Abstract
The latency-related (LR) gene of bovine herpesvirus 1 (BHV-1) is abundantly expressed during latency. A mutant BHV-1 strain that contains three stop codons at the 5′ terminus of the LR gene (LR mutant) does not reactivate from latency. This study demonstrates that the LR mutant does not express open reading frame 2 or an adjacent reading frame that lacks an initiating ATG (reading frame C). Since the LR mutant and wild-type BHV-1 express similar levels of LR RNA, we conclude that LR protein expression plays an important role in regulating the latency reactivation cycle in cattle.
Bovine herpesvirus 1 (BHV-1) belongs to the subfamily Alphaherpesvirinae and shares a number of biological properties with herpes simplex virus types 1 and 2 (HSV-1 and HSV-2) (19). BHV-1 establishes lifelong latency in trigeminal ganglionic neurons of the peripheral nervous system after initial replication in mucosal epithelium. Virus reactivation and spread to other susceptible animals occur after natural or corticosteroid-induced stress (28, 32). Infection can lead to conjunctivitis, pneumonia, genital disorders, abortions, and an upper respiratory infection referred to as “shipping fever” (35), which cost the U.S. cattle industry at least $500 million/year (1). Binding and entry of Pasteurella haemolytica and Pasteurella multocida into bovine cells is enhanced if the cells are infected with BHV-1 (9), supporting other studies concluding that BHV-1 infection can promote secondary bacterial infections (3, 10-12). CD8+-T-cell recognition of infected cells is impaired by repressing expression of major histocompatibility complex class I and the transporter associated with antigen presentation (13, 14, 26). CD4+-T-cell function is impaired during acute infection of calves because BHV-1 infects CD4+ T cells and induces apoptosis (36).
The latency-related (LR) RNA is the only abundant viral transcript detected in latently infected neurons (22, 28, 29). A fraction of LR RNA is polyadenylated and alternatively spliced in trigeminal ganglia, suggesting that LR RNA can be translated into more than one protein (6, 15). LR gene products inhibit S-phase entry, and an LR protein is associated with cyclin-dependent kinase 2 (cdk2) or cdc2/cyclin complexes (15, 18). LR gene products also inhibit apoptosis (4), suggesting that one important function of the LR gene is to promote neuronal survival. A mutant BHV-1 strain that contains three stop codons near the beginning of the LR RNA was constructed to test whether LR proteins play a role in virus growth in cattle (17). Calves infected with the LR mutant exhibit diminished clinical symptoms and reduced virus shedding from the eye or trigeminal ganglionic virus relative to calves infected with wild-type (wt) or the LR rescued virus (16, 17). Conversely, the LR mutant had similar growth properties in productively infected bovine kidney cells and the nasal cavities of calves during acute infection. The LR mutant virus does not reactivate from latency following treatment with dexamethasone, whereas all calves latently infected with wt virus or the LR rescued virus shed infectious virus following dexamethasone treatment. During the transition from acute infection to latency (establishment of latency), higher levels of apoptosis occur in trigeminal ganglionic neurons of calves infected with the LR mutant than in those of calves infected with wt BHV-1 (24). These studies indicate that wt expression of LR gene products is required for the latency-reactivation cycle in cattle.
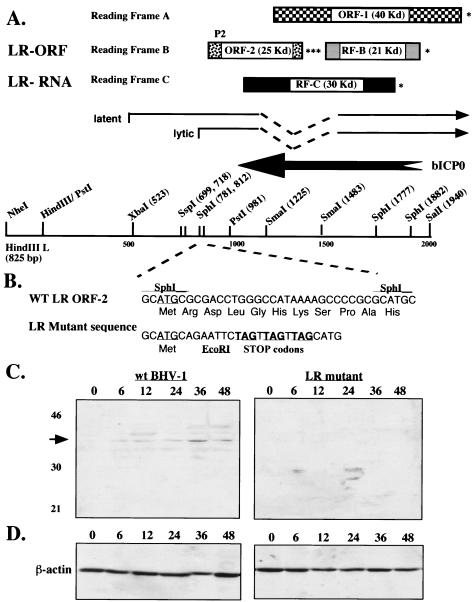
The LR gene has two open reading frames (ORFs), ORF-1 and ORF-2, and two reading frames that lack an initiating ATG (reading frame B [RF-B] and RF-C) (Fig. 1A). The LR mutant that was prepared contains three stop codons adjacent to the first in-frame ATG of ORF-2 (Fig. 1B), suggesting that the LR mutant would not express a protein containing the product of ORF-2. An antibody directed against a peptide located near the amino terminus of the ORF-2 product (P2) recognizes a protein migrating near 40 kDa in productively infected cells or cells transiently transfected with the wt LR gene (4, 15, 18, 30). Since the ORF-2 product is only 25 kDa, we suggested that splicing and/or posttranslational modifications yielded a larger protein. Another peptide antibody directed against the N terminus of the ORF-1 product did not specifically recognize a virus-specific protein in infected or transfected cells (15). Alternative splicing of LR RNA yields a family of transcripts, and one of these transcripts has the potential to be translated into a protein that contains the ORF-2 product fused to the ORF-1 product (6).
FIG. 1.
Schematic of the BHV-1 LR gene and expression of an LR protein in infected cells. (A) Partial restriction map, location of LR RNA, organization of ORFs, and the 3′ terminus of bICP0. The start sites for LR transcription during latency and productive infection were previously described (2, 15). RF-B contains a reading frame after ORF-2 that lacks an initiating Met codon. RF-C also contains a reading frame that lacks an initiating Met codon. The asterisk denotes the position of stop codons that are in frame with the respective ORF or reading frame. The approximate sizes of the ORF and reading frames without an ATG are given. The position of the P2 peptide used to generate peptide-specific antibodies was described previously (15). (B) DNA sequence of the SphI fragment and the mutation in the LR mutant. The first in-frame ATG for ORF-2 is underlined. Stop codons in the mutant oligonucleotide are in all three reading frames (bold and underlined). The EcoRI restriction enzyme site (GAATTC) was incorporated into the mutant oligonucleotide to facilitate screening for the recombinant virus. (C) MDBK cells were infected with virus at a multiplicity of infection of 5, and at the designated times after infection (hours), whole-cell lysate was prepared. A peptide antibody directed against the amino terminus of ORF-2 (P2 antibody, diluted 1:500) was used to probe the Western blot. The arrow indicates the position of the LR-specific protein that migrates with an approximate molecular mass of 40 kDa. One hundred micrograms of protein was used for each lane. (D) The samples (100 μg) used for panel C were electrophoresed, the proteins were transferred to a membrane, and a Western blot was performed with an antibody directed against β-actin (sc-1615; Santa Cruz Biotechnology, Santa Cruz, Calif.).
Bovine kidney cells (MDBK) infected with wt BHV-1, but not the LR mutant, express a protein with an approximate molecular mass of 40 kDa that was recognized by the P2 antibody (Fig. 1C). As previously reported (18), this protein was expressed only at late times after infection. Although the P2 peptide antibody has been useful for certain studies, the 40-kDa protein was difficult to detect in some experiments. We have also observed considerable variability in titers of P2-specific antibodies from rabbit to rabbit, suggesting that the P2 peptide is not a good immunogen. Consequently, we felt that it was necessary to develop a better antibody against ORF-2.
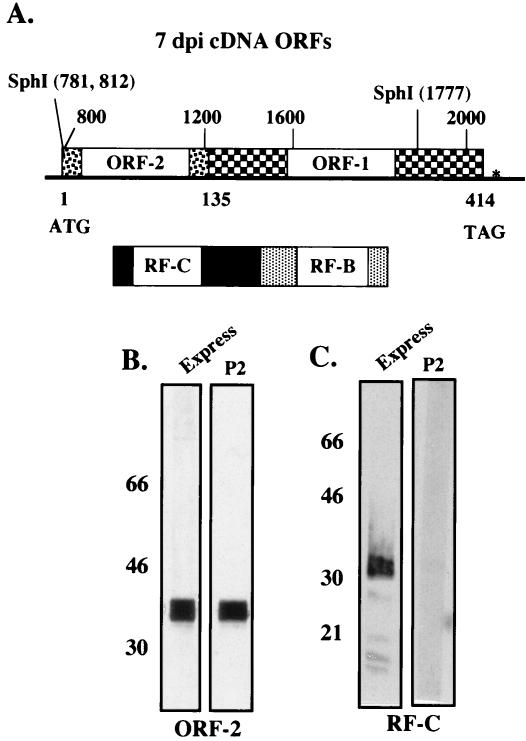
To develop better antibodies directed against ORF-2, an LR cDNA was cloned into a baculovirus expression vector that was identified in trigeminal ganglia of calves infected for 7 days (Fig. 2A). This cDNA contains most of the ORF-2 sequences, and because of splicing, ORF-2 is fused to most of the coding sequences comprising ORF-1 (6). RF-C is also fused to a portion of RF-B because of the unique splicing that occurred to generate the 7-day cDNA. To express RF-C in baculovirus, the 7-day cDNA was released from pcDNA3.1(−) using EcoRI and SalI and cloned into pBlueBacHis2, a baculovirus recombination plasmid (Invitrogen, Carlsbad, Calif.). To generate a baculovirus that would express ORF-2 from the 7-day cDNA, a SphI fragment derived from the 7-day cDNA was cloned into pBlueBacHis2 such that ORF-2 was in frame with the His tag (see the Fig. 2 legend for details of the cloning). Recombinant baculoviruses were prepared by using Bac-N-Blue DNA (Invitrogen), and infectious baculovirus was propagated in insect cells (SF9).
FIG. 2.
Expression of LR ORFs in baculovirus expression vectors. (A) An alternatively spliced LR transcript that was identified in trigeminal ganglia of infected calves at 7 days after infection (dpi) (6) was cloned into pcDNA3.1− (5). This cDNA is spliced at nucleotides 1180 and 1363 such that ORF-2 is fused with ORF-1. The numbers above the ORF-2 schematic are nucleotide numbers of the LR gene, and the numbers below ORF-2 are amino acid numbers. Splicing also generates a fusion protein containing a portion of the RF-C product fused to the RF-B product. The LR cDNA begins at nucleotide 765 and ends at nucleotide 2182. The details of cloning and characterizing the properties of this cDNA were described elsewhere (5). To construct a baculovirus that expressed ORF-2, the SphI fragment containing ORF-2 was cloned into the pGEM-11Zf(−) vector (Promega, Madison, Wis.) into the unique SphI site such that the SphI site at LR nucleotide 810 was adjacent to the unique BamHI site in the vector. This plasmid was digested with HindIII, treated with the Klenow fragment, and then digested with XhoI to release the LR fragment. This LR fragment was cloned into pGEX(GST-5X) (Pharmacia Biotech, Piscataway, N.J.) at unique SmaI and XhoI sites. The resulting fragment was digested to completion with XhoI and then partially digested with BamHI. The resulting BamHI-XhoI fragment was then cloned into pBlueBacHis2 at the unique BamHI-XhoI sites. This method fused ORF-2 in frame with the His fusion sequences and allowed a recombinant baculovirus that expressed the ORF-2 fusion protein to be constructed. (B and C) Recombinant baculovirus strains were constructed and the viruses were grown in SF9 insect cells using procedures described by Invitrogen. Recombinants were characterized by testing for LR protein expression using the Express antibody that recognizes the His tag at the 5′ terminus of LR protein sequences (R910-25; Invitrogen). Large-scale expression was carried out in 2-liter flasks seeded with SF9 cells at a density of 2 × 106/ml in a total volume of 1,000 ml. At 4 days after infection, cells were pelleted by centrifuging for 30 min at 8,000 rpm (Beckman J2-21 centrifuge, JA-10 rotor), suspended in 20 ml of guanidinium lysis buffer (6 M guanidine hydrochloride, 20 mM sodium phosphate, 500 mM NaCl; pH 7.8). Suspended cells were then passed through an 18-gauge needle four times to lyse the cells and shear cellular DNA. (B) His fusion protein purified from SF9 cells infected with the ORF-2 fusion; (C) RF-C fusion protein. The respective protein was loaded at 2 μg/lane, Western blots were performed, and the respective lanes were cut and incubated with the designated antibodies. The P2 antibody was diluted 1:500 for these studies. Western blot analysis was performed with the designated antibodies as previously described (18).
The ORF-2 and RF-C fusion proteins were partially purified by nickel affinity chromatography, using a kit purchased from Invitrogen. With respect to the ORF-2 fusion construct, a His tag-specific antibody (Express) specifically recognized a protein migrating with an approximate molecular mass of 40 kDa (Fig. 2B). The predicted molecular mass of the truncated ORF-2 fusion protein that contains the His tag at its amino terminus is approximately 40 kDa. The P2 antibody also specifically reacted with the ORF-2 fusion protein, as expected. When the RF-C fusion protein was partially purified, the Express antibody specifically recognized a protein with an approximate molecular mass of 30 kDa (Fig. 2C). Splicing of the 7-day cDNA leads to the RF-C protein being fused to the C-terminal sequences in the RF-B product (Fig. 2A), and with the His tag at the N terminus, the RF-C fusion protein was expected to migrate near 30 kDa. The P2 antibody did not recognize the RF-C fusion protein that migrated near 30 kDa, as expected. The two fusion proteins were further purified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the fusion protein bands were excised, eluted, and then injected into rabbits to generate polyclonal antibodies.
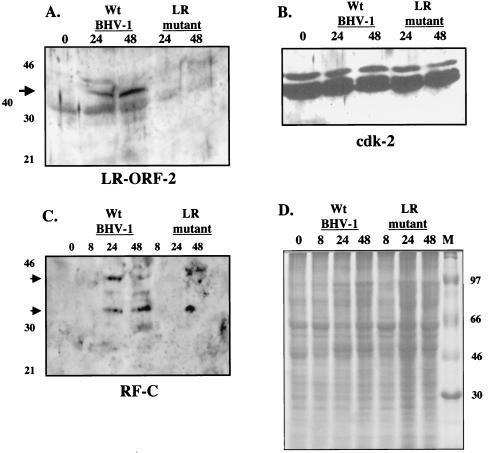
When crude cell lysate was prepared from MDBK cells infected with wt BHV-1 for 24 or 48 h (Fig. 3A), the ORF-2 antibody consistently detected a protein migrating with an approximate molecular mass of 40 kDa. The 40-kDa protein was not detected in MDBK cells infected with the LR mutant even when the blot was overexposed as shown in Fig. 3A, but it was detected when cells were infected with the LR rescued virus (data not shown). In most experiments, the crude serum cross-reacted with a cellular protein migrating around 35 kDa. A polyclonal antibody directed against cdk-2 recognized similar levels of cdk-2 in cells infected with the LR mutant and in those with wt BHV-1 (Fig. 3B).
FIG. 3.
Western blot of MDBK cells infected with wt BHV-1 or the LR mutant. MDBK cells were infected with virus at a multiplicity of infection of 4, and at the designated times after infection (hours), whole-cell lysate prepared. The polyclonal antibody directed against the ORF-2 fusion protein made from a baculovirus vector (A) or cdk2 (sc-748; Santa Cruz Biotechnology) (B) was used for Western blot analysis. All lanes in panels A and B were loaded with 200 μg of protein. The ORF-2 protein-specific antiserum was diluted 1:500 for Western blots, and the cdk2 antibody was diluted as described by the manufacturer. (C) Total cell lysate (2 mg protein) from mock-infected MDBK cells or cells infected with the designated virus for 8, 24, or 48 h after was incubated with 5 μg of IgG from normal rabbit serum for 1 h at 4°C. Immune complexes were collected by using protein A beads. The supernatant was incubated with 3 μg of IgG prepared from the RF-C protein-specific antiserum overnight at 4°C, and immunocomplexes were collected by using protein A beads. The immunoprecipitates were washed as described previously (37). The immunoprecipitates were electrophoresed on a sodium dodecyl sulfate-12% polyacrylamide gel, and Western blot analysis was performed with the RF-C protein-specific IgG (1 μl of a 0.7-mg/ml solution that was diluted 1:500). The details of the immunoprecipitation-Western blot assays were described previously (37). (D) The samples (25 μg of protein) described for panel C were electrophoresed and stained with Coomassie blue. The arrows show the position of the LR-specific proteins detected in the cells infected with wt BHV-1 or the LR rescued virus (data not shown). The results are representative of at least three experiments.
The antiserum directed against RF-C protein reacted with several virus-specific proteins when MDBK cells were infected with wt BHV-1 or the LR mutant and in mock-infected cells (data not shown). This suggested that the RF-C serum cross-reacted with other proteins or that the mutation at the beginning of ORF-2 did not block RF-C protein expression. To test these possibilities, the immunoglobulin G (IgG) present in RF-C antiserum was purified and used to perform immunoprecipitation-Western blot studies as described previously (37). MDBK cells infected with wt BHV-1 (Fig. 3C) or the LR rescued virus (data not shown) contained two proteins, one migrating near 30 kDa and one near 40 to 42 kDa, that were recognized by IgG in the RF-C antiserum at 24 h after infection. At 48 h after infection, the 30-kDa protein was readily detected but the larger protein was not. In contrast, the same IgG preparation did not detect these proteins in crude cell lysate prepared from MDBK cells infected with the LR mutant, even when the Western blot was overexposed. For the immunoprecipitation assays, similar levels of total protein were used (Fig. 3D). In summary, these studies demonstrated that the stop codons in the LR mutant prevented expression of ORF-2 and RF-C.
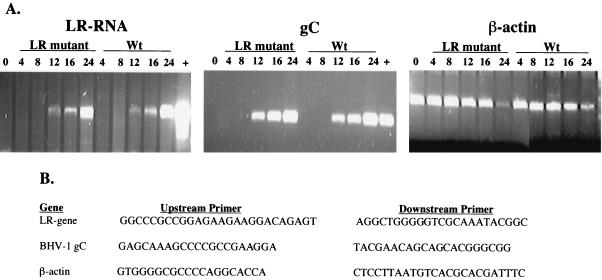
Although LR RNA expression was detected in calves latently infected with the LR mutant by RT-PCR (16), we did not analyze LR RNA expression in bovine cells following infection with the LR mutant or wt BHV-1. The results presented in Fig. 3 suggested that the LR gene encoded two immunologically distinct proteins. However, it was also possible that the LR mutant did not express ORF-2 or the proteins recognized by the RF-C product-specific antibody because the LR mutant virus expressed low levels of LR RNA. To test these possibilities, semiquantitative RT-PCR was performed using primers that specifically amplify the LR RNA family of transcripts, the BHV-1-encoded glycoprotein C mRNA, or β-actin mRNA (Fig. 4B). There was no dramatic difference in the steady-state levels of LR RNA when MDBK cells were infected with the LR mutant or wt BHV-1 (Fig. 4A). Similar levels of gC RNA were also expressed in cells infected with the LR mutant and those infected with wt BHV-1 (Fig. 4A). Since the LR mutant and the wt virus express similar levels of bICP0 and release the same amount of infectious virus during productive infection (17), gC RNA expression was expected to be the same. Finally, no dramatic differences were detected when β-actin levels were compared in cells infected with the LR mutant and those infected with wt BHV-1. In summary, the levels of LR RNA, gC RNA, and β-actin were similar in MDBK cells following infection with wt BHV-1 and with the LR mutant.
FIG. 4.
RNA expression in infected MDBK cells (A) MDBK cells were infected with wt BHV-1 or the LR mutant at a multiplicity of infection of 5. At the designated times after infection (hours), RNA was prepared from infected cells and cDNA synthesis was performed as previously described (16). To detect LR RNA, first-strand cDNA synthesis was performed with the L3A downstream primer, which is antisense to LR RNA. The L3A downstream primer spans nucleotides 1835 to 1815 (5′-GACGAGACCCCCGATTGCCG-3′) and was described previously (15). Oligo(dT) priming was performed to detect gC and β-actin mRNA. +, plasmids containing the LR or gC gene were included as positive controls. Similar results were obtained with the LR rescued virus (data not shown). These results are representative of three studies. (B) DNA sequences of primers used to amplify the cDNA used for panel A. The upstream LR gene primer was originally designated primer 1629, and the downstream primer was primer 872 (15). The gC primers were described previously (31). All primers are listed as 5′-3′.
The three stop codons within the LR mutant were designed to inhibit synthesis of proteins encoded by ORF-2 but not interfere with bICP0 expression and transcription of LR RNA. Previous studies demonstrated that plasmids containing the stop codons express similar levels of LR RNA but not the 40-kDa protein in transiently transfected cells (4) and that LR RNA is expressed in calves latently infected with the LR mutant (16). It is clear that this mutation does not inhibit bICP0 protein expression and that the LR mutant has nearly identical growth properties in cultured bovine cells (17). The results obtained with the P2 antibody are consistent with the results obtained with the ORF-2 protein-specific antiserum, strongly suggesting that a protein containing a portion of the ORF-2 product is expressed.
To our surprise, two distinct proteins recognized by the RF-C protein-specific IgG were expressed in productive infection, but the LR mutant did not express these proteins (Fig. 3). The protein migrating at approximately 30 kDa is the predicted size of the RF-C product (Fig. 1A), suggesting that unspliced LR transcripts produced RF-C protein. The larger protein may be a posttranslationally modified form of the protein migrating near 30 kDa or the result of alternative splicing of the LR RNA. Since the RF-C protein-specific antiserum was made against a RF-C/RF-B fusion protein expressed from baculovirus, it is possible that one of the proteins was derived from RF-C and the other from RF-B. Although reading frames without initiating ATGs can be expressed (8, 21), we cannot rule out the possibility that a small splice occurs near the beginning of RF-C, which brings an adjacent ATG in frame with RF-C. We are in the process of preparing antiserum that recognizes only RF-C, RF-B, and ORF-2 products to test these possibilities.
Insertion of the LR gene into the HSV-1 latency-associated transcript (LAT) locus restores high levels of spontaneous reactivation in the rabbit eye model and in explant-induced reactivation (27), suggesting that the LR gene and the LAT contain common function(s) that promote the latency-reactivation cycle of HSV-1. When the LR gene is mutated and recombined into the HSV-1 LAT locus, this virus behaves like a LAT-minus mutant (25), suggesting that spontaneous reactivation was stimulated by expression of the BHV-1 LR ORF-2 and/or RF-C. LAT contains small ORFs, and there is a report of a latency-associated antigen (7). Overexpression of LAT ORF-2 stimulates productive infection in neuronal cells because it can substitute for ICP0 functions, suggesting that it has functional significance during reactivation from latency (33, 34). An independent study concluded that ORFs within the 2-kb LAT are expressed in transient-transfection assays, but LAT protein expression is not detected in infected cells (23). Interestingly, Lock et al. (23) also concluded that LAT protein expression is regulated at the level of translation. If one compares the ORFs in the LR gene with the LAT, there is no striking similarity, suggesting that a regulatory RNA encoded by the LAT promotes the high-reactivation phenotype in rabbits. The only known shared function of the LAT and the LR gene is inhibiting apoptosis (20), suggesting that an antiapoptosis function plays an important role in the latency reactivation cycle of BHV-1 and HSV-1. In the context of the BHV-1 genome, it will be difficult to make mutations that block expression of RF-C without altering ORF-2 expression or inhibiting bICP0 expression, because the RF-C sequences are contained in the coding sequences of bICP0 and ORF-2. However, such studies are possible if these individual protein coding domains are inserted into an HSV-1 LAT mutant and then assayed in a rabbit or mouse model system.
Acknowledgments
This research was supported by grants from the USDA (2002-02060 and 2003-02213) and NIH (P20RR15635). A faculty development award from the Universidad de Santiago De Compostela supported Nuria Alemañ Posadas.
REFERENCES
- 1.Bowland, S. L., and P. E. Shewen. 2000. Bovine respiratory disease: commercial vaccines currently available in Canada. Can. Vet. J. 41:33-48. [PMC free article] [PubMed] [Google Scholar]
- 2.Bratanich, A. C., N. D. Hanson, and C. J. Jones. 1992. The latency-related gene of bovine herpesvirus 1 inhibits the activity of immediate-early transcription unit 1. Virology 191:988-991. [DOI] [PubMed] [Google Scholar]
- 3.Carter, J. J., A. D. Weinberg, A. Pollard, R. Reeves, J. A. Magnuson, and N. S. Magnuson. 1989. Inhibition of T-lymphocyte mitogenic responses and effects on cell functions by bovine herpesvirus 1. J. Virol. 63:1525-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciacci-Zanella, J., M. Stone, G. Henderson, and C. Jones. 1999. The latency-related gene of bovine herpesvirus 1 inhibits programmed cell death. J. Virol. 73:9734-9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devireddy, L., Y. Zhang, and C. Jones. 2003. Cloning and initial characterization of an alternatively spliced transcript encoded by the bovine herpes virus 1 latency related (LR) gene. J. Neurovirol. 9:612−622. [DOI] [PubMed]
- 6.Devireddy, L. R., and C. Jones. 1998. Alternative splicing of the latency-related transcript of bovine herpesvirus 1 yields RNAs containing unique open reading frames. J. Virol. 72:7294-7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doerig, C., L. I. Pizer, and C. L. Wilcox. 1991. An antigen encoded by the latency-associated transcript in neuronal cell cultures latently infected with herpes simplex virus type 1. J. Virol. 65:2724-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drabkin, H. J., and U. L. RajBhandary. 1998. Initiation of protein synthesis in mammalian cells with codons other than AUG and amino acids other than methionine. Mol. Cell. Biol. 18:5140-5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galdiero, M., M. G. Pisciotta, A. Marinelli, G. Petrillo, E. Galdiero. 2002. Coinfection with BHV-1 modulates cell adhesion and invasion by P. multocida and Mannheima (Pasteurella) haemolytica. New Microbiol. 25:427-436. [PubMed] [Google Scholar]
- 10.Griebel, P. J., H. B. Ohmann, M. J. Lawman, and L. A. Babiuk. 1990. The interaction between bovine herpesvirus type 1 and activated bovine T lymphocytes. J. Gen. Virol. 71:369-377. [DOI] [PubMed] [Google Scholar]
- 11.Griebel, P. J., L. Qualtiere, W. C. Davis, A. Gee, H. Bielefeldt Ohmann, M. J. Lawman, and L. A. Babiuk. 1987. T lymphocyte population dynamics and function following a primary bovine herpesvirus type-1 infection. Viral Immunol. 1:287-304. [DOI] [PubMed] [Google Scholar]
- 12.Griebel, P. J., L. Qualtiere, W. C. Davis, M. J. Lawman, and L. A. Babiuk. 1987. Bovine peripheral blood leukocyte subpopulation dynamics following a primary bovine herpesvirus-1 infection. Viral Immunol. 1:267-286. [DOI] [PubMed] [Google Scholar]
- 13.Hariharan, M. J., C. Nataraj, and S. Srikumaran. 1993. Down regulation of murine MHC class I expression by bovine herpesvirus 1. Viral Immunol. 6:273-284. [DOI] [PubMed] [Google Scholar]
- 14.Hinkley, S., A. B. Hill, and S. Srikumaran. 1998. Bovine herpesvirus-1 infection affects the peptide transport activity in bovine cells. Virus Res. 53:91-96. [DOI] [PubMed] [Google Scholar]
- 15.Hossain, A., L. M. Schang, and C. Jones. 1995. Identification of gene products encoded by the latency-related gene of bovine herpesvirus 1. J. Virol. 69:5345-5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inman, M., L. Lovato, A. Doster, and C. Jones. 2002. A mutation in the latency related gene of bovine herpesvirus 1 interferes with the latency-reactivation cycle of latency in calves. J. Virol. 76:6771-6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inman, M., L. Lovato, A. Doster, and C. Jones. 2001. A mutation in the latency-related gene of bovine herpesvirus 1 leads to impaired ocular shedding in acutely infected calves. J. Virol. 75:8507-8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang, Y., A. Hossain, M. T. Winkler, T. Holt, A. Doster, and C. Jones. 1998. A protein encoded by the latency-related gene of bovine herpesvirus 1 is expressed in trigeminal ganglionic neurons of latently infected cattle and interacts with cyclin-dependent kinase 2 during productive infection. J. Virol. 72:8133-8142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones, C. 1998. Alphaherpesvirus latency: its role in disease and survival of the virus in nature. Adv. Virus Res. 51:81-133. [DOI] [PubMed] [Google Scholar]
- 20.Jones, C. 2003. Herpes simplex virus type 1 and bovine herpesvirus 1 latency. Clin. Microbiol. Rev. 16:79-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozak, M. 2001. New ways of initiating translation in eukaryotes? Mol. Cell. Biol. 21:1899-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kutish, G., T. Mainprize, and D. Rock. 1990. Characterization of the latency-related transcriptionally active region of the bovine herpesvirus 1 genome. J. Virol. 64:5730-5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lock, M., C. Miller, and N. W. Fraser. 2001. Analysis of protein expression from within the region encoding the 2.0-kilobase latency-associated transcript of herpes simplex virus type 1. J. Virol. 75:3413-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lovato, L., M. Inman, G. Henderson, A. Doster, and C. Jones. 2003. Infection of cattle with a bovine herpesvirus 1 (BHV-1) strain that contains a mutation in the latency related gene leads to increased apoptosis in trigeminal ganglia during the transition from acute infection to latency. J. Virol. 77:4848-4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mott, K., N. Osorio, L. Jin, D. Brick, J. Naito, J. Cooper, G. Henderson, M. Inman, C. Jones, S. L. Wechsler, and G.-C. Perng. 2003. The bovine herpesvirus-1 LR ORF2 is crucial for this gene's ability to restore the high reactivation phenotype to a herpes simplex virus-1 LAT null mutant. J. Gen. Virol. 84:2975-2985. [DOI] [PubMed] [Google Scholar]
- 26.Nataraj, C., S. Eidmann, M. J. Hariharan, J. H. Sur, G. A. Perry, and S. Srikumaran. 1997. Bovine herpesvirus 1 downregulates the expression of bovine MHC class I molecules. Viral Immunol. 10:21-34. [DOI] [PubMed] [Google Scholar]
- 27.Perng, G.-C., B. Maguen, L. Jin, K. R. Mott, N. Osorio, S. M. Slanina, A. Yukht, H. Ghiasi, A. B. Nesburn, M. Inman, G. Henderson, C. Jones, and S. L. Wechsler. 2002. A gene capable of blocking apoptosis can substitute for the herpes simplex virus type 1 latency-associated transcript gene and restore wild-type reactivation levels. J. Virol. 76:1224-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rock, D., J. Lokensgard, T. Lewis, and G. Kutish. 1992. Characterization of dexamethasone-induced reactivation of latent bovine herpesvirus 1. J. Virol. 66:2484-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rock, D. L., S. L. Beam, and J. E. Mayfield. 1987. Mapping bovine herpesvirus type 1 latency-related RNA in trigeminal ganglia of latently infected rabbits. J. Virol. 61:3827-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schang, L. M., A. Hossain, and C. Jones. 1996. The latency-related gene of bovine herpesvirus 1 encodes a product which inhibits cell cycle progression. J. Virol. 70:3807-3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schang, L. M., and C. Jones. 1997. Analysis of bovine herpesvirus 1 transcripts during a primary infection of trigeminal ganglia of cattle. J. Virol. 71:6786-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheffy, B. E., and D. H. Davies. 1972. Reactivation of a bovine herpesvirus after corticosteroid treatment. Proc. Soc. Exp. Biol. Med. 140:974-976. [DOI] [PubMed] [Google Scholar]
- 33.Thomas, S. K., G. Gough, D. S. Latchman, and R. S. Coffin. 1999. Herpes simplex virus latency-associated transcript encodes a protein which greatly enhances virus growth, can compensate for deficiencies in immediate-early gene expression, and is likely to function during reactivation from virus latency. J. Virol. 73:6618-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas, S. K., C. E. Lilley, D. S. Latchman, and R. S. Coffin. 2002. A protein encoded by the herpes simplex virus (HSV) type 1 2-kilobase latency-associated transcript is phosphorylated, localized to the nucleus, and overcomes the repression of expression from exogenous promoters when inserted into the quiescent HSV genome. J. Virol. 76:4056-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tikoo, S. K., M. Campos, and L. A. Babiuk. 1995. Bovine herpesvirus 1 (BHV-1): biology, pathogenesis, and control. Adv. Virus Res. 45:191-223. [DOI] [PubMed] [Google Scholar]
- 36.Winkler, M. T., A. Doster, and C. Jones. 1999. Bovine herpesvirus 1 can infect CD4+ T lymphocytes and induce programmed cell death during acute infection of cattle. J. Virol. 73:8657-8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, Y., and C. Jones. 2001. The bovine herpesvirus 1 immediate-early protein (bICP0) associates with histone deacetylase 1 to activate transcription. J. Virol. 75:9571-9578. [DOI] [PMC free article] [PubMed] [Google Scholar]