Abstract
The main heroine traffic from Yunnan province to the Xinjiang Autonomous Region is believed to initiate the transmission of CRF07_BC which is the predominant strain in intravenous drug users (IDUs) in China. However, the great distances between Yunnan and Xinjiang lead to an unclear and elusive diffusion process of CRF07_BC due to the absence of an important middle site such as Sichuan province. Moreover, in recent years the rapidly increasing infection rate among IDUs in the Liangshan region of Sichuan made it necessary to characterize the genetic character of the circulating strain of Sichuan IDUs. In this study, we characterized the genetic character of seven newly isolated CRF07_BC genomes (five from Sichuan and two from Xinjiang) and analyzed the transmission linkage among strains from IDUs in different regions. By conducting Markov chain Monte Carlo (MCMC) analysis and reconstruction of neighbor-joining trees and maximum-likelihood trees, our results revealed the genetic variation and important role of Sichuan-derived CRF07_BC strains during the transmission of CRF07_BC.
A recent large-scaled HIV testing campaign revealed that several regions in China have been heavily devastated by HIV/AIDS among injecting drug users (IDUs), including Dehong Autonomous Municipality in Yunnan province, Liangshan Autonomous Municipality in Sichuan province, and a few regions in Xinjiang Autonomous Region.1–3 Although full-length sequences of the HIV-1 genome isolated from IDUs have been previously reported in Yunnan province and Xinjiang Autonomous Region,4–10 these still remain unavailable for Sichuan province. As Sichuan province is located at the middle between Yunnan and Xinjiang, the sequences from Sichuan will be important to decipher the transmission route of HIV-1. In addition, the full-length genomic sequences will reveal the basis for understanding the genetic evolution in this region. Here we report five near full-length HIV-1 CRF07_BC sequences derived from IDUs in Liangshan Autonomous Municipality in Sichuan province and two near full-length HIV-1 CRF07_BC sequences from IDUs in Ulumuqi of Xinjiang. In addition, the transmission linkage between Sichuan strains and Yunnan, Xinjiang, Guangxi, Liaoning, and Taiwan, which are all severely affected by infection of IDUs, was also analyzed by employing Gag full-length sequences by Bayesian Markov chain Monte Carlo (MCMC) methods and a maximum-likelihood tree. Our data demonstrated that multiple transmission linkage occurred among the strains from Sichuan, Yunnan, and Xinjiang, which suggested that Sichuan is an important secondary epicenter.
Since 2005, there have been numerous new HIV infections in Liangshan of Sichuan and Ulumuqi of Xinjiang every year. Conducted molecular investigations demonstrated that the dominant strain in these regions is the CRF07_BC recombinant strain. Some samples were used for obtaining the near full-length genome of CRF07_BC. Based on a previously analyzed env fragment set from IDUs of Liangshan in Sichuan, two strains among 33 isolates showed the highest homogenicity and another six strains showed the farthest genetic distance from the predicted consensus sequence (Supplementary Fig. S1). (Supplementary Data are available online at www.liebertpub.com/aid). Therefore, these eight study subjects were recruited from IDUs in Liangshan of Sichuan province and two samples were randomly collected from Ulumuqi of Xinjiang Autonomous Region in China, simultaneously. All individuals were antiretroviral therapy naive and signed informed consent to participate in this study. The study was also approved by the Ethics Committee at the Shanghai Public Health Clinical Center. Five milliliters of blood was drawn from each subject into an EDTA prefilled tube and then transported into the laboratory by air; the plasma was separated and used for obtaining the HIV genome.
The spin column from the QIAmp DNA Blood Midi Kit (QIAgen, Germany) and reagents from the QIAmp viral RNA Mini kit were combined for viral RNA extraction.11 Then 1 ml of an HIV-1-positive plasma specimen was used for RNA extraction and the final elution volume was adjusted to 120 μl. RNA extraction and reverse transcriptase polymerase chain reaction (RT-PCR) were performed as previously described. SuperScript III First-Strand Synthesis Supermix (Invitrogen, Carlsbad, CA) was used for cDNA synthesis. Then 25 μl of RNA was mixed with 2.5 μl of 20 mM dNTP and 2.5 μl of 20 mM oligo(dT), or specific primer 1.3MR. The mixture was heated to 65°C for 5 min and rapidly cooled to 0°C. The reverse transcription reaction was performed as previously described and subsequently used for amplification.11
The Expand High Fidelity PCR System (Roche Molecular Biochemicals, Mannheim, Germany) was used to amplify the HIV-1 virion genome from the synthesized cDNA. Three-amplicon strategy was performed as previously described.11 The first amplicon was the gag-pol amplicon (nts 623–4925 HXB2), the second spans pol to env (nts 4743–7668 HXB2), and the third spans env to nef (nts 6996–9635 HXB2).11 A hot-start using DynaWax (Finnzymes, Espoo, Finland) was employed in each PCR, with two layers separated by wax. In the bottom layer were the dNTPs and template cDNA while the top layer contained the PCR buffer, DNA polymerase, and primers.
The PCR product was purified using the gene purification kit (QIAgen, Germany) and was subsequently used to perform the sequencing reaction (BigDye H Terminator v3.1 Cycle Sequencing Assay, Applied Biosystems) in an automated 3730xl sequencer (Applied Biosystems) using a primer-walking approach. The nearly full-length genome sequence was assembled by overlapping the sequences of the three amplicons and merging them into one sequence as long as the two overlapping sequences were greater than 99% homologous.11,12 Seven edited near full-length HIV-1 CRF07_BC genomic sequences were generated, designated as SC006, SC008, SC020, SC025, SC124, 709, and 713, and were ∼9.0 kb in length; the first five were from Sichuan and the last two (709 and 713) were from Xinjiang. Analysis of their genomic organization revealed the presence of nine intact potential open reading frames corresponding to the gag, pol, vif, vpr, tat, rev, vpu, env, and nef genes.
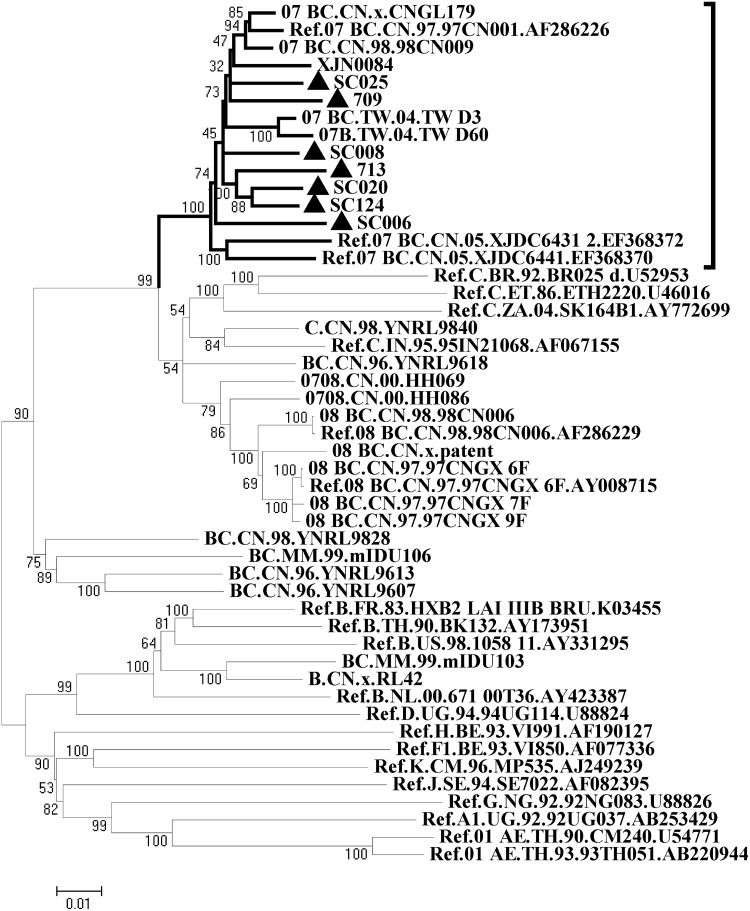
The sequences were aligned with reference sequences (from the Los Alamos HIV database) using Bio-Edit, version 7.0. After managing adjustments, the alignments were used to construct a neighbor-joining tree with 1000 bootstrap replicates, using Mega 5.02 software.12 The neighbor-joining tree shows all seven new full-length sequences clustered with reference sequences of HIV-1 CRF07_BC, supported by 100% of bootstrap trees (Fig. 1). Furthermore, boot scanning analysis was performed by the SimPlot 2.5 program on neighbor-joining trees for a window of 200 nucleotides moving along the alignment in increments of 20 nucleotides,12 in the context of the following reference sequences: A_92UG037, B__RL42, C_95IN21068, D.UG.94UG114, 08_BC.CN. 97CNGX, and 07_BC.CN.97.CN54, which indicated that the seven isolates all shared a C/B′ mosaic structure with similarity to the prototype CRF07_BC reference strains, featured with the genome backbone of subtype C integrated with small fractions of Thailand-B-derived gag, pol, env, and nef genes as well as the first exon of the tat gene (data not shown). Altogether, these data confirmed that the seven sequences are CRF07_BC recombinant strains. Interestingly, Sichuan isolates SC020 and SC124 show a close genetic relationship with Xinjiang strain 713 in the neighbor-joining tree (Fig. 1).
FIG. 1.
Phylogenetic tree analysis of seven near full-length CRF07_BC sequences. A neighbor-joining tree of seven reference sequences (retrieved from the HIV database) was constructed by using Mega 5.2; seven new sequences (▲) were clustered with CRF07_BC reference strains with 100% bootstrap probability. It should be noticed that two Sichuan strains and one Xinjiang strain (713) also formed a clade with 100% bootstrap probability.
Compared with recent isolates, early isolates formed a subcluster supported by a 100% bootstrap branch within the cluster of CRF07_BC in the neighbor-joining tree. Sixteen CRF07_BC full-length sequences (nine from the HIV database) were divided into five groups according to geographic information as follows: early group (98CN009, CNGL179, 97CN001, CN54), Sichuan group (SC006, SC008, SC020, SC025, SC124), Xinjiang group (XJN0084, XJDC6431, XJDC6441, 709, 713), Taiwan strain (TWD3), and Hebei strain (1114). Evolutionary distances between groups were constructed by using neighbor-joining, and calculated with the Kimura two-parameter method. The variance estimation was performed by bootstrap analysis with 500 replicates. For any regional isolate group, the closest genetic distance was always observed in comparison with an early isolate group with a range between 0.03 and 0.041 (Table 1), which suggested that these isolates are likely to share a common ancestor with CRF07_BC.
Table 1.
Genetic Distance Calculated with the Kimura Two-Parameter Method by Mega 5.2
| Sichuan | Xinjiang | Early | Taiwan | Hebei | |
|---|---|---|---|---|---|
| Sichuan | 0.04 | 0.047 | 0.032 | 0.04 | 0.049 |
| Xinjiang | 0.047 | 0.052 | 0.039 | 0.047 | 0.055 |
| Early | 0.032 | 0.039 | 0.016 | 0.03 | 0.041 |
| Taiwan | 0.04 | 0.047 | 0.03 | 0.049 | |
| Hebei | 0.049 | 0.055 | 0.041 | 0.049 |
Bold numbers represent the mean distance in one certain subpopulations. Sixteen CRF07_BC near full-length sequences were collected and divided into five groups, including the Early group (four sequences), Sichuan group (five sequences), Xinjiang group (five sequences), Taiwan group (one sequence), and Hebei group (one sequence). Genetic distances were calculated among different groups.
Similar to a previous report,12 when compared to the early group (genetic distance at 0.016), the dramatically increased genetic distances were observed in the regional strain groups including Sichuan (0.040) and Xinjiang (0.052), respectively, which suggested a dramatically increasing variation of region strains in genetic diversity (Table 1).
Thirty-five HIV-1 CRF07_BC gag sequences (including 28 published sequences in the HIV database) were collected and used for transmission linkage analysis.13 As previously described,14 the general time-reversible model with a proportion of invariant sites and gamma distribution (GTR+γ+R) was selected as the most appropriate analysis model by Modeltest software. The Bayesian phylogenetic inference was performed by employing the MCMC sampling approach in the GTR+γ+R model, as implemented in MrBayes 3.1.15,16 The MCMC search was run for 107 generations with trees sampled every 2000th generation. Burn-in was set at 50% and a posterior consensus tree was generated from 250 sampled trees. The posterior probability of the nodes on the consensus tree was used as a phylogenetic support for clusters.
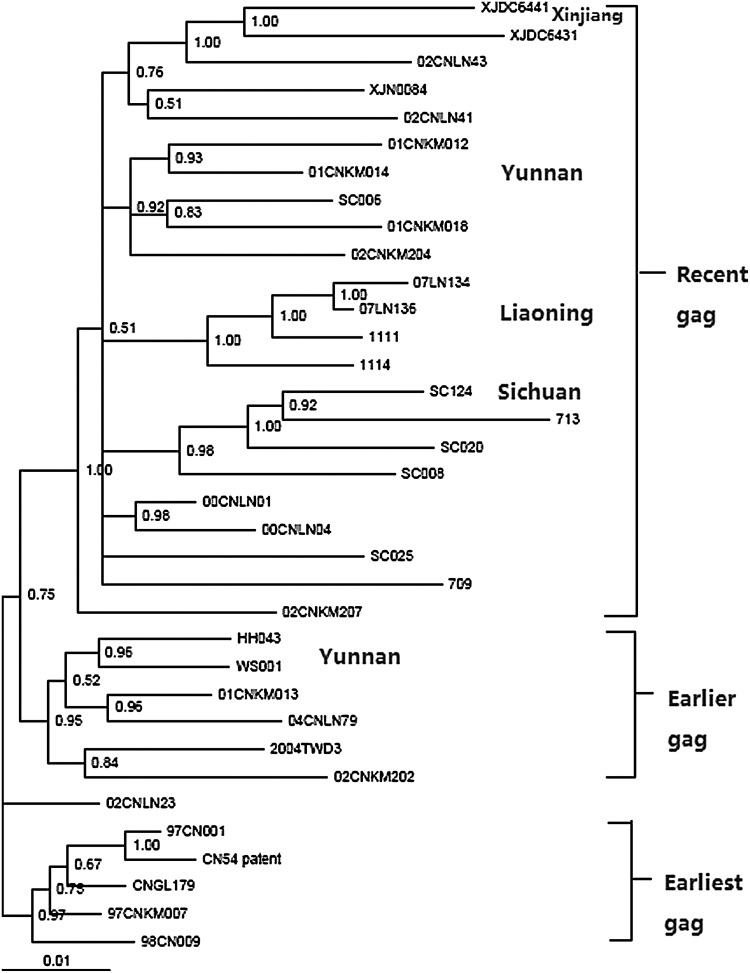
Based on previous reports,15,16 significant linkages in Bayesian phylogenetic inference analysis were considered as those having posterior probability >90% and genetic distances <0.03 nt substitutions per site for gag sequences. As shown in Fig. 2, multiple clusters were formed by geographic strains in the MCMC tree constructed by the Bayes method. Three big clusters were composed of recent gag sequences (sampling time range from 2002 to 2007), earlier gag sequences from Yunnan, Taiwan, and Liaoning provinces (sampling time range from 2000 to 2004), and the earliest gag sequences from Xinjiang, Guangxi, and Yunnan provinces (sampling time in 1997), respectively, with support of >95% posterior probability.
FIG. 2.
Markov chain Monte Carlo (MCMC) tree of 35 CRF07_BC Gag sequences constructed by Bayes 3.1. Significant linkages in Bayesian phylogenetic inference analysis were considered as those having posterior probabilities >90%. Three big clusters were identified and labeled on the right side and they correspond to “Recent gag” sequences, “Earlier gag” sequences, and “earliest gag” sequences. Four regional subclusters are formed by strains from corresponding regions, including Yunnan, Liaoning, Sichuan, and Xinjiang, and are labeled on the right side, neighbored to cluster labels. Note that several regional subclusters are joined by other regional strains.
Within three big clusters in the MCMC tree, several regional subclusters were formed by isolates from their corresponding epidemic region. Isolates from Kunming of Yunnan province (01CNKM012, 01CNKM014, 01CNKM018, and 02CNKM204) formed a subcluster supported by 92% posterior probability. Similarly, Sichuan isolates (SC008, SC020, and SC124) and Liaoning and Hebei isolates (1111, 1114, 07LN134, and 07LN136) also formed subclusters by more than 98% posterior probability. These geographic subclusters suggested on-going relatively independent evolution in different regions, which may have resulted from the long-term history of the CRF07_BC epidemic in those geographic sites.
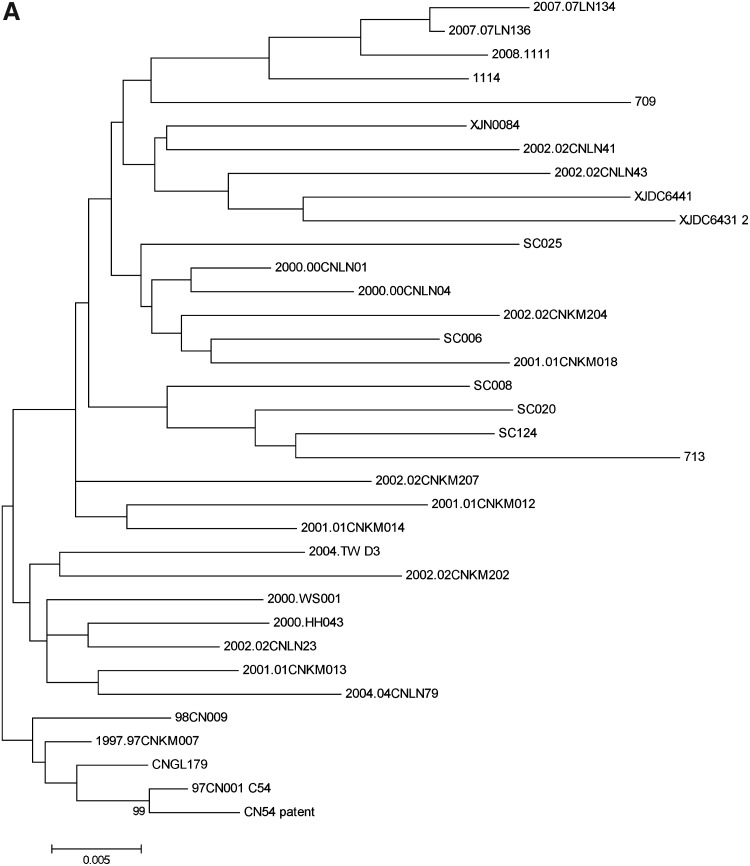
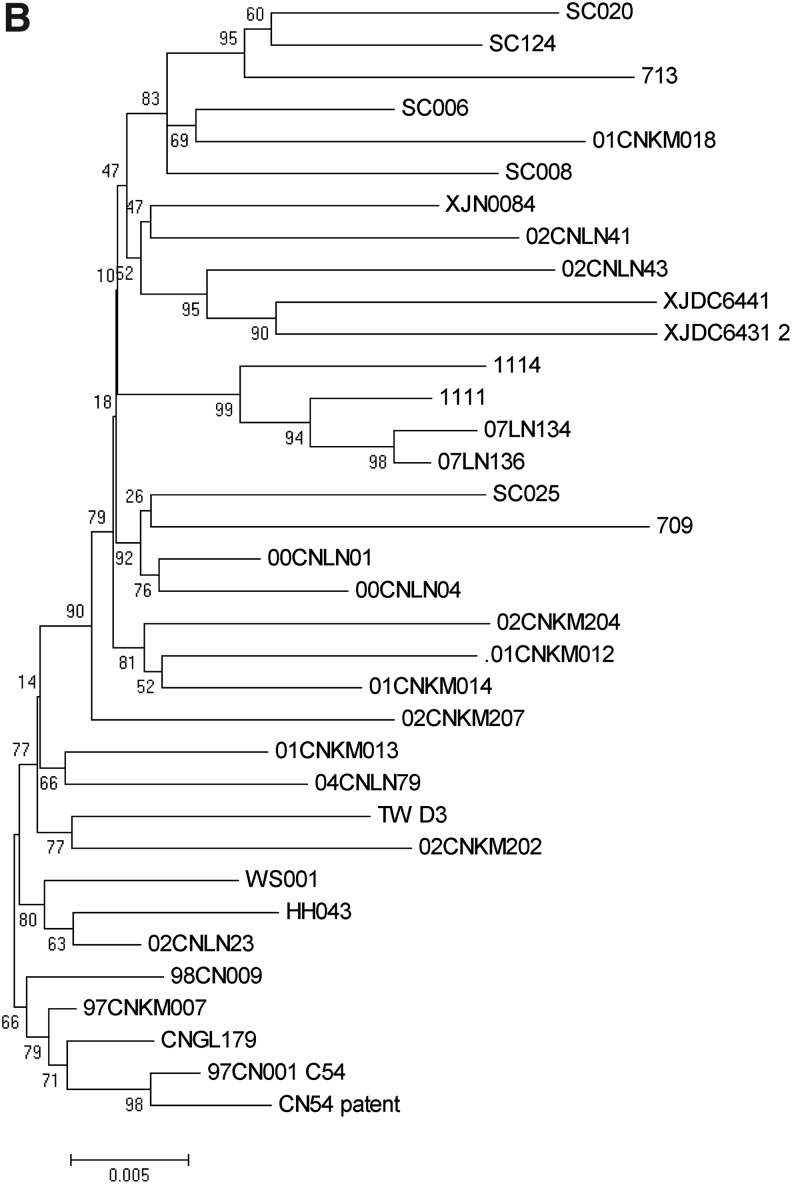
Notably, different regional strains could cluster into one subcluster and significant transmission linkages between two different provinces could also be identified in those subclusters. For instance, in the Kunming subcluster, SC006 from Sichuan had a significant transmission linkage with strains from Kunming of Yunnan province, supported by 92% posterior probability, and Xinjiang strain 713 had significant transmission linkages with three Sichuan isolates (SC008, SC020, and SC124) supported by 100% posterior probability. 02CNLN43 from Liaoning province formed one subcluster with XJDC6431 and XJDC6441 from Xinjiang province with the support of 100% posterior probability. In the earlier cluster, significant transmission linkages were also observed between Taiwan strain TWD3 and Kunming strain 01CNKM013 and between Liaoning strain 04CNLN79 and Kunming strain 01CNKM013 with the support of >90% posterior probability. Furthermore, phylogenetic trees constructed by both the maximum-likelihood method and neighbor-joining method also confirmed the transmission linkage among regional strains identified by Bayesian phylogenetic inference (Fig. 3).
FIG. 3.
A maximum-likelihood tree (a) and a neighbor-joining tree (b) were constructed with the Kimura two-parameter model and the GTR+γ+R model, respectively, by Mega 5.0. The reliability of the branching orders was confirmed by bootstrap analysis with 500 replicates.
To further confirm the transmission linkage, a neighbor-joining tree and maximum-likelihood tree were also constructed with the Kimura two-parameter model and GTR+γ+R model, respectively, by Mega 5.0; the reliability of the branching orders was confirmed by bootstrap analysis with 500 replicates. Furthermore, both the maximum-likelihood tree and neighbor-joining tree confirmed the existence of these three large clusters with high bootstrap value (Fig. 3). Overall, the earliest cluster formed by different regional early strains further indicated that CRF07_BC is likely to share common ancestor strains, whereas the earlier cluster and recent cluster composed of the mixture of different regional strains suggested the existence of a significant transmission linkage among different epidemic regions.
The CRF07_BC recombinant strain is one of the most prevalent Chinese strains and accounts for the majority of Chinese HIV-1 infections in the IDU population.4–10 It was generally accepted that CRF07_BC originated from Yunnan province where many more BC recombinants were identified as URFs,4–10 and then spread to Sichuan, Xinjiang, and other regions. Lines of studies in Yunnan, Guangxi, and Xinjiang observed a close genetic relationship of CRF07_BC strains between Yunnan and Xinjiang or Guangxi.4–7,9,10,12 Surprisingly, the circulating HIV-1 strain among IDUs in Sichuan province received little attention, although Sichuan is neighbored by Yunnan and lies in the transmission route from Yunnan to Xinjiang. Therefore, no direct evidence demonstrated the transmission of CRF07_BC strains from Yunnan to Sichuan up to now.
It was widely accepted that the heroine smuggling routine dictated the transmission of CRF07_BC in IDUs.4–10 Since it was neighbored by Yunnan province,4–10 Sichuan was speculated to be one of the earliest acceptor sites along the heroine routine, and it also played a key role in CRF07_BC transmission.8 In fact, the early HIV-1 epidemic and demographic information supported this hypothesis. First, the earliest report of CRF07_BC in Southwestern China indicated that it had been simultaneously identified in Yunnan and Sichuan in 19988,17; and second, as the main HIV-1 affected population, the local Yi ethnic group in Yunnan frequently communicated with Yi ethnic people living in Liangshan of Sichuan, which could facilitate the rapid transmission of CRF07_BC from Yunnan to Sichuan. In this study, the first isolated five full-length genome sequences identified the circulation of CRF07_BC in IDUs in Liangshan of Sichuan. As expected, the MCMC tree revealed a close genetic relationship between Sichuan and Xinjiang, Yunnan strains, which supported the important role of Sichuan in CRF07_BC transmission. However, it remains unknown whether the earliest CRF07_BC strain in Xinjiang was transmitted from Sichuan or directly from Yunnan province.
Since the initial outbreak was reported in 1997, CRF07_BC has been circulating in IDUs for more than 15 years.4–8 Two observations should be noted in our study. First, different regional strains formed into different subclusters, such as strains from Sichuan, Liaoning, and Yunnan, indicating that upon the early entry of the CRF07_BC strain into one region, viruses have been evolving independently in this region and are imprinted with regional features; second, our analysis revealed that a significant transmission linkage could be established between strains from different regions of Yunnan, Xinjiang, Sichuan, and Liaoning, suggesting the existence of multiple transmission paths during the spread of CRF07_BC. Since the heroine smuggling trade mainly causes one-way spreading in China, our observation is in accord with the explanation that the frequent migration of IDUs has also led to multiple introductions of CRF07_BC into one epidemic region, which undoubtedly resulted in additional complexity in deciphering the CRF07_BC transmission routine and speeded up the expanding diversity of HIV-1. Altogether, our data suggested that the prevalence of the genetic strain is determined not only by the drug traffic, but also by the migration of the population. Indeed, epidemiological information from official reports indicated that IDUs from a few prevalent epidemic regions had introduced CRF07_BC strains into the lowest prevalent regions (personal information exchange), which further emphasized the importance of the migration of IDUs in the transmission of CRF07_BC. These results remind us of the challenge of trying to control the prevalence of HIV.
Taken together, in understanding the countrywide transmission of CRF07_BC, not only Yunnan, but also Guangxi, Xinjiang, and Sichuan played an active role in its diffusion. Therefore, the five newly isolated near full-length CRF07_BC sequences not only provide more data concerning the currently limited CRF07_BC full-length genome sequences in the HIV database, but also facilitate understanding the molecular epidemiology of CRF07_BC.
Nucleotide Sequences Accession Number
The NFLG sequences determined in this study are available in GenBank under accession numbers JX392377– JX392384.
Supplementary Material
Acknowledgments
This study was supported by NSFC Grant 30901257, by the China Grand Program on Key Infectious Disease Control and Prevention (2012ZX10001-006 and 007), and by the China Postdoctoral Science Foundation (Grant 201003233).
Z. Meng conceived the study, carried out the molecular genetic studies, participated in the sequence alignment, and drafted the manuscript. R. Xin, Y. F. Abubakar, J. Sun, and H. Wu participated in the sequence alignment and participated in the design of the study and performed the probability testing of the phylogenetic trees. J. Lv and N. Ya coordinated the study, participated in the experimental design, and helped to draft the manuscript. J. Xu and X. Zhang proposed the concept of the study, designed the study, formulated the major conclusions, and revised this manuscript. All the authors read and approved the final manuscript.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Ministry of Health of the People's Republic of China: China 2010 UNGASS Country Progress Report (2008–2009) Apr 2, 2010.
- 2.National HIV Sentinel Surveillance Collaborative Group. China HIV/AIDS sentinel surveillance during 1995–1998. J China AIDS/STD Prev Cont. 2000;4:242. [Google Scholar]
- 3.Department of Disease Control Minister of Health China National Center for AIDS Prevention Control, Group of National HIV Sentinel Surveillance. National sentinel surveillance of HIV infection in China from 1995 to 1998. Chin J Epidemiol. 2000;21:7–9. [Google Scholar]
- 4.Yu XF. Chen J. Shao Y. Beyrer C. Lai S. Two subtypes of HIV-1 among injection-drug users in southern China. Lancet. 1998;351:1250. doi: 10.1016/S0140-6736(05)79316-8. [DOI] [PubMed] [Google Scholar]
- 5.Yu ES. Xie Q. Zhang K. Lu P. Chan LL. HIV infection and AIDS in China, 1985 through 1994. Am J Public Health. 1996;86:1116–1122. doi: 10.2105/ajph.86.8_pt_1.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tee KK. Pybus OG. Li XJ, et al. Temporal and spatial dynamics of human immunodeficiency virus type 1 circulating recombinant forms 08_BC and 07_BC in Asia. J Virol. 2008;82:9206–9215. doi: 10.1128/JVI.00399-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shao Y. Guan Y. Zhao Q. Zeng Y. Wolf H. Genetic variation and molecular epidemiology of the Ruili HIV-1 strains of Yunan in 1995. Chin J Virol. 1996;12:9–17. [Google Scholar]
- 8.Shao Y. Zhao F. Yang W. The identification of recombinant HIV-1 strains in IDUs in southwest and northwest China. Chin J Exp Clin Virol. 1999;13:109–112. [PubMed] [Google Scholar]
- 9.Chen J. Young NL. Subbarao S, et al. HIV type 1 subtypes in Guangxi Province, China, 1996. AIDS Res Hum Retroviruses. 1999;15:81–84. doi: 10.1089/088922299311754. [DOI] [PubMed] [Google Scholar]
- 10.Jie C. Wei L. Nancy LY. Molecular epidemiological analysis of HIV-1 initial prevalence in Guangxi, China. Chin J Epidemiol. 1999;20:74–77. [PubMed] [Google Scholar]
- 11.Meng ZF. Zhang XY. Xin RL, et al. A new approach for sequencing virion genome of Chinese HIV-1 strains subtype B and BC from plasma. Chin Med J (Eng) 2011;124:302–306. [PubMed] [Google Scholar]
- 12.Meng Z. Xing H. He X. Ma L. Xu W. Shao Y. Genetic characterization of three newly isolated CRF07_BC near full-length genomes in China. AIDS Res Hum Retroviruses. 2007;23:1045–1050. doi: 10.1089/aid.2007.0058. [DOI] [PubMed] [Google Scholar]
- 13.Zhefeng M. Huiliang H. Chao Q, et al. Transmission of new CRF07_BC strains with 7 amino acid deletion in Gag p6. Virol J. 2011;8:60. doi: 10.1186/1743-422X-8-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Posada D. Buckley TR. Model selection and model averaging in phylogenetics: Advantages of the AIC and Bayesian approaches over likelihood ratio tests. Syst Biol. 2004;53:793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- 15.Paraskevis D. Magiorkinis E. Magiorkinis G, et al. Phylogenetic reconstruction of a known HIV-1 CRF04_cpx transmission network using maximum likelihood and Bayesian methods. J Mol Evol. 2004;59:709–717. doi: 10.1007/s00239-004-2651-6. [DOI] [PubMed] [Google Scholar]
- 16.Yang Z. Rananala B. Bayesian phylogenetic inference using DNA sequences: A Markov chain Monte Carlo method. Mol Biol Evol. 1997;14:717–724. doi: 10.1093/oxfordjournals.molbev.a025811. [DOI] [PubMed] [Google Scholar]
- 17.Qin G. Shao Y. Liu G, et al. Subtype and sequence analysis of the C2V3 region of gp120 genes among HIV-1 strains in Sichuan Province. Chin J Epidemiol. 1998;19:39–42. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.