Abstract
Natural antisense transcripts (NATs) are endogenous RNA molecules that are complementary to known RNA transcripts. The functional significance of NATs is poorly understood, but their prevalence in the CNS suggests a role in brain function. Here we investigated a long NAT (antiNOS-2 RNA) associated with the regulation of nitric oxide (NO) production in the CNS of Lymnaea, an established model for molecular analysis of learning and memory. We show the antiNOS-2 RNA is axonally trafficked and demonstrate that this is regulated by classical conditioning. Critically, a single conditioning trial changes the amount of antiNOS-2 RNA transported along the axon. This occurs within the critical time window when neurotransmitter NO is required for memory formation. Our data suggest a role for the antiNOS-2 RNA in establishing memories through the regulation of NO signaling at the synapse.
The recent discovery in higher organisms of a variety of non-coding RNA molecules suggests that the information-content of eukaryotic genomes has been underestimated. Important among non-coding RNAs are those containing regions of significant antisense homology to conventional protein-coding mRNAs. The reported prevalence of these natural antisense transcripts (NATs) in the CNS suggests a role in brain function (for a review, see ref. 1). Generally, NATs include the intensively studied micro RNAs (miRNAs) and longer antisense RNAs, whose precise function remains to be elucidated. Depending on their loci of origin, long NATs can be classified into two major groups: cis- and trans-encoded. Cis-encoded NATs are produced from the same loci as their sense counterparts, whereas trans-encoded NATs are transcribed from different loci (for a review, see ref. 2).
In our previous work on Lymnaea we identified a trans-encoded NAT (antiNOS-2 RNA), which is associated with the negative regulation of the production of the neurotransmitter nitric oxide (NO) by NO synthase (NOS)3. Notably, the antiNOS-2 RNA is transcribed from a NOS pseudogene evolved from a duplicated copy of the NOS gene by an internal DNA inversion. Two mechanisms were indicated for the antiNOS-2-mediated regulation of NO signaling. One involves the formation of an RNA duplex between the antisense region of the NAT and the complementary region of the conventional NOS mRNA. The second mechanism is based on our findings that in addition to the antisense region, antiNOS-2 RNA also has a functional open reading frame. This is translated into a truncated NOS-homologous protein. The antiNOS-2 protein contains the dimerisation domain, but lacks other functional regions essential for NO synthesis3. As the NOS enzyme itself is active only as a homodimer4,5,6 we suggested that by dimerizing with the full-length NOS monomer, the antiNOS-2 protein functions as a dominant negative regulator of NO synthesis by NOS3,7. This conclusion is supported by experiments demonstrating that similarly organized truncated NOS homologues suppress NO production8,9.
NO is known to play an important role in the early stages of learning and memory formation in both invertebrates and vertebrates10,11,12,13. For example, in Lymnaea NO is required for long-term memory (LTM) formation following associative food-reward conditioning14. In this species, a single pairing of two chemosensory stimuli, sucrose (US) and amyl acetate (CS), leads to associative memory formation that can last more than two weeks15. Importantly, there is an obligatory requirement for NO signaling during LTM formation for a continuous period of about 6 hours post-training14. At the neuronal level, the associative conditioning of feeding in Lymnaea is linked with the induction of non-synaptic neuronal plasticity16 and the transient up-regulation of the gene encoding full-length NOS in an identified pair of neurons called the cerebral giant cells (CGCs)17.
Here we focus on the link between NAT-mediated mechanisms controlling NO production and NO-dependent associative memory formation. We show that the antiNOS-2 RNA is: i) co-expressed with the NOS-encoding mRNA in the CGC, a neuron with an established role in memory formation18; ii) subjected to axonal trafficking and iii) exhibits timed learning-induced changes in its peripheral trafficking following a single classical conditioning trial.
Results
AntiNOS-2 RNA and NOS mRNA are co-expressed in the CGC
The CNS of Lymnaea consists of eleven ganglia including the cerebral ganglia that encircle the oesophagus and two smaller ganglia (left and right buccal ganglia) connected to the cerebral ganglia via the cerebral-buccal connective (CBC) (Fig. 1a). The spheroid cell bodies of neurons lie on the periphery of the ganglia and form the neuronal cell body zone. This zone surrounds a central synapse-rich region known as the neuropile19,20,21.
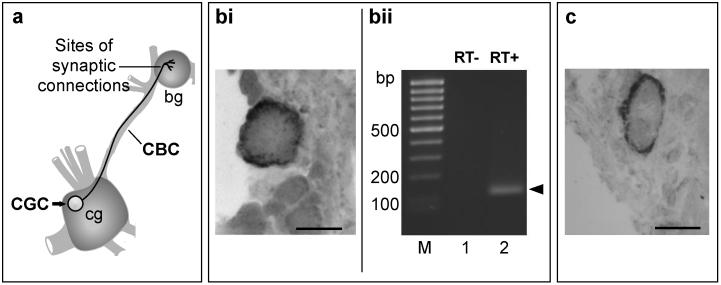
Figure 1. AntiNOS-2 RNA and NOS mRNA are co-expressed in the CGC.
(a) A schematic diagram showing the cerebral (cg) and the buccal (bg) ganglia connected by the CBC. The CGC (arrowed) sends its main axon to the buccal ganglion via the CBC where it makes synaptic connections with neurons of the feeding neural circuit. (bi) In situ hybridization experiments showing the CGC stained with the antiNOS-2 specific probe. (bii) The result of reverse transcription-PCR experiment on RNA extracted from isolated CGCs performed with primers specific for the antiNOS-2 (lane 2). A PCR product of the expected size (119 bp) is shown by arrow. Lane 1 represents the result of control experiment in which reverse transcriptase was omitted. The absence of any PCR products in this lane proves that the RNA preparation was free from DNA contamination. (c) In situ hybridization experiments showing the CGC stained with the NOS specific probe. Scale bars – 50 μm.
We have employed in situ hybridization on frozen sections of Lymnaea CNS to study cellular localization of antiNOS-2 RNA. As antiNOS-2 is expressed at a low level, a protocol designed to detect rare RNAs has been used22,23. These experiments show that the antiNOS-2 RNA is expressed selectively in a subpopulation of neurons in the CNS. Of particular interest is the observation that in the cerebral ganglia the expression of antiNOS-2 is confined to a pair of very large neurons called the CGCs (Fig. 1bi). These neurons are known to have a role in consolidating NO-requiring associative memory. In view of the potential significance of this finding we verified the results of in situ hybridization experiments by using the more powerful reverse transcription-PCR technique conducted on RNA extracted from individually dissected CGCs. Importantly, the results of reverse transcription-PCR confirm in situ hybridization data and unambiguously demonstrate that antiNOS-2 is indeed expressed in the CGC (Fig. 1bii). This is intriguing because the CGC is known to contain also the conventional NOS-encoding mRNA (Fig. 1c)17,24. The discovered cellular co-localization of antiNOS-2 and NOS mRNA suggests that the former can regulate NO production in the CGC by interacting with the latter.
AntiNOS-2 RNA is subjected to peripheral trafficking
Our in situ hybridization experiments show the presence of the antiNOS-2 RNA in a prominent axon in the CBC, the structure linking the cerebral ganglia to the buccal ganglia (Fig. 2a). The axonal hybridization signal has a granular appearance, suggesting the presence of RNA particles known to mediate RNA transport in neurons (for a review, see ref. 25). We also detected the presence of antiNOS-2 RNA within the synapse-rich neuropile of the buccal ganglion (Fig. 2b). Taking together these data indicate strongly that antiNOS-2 RNA is subjected to trafficking from the cerebral ganglion to the synaptic zone of the buccal ganglion.
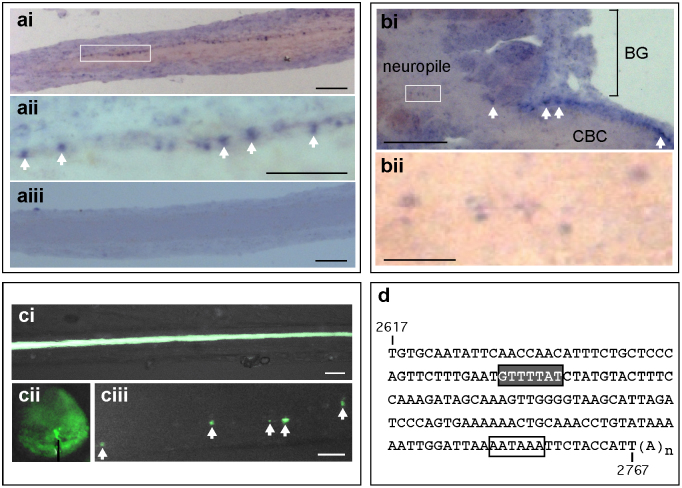
Figure 2. AntiNOS-2 RNA is subjected to peripheral trafficking.
(ai) In situ hybridization with the antiNOS-2 specific probe shows RNA particles within a prominent axon in the CBC. (aii) Higher magnification of boxed area in (ai) (RNA particles are arrowed). (aiii) Control in situ hybridization with ‘sense' probe shows no staining. (bi) In situ hybridization shows RNA particles (arrowed) within a prominent axon entering the buccal ganlion (BG). (bii) Higher magnification of boxed area in (bi) shows the presence of the antiNOS-2 RNA in the synapse-rich neuropile. (ci) A CGC cell body filled with 5,6-carboxyfluorescein to allow clear visualization of the position of the axon in the CBC. (cii) A CGC cell body filled with Alexa Fluor 488-tagged antiNOS-2 RNA. The site of the electrode tip used for injection is indicated in black. (ciii) Putative fluorescent RNA particles (arrowed) in the axon of the CGC shown in (cii). (d) The antiNOS-2 RNA contains a CPE-like motif (grey box). A canonical AATAAA hexamer is boxed. Scale bars: (ai), (aiii) and (bi) – 50 μm; (aii), (ci) and (ciii) – 25 μm; (bii) – 5 μm.
The cell body of the CGC sends its large diameter primary axon to the buccal ganglion via the CBC where it makes synaptic connections with neurons of the feeding circuit (Fig. 1a)26. As the CGCs are the only prominent neurons in the cerebral ganglia containing detectable amount of antiNOS-2 RNA, we can conclude that the axon shown in Fig. 2a belongs to the CGC. To verify if the antiNOS-2 RNA is subjected to peripheral trafficking in the CGC we used imaging techniques in an isolated CNS preparation. Specifically, we produced Alexa Fluor 488 labelled antiNOS-2 RNA and injected this into the neuronal soma of the CGC (Fig. 2ci and 2cii). After injection we examined the preparation using an epifluorescence imaging system. Distinct fluorescently-labeled particles are clearly visible within the CGC's axon in the CBC further supporting the notion that antiNOS-2 RNA is trafficked from the CGC cell body to the buccal ganglia (Fig. 2ciii).
It is well established that RNA transport in neurons depends on the interaction between cis-acting RNA elements and trans-acting RNA binding factors27. With this in mind we have analyzed the primary structure of antiNOS-2 RNA and found the sequence motif GUUUUAU (Fig. 2d), the primary structure and the position of which are typical for the well-characterized cis-acting heptanucleotide called the cytoplasmic polyadenylation element (CPE) (for a review, see ref. 27). The CPE motif is usually located within 100 nt upstream of a nuclear polyadenylation hexanucleotide, has the consensus sequence UUUUUAU and is conserved from invertebrates to mammals. Thus, the presence of the CPE-like motif in antiNOS-2 RNA is in line with its peripheral localization revealed by experiments described above.
Peripheral trafficking of antiNOS-2 RNA is affected by training
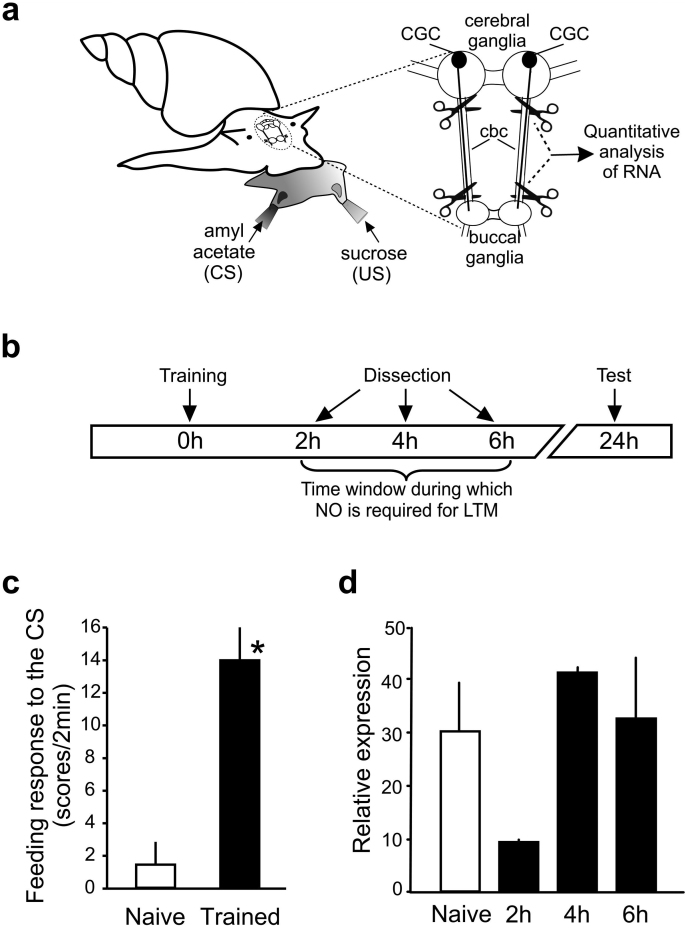
To investigate whether food-reward conditioning is associated with specific changes in the antiNOS-2 RNA peripheral trafficking, the CBCs were subjected to real-time reverse transcription-PCR at 2 hr, 4 hr and 6 hr after one-trial conditioning (Figs 3a, b and c). The amount of the antiNOS-2 RNA was analyzed using a calibrator-normalized relative quantification method. These experiments revealed transient and precisely timed changes in the peripheral trafficking of antiNOS-2 RNA following training. More precisely, our results show a decrease in the amount of the trafficked antiNOS-2 RNA 2 hours after conditioning compared to other time points (Fig. 3d).
Figure 3. AntiNOS-2 RNA is down-regulated in the CBC 2 hours after one-trial appetitive conditioning.
(a) Schematic diagram of the classical single-trial reward conditioning of Lymnaea using amyl acetate as the CS and sucrose as the US. The CBCs, containing the CGC processes, were removed from the brain and were subjected to quantitative reverse transcription PCR analysis. (b) Schematic representation of the experiment to investigate whether food-reward conditioning is associated with specific changes in the antiNOS-2 RNA peripheral trafficking. (c) A typical result of the experimental test of LTM formation at 24 hr post-training. Unpaired t-test showed that the mean feeding response to amyl acetate (the CS) of the trained snails (black bar) is significantly higher than the response of the control animals (white bar) (n = 18, p<0.0001, t = 5.05, df = 34). (d) CBCs dissected from conditioned (2 hours, 4 hours and 6 hours after training) and control animals were subjected to real-time reverse transcription-PCR. The relative level of the antiNOS-2 RNA in the experimental and naive control (NC) groups (n = 10) was calculated using calibrator normalized relative quantification method.
In these experiments a randomly chosen sub-set of each group of animals were retained and behaviorally tested for associative memory formation at 24 hours post-training. This was to confirm that LTM would have occurred in the animals we sacrificed at the earlier time-points at which we measured changes in antiNOS-2 gene expression.
Discussion
That some RNAs are targeted to peripheral destinations is now well established, particularly in cells with complex architecture such as neurons. Coupled with local translation, targeted RNA transport enables neurons to modulate transmission at selected synapses. A number of mRNAs transported along neurites are known (for a review, see ref. 27). In addition some types of non-coding RNAs such as BC1/BC200 RNA and several classes of miRNA have been found at synapses (for reviews, see refs 1, 28). Little is known, however, about peripheral trafficking of the long class of antisense transcripts. Therefore our findings showing the presence of a long NOS-related NAT in the CGC's axon and within the synapse-rich neuropile of the buccal ganglion are of significant importance. They suggest that the antiNOS-2 RNA is delivered to synaptic connections formed between the CGC and target motoneurons located within the buccal ganglion. What could be the functional consequence of this? Considering co-localization of the conventional NOS mRNA and the NAT in the CGCs, as well as the well-established localization of NOS protein in both pre- and post-synaptic terminals29, we hypothesize that the antiNOS-2 RNA is involved in the regulation of NO signaling locally at the synapse.
The most exciting result of our studies is that the peripheral trafficking of the anti-NOS2 RNA is exquisitely sensitive to training – a single conditioning trial only being required to induce a transient decrease in the amount of the NAT delivered to the buccal ganglia via the CBCs. Since this occurs 2 hours after conditioning within a critical time window during which NO is required for memory formation, we propose that precisely timed changes in peripheral trafficking of the antiNOS-2 RNA are a component of the mechanism of associative memory formation. This idea is in line with the results of other studies conducted on invertebrate and vertebrate neurons showing that the peripheral RNA trafficking coupled with local translation underlies long-lasting types of synaptic plasticity (for a review, see ref. 30). Considering the proposed negative regulatory role for the antiNOS-2 RNA, we suggest that the decrease in the amount of this NAT contributes to an increase in the production of NO and consequently facilitates memory formation.
Finally, as the antiNOS-2 RNA is transcribed from a NOS pseudogene, our findings further reinforces the conclusion that the pejorative “junk DNA” may be inappropriately applied to all pseudogenes17,31.
Methods
Experimental animals
Specimens of Lymnaea stagnalis were raised in the breeding facility of the University of Sussex, where they were kept in 20–22°C copper free water under 12 h light and dark cycle. They were fed on lettuce 3 times and a vegetable based fish food twice a week. Snails were housed in aerated aquaria at 20°C, kept under a 12-hour light/dark cycle and fed on a lettuce diet. Animals between 4 and 6 months old were taken for histological studies.
In situ hybridization
The dissected cerebral ganglia were fixed for 1 h at room temperature in 4% paraformaldehyde in phosphate buffered saline (PBS) containing 0.1 M sodium dihydrogen orthophosphate, 0.1 M disodium hydrogen orthophosphate. The tissue was then incubated overnight at 4°C in 30% sucrose in PBS, before being embedded in OCT embedding medium (Raymond Lamb, Eastbourne, UK) and frozen in liquid nitrogen. Fourteen-micrometer frozen sections were cut and mounted on SuperFrost Plus slides (VWR international, Lutterworth, UK). GreenStar DIG-hyperlabeled 48-mer oligonucleotide probes specific for antiNOS-2 RNA were designed and produced by GeneDetect (Auckland, New Zealand). In situ hybridization on frozen sections was performed following the protocol suggested by the probe's manufacturer. Detection procedure was carried out using an anti-DIG antibody conjugated to alkaline phosphatase (Roche, Welwyn Garden City, UK).
Reverse transcription-PCR on isolated identified neurons
The cell bodies of CGCs were identified and individually dissected from the CNS of Lymnaea as described previously17. Total RNA extracted from the pooled neurons by means of the Absolutely RNA Nanoprep Kit (Stratagene, Amsterdam, Netherlands) was copied into cDNA using iScript reverse transcriptase (Bio-Rad, Hemel Hempstead, UK). The produced cDNA was subjected to PCR using the following primers: 5′-GTGAACTATCAAGTCACAATG-3′ and 5′-GAACTCAAGACTCTACGTAG-3′.
One-trial conditioning protocol and surgical procedures
Reward conditioning was carried out using a method based on a previously published protocol15. Snails were randomly assigned to experimental (paired) and control (unpaired) groups to be given a single conditioning and control trial, respectively. Experimental animals were exposed to a solution of amyl acetate (0.004% final concentration) (CS) and 30 sec later to a sucrose solution (0.67%) (US) and stayed in the mixture of solutions for 2 min. Control animals were exposed to the CS and to the US, separated by an interval of 1 hr. A randomly chosen sub-set of 20 animals from each group were retained and tested for LTM formation at 24 hours after the paired and unpaired trials as previously described14. A third group of animals was kept under the same conditions and had the same feeding regime as experimental and unpaired control snails but was not exposed to either the CS or US. This group is referred to as naïve control (NC). At different time points (2 hr, 4 hr, 6 hr) after the treatment a randomly chosen sub-set of animals (usually 10 individuals per time point) were sacrificed and the CNS was removed and used for RNA extraction.
Real-time reverse transcription-PCR
Real-time reverse transcription-PCR was performed on RNAs isolated either from individual CNSs by means of the Absolutely RNA Microprep kit (Stratagene). cDNAs produced from RNA preparations by iScript reverse transcriptase (Bio-Rad) were amplified and analyzed on the Mx3000P real-time cycler (Stratagene) using the FastStart Universal Green Master kit (Roche). We used primers 5′-CTACGTAGAGTCTTGAGTTC-3′ and 5′-TCTATCTCACTCTGCAACTC-3′ for detection of antiNOS-2 and primers 5′-AAGGGACATTACACAGAGG-3′ and 5′-GTGTCAGTTGGAATCCTTG-3′ for detection of β-tubulin. The identity of all PCR products was confirmed by sequencing. The amount of target transcript, normalized to an endogenous reference and relative to a calibrator was calculated as 2−ΔΔCT where ΔΔCT = ΔCT − ΔCT(CAL). ΔCT and ΔCT(CAL) are the differences in threshold cycles for target (antiNOS-2) and reference (β-tubulin) measured in the samples and in the calibrator (CAL) respectively32.
In vitro transcription
The purified plasmid containing the antiNOS-2 cDNA was linearized with BamHI (Fermentas). 1 μg of the resultant plasmid was transcribed in vitro using the Ampliscribe T7 kit (Cambio) in the presence of ChromaTide Alexa Fluor 488-5-UTP (Invitrogen). The synthesized RNA was then purified on Micro Bio-Spin P30 columns (Bio-Rad). An aliquot of purified fluorescently labeled RNA was analyzed in formaldehyde-containing agarose gel to verify its integrity.
Microinjection of RNA into the CGC
Microinjection of the fluorescently labeled RNA was performed by the application of repetitive, short pressure pulses with a Picospritzer (General Valve Corp.) to the microelectrode inserted into the CGC soma. The injection protocol consisted of 15 single pulses, 10 ms long each, at a pressure of 15 psi with 15 s intervals between pulses.
Imaging
Live fluorescence imaging was carried out using an Olympus BX61WI microscope, Cell-M light source and XM10 CCD camera. Injection of 5,6-carboxyfluorescein was used to localize the CGC axon in the cerebro-buccal connective and Alexa-488-tagged antiNOS-2 RNA to visualize RNA puncta. Excitation and emission were: 480/20 nm and 520/35 nm, respectively.
Author Contributions
Experiments were conceived by S.A.K., G.K. and K.S. and carried out by S.A.K., I.K., N.B. and K.S. Results were analysed and interpreted by S.A.K., G.K., K.S., P.B. and M.O. The manuscript was written by S.A.K. and M.O. Infrastructure and reagents were provided by S.A.K., G.K., K.S., P.B. and M.O.
References
- Cao X., Yeo G., Muotri A. R., Kuwabara T. & Gage F. H. Noncoding RNAs in the mammalian central nervous system. Annu. Rev. Neurosci. 29, 77–103 (2006). [DOI] [PubMed] [Google Scholar]
- Korneev S. A. & O'Shea M. Natural antisense RNAs in the nervous system. Rev. Neurosci. 16, 213–222 (2005). [DOI] [PubMed] [Google Scholar]
- Korneev S. A. & O'Shea M. Evolution of nitric oxide synthase regulatory genes by DNA inversion. Mol. Biol. Evol. 19, 1228–1233 (2002). [DOI] [PubMed] [Google Scholar]
- Abu-Soud H. M., Loftus M. & Stuehr D. J. Subunit dissociation and unfolding of macrophage NO synthase: relationship between enzyme structure, prosthetic group binding, and catalytic function. Biochemistry 34, 11167–11175 (1995). [DOI] [PubMed] [Google Scholar]
- Klatt P. et al. Characterization of heme-deficient neuronal nitric oxide synthase reveals a role for heme in subunit dimerization and binding of the amino acid substrate and tetrahydrobiopterin. J. Biol. Chem. 271, 7336–7342 (1996). [DOI] [PubMed] [Google Scholar]
- Crane B. R. et al. (1998) Structure of nitric oxide synthase oxygenase dimer with pterin and substrate. Science 279, 2122–2126 (1998). [DOI] [PubMed] [Google Scholar]
- Korneev S. A. & O'Shea M. Regulatory role and evolution of unconventional NOS-related RNAs. Adv. Exp. Biol. 1, 181–197 (2007). [Google Scholar]
- Lee C. M., Robinson L. J. & Michel T. Oligomerization of endothelial nitric oxide synthase. Evidence for a dominant negative effect of truncation mutants. J. Biol. Chem. 270, 27403–27406 (1995). [DOI] [PubMed] [Google Scholar]
- Stasiv Y., Regulski M., Kuzin B., Tully T. & Enikolopov G. The Drosophila nitric-oxide synthase gene (dNOS) encodes a family of proteins that can modulate NOS activity by acting as dominant negative regulators. J. Biol. Chem. 276, 42241–42251 (2001). [DOI] [PubMed] [Google Scholar]
- Lu Y. F., Kandel E. R. & Hawkins R. D. Nitric oxide signaling contributes to late-phase LTP and CREB phosphorylation in the hippocampus. J. Neurosci. 19, 10250–10261 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose S. P. God's organism? The chick as a model system for memory studies. Learn. Mem. 7, 1–17 (2000). [DOI] [PubMed] [Google Scholar]
- Schweighofer N. & Ferriol G. Diffusion of nitric oxide can facilitate cerebellar learning: a simulation study. Proc. Natl. Acad. Sci. USA 97, 10661–10665 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonov I., Ha T., Antonova I., Moroz L. L. & Hawkins R. D. Role of nitric oxide in classical conditioning of siphon withdrawal in Aplysia. J. Neurosci. 27, 10993–11002 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemenes I., Kemenes G., Andrew R. J., Benjamin P. R. & O'Shea M. Critical time-window for NO-cGMP dependent long-term memory formation after one-trial appetitive conditioning. J. Neurosci. 22, 1414–1425 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander J. Jr., Audesirk T. E. & Audesirk G. J. One-trial reward learning in the snail. Lymnaea stagnalis. J. Neurobiol. 15, 67–72 (1984). [DOI] [PubMed] [Google Scholar]
- Kemenes I. et al. Curr. Biol. 16, 1269–1279 (2006). [DOI] [PubMed] [Google Scholar]
- Korneev S. A. et al. Timed and targeted differential regulation of NOS and antiNOS genes by reward conditioning leading to long-term memory formation. J. Neurosci. 25, 1188–1192 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemenes I. et al. Role of delayed nonsynaptic neuronal plasticity in long-term associative memory. Curr. Biol. 16, 1269–1279 (2006). [DOI] [PubMed] [Google Scholar]
- Chase R. & Tolloczko B. Synaptic innervation of the giant cerebral neuron in sated and hungry snails. J. Comp. Neurol. 318, 93–102 (1992). [DOI] [PubMed] [Google Scholar]
- Nagy T. & Elekes K. Embryogenesis of the central nervous system of the pond snail Lymnaea stagnalis L. An ultrastructural study. J. Neurocytol. 29, 43–60 (2000). [DOI] [PubMed] [Google Scholar]
- van Nierop P. et al. Identification and functional expression of a family of nicotinic acetylcholine receptor subunits in the central nervous system of the mollusc Lymnaea stagnalis. J. Biol. Chem. 281, 1680–1691 (2006). [DOI] [PubMed] [Google Scholar]
- Ribeiro M. et al. Characterization of NO-sensitive guanylyl cyclase: expression in an identified interneuron involved in NO-cGMP-dependent memory formation. Eur. J. Neurosci. 28, 1157–1165 (2008). [DOI] [PubMed] [Google Scholar]
- Ribeiro M., Schofield M., Kemenes I., Benjamin P. R., O'Shea M. & Korneev S. A. Atypical guanylyl cyclase from the pond snail Lymnaea stagnalis: clonining, sequence analysis and characterization of expression. Neuroscience 165, 794–800 (2010). [DOI] [PubMed] [Google Scholar]
- Korneev S. A., Park J.-H. & O'Shea M. Neuronal expression of neural nitric oxide synthase (nNOS) protein is supressed by an antisense RNA transcribed from a NOS pseudogene. J. Neurosci. 19, 7711–7720 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramham C. R. & Wells D. G. Dendritic mRNA: transport, translation and function. Nature Reviews 8, 776–789 (2007). [DOI] [PubMed] [Google Scholar]
- McCrohan C. R. & Benjamin P. R. Patterns of activity and axonal projections of the cerebral giant cells of the snail, Lymnaea stagnalis. J. Exp. Biol. 85, 149–168 (1980). [DOI] [PubMed] [Google Scholar]
- Kindler S., Wang H., Richter D. & Tiedge H. RNA transport and local control of translation. Annu. Rev. Cell Dev. Biol. 21, 223–245 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldie B. J. & Cairns M. J. Post-transcriptional trafficking and regulation of neuronal gene expression. Mol. Neurobiol. 45, 99–108 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson L., Batten T. F. C., Corbett E. K. A., Sinfield J. K. & Deuchars J. Subcellular localization of neuronal nitric oxide synthase in the rat nucleus of the solitary tract in relation to vagal afferent inputs. Neuroscience 118, 115–122 (2003). [DOI] [PubMed] [Google Scholar]
- Siegel G., Saba R. & Schratt G. microRNAs in neurons: manifold regulatory roles at the synapse. Curr. Opin. Genet. Dev. 21, 491–497 (2011). [DOI] [PubMed] [Google Scholar]
- Guo X., Zhang Z., Gerstein M. B. & Zheng D. Small RNAs originated from pseudogenes: cis- or trans-acting? PLoS Comput. Biol. 5, e1000449 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucl. Acids Res. 29, 2002–2007 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]