent experiments. *P<0.05; **P<0.01; ***P<0.001; Student’s t-test.
To directly assess the role of PtdIns5P in cell migration, we performed rescue experiments with PtdIns5P by two alternative approaches. To increase cellular PtdIns5P levels in MTMR3 or PIKfyve-depleted cells, we added exogenous PtdIns5P di-C16 (and PtdIns(3,5)P2 di-C16 in the case of PIKfyve depletion) in combination with a carrier to cells. We observed a significant increase in cell migration velocity compared with depleted cells (Fig 4C,D). The knockdown phenotype was not fully restored to control levels, which could be due to a suboptimal spatial distribution of externally added PtdIns5P. Addition of other phosphoinositides did not cause any differences in velocity compared with knockdown cells, demonstrating the specific ability of PtdIns5P to rescue cell velocity after depletion of MTMR3 or PIKfyve. Addition of PtdIns5P to control cells did not affect cell migration (Fig 4C). To confirm a possible role for endogenous PtdIns5P in cell migration, we transfected MTMR3-depleted cells with IpgD, a bacterial PtdIns(4,5)P2 4-phosphatase that increases the level of PtdIns5P in the cell [15]. Compared with the MTMR3-depleted cells, IpgD-transfected cells showed an increase in migration velocity, whereas the inactive mutant IpgD C438S did not (Fig 4E). Expression of IpgD in control cells did not significantly change cell velocity. Taken together, these experiments provide direct evidence that PtdIns5P promotes cell migration.
In conclusion, we have shown here that all members of a phosphoinositide pathway for cellular PtdIns5P production (Fig 4F), starting with PI3KIII, followed by PIKfyve and MTMR3 catalytic activities, are involved in cell migration. These data, combined with the effects of exogenous PtdIns5P and IpgD, therefore, suggest a new role for the recently discovered phosphoinositide PtdIns5P in cell migration. Interestingly, MTMR3 binds to PtdIns5P through its PH-GRAM domain [29] and thus creates a positive feedback loop [30], possibly producing local PtdIns5P domains on membranes that, in turn, could recruit other PtdIns5P effectors regulating cell migration. The precise localization of PtdIns5P is at present not clear due to the lack of specific probes. Using a cell fractionation approach, it was previously shown to be most abundant in the plasma membrane [5], and it is therefore possible that MTMR3 and PIKfyve modulate PtdIns5P levels there to regulate cell migration.
Methods
Time-lapse live-cell imaging and cell tracking. BJ cells plated on Glass Bottom Dishes (MatTek Corporation) were observed with a BioStation IM Live Cell Recorder (Nikon Instruments Inc.). In all experiments, cells were stimulated with FGF1 (200 ng ml−1; prepared as previously described [31]) and heparin (20 U ml−1; Sigma-Aldrich). Image acquisition was done every 10 min. For comparison of maximum velocities, we analysed for the period after stimulation with FGF1 for 7 h, that is, the last 3 h. Images were analysed with ImageJ software with Manual Tracking and Chemotaxis and Migration Tool (ibidi GmbH) plugins. Persistence is calculated by dividing the euclidean distance by the accumulated distance of the cell trajectories.
Border cell migration assay during D. melanogaster oogenesis. For RNAi-mediated gene silencing in border cells, the GAL4-UAS system was used to express RNAi transgenes. The GAL4 strain used was slbo-GAL4, UAS-GFP.
Metabolic labelling and analysis of phosphoinositides. BJ cells were metabolically labelled with 10 μCi ml−1 [3H]inositol (American Radiolabeled Chemicals Inc.) for 48 h in inositol-free DMEM (MP Biomedicals, LLC) supplemented with dialysed fetal bovine serum (GIBCO) and 200 mM L-glutamine (Invitrogen). Before extracting cellular phosphoinositides, cells were incubated with heparin alone (20 U ml−1) or FGF1 (200 ng ml−1) and heparin (20 U ml−1) for 2 h. Extraction, deacylation and separation by anion-exchange HPLC were done as previously described [5].
Addition of exogenous PtdIns5P to cells. The PtdIns5P/PtdIns3P/PtdIns4P or PtdIns(3,5)P2 Shuttle PIP Kit (Echelon Biosciences) were used. The final concentration of the lipid was 50 μM
A description of extra reagents and detailed protocols are available in the supplementary information online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank M.J. Clague and B. Payrastre for providing plasmids. J. Jensen and P. Shepherd are acknowledged for providing the PIKfyve inhibitor. We acknowledge the Vienna Drosophila RNAi Center (VDRC), Fly stocks of National Institute of Genetics (NIG-Fly) and Bloomington Stock Centre for Drosophila stocks. A.O. was supported by a grant from Norway through the Norwegian Financial Mechanism (PNRF-87-AI-1/07). J.W and K.H. hold senior scientist grants from the South-Eastern Norway Regional Health Authority. H.S. holds an Advanced Grant from the European Research Council. This work was supported by the Norwegian Cancer Society.
Author contributions: A.O. and J.W. conceived and designed the experiments with input from the co-authors. A.O. executed the siRNA screen with the help of N.M.P. A.O. did siRNA knockdown, pull-down, western blotting, rescue experiments and live-cell microscopy. V.H.L. performed immunofluorescence and confocal microscopy for polarization and actin studies, spreading assays and integrin recycling experiments. K.H. performed the Drosophila experiments. A.M.M. and L.E.R. performed phosphoinositide analyses on samples prepared by A.O. K.L. performed statistical analysis. K.O.S. did structured illumination microscopy. A.B. performed electron microscopy. T.E.R., E.M.W. and E.M.H. contributed to the experiments. A.O., V.H.L., K.H., H.S. and J.W. wrote the manuscript with input from the co-authors.
Footnotes
The authors declare that they have no conflict of interest.
References
- Di Paolo G, De Camilli P (2006) Phosphoinositides in cell regulation and membrane dynamics. Nature 443: 651–657 [DOI] [PubMed] [Google Scholar]
- Di Paolo G et al. (2002) Recruitment and regulation of phosphatidylinositol phosphate kinase type 1 gamma by the FERM domain of talin. Nature 420: 85–89 [DOI] [PubMed] [Google Scholar]
- Ling K, Doughman RL, Firestone AJ, Bunce MW, Anderson RA (2002) Type I gamma phosphatidylinositol phosphate kinase targets and regulates focal adhesions. Nature 420: 89–93 [DOI] [PubMed] [Google Scholar]
- Rameh LE, Tolias KF, Duckworth BC, Cantley LC (1997) A new pathway for synthesis of phosphatidylinositol-4,5-bisphosphate. Nature 390: 192–196 [DOI] [PubMed] [Google Scholar]
- Sarkes D, Rameh LE (2010) A novel HPLC-based approach makes possible the spatial characterization of cellular PtdIns5P and other phosphoinositides. Biochem J 428: 375–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JB, Hinchliffe KA, Ciruela A, Letcher AJ, Irvine RF (2000) Thrombin stimulation of platelets causes an increase in phosphatidylinositol 5-phosphate revealed by mass assay. FEBS Lett 475: 57–60 [DOI] [PubMed] [Google Scholar]
- Sbrissa D, Ikonomov OC, Deeb R, Shisheva A (2002) Phosphatidylinositol 5-phosphate biosynthesis is linked to PIKfyve and is involved in osmotic response pathway in mammalian cells. J Biol Chem 277: 47276–47284 [DOI] [PubMed] [Google Scholar]
- Guittard G et al. (2009) Cutting edge: Dok-1 and Dok-2 adaptor molecules are regulated by phosphatidylinositol 5-phosphate production in T cells. J Immunol 182: 3974–3978 [DOI] [PubMed] [Google Scholar]
- Sbrissa D, Ikonomov OC, Strakova J, Shisheva A (2004) Role for a novel signaling intermediate, phosphatidylinositol 5-phosphate, in insulin-regulated F-actin stress fiber breakdown and GLUT4 translocation. Endocrinology 145: 4853–4865 [DOI] [PubMed] [Google Scholar]
- Clarke JH, Letcher AJ, D’Santos CS, Halstead JR, Irvine RF, Divecha N (2001) Inositol lipids are regulated during cell cycle progression in the nuclei of murine erythroleukaemia cells. Biochem J 357: 905–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DR et al. (2006) Nuclear PtdIns5P as a transducer of stress signaling: an in vivo role for PIP4Kbeta. Mol Cell 23: 685–695 [DOI] [PubMed] [Google Scholar]
- Grainger DL, Tavelis C, Ryan AJ, Hinchliffe KA (2011) Involvement of phosphatidylinositol 5-phosphate in insulin-stimulated glucose uptake in the L6 myotube model of skeletal muscle. Pflugers Arch 462: 723–732 [DOI] [PubMed] [Google Scholar]
- Gozani O et al. (2003) The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell 114: 99–111 [DOI] [PubMed] [Google Scholar]
- Coronas S, Lagarrigue F, Ramel D, Chicanne G, Delsol G, Payrastre B, Tronchère H (2008) Elevated levels of PtdIns5P in NPM-ALK transformed cells: implication of PIKfyve. Biochem Biophys Res Commun 372: 351–355 [DOI] [PubMed] [Google Scholar]
- Niebuhr K, Giuriato S, Pedron T, Philpott DJ, Gaits F, Sable J, Sheetz MP, Parsot C, Sansonetti PJ, Payrastre B (2002) Conversion of PtdIns(4,5)P(2) into PtdIns(5)P by the S.flexneri effector IpgD reorganizes host cell morphology. EMBO J 21: 5069–5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendaries C, Tronchere H, Arbibe L, Mounier J, Gozani O, Cantley L, Fry MJ, Gaits-Iacovoni F, Sansonetti PJ, Payrastre B (2006) PtdIns5P activates the host cell PI3-kinase/Akt pathway during Shigella flexneri infection. EMBO J 25: 1024–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason D, Mallo GV, Terebiznik MR, Payrastre B, Finlay BB, Brumell JH, Rameh L, Grinstein S (2007) Alteration of epithelial structure and function associated with PtdIns(4,5)P2 degradation by a bacterial phosphatase. J Gen Physiol 129: 267–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel D, Lagarrigue F, Pons V, Mounier J, Dupuis-Coronas S, Chicanne G, Sansonetti PJ, Gaits-Iacovoni F, Tronchère H, Payrastre B (2011) Shigella flexneri infection generates the lipid PI5P to alter endocytosis and prevent termination of EGFR signaling. Sci Signal 4: ra61. [DOI] [PubMed] [Google Scholar]
- Sagona AP, Nezis IP, Pedersen NM, Liestol K, Poulton J, Rusten TE, Skotheim RI, Raiborg C, Stenmark H (2010) PtdIns(3)P controls cytokinesis through KIF13A-mediated recruitment of FYVE-CENT to the midbody. Nat Cell Biol 12: 362–371 [DOI] [PubMed] [Google Scholar]
- Lobert VH, Brech A, Pedersen NM, Wesche J, Oppelt A, Malerod L, Stenmark H (2010) Ubiquitination of alpha 5 beta 1 integrin controls fibroblast migration through lysosomal degradation of fibronectin-integrin complexes. Dev Cell 19: 148–159 [DOI] [PubMed] [Google Scholar]
- Ayala I, Giacchetti G, Caldieri G, Attanasio F, Mariggio S, Tete S, Polishchuk R, Castronovo V, Buccione R (2009) Faciogenital dysplasia protein Fgd1 regulates invadopodia biogenesis and extracellular matrix degradation and is up-regulated in prostate and breast cancer. Cancer Res 69: 747–752 [DOI] [PubMed] [Google Scholar]
- Hayakawa M, Matsushima M, Hagiwara H, Oshima T, Fujino T, Ando K, Kikugawa K, Tanaka H, Miyazawa K, Kitagawa M (2008) Novel insights into FGD3, a putative GEF for Cdc42, that undergoes SCF(FWD1/beta-TrCP)-mediated proteasomal degradation analogous to that of its homologue FGD1 but regulates cell morphology and motility differently from FGD1. Genes Cells 13: 329–342 [DOI] [PubMed] [Google Scholar]
- Sankaran VG, Klein DE, Sachdeva MM, Lemmon MA (2001) High-affinity binding of a FYVE domain to phosphatidylinositol 3-phosphate requires intact phospholipid but not FYVE domain oligomerization. Biochemistry 40: 8581–8587 [DOI] [PubMed] [Google Scholar]
- Walker DM, Urbe S, Dove SK, Tenza D, Raposo G, Clague MJ (2001) Characterization of MTMR3. an inositol lipid 3-phosphatase with novel substrate specificity. Curr Biol 11: 1600–1605 [DOI] [PubMed] [Google Scholar]
- Lecompte O, Poch O, Laporte J (2008) PtdIns5P regulation through evolution: roles in membrane trafficking? Trends Biochem Sci 33: 453–460 [DOI] [PubMed] [Google Scholar]
- Rutherford AC, Traer C, Wassmer T, Pattni K, Bujny MV, Carlton JG, Stenmark H, Cullen PJ (2006) The mammalian phosphatidylinositol 3-phosphate 5-kinase (PIKfyve) regulates endosome-to-TGN retrograde transport. J Cell Sci 119: 3944–3957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupfer A, Louvard D, Singer SJ (1982) Polarization of the Golgi apparatus and the microtubule-organizing center in cultured fibroblasts at the edge of an experimental wound. Proc Natl Acad Sci USA 79: 2603–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchek P, Somogyi K, Jekely G, Beccari S, Rorth P (2001) Guidance of cell migration by the Drosophila PDGF/VEGF receptor. Cell 107: 17–26 [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Urbe S, Clague MJ (2005) Analysis of phosphoinositide binding domain properties within the myotubularin-related protein MTMR3. J Cell Sci 118: 2005–2012 [DOI] [PubMed] [Google Scholar]
- Schaletzky J, Dove SK, Short B, Lorenzo O, Clague MJ, Barr FA (2003) Phosphatidylinositol-5-phosphate activation and conserved substrate specificity of the myotubularin phosphatidylinositol 3-phosphatases. Curr Biol 13: 504–509 [DOI] [PubMed] [Google Scholar]
- Nilsen T, Rosendal KR, Sorensen V, Wesche J, Olsnes S, Wiedlocha A (2007) A nuclear export sequence located on a beta-strand in fibroblast growth factor-1. J Biol Chem 282: 26245–26256 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.