Abstract
Unlike other Salmonella, which can infect a broad range of hosts causing self-limiting infection, Salmonella Typhi is an exclusively human pathogen that causes typhoid fever, a life-threatening systemic disease. Typhoid toxin is a unique virulence factor of Salmonella Typhi, which is expressed when the bacteria are within mammalian cells. Here, we report that an N-acetyl-β-D-muramidase similar to phage endolysins encoded within the same pathogenicity islet as the toxin is required for typhoid toxin secretion. Genetic and functional analysis of TtsA revealed unique amino acids at its predicted peptidoglycan-binding domain that are essential for protein secretion and that distinguishes this protein from other homologues. We propose that TtsA defines a new protein secretion mechanism recently evolved from the machine that mediates phage release.
Keywords: bacterial toxins, protein secretion, typhoid fever, pathogen evolution, holins
Introduction
Salmonella enterica serovar Typhi (S. Typhi) is the cause of typhoid fever, which kills an estimated 200,000 people every year 1], [2, 3]. In contrast to other S. enterica serovars, S. Typhi can only infect humans causing a life-threatening systemic illness [1, 4]. The molecular bases for these remarkable differences in the disease presentation and host range are poorly understood although it is believed that they are the result of genome reduction and the acquisition of unique genes [5, 6]. One of the few virulence factors uniquely present in S. Typhi is typhoid toxin, a remarkable AB5 toxin with similarities to pertussis and cytolethal distending toxins [7, 8, 9]. Typhoid toxin has a unique biology in that it is only expressed when S. Typhi is within mammalian host cells [8, 10]. After its synthesis and secretion from the bacteria, the toxin is transported by carrier intermediates from the Salmonella-containing vacuole to the extracellular space, from where it gains access to target cells via autocrine or paracrine routes [7, 11]. The mechanisms by which the toxin is secreted from the bacteria are unknown. The three components of typhoid toxin have canonical secretion signals, implying that, like other AB5 toxins, the Sec machinery exports the toxin components to the periplasm where they assemble into a holotoxin. However, there is no information on how the holotoxin gets out of the bacteria before its packaging into transport carriers. Here, we report that an N-acetyl-β-D-muramidase similar to phage endolysins encoded within the same pathogenicity islet as the toxin is required for typhoid toxin secretion. We identified a critical region at the carboxy-terminal peptidoglycan (PG)-binding region of this enzyme that is necessary to carry out protein secretion functions. We propose that this muramidase is the result of the evolutionary adaptation of a phage enzyme to conform a new protein secretion mechanism. Finally, we present bioinformatics evidence that suggests that the protein secretion mechanism described here is widespread in bacteria.
Results and discussion
ttsA is required for typhoid toxin secretion
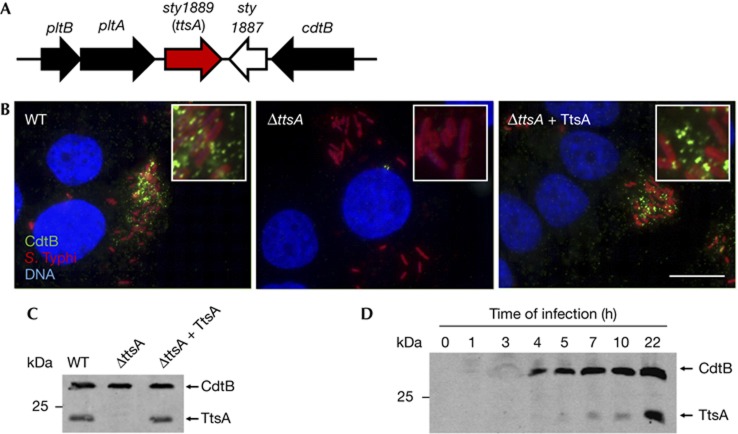
The genomic islet that encodes typhoid toxin also contains two other uncharacterized genes, sty1887 and sty1889 (Fig 1A). As functionally related genes are often encoded in close proximity to one another, we investigated the potential involvement of these genes in typhoid toxin secretion. Typhoid toxin secretion cannot be studied in vitro because its components are not expressed [7, 8, 9]. Therefore, we investigated toxin secretion by immunofluorescence staining of CdtB, a typhoid toxin subunit, in S. Typhi-infected cultured epithelial cells. Previous studies have shown that several hours after infection, typhoid toxin can be seen in puncta radiating from the S. Typhi-containing vacuole and spreading throughout the entire cell [7]. Therefore, we used the appearance of these puncta as an assay to measure typhoid toxin secretion from the bacterial cell. Cells infected with a S. Typhi mutant strain carrying a deletion of sty1887 showed a staining pattern that was indistinguishable from that of cells infected with wild type (supplementary Fig S1 online). In contrast, cells infected with an S. Typhi strain carrying a deletion in sty1889 showed no CdtB puncta staining (Fig 1B and supplementary Figs S2 and S3 online), although the CdtB expression levels in the mutant were indistinguishable from those of wild type (Fig 1C). Introduction of a plasmid encoding sty1889 effectively restored puncta staining (Fig 1B,C and supplementary Fig S3 online). Expression of the periplasmic protein MalE in intracellular S. Typhi did not result in its detection on either the bacterial surface or in puncta after equally processing the samples for staining, indicating that the detection of CdtB in wild-type intracellular bacteria was not owing to nonspecific leakage of periplasmic proteins or bacterial lysis (supplementary Fig S4 online). These results indicate that Sty1889 is required for toxin secretion from the bacterial cell, and therefore we renamed it ttsA (for typhoid toxin secretion A). To examine whether ttsA was co-regulated with the typhoid toxin genes, we tested its pattern of expression during infection. We found that, like typhoid toxin [7, 8], TtsA was not detected when S. Typhi was grown in L-broth. However, TtsA was detected within cultured epithelial cells (Fig 1D) although it required slightly longer infection times than those required to detect typhoid toxin (Fig 1D). Taken together, these results indicate that TtsA is required for toxin secretion, and that, consistent with this function, its expression correlated with that of typhoid toxin.
Figure 1.
TtsA is required for typhoid toxin secretion. (A) Schematic representation of the S. Typhi pathogenicity islet encoding typhoid toxin. (B) Sty1889 (TtsA) is required for typhoid toxin secretion. Henle-407 cells were infected with S. Typhi expressing chromosomally encoded 3 × FLAG-epitope-tagged CdtB, a ΔttsA isogenic mutant, or the complemented ΔttsA mutant. Twenty-two hours after infection, cells were stained with an antibody directed to the FLAG epitope (green) (to visualize CdtB), a rabbit antibody directed to S. Typhi LPS (red) and DAPI for DNA detection (blue). The bar represents 10 μm. (C) Western blot analysis of the strains used in panel (B). (D) TtsA expression profile during bacterial infection of cultured cells. At the indicated times after infection, cells were lysed and the levels of CdtB and TtsA were examined in CFU standardized samples by western blot analysis. CFU, colony-forming units; DAPI, 4,6-diamidino-2-phenylindole; LPS, lipopolysaccharide; WT, wild-type.
TtsA belongs to a family of bacteriophage muramidases
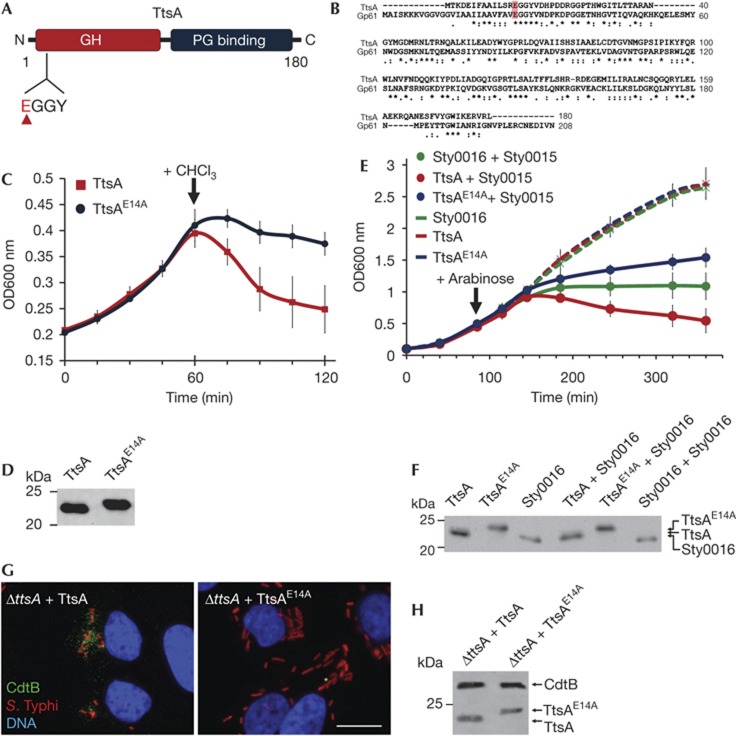
Stojkovic and Rothman–Denes have previously reported that the coliphage N4 Gp61 N-acetylmuramidase defines a new family of bacteriophage muramidases and noticed the presence of homologues of this protein family in S. enterica including sty1889 (which we now have renamed ttsA) [12]. This protein family is composed of a amino-terminal lysozyme-like domain belonging to glycoside hydrolase family 108, and a C-terminal putative PG-binding domain (Fig 2A,B) [13]. Most members of this family, including ttsA, lack an identifiable N-terminal sec-dependent secretion signal and are delivered to the periplasm by holins, a diverse group of small membrane proteins that are most often encoded immediately adjacent to the muramidases [14]. In a regulated fashion, holins form a pore in the inner membrane that allows the passage of the murein-lytic enzymes. Interestingly, in the vicinity of ttsA there are no genes that could encode a protein with features similar to those expected in a holin (a small protein with one or two transmembrane domains). Despite repeated efforts, we were not able to demonstrate in vitro muramidase activity with purified TtsA using standard zymography. Similar observations have been previously made for other members of this protein family [12]. Therefore, we investigated its putative muramidase activity using alternative indirect approaches. It has been observed that in the absence of holins (or in the absence of an activating signal that would trigger their activity), this family of muramidases is still capable of inducing bacterial lysis when overexpressed and after addition of low concentrations of chloroform [12]. It is thought that chloroform destabilizes the inner membrane allowing the ‘leakage’ of the cytoplasmic muramidases into the periplasmic space. Consistent with its putative muramidase activity, TtsA was capable of inducing bacterial lysis under these conditions (Fig 2C,D). Although each muramidase exerts its function together with a holin encoded in its immediate vicinity, some holins show little specificity and are indeed capable of transporting unrelated muramidases [15]. Therefore to further test TtsA’s putative muramidase activity, we co-expressed it with Sty0015, a holin for the adjacently encoded muramidase Sty0016, both encoded distantly from the typhoid toxin locus [12]. We found that co-expression of TtsA and Sty0015 led to rapid bacterial lysis (Fig 2E,F). In contrast, expression of TtsA alone did not. These results further demonstrate that TtsA exhibits muramidase activity and suggest that TtsA is likely to exert its activity in conjunction with a co-regulated holin encoded elsewhere in the S. Typhi genome. Such holin is not Sty0015 as we found wild-type toxin secretion in a Δsty0015 S. Typhi mutant (supplementary Figure S5 online).
Figure 2.
The TtsA enzymatic activity is required for toxin secretion. (A) TtsA domain organization. Indicated are the locations of the GH and PG-binding domains, as well as the catalytic site (EGGY). (B) Amino-acid sequence alignment of TtsA and coliphage N4 Gp61 muramidase. The conserved catalytic glutamate is highlighted. (C) Bacteriolytic activity of TtsA. FLAG-epitope tagged TtsA or TtsAE14A were overexpressed in S. Typhi ΔttsA mutant and at the indicated time 0.3% chloroform (CHCl3). Bacterial lysis was monitored by measuring the OD600 nm of the bacterial cultures. The graph shows the average and s.d.’s of six independent assays. (D) Western blot analysis of the expression levels of the indicated proteins in the strains used in panel (C) in aliquots obtained immediately before adding CHCl3. (E) Holin-assisted bacteriolysis by TtsA. FLAG-tagged TtsA, TtsAE14A or Sty0016 were expressed in a ΔttsA S. Typhi mutant from an arabinose-inducible promoter either by themselves, or along with the Sty0015 holin. At the indicated time, arabinose was added to induce holin/endolysin expression and bacterial lysis monitored by measuring the OD600 nm of the different cultures. The graph shows the average and s.d.’s of six independent assays. (F) Western blot analysis of the expression levels of the different endolysins in the strains used in panel (E) 20 min following the induction with arabinose. (G) The TtsA catalytic activity is required for toxin secretion. Henle-407 cells were infected with a ΔttsA S. Typhi mutant expressing chromosomally encoded FLAG-tagged cdtB complemented with either wild-type TtsA or the catalytic mutant TtsAE14A. Twenty-two hours post infection, infected cells were fixed and stained with a mouse monoclonal antibody directed to the FLAG epitope (to visualize CdtB), a rabbit antibody directed to S. Typhi LPS and DAPI for DNA detection. The bar represents 10 μm. (H) Protein levels in CFU standardized lysates of strains used in panel (G) 22 h after infection of Henle-407 cells. CFU, colony forming units; DAPI, 4,6-diamidino-2-phenylindole; GH, glycosyl hydrolase; LPS, lipopolysaccharide; PG, peptidoglycan; s.d., standard deviation.
It has been previously reported that the muramidase activity of the TtsA protein family is associated with a catalytic glutamic acid located within a conserved EGGY motif located close to their N terminus [12]. Mutation in TtsA of a glutamic acid residue within this motif (TtsAE14A) (Fig 2A,B) abolished its ability to induce bacterial lysis either on addition of chloroform (Fig 2C,D) or after its co-expression with the Sty0015 holin (Fig 2E,F). These results further support the notion that TtsA is a muramidase. Inducible expression of a mutant TtsA containing a sec signal from DsbA at its N terminus also led to bacterial lysis. However, we were not able to establish experimental conditions with which we could consistently observe differences between wild-type and the catalytic mutant. We then tested the requirement of the predicted TtsA muramidase activity for typhoid toxin secretion. Consistent with the requirement of its predicted enzymatic activity for toxin secretion, no typhoid toxin puncta were detected in cells infected with a S. Typhi strain expressing TtsAE14A (Fig 2G,H). Taken together, these results indicate that TtsA is a muramidase and that this enzymatic activity is required for typhoid toxin secretion or release.
Functional specificity of the TtsA protein family
To investigate whether the amino-acid sequence of TtsA could be correlated with its specific adaptation to mediate typhoid toxin secretion, we generated a phylogenetic tree of TtsA homologues presenting at least 50% identity (supplementary Fig S6 online). In addition, we use the PHAge Search Tool (PHAST) to analyse the loci of all the homologues represented in the tree to identify possible prophages. The different muramidases clustered into three large groups in the phylogenetic tree, which we arbitrarily refer to as group 1 (which harbours TtsA), 2 and 3. As expected for this family of muramidases, many of the TtsA homologues were encoded within the context of complete or incomplete phage genomes (supplementary Fig S6 online). However, no phage-related genes were detected in the vicinity of a significant number of family members suggesting a much more diverse set of function for this protein family. Although muramidases associated with phage genomes were found in the three phylogenetic groups, there were fewer phage-associated muramidases within group 2. Whether this observation suggests a specific function associated with this subgroup of muramidases is unknown. Nevertheless, TtsA, which most likely does not have a phage-associated function, did not cluster in this group, although Zymomonas mobilis ZlyS, which has been proposed to activate the secretion of an extracellular levansucrase and invertase [16], did.
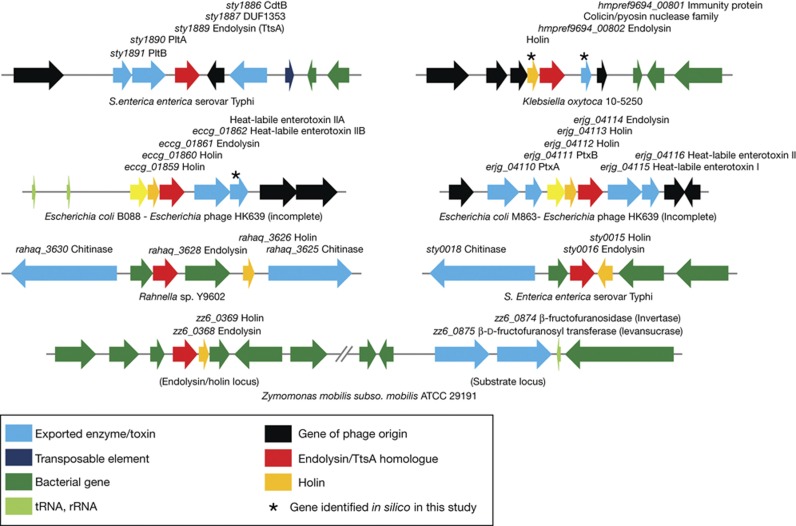
Consistent with the most common mechanism associated with the transport of these cytoplasmic muramidases through the inner membrane, we were able to identify holins encoded immediately adjacent to most of the muramidases we analysed (supplementary Table S1 online). Interestingly, an exception was ttsA, as we were not able to identify in its vicinity any protein that shows the expected features of a putative holin, suggesting that TtsA might be transported by a holin encoded elsewhere in the chromosome that is co-expressed with typhoid toxin. Of note, in several instances, we found toxins or predicted extracellular enzymes encoded immediately adjacent to muramidase/holin pairs, including a chitinase encoded in the vicinity of the S. Typhi sty0015/sty0016 holin/endolysin pair (Fig 3 and supplementary Table S1 online). This finding suggests that the mechanism of secretion described here might not be unique to typhoid toxin but, rather, widespread in bacteria. In most of these cases, no phage sequences were detected in these loci suggesting that, like in the case of TtsA, the holin/muramidase pairs have been coopted from their phage origins to carry out protein secretion functions.
Figure 3.
Genomic organization of loci showing the presence of toxin or extracellular enzymes in the vicinity endolysin/holin pairs. rRNA, ribosomal RNA; tRNA, transfer RNA.
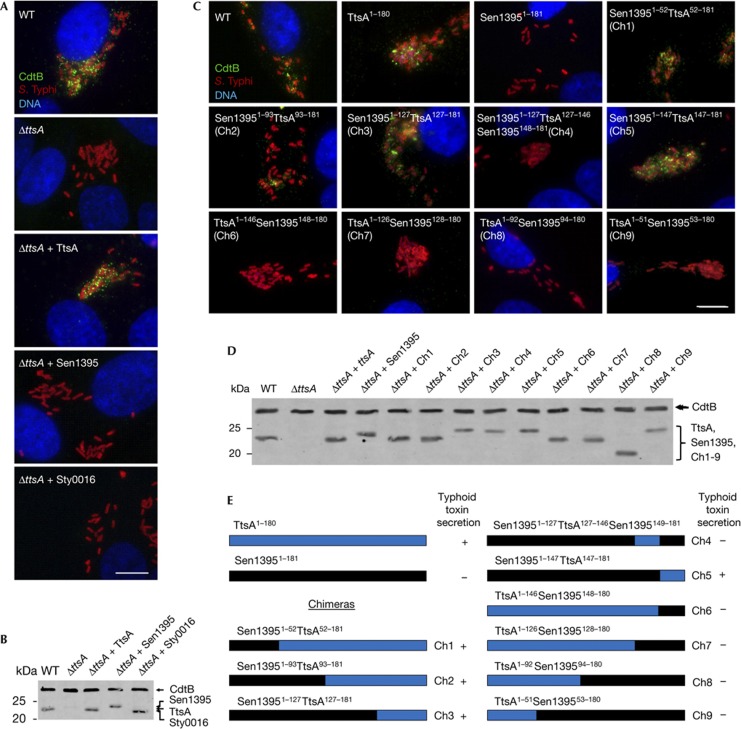
To gain insight into possible functional specificity determinants within this protein family, we tested whether members of other phylogenetic groups could complement a ΔttsA S. Typhi mutant. We chose a representative member of the phylogenetic groups encoded by S. enterica to avoid potential expression and/or codon usage issues. We found that neither S. Typhi Sty0016 nor S. Enteritidis Sen1395 belonging to groups 2 and 3, respectively, were able to complement a ΔttsA S. Typhi mutant for toxin secretion (Fig 4A,B), despite the fact that both proteins are likely to be enzymatically active as, like TtsA, their overexpression resulted in bacterial lysis (supplementary Fig S7 online). These results indicate that despite the high structural similarity, these muramidases are adapted to carry out specific functions.
Figure 4.
Identification of functional specificity determinants in TtsA. (A) Functional complementation analysis of TtsA homologues. Henle-407 cells were infected with WT S. Typhi or a ΔttsA mutant derivative expressing chromosomally encoded FLAG-tagged cdtB and carrying plasmids encoding TtsA, Sty0016 or Sen1395. Twenty-two hours post infection, infected cells were fixed and stained with a mouse monoclonal antibody directed to the FLAG epitope (to visualize CdtB), a rabbit antibody directed to S. Typhi LPS and DAPI for DNA detection. The bar represents 10 μm. (B) Western blot analysis of the expression levels of the indicated proteins in the strains used in panel (A). (C) Ability of TtsA-Sen1395 chimeras to complement a S. Typhi ΔttsA mutant for typhoid toxin secretion. Henle-407 cells were infected with wild-type (WT) S. Typhi or a ΔttsA mutant derivative expressing chromosomally encoded FLAG-tagged cdtB and carrying a plasmid encoding the indicated chimeric proteins. Twenty-two hours post infection, infected cells were fixed and stained with a mouse monoclonal antibody directed to the FLAG epitope (to visualize CdtB), a rabbit antibody directed to S. Typhi LPS and DAPI for DNA detection. The bar represents 10 μm. (D) Western blot analysis of the expression levels of the indicated proteins in the strains used in (C). (E) Schematic representation and summary of the typhoid toxin secretion phenotype of the chimeras analysed in panels (C) and (D). DAPI, 4,6-diamidino-2-phenylindole; LPS, lipopolysaccharide; WT, wild-type.
The C-terminus of TtsA determines secretion function
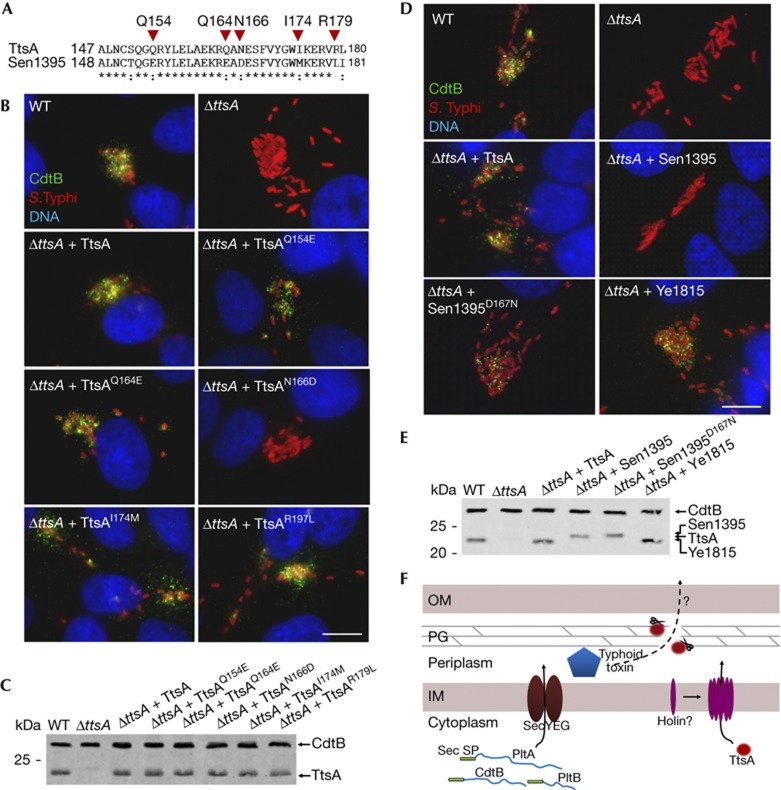
To identify determinants within TtsA that are important to carry out protein secretion functions, we examined the ability of a series of chimeras between TtsA and the non-complementing muramidase Sen1395 to complement a S. Typhi ΔttsA mutant for typhoid toxin secretion (Fig 4C,D). This protein family is composed of a glycosyl hydrolase (GH) domain at the N terminus and a PG-binding domain at the C terminus (supplementary Fig S8 online). We replaced the GH domain of TtsA for an equivalent domain from Sen1395. The resulting chimera, as well as other chimeras carrying smaller segments of this domain, was able to complement the S. Typhi ΔttsA mutant (Fig 4C–E). These results indicate that the specificity determinants that allow TtsA to carry out its specialized function must lie within the PG-binding domain. Consistent with this hypothesis, a chimera consisting of the GH domain from TtsA and the PG-binding domain from Sen1395 did not complement the S. Typhi ΔttsA mutant despite the fact that it was enzymatically active (supplementary Fig S9 online) and exhibited wild-type levels of chimeric protein expression (Fig 4C–E and supplementary Fig S9 online). To narrow down the specificity region, we constructed more chimeras with an increasing proportion of the N terminus of Sen1395 including portions of its PG-binding domain. We found that a chimera consisting of all but the last 32 amino acids of Sen1395 was able to complement the S. Typhi ΔttsA mutant (Fig 4C–E and supplementary Fig S9 online), indicating that the specificity determinants that confer TtsA its ability to carry out typhoid toxin secretion function must lie within its last 32 amino acids.
We compared the amino-acid sequence of this region of TtsA with the equivalent region of Sen1395 and identified several amino acids that were not conserved between these two muramidases and therefore potentially important for TtsA function (Fig 5A). We tested this hypothesis by changing these residues in TtsA into the equivalent residues present in Sen1395. We found that changing Q154, Q164, I174 and R179 into the corresponding amino acids present in Sen1395 did not affect TtsA function (Fig 5B,C). However, a TtsA mutant, in which asparagine 166 was changed into aspartic acid (present in Sen1395), was unable to complement the S. Typhi ΔttsA mutant for typhoid toxin secretion (Fig 5B,C). Furthermore, we found that, remarkably, simply changing the aspartic acid residue in Sen1395 into asparagine (present in TtsA at that position) allowed this muramidase to restore typhoid toxin secretion to a S. Typhi ΔttsA mutant (Fig 4D,E). Overexpression of TtsAN166D (unable to complement the ΔttsA mutant for typhoid toxin secretion) led to S. Typhi lysis after chloroform treatment (supplementary Fig S10 online), indicating that introduction of these mutations did not affect its enzymatic activity. These results not only indicate that N166 is critical for TtsA function but suggest that remarkably minor changes in the PG-binding domain of members of this muramidase family can result in significant changes in their function and specificity, presumably by altering their targeting mechanisms. We examined other muramidases for the presence of N166 and found that this amino acid is present in many members of the family. We expressed one of these members, the Yersinia enterocolitica Ye1815, in the S. Typhi ΔttsA mutant strain and found that it was able to complement this strain for typhoid toxin secretion (Fig 5D,E), consistent with the importance of N166 in conferring these muramidases the ability to function in protein secretion. However, Sty0016, which has the N166 residue, was unable to complement the S. Typhi ΔttsA mutant strain for typhoid toxin secretion (Fig 4A) despite the fact that its overexpression in S. Typhi indicated that it is enzymatically active (supplementary Fig S10 online). This finding indicates that in addition to N166, other residues must contribute to the TtsA specificity. Indeed, structure modelling of this region predicts that N166 is located within a loop bounded by two well-organized helixes (supplementary Fig S11 online). It is therefore possible that the overall configuration of the loop might be important for targeting. Interestingly, phylogenetic analysis of the last 36 amino acids of TtsA family members clustered all the muramidases capable of complementing typhoid toxin within the same group, indicating the presence of common features within this domain in these protein homologues (supplementary Fig S12 online). Taken together, these results indicate that information located within the C terminus of this enzyme family determines functional specificity, perhaps by targeting the enzymatic activity so as to disrupt the PG layer in close proximity to a periplasmic pool of the protein or complex of proteins (for example, typhoid toxin) destined to be secreted by this pathway.
Figure 5.
A C-terminal region of TtsA determines its ability to secrete typhoid toxin. (A) Amino-acid sequence comparison of the C terminus of TtsA and Sen1395. The residues mutated in this analysis are indicated. (B) Mutagenesis analysis of the C-terminal region of TtsA. Henle-407 cells were infected with WT S. Typhi or a ΔttsA mutant derivative expressing chromosomally encoded FLAG-tagged cdtB and carrying plasmids encoding the indicated TtsA mutant proteins. Twenty-two hours post infection, infected cells were fixed and stained with a mouse monoclonal antibody directed to the FLAG epitope (to visualize CdtB), a rabbit antibody directed to S. Typhi LPS and DAPI for DNA detection. The bar represents 10 μm. (C) Western blot analysis of the expression levels of the indicated proteins in the strains used in panel (B). (D) The N166 residue of TtsA is crucial for typhoid toxin secretion. Henle-407 cells were infected with WT S. Typhi or a ΔttsA mutant derivative expressing chromosomally encoded FLAG-tagged cdtB and carrying plasmids encoding the indicated proteins. Twenty-two hours post infection, infected cells were fixed and stained with a mouse monoclonal antibody directed to the FLAG epitope (to visualize CdtB), a rabbit antibody directed to S. Typhi LPS and DAPI for DNA detection. The bar represents 10 μm. (E) Western blot analysis of the expression levels of the indicated proteins in the strains used in panel (D). (F) Typhoid toxin secretion model. Typhoid toxin components PltA, PltB and CdtB have an N-terminal Sec signal peptide, which mediates their export to the periplasm through the Sec machinery. Further protein export requires enzymatically active TtsA, which lacks a canonical N-terminal signal peptide and therefore must be exported to the periplasm by a yet unidentified holin. Sec signal peptides (Sec SP), PG, IM and OM are indicated. DAPI, 4,6-diamidino-2-phenylindole; IM, inner membrane; LPS, lipopolysaccharide; OM, outer membrane; PG, peptidoglycan; WT, wild-type.
Concluding remarks
We have identified a muramidase belonging to a widely distributed protein family that is required for S. Typhi’s typhoid toxin secretion. We propose that these muramidases represent a recent adaptation of phage-associated enzymes to carry out a different function. Consistent with this hypothesis, we note that Sty1887, located immediately adjacent to ttsA, is predicted to encode a homologue of phage tail proteins. In addition, we propose that these enzymes define a new protein secretion system (Fig 5F) that is likely to be widespread in bacteria as we found several instances in which toxins or predicted extracellular enzymes are encoded immediately adjacent to holin/muramidase pairs.
Methods
Bacterial strains were constructed as previously described [17]. Immunofluorescence microscopy and cultured cell infection assays, and western blot analysis were performed as previously described [7]. Bacteriolytic assays after addition of 0.3% CHCl3 were carried out as previously described [12] with some modifications. Briefly, overnight grown bacteria were diluted to an OD600 nm of 0.1 in fresh LB medium containing 0.2% arabinose. After 60 min of induction, 0.3% CHCl3 was added to the cultures and lysis was monitored for additional 60 min by measuring their OD600 nm. For holin-assisted bacteriolysis, overnight cultures were diluted in fresh LB medium to an OD600 nm of 0.1 and further grown to an OD600 nm of 0.5. At that time, 0.002% arabinose was added to induce holin and/or endolysin expression, and bacterial lysis was monitored for additional 280 min by measuring the OD600 nm of the bacterial cultures. Phylogenetic trees were generated using ClustalW2, homologues of Sty1889 were identified using BLASTP and phage genes were detected using the PHAST. A more detailed description of the Methods appears in supplementary materials.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank members of the Galán laboratory for critical reading of this manuscript. This work was supported by NIAID Grant AI079022 to J.E.G.
Author contributions: H.H. designed, interpreted and conducted experiments; wrote the paper. J.E.G. designed and interpreted experiments; wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Parry C, Hien TT, Dougan G, White N, Farrar J (2002) Typhoid fever. N Engl J Med 347: 1770–1782 [DOI] [PubMed] [Google Scholar]
- Crump J, Mintz E (2010) Global trends in typhoid and paratyphoid fever. Clin Infect Dis 50: 241–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffatellu M, Wilson R, Winter S, Bäumler A (2008) Clinical pathogenesis of typhoid fever. J Infect Dev Ctries 2: 260–266 [DOI] [PubMed] [Google Scholar]
- House D, Bishop A, Parry C, Dougan G, Wain J (2001) Typhoid fever: pathogenesis and disease. Curr Opin Infect Dis 14: 573–578 [DOI] [PubMed] [Google Scholar]
- Baker S, Dougan G (2007) The genome of Salmonella enterica serovar Typhi. Clin Infect Dis 45: Suppl 1S29–S33 [DOI] [PubMed] [Google Scholar]
- Parkhill J et al. (2001) Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413: 848–852 [DOI] [PubMed] [Google Scholar]
- Spano S, Ugalde JE, Galan JE (2008) Delivery of a Salmonella Typhi exotoxin from a host intracellular compartment. Cell Host Microbe 3: 30–38 [DOI] [PubMed] [Google Scholar]
- Haghjoo E, Galan JE (2004) Salmonella Typhi encodes a functional cytolethal distending toxin that is delivered into host cells by a bacterial-internalization pathway. Proc Natl Acad Sci USA 101: 4614–4619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spano S, Galan JE (2008) A novel pathway for exotoxin delivery by an intracellular pathogen. Curr Opin Microbiol 11: 15–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghjoo E, Galan JE (2007) Identification of a transcriptional regulator that controls intracellular gene expression in Salmonella Typhi. Mol Microbiol 64: 1549–1561 [DOI] [PubMed] [Google Scholar]
- Spano S, Liu X, Galan JE (2011) Proteolytic targeting of Rab29 by an effector protein distinguishes the intracellular compartments of human-adapted and broad-host Salmonella. Proc Natl Acad Sci USA 108: 18418–18423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojković E, Rothman-Denes L (2007) Coliphage N4 N-acetylmuramidase defines a new family of murein hydrolases. J Mol Biol 366: 406–419 [DOI] [PubMed] [Google Scholar]
- Pei J, Grishin NV (2005) COG3926 and COG5526: a tale of two new lysozyme-like protein families. Protein Sci 14: 2574–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R (2002) Bacteriophage holins: deadly diversity. J Mol Microbiol Biotechnol 4: 21–36 [PubMed] [Google Scholar]
- Wang I, Smith D, Young R (2000) Holins: the protein clocks of bacteriophage infections. Annu Rev Microbiol 54: 799–825 [DOI] [PubMed] [Google Scholar]
- Kondo Y, Toyoda A, Fukushi H, Yanase H, Tonomura K, Kawasaki H, Sakai T (1994) Cloning and characterization of a pair of genes that stimulate the production and secretion of Zymomonas mobilis extracellular levansucrase and invertase. Biosci Biotechnol Biochem 58: 526–530 [DOI] [PubMed] [Google Scholar]
- Kaniga K, Bossio JC, Galan JE (1994) The Salmonella typhimurium invasion genes invF and invG encode homologues of the AraC and PulD family of proteins. Mol Microbiol 13: 555–568 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.