Abstract
The human monoclonal antibody 2F5 neutralizes primary human immunodeficiency virus type 1 (HIV-1) with rare breadth and potency. A crystal structure of a complex of 2F5 and a peptide corresponding to its core epitope on gp41, ELDKWAS, revealed that the peptide interacts with residues at the base of the unusually long (22-residue) third complementarity-determining region of the heavy chain (CDR H3) but not the apex. Here, we perform alanine-scanning mutagenesis across CDR H3 and make additional substitutions of selected residues to map the paratope of Fab 2F5. Substitution of residues from the base of the H3 loop or from CDRs H1, H2, and L3, which are proximal to the peptide, significantly diminished the affinity of Fab 2F5 for gp41 and a short peptide containing the 2F5 core motif. However, nonconservative substitutions to a phenylalanine residue at the apex of the H3 loop also markedly decreased 2F5 binding to both gp41 and the peptide, suggesting that recognition of the core epitope is crucially dependent on features at the apex of the H3 loop. Furthermore, substitution at the apex of the H3 loop had an even more pronounced effect on the neutralizing activity of 2F5 against three sensitive HIV-1. These observations present a challenge to vaccine strategies based on peptide mimics of the linear epitope.
The target of human immunodeficiency virus type 1 (HIV-1) neutralizing antibodies (Abs) is the putative trimer of gp120-gp41 heterodimers that decorates the surface of HIV-1 (10, 28, 43, 52, 54). In the case of gp41, it appears that antibody access to neutralizing epitopes may be more restricted than access to those on gp120, since the relevant epitopes on gp41 probably become fully exposed only during HIV-1 envelope-mediated virus-cell membrane fusion (4, 19, 20, 46). The two anti-gp41 monoclonal Abs (MAbs) that are the most potent and broadly neutralizing are the human immunoglobulin G (IgG) MAbs 2F5 and 4E10 (12, 14, 16, 21, 47, 49, 58). The core epitope of 2F5, the most studied of the two MAbs, has been defined conveniently by a short linear sequence, ELDKWA, which is found at the extreme C-terminal end of the C-heptad repeat region on the ectodomain of gp41 (37). MAb 4E10 appears to recognize an epitope immediately C-terminal to the 2F5 epitope. The 4E10 epitope has been defined by the sequence NWFDIT from mapping with a phage display expression library of gp160 gene fragments as well as overlapping peptides (47, 58).
2F5 not only neutralizes primary HIV-1 from several different subtypes but confers protection against challenge by immunodeficiency virus by passive transfer in animal models (3, 23, 31, 32). Thus, a justifiable interest has arisen in developing immunogens capable of eliciting 2F5-like Abs by immunization. Unfortunately, despite many attempts, the neutralizing activity of 2F5 has not been recapitulated by immunizing with either gp41, gp160, or a variety of immunogens bearing the 2F5 core epitope in different contexts (11, 17, 25, 29, 34, 36). The importance of residues flanking the ELDKWA sequence in binding 2F5 has been revealed (25, 34, 39, 48, 58), which could explain the inability of some, but not all, of the immunogens tested to elicit neutralizing Abs. A crystal structure of the peptide ELDKWAS in complex with Fab 2F5 revealed a β-turn conformation in the peptide (38), which led to design of a constrained β-lactam bridge that enhanced reactivity of 2F5 to a 13-mer peptide (34). However, this constrained peptide was still unable to elicit neutralizing antisera in guinea pigs despite high antipeptide Ab titers.
One interpretation of the inability to elicit 2F5-like Abs in animals is that it may be difficult to elicit an Ab with a very long third complementarity-determining region of the heavy chain (CDR H3), as is found in 2F5. This 22-residue CDR H3 loop in 2F5 is much longer than the average length of CDR H3s in humans, rabbits, and mice, ∼13, ∼11 to 12, and ∼9 to 10 residues, respectively (53). (The average length of H3 loops for guinea pigs has not been determined.) Although H3 loops of around 20 or more residues are not rare in humans, the immunization experiments cited above were done mostly with mice, rabbits, and guinea pigs. However, it is not clear whether a long H3 loop is actually a requirement for Abs to interact with the 2F5 epitope and neutralize HIV-1. In the crystal structure of Fab 2F5 in complex with ELDKWAS, the apex of the H3 loop is distant from the bound peptide (38). This lack of apparent interaction between the apex of H3 and the peptide caused us to reconsider whether the length per se of the H3 loop of 2F5 is relevant for its neutralizing activity. On the other hand, the tip of the CDR H3 of 2F5 might be involved in a crucial interaction with an as yet unidentified determinant on the virion that might need to be incorporated in an immunogen designed to elicit 2F5-like Abs.
Since eliciting an Ab with 100% sequence identity to 2F5 is extremely unlikely, it is beneficial to delineate more precisely the molecular requirements of 2F5 for binding to its epitope on gp41. Recently, we reported on an extensive study in which alanine-scanning mutagenesis was used to map the paratope of the broadly neutralizing MAb b12 (59) by identifying residues that are key for recognition of gp120. In that study, the entire H3 loop of Fab b12 was “scanned” using Ala mutagenesis. Furthermore, residues outside CDR H3 of b12 were also mutated; many were “back mutated” to the residue encoded by the closest germ line gene. Here, we took a similar approach to mapping the paratope of Fab 2F5. As a first step, the amino acid sequence of 2F5 (38) was used as the template to engineer a Fab 2F5 that could be expressed in Escherichia coli. Overlapping oligonucleotide primers spanning the VH and Vκ regions were assembled, in equimolar ratio, by overlap extension PCR (24). The PCR gene products were cloned sequentially into the pComb3H vector (56), and sequence errors were corrected with site-directed mutagenesis primers using the QuikChange mutagenesis kit (Stratagene). The entire synthetic Vκ and VH as well as the CL and CH1 gene segments of 2F5 were verified by DNA sequencing.
The sequences of VL and VH of 2F5 (Fig. 1) were aligned with the closest matching germ line genes, VK-1 (L4/18a) (41) and VH2-5 (33), respectively, as determined using IgG BLAST (1) and a previous report (27). Substitutions were introduced into the 2F5 Fab (see Fig. 1), employing the QuikChange mutagenesis kit, using pComb3H DNA containing the wild-type 2F5 VL and VH genes as templates. The sequences of the mutant clones were verified by DNA sequencing within the variable regions.
FIG. 1.
Alignment of 2F5 variable regions with the sequences encoded by the closest germ line genes. The germ line genes for 2F5 are VH2-5 and JH6 for the heavy chain (D segment is not defined) and L4/18a (VK1) and JK4 for the light chain (27). The residues that were chosen for individual substitution are shown immediately below the alignment (†), and their position numbers are indicated directly underneath. The CDR H3 of 2F5 contains a 14-residue insertion. In numbering by Kabat and Wu (26), these residues are designated 100A, 100B,…, and 100N. For clarity, these inserted residues are denoted with a subscript letter after the number of the residue position so as to avoid confusion with the replaced residue when referring to substitutions (e.g., F100BA). Residues in CDRs H3 and L3 were replaced by Ala, whereas the other targeted residues in the variable regions were replaced by germ line-encoded residues.
The 2F5 Fab mutants were compared in an enzyme-linked immunosorbent assay (ELISA) for their ability to bind to a short peptide, GELDKWASLC, and to a fusion protein of the ectodomain of gp41JR-FL linked to the C terminus of the maltose binding protein, designated M41xt. To construct M41xt, the gene segment encoding residues 535 to 681 (MTLTVQARLLLSGIV QQQNNLLRAIEAQQRMLQLTVWGIKQLQARVLAVERYLGDQQLLGIWGCSGKLICTTAVPWNASWSNKSLDRIWNNMTWMEWEREIDNYTSEIYTLIEESQNQQEKNEQELLELDKWASLWNWFDITKWLWY) of gp41JR-FL was amplified by PCR using the template pSyngp140JR-FL (obtained from the National Institutes of Health Research and Reference Reagent Program and contributed by Eun-Chung Park and Brian Seed [2, 22]). For the PCR, 5′- and 3′-specific oligonucleotide primers bearing overhanging BamHI and PstI sites, respectively, were used. The gene fragment was cloned into the multiple cloning site of the pMal-p2X vector (New England Biolabs) as a BamHI-PstI fragment in such a way that the 3′ PstI site was eliminated and a unique AatII site was introduced immediately 3′ to the eliminated PstI site. M41xt was produced in E. coli and purified (∼90% pure by sodium dodecyl sulfate-polyacrylamide gel electrophoresis) on an amylose column (New England Biolabs). Crude Fab supernatants were prepared as described previously (59). For the Fab ELISA, apparent affinities were calculated as outlined in Fig. 2.
FIG. 2.
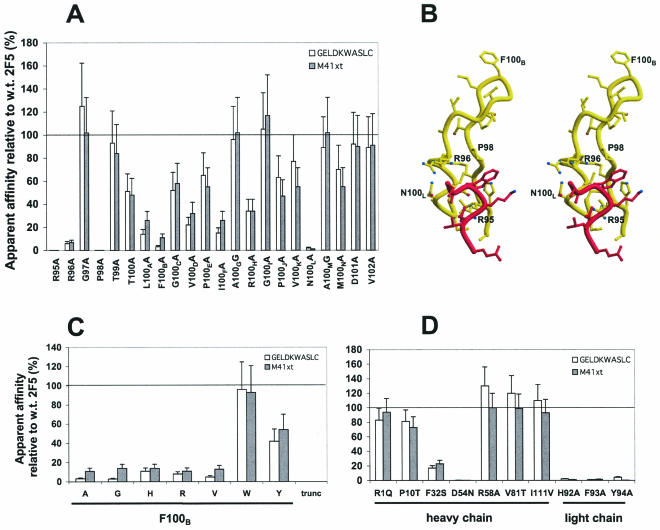
Mutagenesis of the paratope of 2F5 and the effect on binding to gp41 and GELDKWASLC peptide. (A) Alanine-scanning mutagenesis of the CDR H3 of 2F5. Bars indicate the apparent affinities of Fab mutants relative to wild-type Fab 2F5 for M41xt (residues 535 to 681 of gp41JR-FL fused to maltose binding protein) and the peptide GELDKWASLC. For the Fab ELISA, the concentration of Fab was determined with an anti-Fab ELISA (full curve, threefold dilution series) using simple linear regression; the concentrations of Fab in the samples were generally within about two- to threefold those of wild-type Fab 2F5. A full ELISA binding curve was also generated for groups of Fab mutants, alongside wild-type 2F5 and a negative Fab control (i.e., b12) against M41xt or GELDKWASLC peptide. Apparent affinities were calculated as the antibody concentration at half-maximal binding, as described previously (59). Apparent affinities relative to those of wild-type Fab 2F5 were calculated using the following formula: [(apparent affinity wild-type)/(apparent affinity mutant)] × 100%. Each mutant Fab was prepared in duplicate and tested at least twice; the mean was taken as the final reported value. (B) Tube representation of the crystal structure of the 2F5 CDR H3 loop with bound peptide, ELDKWAS, using the coordinates from the published patent (38). The H3 loop (yellow) and peptide (red) are shown, with H3 residues for which substitution by Ala diminished binding by ≥10-fold labeled in black. (C) Further substitution analysis of residues at position 100B in the CDR H3 of 2F5 and effect of deleting residues from the H3 loop. The CDR H3 truncation mutant, trunc, is one in which residues T99 to A100G of the CDR H3 loop of 2F5 were replaced with the sequence GSG. (D) Analysis of selected non-H3 substitutions in the paratope of 2F5. The apparent affinities shown in panels C and D were determined as described in panel A. w.t., wild type.
The effects of Ala substitutions in the H3 loop of 2F5 are shown in Fig. 2A together with the structure of CDR H3 of 2F5 and bound peptide, ELDKWAS (Fig. 2B); the residues for which Ala substitution diminished apparent binding affinity by ≥10-fold are also indicated (Fig. 2B). Substitutions at the base of the H3 loop were expected to affect 2F5 activity, since the base of the CDR H3 is proximal to the peptide in the crystal structure (38). Indeed, four substitutions near the base of the H3 loop, R95A, R96A, P98A, and N100LA, diminished the apparent affinities of 2F5 for both gp41 and the peptide by about 10-fold. Surprisingly, several residues towards the tip of H3 also significantly diminished 2F5 affinity, particularly F100BA (∼10% wild-type), but also L100AA, V100DA, I100FA, and R100HA (15 to 35% wild-type). Additional substitutions to F100B were also made (see below). Several substitutions in H3 had only a moderate effect (≤2-fold) on 2F5 affinity, including T100A, G100CA, P100EA, P100JA, V100KA, and M100NA. Still other substitutions had no effect on 2F5 affinity, including G97A, T99A, A100GG, G100IA, A100MG, D101A, and V102A.
The most unexpected result of the Ala scan of H3 was that replacement of F100B at the apex of the H3 loop by Ala produced such a large decrease in affinity of 2F5 for peptide despite the relatively long distance between F100B and any peptide residues in the crystal structure. We decided to investigate the effect of further replacement of F100B on 2F5 activity. Residues G, H, R, V, W, and Y were substituted at this position, and the effects on 2F5 binding were assessed (Fig. 2C). All five nonaromatic substitutions diminished 2F5 binding by roughly 10-fold, whereas the aromatic substitutions either had no effect (F100BW) or diminished 2F5 activity by about twofold (F100BY). The F100BW mutant was subsequently purified on an anti-Fab column, according to methods described elsewhere (6), and tested in an ELISA alongside similarly purified wild-type Fab 2F5. The ELISA confirmed that Fab F100BW had affinity for both gp41 and the peptide that was indistinguishable from that of wild-type 2F5 (data not shown). A truncated H3-loop mutant of 2F5 (trunc) was also engineered, in which residues T99 to A100G were replaced with the sequence GSG to ascertain whether a shortened loop would have any binding activity. The H3 truncation mutant of 2F5 was unable to bind gp41 or the 2F5 peptide in our assay format (Fig. 2C).
A number of residues outside the H3 loop were also chosen for mutagenesis, either by virtue of predicted proximity to the peptide or due to the rarity in occurrence of a certain residue in a particular position among Abs in the Kabat database (30). When we began the project, only the coordinates of the CDR H3 loop and bound peptide were available in the published patent, so we created a model in which we grafted the H3 loop of 2F5 onto a preexisting Fab structure (1HYS) that bore good sequence homology to the 2F5 heavy chain (Fab-28) (45). In the grafted model, the original relationship between H3 and the peptide was preserved. At the same time, CDR residues in the preexisting Fab structure were replaced by residues of the CDRs of 2F5. The resulting model of 2F5 (see below) allowed prediction of which residues in the CDRs might be important for 2F5 binding to the peptide (and, therefore, to gp41 and to HIV-1). The residues selected for replacement, and the effects of changing these residues to Ala for CDR L3 and to those encoded by the closest germ line gene for all others, are shown in Fig. 2D. All three of the substitutions in L3 (i.e., H92A, F93A, and Y94A) diminished 2F5 activity by at least 20-fold, revealing the importance of this CDR in 2F5 epitope recognition. Similarly, changing F32 of CDR H1 to an S residue and changing D54 of CDR H2 to an N residue diminished binding of 2F5 to gp41 and to the 2F5 peptide by about 4-fold and >30-fold, respectively. In contrast, the substitution R58A in H2 had no significant effect on 2F5 activity. The residues R1, P10, V81, and I111 in the heavy chain of 2F5 rarely occur (<0.5%) at these positions in Abs, as determined using the AbCheck webtool (http://www.rubic.rdg.ac.uk/abs/seqtest.html). The corresponding replacements, back to germ line-encoded residues, namely, R1Q and P10T in framework region 1 (FR1), V81T in FR3, and I111V in FR4, had no significant effect on 2F5 binding, indicating that the somatic mutations that produced these unconventional residues were incidental and were not required for 2F5 activity.
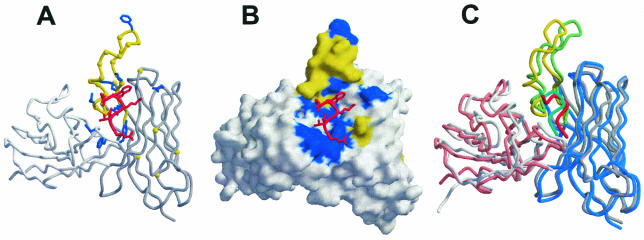
The model of 2F5, in which the CDR H3 loop of 2F5 was grafted onto an existing Fab structure together with replacement of the scaffold residues by residues from the other CDRs of 2F5, is shown in Fig. 3A. The peptide appears to lie between the base of H3 and L3, distant from L1 and L2, and with the crucial Asp, Trp, and Lys residues of the peptide pointing in the direction of R95 of H3, F32 of H1, and D54 of H2, respectively. A surface rendering of the model complex helps depict the key interactions as determined by our mutational analysis (Fig. 3B). At the time of submission, the full coordinates of the Fab 2F5-peptide complex became available (2F5B.PDB). The actual crystal structure of the Fab 2F5-peptide complex is shown superimposed on the model in tube format (Fig. 3C). Reassuringly, the model very closely matches the structure with root-mean square deviations of 0.87 Å for L1 to L107 and 0.84 Å for H1 to H113. Note that without the H3-peptide coordinates from the patent, the model would probably not have been so accurate.
FIG. 3.
Model and crystal structure of the 2F5 variable domain with bound peptide. The 2F5 model that was used to predict residues important for peptide recognition is shown in tube (A) and molecular surface (B) representations. The light and heavy chains of the model are shown in gray with H3 in yellow, peptide in red, and CDRs labeled. The 2F5 residues for which Ala substitution reduced binding by ≥10-fold are shown in blue, and all other residues that were substituted in this study are represented by a yellow Cα atom on the 2F5 backbone. To construct the model of the 2F5 variable domain, coordinates for H3 residues H93 to H101 and peptide residues ELDKWAS were first obtained from Patent WO-00/61618 (38). The 2F5 light and heavy chain variable sequences were grafted onto template Fab-28 (1HYX.PDB [45]) with SWISS-MODEL (42). The H3 region of the model was then replaced with that from 2F5. Minor adjustments were made to the model to eliminate close contacts, and the model was then conjugate-gradient energy minimized with the Crystallography & NMR System (9). (C) Superimposition of the model (gray and yellow) and the actual 2F5 Fab peptide crystal structure (2F5B.PDB) in tube representation. All Cα atoms were used (L1 to L107 and H1 to H113) for the overlap. The red and turquoise peptides are from the crystal structure and model, respectively. The light and heavy variable domains from the crystal structure are depicted in pink and blue, respectively.
We wanted to compare the abilities of the mutant Fabs to neutralize HIV-1, but it was impractical to make this comparison for all of the 2F5 mutants. To limit the search, we reasoned (although we do not prove) that the mutants that significantly diminish 2F5 affinity for gp41 would have a similar effect on 2F5-mediated neutralization of HIV-1. In addition, the most obvious hurdle for vaccine design involving 2F5 may be the difficulty of eliciting Abs with H3s as long as 2F5. Therefore, we focused on the F100BW mutant, which binds at wild-type levels to gp41 but bears a mutation at the very tip of the long H3 loop of 2F5. If a highly specific interaction between F100B and an unidentified determinant on HIV-1 was crucial for neutralization of HIV-1 by 2F5, then F100BW may not neutralize virus. We tested F100BW and wild-type Fab 2F5 in two neutralization assay formats. In the first format, p24 was used as the endpoint (60), and the T-cell-line-adapted strain, HIV-1MN, and H9 cells were used as the virus and target cells, respectively. The second format was a single-round infectivity assay using the T-cell-line-adapted strain, HIV-1HxB2, or a 2F5-sensitive mutant of HIV-1JR-FL, which we designate HIV-1JR2 and will describe elsewhere (M. B. Zwick and D. R. Burton, unpublished data). The viruses were pseudotyped using pNL4-3.luc.R−E−(13, 60), and U87.CD4.CCR5 (or CXCR4) cells were used as target cells (8). Equivalent neutralizing activity for F100BW and wild-type 2F5 Fab was observed against all three viruses, demonstrating no special requirement for an F residue at position 100B for HIV-1 neutralization (Fig. 4A, B, and C).
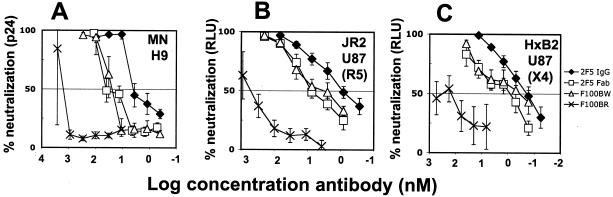
FIG. 4.
Neutralization of HIV-1 and HIV-1 pseudoviruses by Fab and IgG 2F5 and 2F5-Fab mutants, F100BW and F100BR. (A) Ability of IgG and Fab 2F5, as well as 2F5-Fab mutants, F100BW and F100BR, to neutralize HIV-1MN by using p24 as the endpoint and H9 cells as target cells (60). (B and C) Ability of the MAbs in panel A to neutralize the pseudovirions, HIV-1JR2 (B) and HIV-1HxB2 (C), using relative light units (RLU) produced by luciferase activity following a single round of infection of U87.CD4.CCR5 or CXCR4 cells (8) as the endpoint (13), as described previously (60). Neutralizing activity is expressed as a percentage of inhibition of infection. The error bars indicate standard deviations.
It may be argued that because W and F are both chemically similar residues, W100B might substitute for F100B even for a fairly specific interaction, such as that involving burying an F or W residue in a hydrophobic cavity. We therefore also tested F100BR, a mutant bearing an extended, positively charged side chain at position 100B. This mutant was shown to bind to gp41 and the peptide with ∼10-fold-lower affinity (Fig. 2C). The mutant was then evaluated in both neutralization assay formats and shown to neutralize the three standard viruses, but the 50% inhibitory concentrations were ∼42-, 67- and 160-fold lower than with the wild-type Fab 2F5 for MN, JR2, and HxB2, respectively (Fig. 4A, B, and C). The loss in neutralization potency of F100BR can be partially explained by the ∼10-fold decrease in binding to the epitope peptide and gp41, but its magnitude suggests additional factors may be involved. Thus, it might be that the epitope of 2F5 is larger on the virus than hitherto appreciated from prior peptide mapping studies. Alternatively, the F100BR Fab might be restricted in some way from accessing the 2F5 epitope on HIV-1. The neutralization assay results with F100BR can be considered in light of studies that show that neutralization of HIV-1, as well as of other viruses, does not necessarily correlate with antibody affinity to isolated (glyco)proteins from the virus but is considered rather a function of antibody affinity to the relevant determinants on the surface of the infectious virion particle (40). In the case of 2F5, this situation is made more complex by the fact that the antibody likely recognizes an intermediate in the fusion process (4, 18, 19, 21). Whether Abs nonhomologous to 2F5 would need to interact with HIV-1 in a way similar to that of 2F5 in order to neutralize HIV-1 remains unknown but should be addressed in light of HIV-1 vaccine development strategies involving the 2F5 epitope.
In the present study, the reactivity profile of the 2F5 epitope peptide was similar to that observed for gp41 for all of the mutant Fabs tested. In a related study involving mutants of Fab b12, the reactivity profile of a mimotope peptide, B2.1, had significant differences from that observed for gp120 (57). The contrasting results of the two studies can be explained in that the 2F5 peptide and gp41 share similar contacts with 2F5, whereas the B2.1 mimotope peptide and gp120 each make quite different contacts with b12. The 2F5 epitope peptides would be expected to more closely “mimic” the corresponding epitope on gp41, which is at least somewhat linear in nature, whereas the B2.1 peptide was selected using phage display to bind to an Ab that targets a discontinuous epitope that overlaps the CD4 binding site on gp120 (57).
Perhaps the most puzzling result from this study was the large negative effect on 2F5 binding to peptide of so many of the substitutions at F100B, despite a minimum distance of around 18 Å between the peptide and F100B in the crystal structure (38). This distance is inconsistent with a specific interaction (i.e., H bonding or hydrophobic interaction) between F100B and the peptide. The F100BW mutant retained full binding activity, suggesting a preference for aromatic residues at position 100B, but a Y substitution still diminished 2F5 activity by twofold. The interpretation of these results is not readily forthcoming, nor is the interpretation of some other functional hot spots, as discovered by alanine scanning and described in the literature (15). Indeed, it has been shown that somatic mutations that increase the affinity of an antibody to an antigen quite often are not involved in direct contact with the antigen (50, 51, 55). It might be that certain substitutions to 100B can somehow perturb the orientation of key residues at the base of the H3 loop or reduce the free energy associated with the ground state of 2F5. Finally, F100B might be involved in initial hydrophobic interactions that draw the core of the epitope into the binding site. The importance of the tip of the H3 loop is reinforced by the complete lack of gp41-binding affinity of the 2F5 truncation mutant (trunc) in our assay format. We note here that the 2F5 trunc mutant was found to be somewhat poorly produced in crude bacterial supernatants relative to the wild type (∼10-fold reduction; data not shown), suggesting a possible effect on folding or stability for this Fab.
It is interesting that other potent and broadly neutralizing MAbs against HIV-1, including IgG1 b12 (44, 59) and Fab X5 (35), also have H3 loops of above-average length (18 and 22 amino acids, respectively), although there are nonneutralizing anti-HIV-1 antibodies with long H3s as well (5, 7). Whether or not long H3s will be a general trend found in HIV-1-neutralizing antibodies will await the discovery of more such antibodies. However, it might be that the long H3 loop is a common feature contributing to the difficulty of eliciting neutralizing antibodies to HIV-1.
With respect to vaccine development, eliciting neutralizing Abs against the elusive 2F5 target will require increased efforts to precisely determine the 2F5 epitope on gp41. A crystal structure of a complex of 2F5 with a longer peptide, or even gp41 itself, would provide a more precise structural context for designing structurally constrained immunogens. Since 2F5 can neutralize HIV-1 following its attachment to host cells, it is therefore likely that the preponderance of Abs raised against recombinant gp41 or the ELDKWAS-displaying immunogens cannot access, or bind poorly to, the relevant epitope on the gp41 fusion-intermediate, which is poorly immunogenic. We have now identified paratope residues that are key determinants of the neutralizing activity of 2F5 and show that the long CDR H3 loop appears to contribute to its ability to neutralize HIV-1. Hence, it would seem imprudent to pursue efforts to elicit 2F5-like Abs in mice, which produce Abs with much shorter H3s. The paratope map of 2F5 adds to our understanding of the interaction 2F5 makes with HIV-1 and may facilitate the design of molecules that are able to elicit 2F5-like activities.
Acknowledgments
We thank Dawn Slifka for excellent technical assistance and Erica Saphire for helpful discussions.
We acknowledge support from the Elizabeth Glaser Pediatric AIDS Foundation (M.B.Z.) and grants from the National Institutes of Health, AI40377 (P.W.H.I.P.), GM46192 (I.A.W.), AI33292 (D.R.B.), the International AIDS Vaccine Initiative through the Neutralizing Antibody Consortium (D.R.B. and I.A.W.), and the Pendleton Trust.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andre, S., B. Seed, J. Eberle, W. Schraut, A. Bultmann, and J. Haas. 1998. Increased immune response elicited by DNA vaccination with a synthetic gp120 sequence with optimized codon usage. J. Virol. 72:1497-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baba, T. W., V. Liska, R. Hofmann-Lehmann, J. Vlasak, W. Xu, S. Ayehunie, L. A. Cavacini, M. R. Posner, H. Katinger, G. Stiegler, B. J. Bernacky, T. A. Rizvi, R. Schmidt, L. R. Hill, M. E. Keeling, Y. Lu, J. E. Wright, T. C. Chou, and R. M. Ruprecht. 2000. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat. Med. 6:200-206. [DOI] [PubMed] [Google Scholar]
- 4.Barbas, C. F., III, D. R. Burton, J. K. Scott, and G. J. Silverman. 2001. Phage display: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 5.Barbas, C. F., III, T. A. Collet, W. Amberg, P. Roben, J. M. Binley, D. Hoekstra, D. Cababa, T. M. Jones, R. A. Williamson, G. R. Pilkington, et al. 1993. Molecular profile of an antibody response to HIV-1 as probed by combinatorial libraries. J. Mol. Biol. 230:812-823. [DOI] [PubMed] [Google Scholar]
- 6.Binley, J. M., C. S. Cayanan, C. Wiley, N. Schulke, W. C. Olson, and D. R. Burton. 2003. Redox-triggered infection by disulfide-shackled human immunodeficiency virus type 1 pseudovirions. J. Virol. 77:5678-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Binley, J. M., H. J. Ditzel, C. F. Barbas III, N. Sullivan, J. Sodroski, P. W. Parren, and D. R. Burton. 1996. Human antibody responses to HIV type 1 glycoprotein 41 cloned in phage display libraries suggest three major epitopes are recognized and give evidence for conserved antibody motifs in antigen binding. AIDS Res. Hum. Retrovir. 12:911-924. [DOI] [PubMed] [Google Scholar]
- 8.Bjorndal, A., H. Deng, M. Jansson, J. R. Fiore, C. Colognesi, A. Karlsson, J. Albert, G. Scarlatti, D. R. Littman, and E. M. Fenyo. 1997. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J. Virol. 71:7478-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR System: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905-921. [DOI] [PubMed] [Google Scholar]
- 10.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 11.Coeffier, E., J. M. Clement, V. Cussac, N. Khodaei-Boorane, M. Jehanno, M. Rojas, A. Dridi, M. Latour, R. El Habib, F. Barre-Sinoussi, M. Hofnung, and C. Leclerc. 2000. Antigenicity and immunogenicity of the HIV-1 gp41 epitope ELDKWA inserted into permissive sites of the MalE protein. Vaccine 19:684-693. [DOI] [PubMed] [Google Scholar]
- 12.Conley, A. J., J. A. Kessler II, L. J. Boots, J. S. Tung, B. A. Arnold, P. M. Keller, A. R. Shaw, and E. A. Emini. 1994. Neutralization of divergent human immunodeficiency virus type 1 variants and primary isolates by IAM-41-2F5, an anti-gp41 human monoclonal antibody. Proc. Natl. Acad. Sci. USA 91:3348-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 14.Cotropia, J., K. E. Ugen, S. Kliks, K. Broliden, P. A. Broliden, J. A. Hoxie, V. Srikantan, W. V. Williams, and D. B. Weiner. 1996. A human monoclonal antibody to HIV-1 gp41 with neutralizing activity against diverse laboratory isolates. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 12:221-232. [DOI] [PubMed] [Google Scholar]
- 15.DeLano, W. L. 2002. Unraveling hot spots in binding interfaces: progress and challenges. Curr. Opin. Struct. Biol. 12:14-20. [DOI] [PubMed] [Google Scholar]
- 16.D'Souza, M. P., D. Livnat, J. A. Bradac, and S. H. Bridges. 1997. Evaluation of monoclonal antibodies to human immunodeficiency virus type 1 primary isolates by neutralization assays: performance criteria for selecting candidate antibodies for clinical trials. AIDS Clinical Trials Group Antibody Selection Working Group. J. Infect. Dis. 175:1056-1062. [DOI] [PubMed] [Google Scholar]
- 17.Eckhart, L., W. Raffelsberger, B. Ferko, A. Klima, M. Purtscher, H. Katinger, and F. Ruker. 1996. Immunogenic presentation of a conserved gp41 epitope of human immunodeficiency virus type 1 on recombinant surface antigen of hepatitis B virus. J. Gen. Virol. 77:2001-2008. [DOI] [PubMed] [Google Scholar]
- 18.Finnegan, C. M., W. Berg, G. K. Lewis, and A. L. DeVico. 2001. Antigenic properties of the human immunodeficiency virus envelope during cell-cell fusion. J. Virol. 75:11096-11105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Follis, K. E., S. J. Larson, M. Lu, and J. H. Nunberg. 2002. Genetic evidence that interhelical packing interactions in the gp41 core are critical for transition of the human immunodeficiency virus type 1 envelope glycoprotein to the fusion-active state. J. Virol. 76:7356-7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Golding, H., M. Zaitseva, E. de Rosny, L. R. King, J. Manischewitz, I. Sidorov, M. K. Gorny, S. Zolla-Pazner, D. S. Dimitrov, and C. D. Weiss. 2002. Dissection of human immunodeficiency virus type 1 entry with neutralizing antibodies to gp41 fusion intermediates. J. Virol. 76:6780-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorny, M. K., and S. Zolla-Pazner. 2000. Recognition by human monoclonal antibodies of free and complexed peptides representing the prefusogenic and fusogenic forms of human immunodeficiency virus type 1 gp41. J. Virol. 74:6186-6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haas, J., E. C. Park, and B. Seed. 1996. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr. Biol. 6:315-324. [DOI] [PubMed] [Google Scholar]
- 23.Hofmann-Lehmann, R., J. Vlasak, R. A. Rasmussen, S. Jiang, P. L. Li, T. W. Baba, D. C. Montefiori, B. J. Bernacky, T. A. Rizvi, R. Schmidt, L. R. Hill, M. E. Keeling, H. Katinger, G. Stiegler, L. A. Cavacini, M. R. Posner, and R. M. Ruprecht. 2002. Postnatal pre- and postexposure passive immunization strategies: protection of neonatal macaques against oral simian-human immunodeficiency virus challenge. J. Med. Primatol. 31:109-119. [DOI] [PubMed] [Google Scholar]
- 24.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 25.Joyce, J. G., W. M. Hurni, M. J. Bogusky, V. M. Garsky, X. Liang, M. P. Citron, R. C. Danzeisen, M. D. Miller, J. W. Shiver, and P. M. Keller. 2002. Enhancement of alpha-helicity in the HIV-1 inhibitory peptide DP178 leads to an increased affinity for human monoclonal antibody 2F5 but does not elicit neutralizing responses in vitro. Implications for vaccine design. J. Biol. Chem. 277:45811-45820. [DOI] [PubMed] [Google Scholar]
- 26.Kabat, E. A., T. T. Wu, H. M. Perry, K. S. Gottesman, and C. Foeller. 1991. Sequences of proteins of immunological interest. Department of Health and Human Services, Washington, D.C.
- 27.Kunert, R., F. Ruker, and H. Katinger. 1998. Molecular characterization of five neutralizing anti-HIV type 1 antibodies: identification of nonconventional D segments in the human monoclonal antibodies 2G12 and 2F5. AIDS Res. Hum. Retrovir. 14:1115-1128. [DOI] [PubMed] [Google Scholar]
- 28.Kwong, P. D., R. Wyatt, Q. J. Sattentau, J. Sodroski, and W. A. Hendrickson. 2000. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J. Virol. 74:1961-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang, X., S. Munshi, J. Shendure, G. Mark III, M. E. Davies, D. C. Freed, D. C. Montefiori, and J. W. Shiver. 1999. Epitope insertion into variable loops of HIV-1 gp120 as a potential means to improve immunogenicity of viral envelope protein. Vaccine 17:2862-2872. [DOI] [PubMed] [Google Scholar]
- 30.Martin, A. C. 1996. Accessing the Kabat antibody sequence database by computer. Proteins 25:130-133. [DOI] [PubMed] [Google Scholar]
- 31.Mascola, J. R., M. G. Lewis, G. Stiegler, D. Harris, T. C. VanCott, D. Hayes, M. K. Louder, C. R. Brown, C. V. Sapan, S. S. Frankel, Y. Lu, M. L. Robb, H. Katinger, and D. L. Birx. 1999. Protection of macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J. Virol. 73:4009-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mascola, J. R., G. Stiegler, T. C. VanCott, H. Katinger, C. B. Carpenter, C. E. Hanson, H. Beary, D. Hayes, S. S. Frankel, D. L. Birx, and M. G. Lewis. 2000. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med. 6:207-210. [DOI] [PubMed] [Google Scholar]
- 33.Matsuda, F., K. Ishii, P. Bourvagnet, K. Kuma, H. Hayashida, T. Miyata, and T. Honjo. 1998. The complete nucleotide sequence of the human immunoglobulin heavy chain variable region locus. J. Exp. Med. 188:2151-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGaughey, G. B., M. Citron, R. C. Danzeisen, R. M. Freidinger, V. M. Garsky, W. M. Hurni, J. G. Joyce, X. Liang, M. Miller, J. Shiver, and M. J. Bogusky. 2003. HIV-1 vaccine development: constrained peptide immunogens show improved binding to the anti-HIV-1 gp41 MAb. Biochemistry 42:3214-3223. [DOI] [PubMed] [Google Scholar]
- 35.Moulard, M., S. K. Phogat, Y. Shu, A. F. Labrijn, X. Xiao, J. M. Binley, M. Y. Zhang, I. A. Sidorov, C. C. Broder, J. Robinson, P. W. Parren, D. R. Burton, and D. S. Dimitrov. 2002. Broadly cross-reactive HIV-1-neutralizing human monoclonal Fab selected for binding to gp120-CD4-CCR5 complexes. Proc. Natl. Acad. Sci. USA 99:6913-6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muster, T., R. Guinea, A. Trkola, M. Purtscher, A. Klima, F. Steindl, P. Palese, and H. Katinger. 1994. Cross-neutralizing activity against divergent human immunodeficiency virus type 1 isolates induced by the gp41 sequence ELDKWAS. J. Virol. 68:4031-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muster, T., F. Steindl, M. Purtscher, A. Trkola, A. Klima, G. Himmler, F. Ruker, and H. Katinger. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 67:6642-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pai, E. F., M. H. Klein, P. Chong, and A. Pedyczak. October2000. World Intellectual Property Organization patent WO-00/61618.
- 39.Parker, C. E., L. J. Deterding, C. Hager-Braun, J. M. Binley, N. Schulke, H. Katinger, J. P. Moore, and K. B. Tomer. 2001. Fine definition of the epitope on the gp41 glycoprotein of human immunodeficiency virus type 1 for the neutralizing monoclonal antibody 2F5. J. Virol. 75:10906-10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parren, P. W., and D. R. Burton. 2001. The antiviral activity of antibodies in vitro and in vivo. Adv. Immunol. 77:195-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pech, M., H. R. Jaenichen, H. D. Pohlenz, P. S. Neumaier, H. G. Klobeck, and H. G. Zachau. 1984. Organization and evolution of a gene cluster for human immunoglobulin variable regions of the kappa type. J. Mol. Biol. 176:189-204. [DOI] [PubMed] [Google Scholar]
- 42.Peitsch, M. C. 1995. Protein modelling by E-Mail. Bio/Technology 13:658-660. [Google Scholar]
- 43.Poignard, P., E. O. Saphire, P. W. Parren, and D. R. Burton. 2001. gp120: biologic aspects of structural features. Annu. Rev. Immunol. 19:253-274. [DOI] [PubMed] [Google Scholar]
- 44.Saphire, E. O., P. W. Parren, R. Pantophlet, M. B. Zwick, G. M. Morris, P. M. Rudd, R. A. Dwek, R. L. Stanfield, D. R. Burton, and I. A. Wilson. 2001. Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science 293:1155-1159. [DOI] [PubMed] [Google Scholar]
- 45.Sarafianos, S. G., K. Das, C. Tantillo, A. D. Clark, Jr., J. Ding, J. M. Whitcomb, P. L. Boyer, S. H. Hughes, and E. Arnold. 2001. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 20:1449-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sattentau, Q. J., S. Zolla-Pazner, and P. Poignard. 1995. Epitope exposure on functional, oligomeric HIV-1 gp41 molecules. Virology 206:713-717. [DOI] [PubMed] [Google Scholar]
- 47.Stiegler, G., R. Kunert, M. Purtscher, S. Wolbank, R. Voglauer, F. Steindl, and H. Katinger. 2001. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 17:1757-1765. [DOI] [PubMed] [Google Scholar]
- 48.Tian, Y., C. V. Ramesh, X. Ma, S. Naqvi, T. Patel, T. Cenizal, M. Tiscione, K. Diaz, T. Crea, E. Arnold, G. F. Arnold, and J. W. Taylor. 2002. Structure-affinity relationships in the gp41 ELDKWA epitope for the HIV-1 neutralizing monoclonal antibody 2F5: effects of side-chain and backbone modifications and conformational constraints. J. Peptide Res. 59:264-276. [DOI] [PubMed] [Google Scholar]
- 49.Trkola, A., A. B. Pomales, H. Yuan, B. Korber, P. J. Maddon, G. P. Allaway, H. Katinger, C. F. Barbas III, D. R. Burton, D. D. Ho, et al. 1995. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. J. Virol. 69:6609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Valjakka, J., A. Hemminki, S. Niemi, H. Soderlund, K. Takkinen, and J. Rouvinen. 2002. Crystal structure of an in vitro affinity- and specificity-matured anti-testosterone Fab in complex with testosterone. Improved affinity results from small structural changes within the variable domains. J. Biol. Chem. 277:44021-44027. [DOI] [PubMed] [Google Scholar]
- 51.Wedemayer, G. J., P. A. Patten, L. H. Wang, P. G. Schultz, and R. C. Stevens. 1997. Structural insights into the evolution of an antibody combining site. Science 276:1665-1669. [DOI] [PubMed] [Google Scholar]
- 52.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 53.Wu, T. T., G. Johnson, and E. A. Kabat. 1993. Length distribution of CDRH3 in antibodies. Proteins 16:1-7. [DOI] [PubMed] [Google Scholar]
- 54.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 55.Yang, P. L., and P. G. Schultz. 1999. Mutational analysis of the affinity maturation of antibody 48G7. J. Mol. Biol. 294:1191-1201. [DOI] [PubMed] [Google Scholar]
- 56.Yang, W. P., K. Green, S. Pinz-Sweeney, A. T. Briones, D. R. Burton, and C. F. Barbas III. 1995. CDR walking mutagenesis for the affinity maturation of a potent human anti-HIV-1 antibody into the picomolar range. J. Mol. Biol. 254:392-403. [DOI] [PubMed] [Google Scholar]
- 57.Zwick, M. B., L. L. Bonnycastle, A. Menendez, M. B. Irving, C. F. Barbas III, P. W. Parren, D. R. Burton, and J. K. Scott. 2001. Identification and characterization of a peptide that specifically binds the human, broadly neutralizing anti-human immunodeficiency virus type 1 antibody b12. J. Virol. 75:6692-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zwick, M. B., A. F. Labrijn, M. Wang, C. Spenlehauer, E. O. Saphire, J. M. Binley, J. P. Moore, G. Stiegler, H. Katinger, D. R. Burton, and P. W. Parren. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 75:10892-10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zwick, M. B., P. W. Parren, E. O. Saphire, S. Church, M. Wang, J. K. Scott, P. E. Dawson, I. A. Wilson, and D. R. Burton. 2003. Molecular features of the broadly neutralizing immunoglobulin G1 b12 required for recognition of human immunodeficiency virus type 1 gp120. J. Virol. 77:5863-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zwick, M. B., M. Wang, P. Poignard, G. Stiegler, H. Katinger, D. R. Burton, and P. W. Parren. 2001. Neutralization synergy of human immunodeficiency virus type 1 primary isolates by cocktails of broadly neutralizing antibodies. J. Virol. 75:12198-12208. [DOI] [PMC free article] [PubMed] [Google Scholar]