Abstract
Atlantic cod (Gadus morhua) is a large, cold-adapted teleost that sustains long-standing commercial fisheries and incipient aquaculture1,2. Here we present the genome sequence of Atlantic cod, showing evidence for complex thermal adaptations in its haemoglobin gene cluster and an unusual immune architecture compared to other sequenced vertebrates. The genome assembly was obtained exclusively by 454 sequencing of shotgun and paired-end libraries, and automated annotation identified 22,154 genes. The major histocompatibility complex (MHC) II is a conserved feature of the adaptive immune system of jawed vertebrates3,4, but we show that Atlantic cod has lost the genes for MHCII, CD4 and Ii that are essential for the function of this pathway. Nevertheless, Atlantic cod is not exceptionally susceptible to disease under natural conditions5. We find a highly expanded number of MHCI genes and a unique composition of its Toll-like receptor (TLR) families. This suggests how the Atlantic cod immune system has evolved compensatory mechanisms within both adaptive and innate immunity in the absence of MHCII. These observations affect fundamental assumptions about the evolution of the adaptive immune system and its components in vertebrates.
We sequenced the genome of a heterozygous male Atlantic cod (NEAC_001, Supplementary Note 1 and 2) applying a whole-genome shotgun (WGS) approach to 40x coverage (estimated genome size 830Mb, Supplementary Note 4, Supplementary Figure 2) using 454 technology (Supplementary Note 3). Two programs (Newbler6 and Celera7, Supplementary Note 5 and 6) produced assemblies with short contigs, yet scaffolds of comparable size to those of Sanger sequenced teleost genomes (Supplementary Note 10, Supplementary Figure 8). While fragmentation due to short tandem repeats is difficult to address (Supplementary Note 7), we resolved numerous gaps due to heterozygosity (Supplementary Note 8). The assemblies differ in scaffold and contig length (Table 1) though their scaffolds align to large extent (Supplementary Note 9, Supplementary Figure 7). We obtained ~1 million SNPs by mapping 454 and Illumina reads from the sequenced individual to the Newbler assembly (Supplementary Note 11). Both assemblies cover over 98% of the reads from an extensive transcriptome dataset, indicating that the proteome is well represented (Supplementary Note 13). The assemblies are consistent with four independently assembled BAC insert clones (Supplementary Note 14, Supplementary Figure 9) and with the expected insert size of paired BAC-end reads (Supplementary Note 15, Supplementary Figure 10).
Table 1.
Assembly statistics
| Number | Bases (Mb) | N50L (bp)a | N50 (n)b | MLc (bp) | |
|---|---|---|---|---|---|
| Newbler | |||||
| Contigsd | 284,239 | 536 | 2,778 | 50,237 | 76,504 |
| Scaffolds | 6,467 | 611 | 687,709 | 218 | 4,999,318 |
| Entire assemblye | 157,887 | 753 | 459,495 | 344 | 4,999,318 |
| Celera | |||||
| Contigsd | 135,024 | 555 | 7,128 | 19,938 | 117,463 |
| Scaffolds | 3,832 | 608 | 488,312 | 373 | 2,810,583 |
| Entire assemblye | 17,039 | 629 | 469,840 | 395 | 2,810,583 |
Minimum sequence length in which half of the assembled bases occur.
Number of sequences with N50L or longer.
Maximum Length.
Contigs greater than 500bp.
Scaffolds and unplaced contigs.
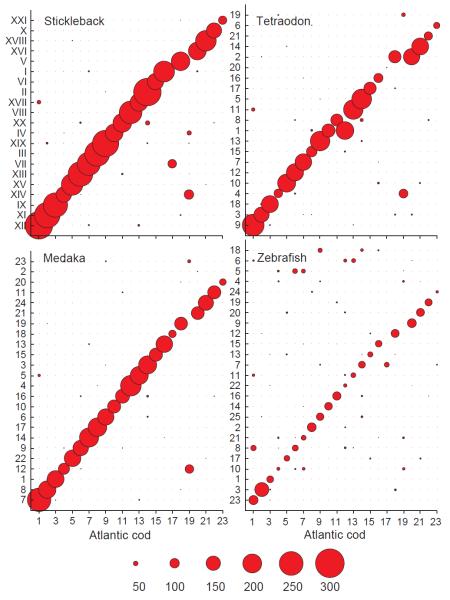
A standard protein-evidence based annotation approach was complemented with a whole genome alignment with stickleback (Gasterosteus aculeatus), after repeatmasking 25.4% of the Newbler assembly (Supplementary Note 16, Supplementary Table 6). In this way, 17,920 out of 20,787 protein-coding stickleback genes were mapped onto reorganized scaffolds (Supplementary Note 17). Additional protein-coding genes, pseudogenes and non-coding RNAs were annotated using the standard Ensembl pipeline. These approaches resulted in a final gene set of 22,154 genes (Supplementary Table 7). Comparative analysis of Gene Ontology (GO) classes indicates that the major functional pathways are represented in the annotated gene set (Supplementary Note 18, Supplementary Figure 11). We anchored 332Mb of the Newbler assembly to 23 linkage groups of an existing Atlantic cod linkage map using 924 SNPs8 (Supplementary Note 19, Supplementary Table 8). These linkage groups have distinct orthology to chromosomes of other teleosts based on the number of co-occuring genes showing that the WGS assembly reflects the expected chromosomal ancestry (Figure 1, Supplementary Note 20, Supplementary Table 9).
Figure 1.
Synteny between Atlantic cod and selected teleosts. The co-occurrence of orthologous genes (with a minimum of 50% sequence identity over 50% of the alignment) located on 23 Atlantic cod linkage groups8 reveals synteny with the chromosomes of four teleosts. Multiple genes located on the stickleback XIV, tetraodon 4 and medaka 12 chromosomes indicate a lineage-specific chromosomal rearrangement in Atlantic cod.
Well-studied haemoglobin polymorphisms are indicative of functional molecular adaptation to thermal variation in Atlantic cod9,10,11,12. Its genome contains nine α- and β- globin genes that are organized in two unlinked clusters, β5-α1-β1-α4 and β3-β4-α2-α3-β213,14. We discovered a 73 bp indel polymorphism in the intergenic promoter region of the α1-β1 globin pair (Figure 2a, Supplementary Note 21). This promoter polymorphism occurs in highly significant linkage disequilibrium with two known polymorphic sites, the Val55β1Met and Ala62β1Lys substitutions11, in eight Atlantic cod populations (Supplementary Note 22, Supplementary Figure 12). In fact, in the three most northern Atlantic and both Baltic populations, cod β1 globin predominantly occurs as a single homozygous genotype consisting of the long promoter and the Val55—Ala62 allele (Supplementary Table 10). By placing the two promoter variants in front of a luciferase reporter gene and transfecting the constructs into salmon kidney cells (Supplementary Note 23), we found that temperature and promoter type have a significant interaction effect (GLM, F2,36 = 7.85, P = 0.007, Figure 2b) and that the long promoter has a two-fold higher transcriptional activity compared to the short promoter at 15 °C and 20 °C. Increased globin synthesis of the Val—Ala allele would compensate for its lower affinity10,11 at high temperatures. Thus, the promoter polymorphism provides a molecular compensatory mechanism that helps maintain the total oxygen carrying capacity15. The tight linkage between the two types of polymorphism provides a compelling example of the coevolution of structural and regulatory adaptation and highlights the relationship between temperature and functional molecular variation in the haemoglobin system16.
Figure 2.
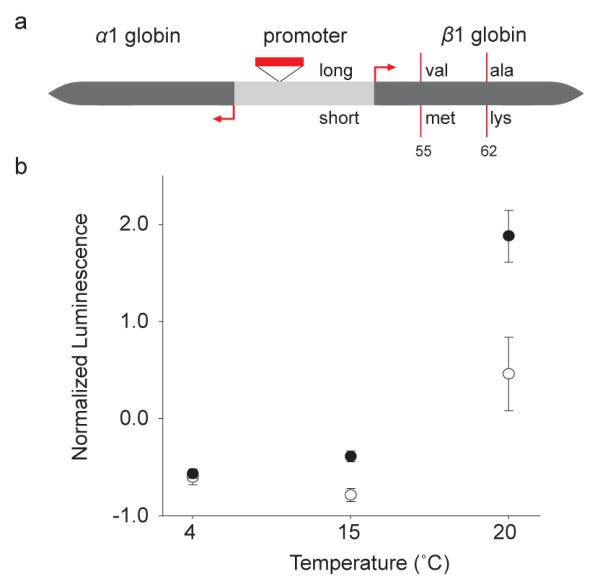
Functional haemoglobin polymorphisms in Atlantic cod (a) Schematic of the head-to-head organized α1 and β1 globin genes, the intergenic promoter region and transcription start sites (red arrows). A 73 bp insert promoter polymorphism (red box) segregates in linkage disequilibrium with two amino acid substitution polymorphisms at position 55 and 62 (vertical lines), which affect oxygen-binding affinity. This linkage disequilibrium results in two predominant haplotypes, Long-Val-Ala and Short-Met-Lys. (b) Normalized luciferase luminescence ratios in salmon kidney cells. Cells were transfected using the long (black symbol) or short (white symbol) promoter and incubated at 4, 15 or 20 °C (n = 3 for each treatment level). Error bars show 95% confidence intervals.
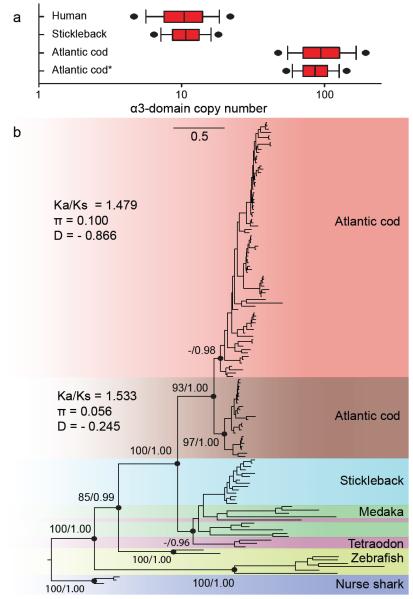
The Atlantic cod immune system has unusual properties that set it apart from other teleosts, i.e. high levels of IgM17, a minimal antibody response after pathogen exposure5,17,18 and abundant phagocytic neutrophils in its peripheral blood19,20. Despite speculations, the exact causes for these differences remain unknown5. We found that most genes involved in the vertebrate immune response are present in Atlantic cod (Supplementary Note 24, Supplementary Figure 13, Supplementary Table 11). Nevertheless, we did not find the major histocompatibility complex (MHC) II isoforms, their assembly and trafficking chaperone Invariant chain (Ii)21 and the MHCII interacting protein CD4, essential for helper T cell activation. By comparing a comprehensive set of vertebrate MHCII, CD4 and Ii sequences to the genome assemblies and all unassembled 454 and Illumina sequencing reads (a dataset of ~49.5 Gb), we detected a truncated pseudogene for CD4 (Supplementary Note 25), which is located in a region of conserved synteny (Supplementary Note 27, Supplementary Figure 18). No traces of MHCII and Ii were found in syntenous regions (Supplementary Note 27, Supplementary Figures 16, 17, 19 and 20) and qPCR targeting a conserved domain in MHCII did not amplify the target sequence (Supplementary Note 26, Supplementary Figure 15). The absence of MHCII and Ii and the pseudogenic nature of CD4 show that Atlantic cod has lost function of the classical pathway for adaptive immunity against bacterial and parasitic infections. Nevertheless, Atlantic cod deals adequately with its prevailing pathogen load in its natural ecological settings5. Earlier transcriptional (cDNA) studies in Atlantic cod have indicated an expansion of the number of MHCI loci22,23. By targeting the conserved MHCI α3 domain in genomic DNA using qPCR, we more accurately quantified the number of loci belonging to the teleost U-lineage24 (Supplementary Note 28). Remarkably, Atlantic cod has ~100 classical MHCI loci, which is a highly expanded number compared to other teleosts (Figure 3a). A phylogenetic analysis of teleost MHCI sequences supports two clades in cod (Figure 3b, Supplementary Note 29). Within each clade, the mutation patterns display statistically significant signs of positive selection that are indicative of subfunctionalization. These findings suggest that loss of MHCII functionality has coincided with a more versatile usage of the cytosolic pathway of MHCI. Two different MHCI antigen presentation pathways – the classical and the alternative cross-presentation pathway – can initiate immune responses in mammals25. The cross-presentation pathway represents a structural and cellular modification of the MHCI machinery that allows activation of CD8+ T cells upon bacterial infection. The Atlantic cod cytokine gene profile (Supplementary Table 11) supports the possibility of generating different CD8+ T cell subsets that provide either direct protection or regulate other immune cells and thus compensate for the loss of CD4+ T cells.
Figure 3.
Major histocompatibility complex (MHC) I diversity in Atlantic cod (a) MHCI α3 domain copy number estimates. Estimates are based on qPCR Cp ratios (Supplementary Note 28) of the MHCI α3 domain and a single copy reference gene. For Atlantic cod, β-2-microglobulin and topoisomerase-III-α (*) are used as single copy reference gene, for human and stickleback β-2-microglobulin is used. 95% bootstrap confidence intervals (black dots) were calculated by bootstrapping (n = 50,000). The human and stickleback estimates agree with the expected number of α3 domains found in both reference genomes (Supplementary Table 15) (b) Phylogeny of teleost MHCI α1-3 domain amino acid sequences. The Atlantic cod sequences are derived from cDNA and comprise classical U-lineage MHCI only. The other teleost sequences were obtained from Ensembl and NCBI and contain classical and non-classical U-lineage MHCI. Alignments were visually inspected and corrected where necessary. Maximum Likelihood (ML) values and bayesian posterior probabilities (dots) support the main branches on the ML topology. Distance represents the average number of substitutions per site (scale). The ratio of non-synonymous over synonymous variable sites (Ka/Ks), average nucleotide diversity per site (π) and Tajimas D (D) were calculated for the two main clades in Atlantic cod.
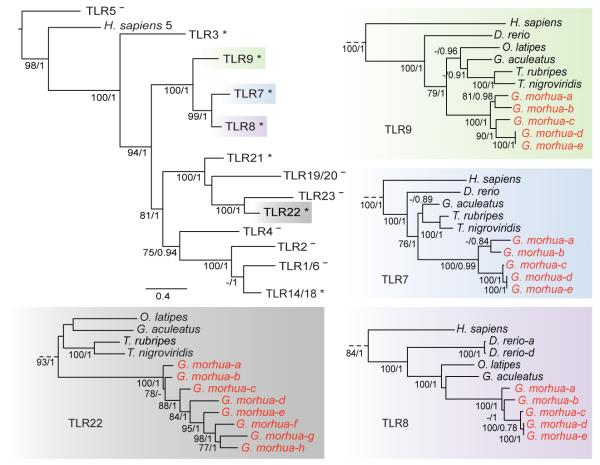
In addition to the MHCI expansion, we find an unusual composition of the highly conserved Toll-like receptor (TLR) families that play a fundamental role in the innate immune response and the initial detection of pathogens. Teleost TLR genes occur in well-supported phylogenetic clusters most of which share functional properties with mammalian orthologs though some are fish-specific26. The Atlantic cod TLR genes form monophyletic groups within the known teleost functional groups (Figure 4, Supplementary Note 30, Supplementary Figure 22). Several bacterial surface antigen-recognizing TLRs (TLR1, -2 and -5) are absent however, leaving only the teleost-specific TLR14/18 as members of the TLR1 family in Atlantic cod. Moreover, multiple families of nucleic acid-recognizing TLRs (TLR7, -8, -9 and -22) have markedly expanded, resulting in the highest number of TLRs found in a teleost so far. This TLR repertoire suggests that the Atlantic cod immune system relies relatively more on nucleic acid-detecting TLRs in order to recognize bacterial pathogens. Notably, the gene expansion of TLR9 coincides with an expansion of interleukin 8 genes (IL-8, Supplementary Table 11). IL-8 is an important chemokine in the innate immune response and is directly induced by TLR9 in human neutrophils27. The corresponding expansions of IL-8 and TLR9 indicate that this signalling cascade is particularly important in Atlantic cod.
Figure 4.
Phylogeny of Toll-like receptor (TLR) families in Atlantic cod. TLR protein sequences were selected based on the conserved TIR domain for Atlantic cod, including known sequences from stickleback, zebrafish, tetraodon, fugu, medaka and human as reference. TLR clades with (*) or without (−) Atlantic cod sequences are denoted according to human or teleost orthologs (summary tree topology, top left). Distance represents the average number of substitutions per site (scale). Maximum Likelihood (ML) values and bayesian posterior probabilities over 75/0.75 support the ML topology. Detailed topologies of TLR 7 (blue) -8 (purple) -9 (green) -22 (grey) show gene expansions for Atlantic cod (red).
The loss of MHCII function and lack of CD4+ T cell response represent a fundamental change in how the adaptive immune system is initiated and regulated in Atlantic cod. The marked expansion of MHCI genes and unusual TLR composition signify a shift of its immune system in handling microbial pathogens. An expanded MHCI repertoire in the presence of a non-polymorphic MHCII is found in a distant vertebrate, the axolotl (Ambystoma mexicanum)28,29. These observations suggest that anomalous immune systems (possibly analogous to that of Atlantic cod) have evolved independently. Additionally, we did not recover evidence for expressed MHCII, CD4 and Ii in the transcriptomes of three other gadoids, indicating that the unusual immune system is a derived characteristic of the gadoid lineage (Supplementary Table 18 and 19).
We have provided the first annotated genome of a species that supports extensive fisheries and is on the verge of becoming an important aquaculture species. This work provides a major foundation for addressing key issues related to the management of natural Atlantic cod populations, such as the concept of fisheries-induced evolution, which dictates that selective harvesting changes the evolutionary trajectory in major life history traits of natural populations30. Moreover, our novel immune findings allow for more targeted vaccine development aiding disease management and aid the process of domestication of Atlantic cod. Importantly, these findings change fundamental assumptions regarding the evolution of the vertebrate immune system.
METHODS SUMMARY
Detailed methods on the sequencing and assembly of data from genomic and transcriptomic origin, annotation, syntheny analyses, transfection experiments, bioinformatic analyses and phylogenetic analyses presented in this manuscript are described in the Supplementary Information.
Supplementary Material
Acknowledgements
This work was supported by a grant From the Research Council of Norway (FUGE program) to KSJ. The authors wish to thank the following people and organisations: the 454 Life Science Sequencing Center (Branford, USA); the 454 and Illumina nodes of the Norwegian Sequencing Centre (Univ. of Oslo); Michael Egholm (formerly 454 Life Science); the Norwegian Metacenter for Computational Science (Notur) and the Norwegian Storage Infrastructure (Norstore); the Research Computing Services (RCS) group, especially Bjørn-Helge Mevik, at the Center for Information Technology (USIT, Univ. Oslo); Brian Walenz from Celera (San Francisco, USA); the Canadian Cod Genomics and Broodstock Development Consortium; Pål Olsvik, Kai Lie and Elisabeth Holen at the Norwegian National Institute of Nutrition and Seafood Research (NIFES); Junita Gaup and Hege Bakke (CEES, Univ. of Oslo); Matthew Kent (CIGENE, Norwegian University of Life Sciences); Sharen Bowman (Genome Atlantic, Canada); the FUGE bioinformatics platforms’ group, especially Svenn Grindhaug, at CBU (Univ. Bergen, Norway); Inger Sandlie and Ole B. Landsverk (Centre for Immune Regulation, Univ. of Oslo); Roche Norway.
This manuscript is dedicated to the memory of Prof. Lars Pilström and Prof. Rene J.M. Stet. Their research inspired our work to further understand the Atlantic cod immune system.
Footnotes
Author Contributions DNA and RNA isolation, library construction and sequencing: A.T.K., M.S., M.H.S., T.B.R., M.M., M.E., B.S., A.J.N. and J.T. Sanger BAC (end-) sequencing: H.K. and R.R. Assembly: A.J.N., B.S., A.S. and A.L. Linkage map analyses: K.G.T. and B.S. SNP analyses: K.G.T., P.R.B, S.L. and A.J.N. Annotation: J.H.V., B.A. and S.S. Repeat analyses: B.S. Synteny analyses: J.P. and B.S. Haemoglobin analyses: Ø.A., O.F.W., B.S. and T.G. Bioinformatics: A.J.N., B.S., A.S., T.B.R., J.P., C.P., C.N., R.B.E., R.W., J.K., K.L., A.L., I.J., M.M, K.M., P.R.B., K.G.T. and M.H.S. Immune analyses: U.G., M.M., M.H.S., M.E., B.S., B.O.K., T.M., K.L., S.D.J. and T.B.R. Interpretation of immune results: U.G., T.F.G., S.J., B.S. and K.S.J. 454 contributions: L.D. Revisions: Ø.A., T.M., S.D.J, F.N., I.J., S.J., N.C.S and S.W.O. Project initiation: S.W.O, I.J., F.N., S.L., N.C.S. and K.S.J. Project coordination: S.J. Consortium leader: K.S.J. The unassembled sequencing reads and Newbler assembly have been deposited at ENA-EMBL under the accession numbers CAEA01000001 through CAEA01554869. The annotation is available through Ensembl at http://www.ensembl.org/index.html. These and more resources are additionally available through http://codgenome.no.
The authors declare no competing financial interests.
References
- 1.Kurlansky M. Cod: a biography of the fish that changed the world. Penguin Books Ltd.; 1998. [Google Scholar]
- 2.Johansen SD, et al. Large-scale sequence analyses of Atlantic cod. New Biotech. 2009;25:263–271. doi: 10.1016/j.nbt.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 3.Flajnik MF, Kasahara M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nature Reviews Genetics. 2010;11:47–59. doi: 10.1038/nrg2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Litman GW, Rast JP, Fugmann SD. The origins of vertebrate adaptive immunity. Nature Reviews Immunology. 2010;10:543–553. doi: 10.1038/nri2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pilstrom L, Warr GW, Stromberg S. Why is the antibody response of Atlantic cod so poor? The search for a genetic explanation. Fisheries Sci. 2005;71:961–971. [Google Scholar]
- 6.Miller JR, Koren S, Sutton G. Assembly algorithms for next-generation sequencing data. Genomics. 2010;95:315–327. doi: 10.1016/j.ygeno.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller JR, et al. Aggressive assembly of pyrosequencing reads with mates. Bioinformatics. 2008;24:2818–2824. doi: 10.1093/bioinformatics/btn548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hubert S, Higgins B, Borza T, Bowman S. Development of a SNP resource and a genetic linkage map for Atlantic cod (Gadus morhua) BMC Genomics. 2010;11 doi: 10.1186/1471-2164-11-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sick K. Haemoglobin polymorphism in fishes. Nature. 1961;192:894–896. doi: 10.1038/192894a0. [DOI] [PubMed] [Google Scholar]
- 10.Brix O, Thorkildsen S, Colosimo A. Temperature acclimation modulates the oxygen binding properties of the Atlantic cod (Gadus morhua L.) genotypes HbI*1/1, HbI*1/2, and HbI*2/2 by changing the concentrations of their major hemoglobin components (results from growth studies at different temperatures) Comp Biochem Phys A. 2004;138:241–251. doi: 10.1016/j.cbpb.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Andersen O, et al. Haemoglobin polymorphisms affect the oxygen-binding properties in Atlantic cod populations. Proc. R. Soc. B. 2009;276:833–841. doi: 10.1098/rspb.2008.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petersen MF, Steffensen JF. Preferred temperature of juvenile Atlantic cod Gadus morhua with different haemoglobin genotypes at normoxia and moderate hypoxia. J. of Exper. Biol. 2003;206:359–364. doi: 10.1242/jeb.00111. [DOI] [PubMed] [Google Scholar]
- 13.Borza T, Stone C, Gamperl AK, Bowman S. Atlantic cod (Gadus morhua) hemoglobin genes: multiplicity and polymorphism. BMC Genet. 2009;10 doi: 10.1186/1471-2156-10-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wetten O, et al. Genomic organization and gene expression of the multiple globins in Atlantic cod: conservation of globin-flanking genes in chordates infers the origin of the vertebrate globin clusters. BMC Evol. Biol. 2010;10:315. doi: 10.1186/1471-2148-10-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gamperl AK, Busby CD, Hori TSF, Afonso LOB, Hall JR. Hemoglobin genotype has minimal influence on the physiological response of juvenile Atlantic cod (Gadus morhua) to environmental challenges. Physiol Biochem Zool. 2009;82:483–494. doi: 10.1086/603636. [DOI] [PubMed] [Google Scholar]
- 16.Weber RE, Fago A. Functional adaptation and its molecular basis in vertebrate hemoglobins, neuroglobins and cytoglobins. Respir. Physiol. Neuro. 2004;144:141–159. doi: 10.1016/j.resp.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 17.Magnadottir B, Gudmundsdottir S, Gudmundsdottir BK, Helgason S. Natural antibodies of cod (Gadus morhua L. Specificity, activity and affinity. Comp Biochem Phys B. 2009;154:309–316. doi: 10.1016/j.cbpb.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 18.Solem ST, Stenvik J. Antibody repertoire development in teleosts -a review with emphasis on salmonids and Gadus morhua L. Dev Comp Immunol. 2006;30:57–76. doi: 10.1016/j.dci.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 19.Rønneseth A, Wergeland HI, Pettersen EF. Neutrophils and B-cells in Atlantic cod (Gadus morhua L. Fish Shellfish Immun. 2007;23:493–503. doi: 10.1016/j.fsi.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 20.Øverland HS, Pettersen EF, Rønneseth A, Wergeland HI. Phagocytosis by B-cells and neutrophils in Atlantic salmon (Salmo salar L.) and Atlantic cod (Gadus morhua L.) Fish Shellfish Immun. 2010;28:193–204. doi: 10.1016/j.fsi.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 21.Landsverk OJ, Bakke O, Gregers TF. MHC II and the endocytic pathway: regulation by invariant chain. Scand J Immunol. 2009;70:184–193. doi: 10.1111/j.1365-3083.2009.02301.x. [DOI] [PubMed] [Google Scholar]
- 22.Persson AC, Stet RJM, Pilstrom L. Characterization of MHC class I and beta(2)-microglobulin sequences in Atlantic cod reveals an unusually high number of expressed class I genes. Immunogenetics. 1999;50:49–59. doi: 10.1007/s002510050685. [DOI] [PubMed] [Google Scholar]
- 23.Miller KM, Kaukinen KH, Schulze AD. Expansion and contraction of major histocompatibility complex genes: a teleostean example. Immunogenetics. 2002;53:941–963. doi: 10.1007/s00251-001-0398-4. [DOI] [PubMed] [Google Scholar]
- 24.Dijkstra JM, et al. A third broad lineage of major histocompatibility complex (MHC) class I in teleost fish; MHC class II linkage and processed genes. Immunogenetics. 2007;59:305–321. doi: 10.1007/s00251-007-0198-6. [DOI] [PubMed] [Google Scholar]
- 25.Amigorena S, Savina A. Intracellular mechanisms of antigen cross presentation in dendritic cells. Curr Opin Immunol. 2010;22:109–117. doi: 10.1016/j.coi.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 26.Rebl A, Goldammer T, Seyfert HM. Toll-like receptor signaling in bony fish. Vet. Immunol. Immunopathol. 2010;134:139–150. doi: 10.1016/j.vetimm.2009.09.021. [DOI] [PubMed] [Google Scholar]
- 27.József L, Khreiss T, El Kebir D, Filep JG. Activation of TLR-9 induces IL-8 secretion through peroxynitrite signaling in human neutrophils. J Immunol. 2006;176:1195–1202. doi: 10.4049/jimmunol.176.2.1195. [DOI] [PubMed] [Google Scholar]
- 28.Tournefier A, et al. Structure of MHC class I and class II cDNAs and possible immunodeficiency linked to class II expression in the Mexican axolotl. Immunol Rev. 1998;166:259–277. doi: 10.1111/j.1600-065x.1998.tb01268.x. [DOI] [PubMed] [Google Scholar]
- 29.Kaufman J, Volk H, Wallny HJ. A “minimal essential Mhc” and an “unrecognized Mhc”: two extremes in selection for polymorphism. Immunol Rev. 1995;143:63–88. doi: 10.1111/j.1600-065x.1995.tb00670.x. [DOI] [PubMed] [Google Scholar]
- 30.Stenseth NC, Dunlop ES. Evolution: Unnatural selection. Nature. 2009;457:803–804. doi: 10.1038/457803a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.