Abstract
Geminiviruses are small DNA viruses that replicate in nuclei of infected plant cells after accumulation of host replication machinery. Tomato golden mosaic virus (TGMV) and Tomato yellow leaf curl Sardinia virus (TYLCSV) encode a protein, RepAC1 (or Rep), that is essential for viral replication. Rep/RepAC1 is an oligomeric protein that binds to double-stranded DNA, catalyzes cleavage and ligation of single-stranded DNA, and is sufficient for host induction. It also interacts with several host proteins, including the cell cycle regulator, retinoblastoma, and essential components of the cell DNA replication machinery, like proliferating nuclear cell antigen (PCNA) and RFC-1. To identify other cellular proteins that interact with Rep/RepAC1 protein, a Nicotiana benthamiana cDNA library was screened with a yeast two-hybrid assay. The host cell sumoylation enzyme, NbSCE1 (N. benthamiana SUMO-conjugating enzyme, homolog to Saccharomyces cerevisiae UBC9), was found to interact specifically with RepAC1. Mapping studies localized the interaction to the N-terminal half of RepAC1. Effects on geminivirus replication were observed in transgenic plants with altered levels of SUMO, the substrate for UBC9.
Geminiviruses are a large family of plant viruses with circular, single-stranded DNA (ssDNA) genomes packaged within geminate particles. The Geminiviridae family is divided into four genera according to their genome organizations and biological properties (15, 16). The Begomovirus genus includes viruses that are transmitted by whiteflies and infect only dicotyledonous plants. Begomoviruses can have bipartite genomes (A and B components), like Tomato golden mosaic virus (TGMV), or monopartite genomes, like Tomato yellow leaf curl Sardinia virus (TYLCSV) (40). The TGMV genome consists of two circular DNA components of about 2.5 kb in size, which together encode at least six proteins. TYLCSV has a single 2.8-kb circular ssDNA genome that contains six open reading frames (ORFs). The arrangement of TYLCSV ORFs is similar to that of the DNA-A component of bipartite begomoviruses. The ORFs encoding Rep, TrAP, and REn partially overlap, and a small ORF, C4, is located within the Rep ORF, but in a different reading frame (other names for these genes in TGMV are AL1 [or AC1], AL2 [or AC2], AL3 [or AC3], and AL4 [or AC4], respectively) (25). Two of these proteins, Rep and REn, are required for efficient viral DNA replication. Rep is essential for replication, whereas REn enhances viral DNA accumulation by an unknown mechanism. Rep/RepAC1 is a multifunctional protein that initiates and terminates plus strand replication (27, 34, 43) and specifically binds to double-stranded DNA during origin recognition (2, 8, 19). Rep/RepAC1 can also hydrolyze ATP and interacts with itself (44) and with the viral replication enhancer REn (54).
Geminiviruses do not encode their own DNA polymerases and instead rely on the nuclear DNA replication machinery of their host, like many mammalian DNA tumor viruses do. They replicate their genomes in nuclei of mature cells, which are not competent for DNA replication, so an early step in geminivirus infection may be the induction of host DNA replication enzymes (reviewed in references 23 and 25). This idea is supported strongly by the accumulation of proliferating cell nuclear antigen (PCNA) in differentiated cells of TGMV-infected plants (42) and in transgenic plants expressing TGMV RepAC1. TGMV RepAC1 (1, 31) binds to plant homologs of the cell cycle regulator, retinoblastoma (pRb). By analogy with mammalian DNA viruses, these interactions may bypass a pRb phosphorylation requirement for the G1/S transition and S-phase progression (39, 57) during geminivirus infection. Recently, new interactions between geminivirus Rep/RepAC1 and plant cellular proteins have been described. It was demonstrated that a serine/threonine protein kinase, a kinesin, and histone H3 are partners of TGMV and CbLCV (Cabbage leaf curl virus) RepAC1 (32). In addition, the replication factor C complex from wheat (TmRFC-1) interacts with Rep from Wheat dwarf virus (WDV) (37), and tomato PCNA interacts with Rep from TYLCSV (9).
Here, we show that Rep/RepAC1 interacts with a plant homolog of those proteins acting as SUMO-conjugating enzymes in other organisms. Sumoylation is a posttranslational process that modifies function, activity, or localization of the target protein by the covalent attachment of a ubiquitin (Ub)-like polypeptide (Ubl) called SUMO (also known as sentrin, Smt3, UPL, and PIC1) (38, 53). The posttranslational modifications by ubiquitin or SUMO have mechanistically similar ATP-dependent reaction cascades, involving activation and conjugation. The two reactions are catalyzed by different enzymes: the activating enzyme (E1) and conjugating enzyme (E2), respectively. Sometimes an additional enzyme (E3) participates in target recognition and ligation (53). Contrary to ubiquitination, SUMO conjugation is mediated by a single E2 enzyme (UBC9) that is essential for cell viability and sumoylation in yeast and animals (26, 56). In yeast, there is a single gene that encodes SUMO (SMT3), but in metazoans, SUMO proteins can be divided in two families: SUMO1 and SUMO2/SUMO3. Even though SUMO2 and SUMO3 share 50% sequence identity with SUMO1, they are functionally different (50, 58).
The general consequence for SUMO modification remains unclear and varies, depending on the target. SUMO conjugation has been implicated in cellular responses to environmental stress (50), subcellular protein translocation (48), nuclear body formation, centromere segregation, protection from ubiquitin-mediated proteolysis (38), and regulation of transcriptional activity (59). Several mammalian viral proteins, including cytomegalovirus IE1 and IE2, Epstein-Barr virus BZLF1, and papillomavirus E1, interact with UBC9 and/or SUMO (60). As with sumoylated cellular proteins, the biological effect ensuing from viral protein sumoylation is target specific.
In plants, little is known about the SUMO pathway or the nature of its targets. Many of the core components for sumoylation have been identified in Arabidopsis thaliana (33, 35). The Arabidopsis genome contains eight full-length SUMO genes (AtSUMOs) and a single gene that encodes a SUMO-conjugating enzyme homolog to UBC9 (AtSCE1a, for A. thaliana SUMO-conjugating enzyme). Phylogenetic analysis clustered AtSUMO into five subfamilies. Sequence comparison and expression profiles indicated that AtSUMO1, AtSUMO2, and AtSUMO3 are SUMO orthologs of human HsSUMO2, HsSUMO3, and HsSUMO1, respectively. Like human cells, the accumulation of SUMO1/2 conjugates in A. thaliana is increased by stress (heat, H2O2, ethanol, and canavanine) (33). Sumoylation also contributes to the regulation of ABA signaling that mediates plant responses to several environmental stresses (35). SUMO may also play an important role in pathogen plant defense responses. The tomato SUMO ortholog (LeSUMO) was isolated in a yeast two-hybrid screen by its interaction with ethylene-inducing xilanase (EIX) from the fungus Trichoderma viridae (24). The expression of LeSUMO in transgenic tobacco plants suppressed the induction of the defense response by EIX (a strong elicitor of the rapid defense response in tomato). A virulence factor (AvrBsT) from the plant pathogen Xanthomonas campestris has SUMO protease activity that interferes with the plant defense response, probably by desumolating a key defense regulator (47).
Here we describe the isolation of an ortholog to the SUMO-conjugating enzyme UBC9 from Nicotiana bethamiana (NbSCE1). This protein interacts with the Rep/RepAC1 proteins from several begomoviruses. These results and infection analyses of transgenic tobacco plants with altered SUMO levels suggested that posttranslational protein modification by SUMO plays an important role in geminivirus infection. This is the first time that an interaction between a protein and the E2 component of the SUMO pathway has been described in plants.
MATERIALS AND METHODS
Microorganisms and general methods.
Manipulations of Escherichia coli and Saccharomyces cerevisiae strains and nucleic acids were performed according to standard methods (3, 51). E. coli strains DH5-α and JA226 were used for subcloning. All PCR-amplified fragments cloned in this work were fully sequenced. E. coli strains LE392 and BNN132 were used for titration and conversion of N. benthamiana cDNA library from lambda to two-hybrid plasmid vectors, respectively. S. cerevisiae strain PJ696 (MATa trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ), a derivative of PJ69-4A (29), was used for the two-hybrid experiments.
For DNA gel blots, total DNA was extracted from N. benthamiana leaves as described previously (11). DNA samples (10 μg) were digested, separated on 0.8% agarose gels, and blotted onto Hybond-N membranes (Amersham Pharmacia Biotech AB) according to the manufacturer's instructions. The membrane was hybridized in Church buffer (10) at 65°C with radiolabeled NbSCE1 ORF probe. The probe was an [α-32P]dCTP-labeled, 500-bp PCR fragment generated by random priming with the Megaprime DNA labeling system (Amersham Pharmacia Biotech AB) and the primers UPNBUB and LWNBUB. (All primers used in this work are described in Table 1.) The filter was washed two times in a mixture of 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) plus 0.1% sodium dodecyl sulfate (SDS) at 65°C and autoradiographed.
TABLE 1.
Primers used in this study
| Primer | Oligonucleotide sequence (5′→3′) |
|---|---|
| UPNBUB | GAGAAGGATCCTAATGTCAGGAGGTATAGC |
| LWNBUB | GCATTAGAACCAGGATCCAAACATACGACC |
| O5REPACK | TGACTTGATATCCATGAGAACTCCTCG |
| O3REPACK | TCCACAGACGATATCCACTCTCCTACG |
| O5REPTG | GCCTCTTGATATCAAAAATGCCATCGC |
| O3REPTG | CGCCTTCTGATATCTCTTCGTTTAGC |
| O3NRepTG | TGGAACGGATATCACCATGGTTCAGGAGTC |
| O5CAL1 | CTTTGACGATATCATGGCTTCCTCCGTTCC |
| O3CAL1 | CGCCTTCTGATATCTCTTCGTTTAGCTGC |
| O3RepTG130 | AGCGTCGATATCTGTTTGGCAACCTCCTC |
| Forward | AGCGAATTCACCATGGATTTACGCACAGG |
| Reverse | GATCTGCAGGCGTTGTTGTTGTATATGGC |
| O5HISUBC | GAGAAAGACACATATGTCAGGAGGTATAGCC |
| O3HISUBC | CGACCATTATGAATTCTACACCAGAGGAGGATAC |
For RNA gel blots, total RNA was extracted from frozen leaves, flowers, stems, and veins of N. benthamiana as described previously (28) and fractionated by 1% agarose gel electrophoresis. Just before loading the gel, formamide, formaldehyde, and ethidium bromide were added to each RNA sample (10 μg). RNA was blotted onto Hybond N+ membrane (Amersham Pharmacia Biotech AB) according to the manufacturer's instructions, and the membrane was hybridized in Church buffer at 65°C with the radiolabeled NbSCE1 ORF probe described above. The filter was washed two times in 0.2× SSC-0.2% SDS at 65°C and autoradiographed.
Plasmids and cloning.
The two-hybrid plasmid pBD-GAL4 (Stratagene) was used for the fusion of the bait protein to the GAL4 DNA binding domain (DB). The bait plasmid pDBRep was constructed as follows. The plasmid pTYC—KpnI fragment from position 839 to 2634 of TYLCSV-ES[2] (accession no. L27708) cloned into the KpnI site of pBluescriptIIKS+—was digested with KpnI and BglII and repaired with Klenow enzyme. This fragment was then cloned into the SmaI site of pBDGal4 to yield pDBRep.
Complete Rep/AC1 ORFs from different begomoviruses were amplified with Pfu by using the primers (Table 1) O5REPACK and O3REPACK for ACMV-KE (positions 2598 through 2779 to nucleotide 1539; accession no. J02057) and O5REPTG and O3REPTG for TGMV (positions 2461 through 2588 to nucleotide 1416; accession no. M73794). Amplified fragments were digested with EcoRV and cloned into the EcoRI site of pBD-GAL4, previously repaired with Klenow enzyme, to yield pDBRepAC and pDBRepTG, respectively.
The yeast two-hybrid plasmid containing full length AC1 from TGMV in pACT2 (pNSB809) was described elsewhere (46). Yeast two-hybrid plasmids containing truncated forms of TGMV AC1 in pBD-GAL4 were constructed as follows. The N-terminal half of TGMV RepAC1 (amino acids 1 to 181) was PCR amplified with the primers O5REPTG and O3NRepTG; the C-terminal half of RepAC1 (amino acids 182 to 352) was amplified with the primers O5CAL1 and O3CAL1; and the truncated form, RepAC1N1-130, was amplified with the primers O5RepTG and O3RepTG130 (Table 1). Amplified fragments were digested with EcoRV and cloned into the EcoRI site of pBD-GAL4, previously repaired with Klenow enzyme, to yield pDBRepAC1N1-180, pDBRepAC1C181-352, and pDBRepAC1N1-130, respectively.
Bait plasmids used as negative controls for the interactions were pDBCP (CP from TYLCSV in pBDGal4; S. Ohnesorge and E. R. Bejarano, unpublished data) and pDBREn. pDBREn was constructed as follows. The coding region from REn of TYLCSV-ES[2] was amplified by PCR from pTYA50—a plasmid containing the EcoRI full genome of TYLCSV-ES2 (accession no. L27708) cloned into EcoRI pBluescriptIIKS+—by using the forward and reverse primers. The amplified fragment was digested with the restriction enzymes EcoRI and PstI and cloned into the EcoRI-PstI sites of pBD-GAL4 to yield pDBREn.
Yeast two-hybrid screen.
The N. benthamiana cDNA library was kindly provided by L. Jongejan (Leiden University, Leiden, The Netherlands). It was constructed in the EcoRI-XhoI sites of λACTII as described by J. Memelink on the Technical Tips Online site (T01111; http://www.elsevier.com/locate/tto) and contains 1.2 × 106 primary transformants. Automatic subcloning conversion of the cDNA library from lambda to plasmid vectors was done as described by Elledge et al. (13).
The yeast strain PJ696, which contains the three reporter genes lacZ, HIS3, and ADE2, was used in the two-hybrid screens (17). For the screens, yeast cells were cotransformed with pDBRep and the N. benthamiana cDNA library in pACTII (AD-Gal4; LEU2), as described by Gietz and Schiestl (21). For medium-stringency selection, the transformation mixture was plated on yeast selection medium SD/−Trp −Leu −His (Yeast Protocols Handbook, CLONTECH Laboratories, Inc., 2001; www.clontech.com) supplemented with 2 mM 3-amino-1, 2, 4-triazole (3-AT) (4) to reduce the appearance of false-positive colonies. Transformants were routinely recovered during a period of 5 to 10 days and checked for growth on the alternative selection medium. For a higher-stringency selection, the transformation mixture was plated on two selection media: SD/−Trp −Leu −Ade or SD/−Trp −Leu −His −Ade + 3-AT. To corroborate the interaction between the two fusion proteins, β-galactosidase activity was assayed in agar plates (12) or on filters (Yeast Protocols Handbook). Plasmid DNA was recovered from positive colonies by transformation into E. coli JA226, which is leuB, and can be complemented by the LEU2 gene in the pACT2 plasmid.
Quantitative β-galactosidase assays from liquid cultures and immunoblot analysis of bait plasmids were performed as described by Castillo et al. (9). A TGMV polyclonal anti-RepAC1 antibody (54) was used for the immunoblotting.
Yeast complementation assay.
S. cerevisiae mutant strain YWO98/ ubc9-2 is isogenic to YWO2 (MATα his3-Δ200 leu2-3,112 lys2-801 trp1-1 ura3-52) (55), but has a deletion of UBC9 gene that is complemented with the plasmid pSE362-ubc9-2 (YWO98: MATα, ubc9Δ1::TRP1 pSE362-ubc9-2) (7). YWO2 and YWO98 were used for the complementation assay with UBC9 from N. benthamiana (NbSCE1). NbSCE1 was amplified from pACTII-NbSCE1 (clone isolated from the two-hybrid screenings) with Pfu by using the oligonucleotides UPNBUB and LWNBUB (Table 1). The amplified fragment was digested with BamHI and cloned into BamHI sites of pCM190 (multicopy plasmid) and pCM188 (centromeric plasmid) (20) to yield pCM190-NbSCE1 and pCM188-NbSCE1, respectively. Sense clones were selected, and the S. cerevisiae strain YW098/ubc9-2 was transformed (21) with both constructions and plated at 25°C on minimal medium (SD) supplemented with leucine, lysine, and doxycycline (10 μg/ml). Both vectors express NbSCE1 from a tetracycline-regulatable promoter, so the tetracycline analog doxycycline (Sigma), was added to the plates to inhibit NbSCE1 expression (20). Two colonies were selected for each transformant and plated on minimal medium (SD) supplemented with leucine, lysine, and several doses of doxycycline (0, 1, 5, and 10 μg/ml). The plates were incubated at 25 or 37°C for 5 days.
Recombinant baculoviruses.
Baculovirus transfer vectors containing the polyhedrin promoter and the simian virus 40 poly(A) site were constructed for the full-length and truncated forms of TGMV AC1, NbSCE1, and chloramphenicol acetyltransferase (CAT). Baculovirus DNA corresponding to each of the transfer vectors was generated by Tn7-mediated transposition with the bacmid plasmid bMON14242, transfected into Spodoptera frugiperda Sf9 cells, and screened for recombinant protein expression as described previously (36, 44). The baculovirus transfer vector pNSB910 contains a HIS-tagged maize retinoblastoma gene downstream of the polyhedrin promoter in pMON27025 (36). NbSCE1 was amplified by PCR with Vent Taq polymerase (New England Biolabs) and the oligonucletides O5HISUBC and O3HISUBC. The amplified fragment was digested with NdeI and EcoRI and cloned into pNSB910 NdeI and EcoRI sites, after removing the maize retinoblastoma gene by using Geneclean. The resulting vector, pNSB910-NbSCE1, was used to obtain recombinant baculoviruses expressing HIS-NbSCE1. Recombinant baculoviruses corresponding to full-length TGMV RepAC11-352 (pNSB244) and the truncated proteins RepAC1N1-180 (pNSB517) and RepAC1C181-352 (pNSB469) were described elsewhere (31). Recombinant baculovirus corresponding to the HIS-CAT fusion was described by Ach et al. (1).
Protein expression, extraction, and coimmunoprecipitation.
Sf9 cells (106/ml) were coinfected with recombinant baculoviruses (either HIS-NbSCE1 and any of the full-length or truncated forms of TGMV RepAC1 or HIS-CAT and any of the full-length or truncated forms of RepAC1) in a 25-mm2 T flask. Cells were harvested 60 h postinfection and lysed in NIB buffer (25 mM Tris-HCl [pH 8], 1 mM EDTA, 0.014 mM β-mercaptoethanol, 0.01% Triton X-100) with protease inhibitors (10-μg/ml pepstatin, 50-μg/ml leupeptin, 1 mM phenylmethylsulfonyl fluoride). Immunoprecipitations were performed by incubating protein extract with either an anti-HIS monoclonal antibody (6xHIS; CLONTECH) or TGMV anti-RepAC1 antisera (54). Protein-antibody complexes were mixed with protein A-Sepharose CL-4B (Amersham Pharmacia Biotech AB) in Tris-buffered saline-Tween (TBST) at 4°C, mixed for 2 h, and then washed with TBST four times. Bound proteins were released by boiling in SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer for 5 min. Coimmunoprecipitation was monitored by SDS-PAGE followed by transfer to nitrocellulose membrane (Hybond ECL; Amersham Pharmacia Biotech AB) and immunoblotting with the ECL enhanced chemiluminescence detection system (Amersham Pharmacia Biotech AB). The primary antibodies were monoclonal anti-HIS antibody (6xHIS monoclonal antibody), TGMV polyclonal anti-RepAC1 antibody (54), or TGMV monoclonal anti-RepAC1 antibody (18).
Geminivirus replication assays.
Untransformed or T2 seeds from transgenic Nicotiana tabacum cultivar Samsun plants were used for the geminivirus infection assays. The transgenic plants express LeSUMO in a sense or antisense orientation under the control of the 35S-Ω promoter containing the translation enhancer signal and the Nos terminator (24). Both transgenic lines contain several copies of the LeSUMO construction. Six to eight T2 kanamycin-resistant plants of each type and a similar number of untransformed plants were agroinoculated with TGMV (5) in two independent experiments. Total DNA was extracted from N. tabacum leaves (16, 24, and 45 days postinfection [dpi]) as previously described (11) and analyzed by DNA gel blotting with a radiolabeled TGMV A component probe radiolabeled by random priming, as described above. A PhosphorImager (FUJIFILM BAS-1500) was used to quantify the radioactivity signal on each sample.
Leaf disks from untransformed N. tabacum cultivar Samsun or transgenic plants overexpressing LeSUMO in a sense or antisense orientation (24) were infected with the TGMV A component according to the method described by Elmer et al. (14). DNA extraction and DNA gel blots were done as described above.
Nucleotide sequence accession number.
The database accession number for NbSCE1 is AJ580839.
RESULTS
Isolation of a novel geminivirus Rep-interacting protein.
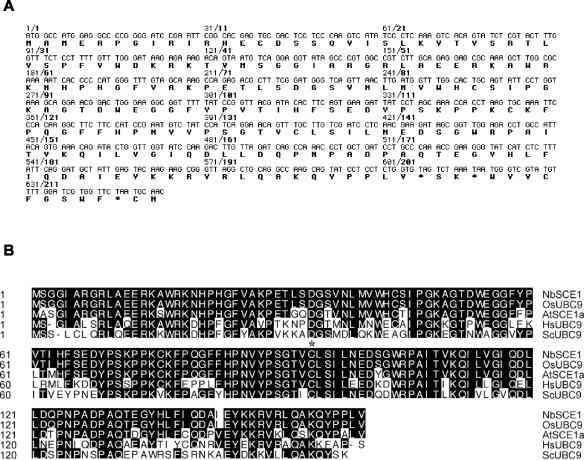
To identify cDNAs encoding polypeptides that interact with TYLSCV Rep protein, we used the yeast two-hybrid system (17). The Rep gene from TYLSCV was fused to the DB of the yeast transcription factor GAL4. The fusion protein did not activate the reporter genes HIS3, ADE2, and lacZ in yeast by itself (data not shown). Yeast strain PJ696 was then cotransformed with the Rep bait plasmid and the cDNA N. benthamiana prey library (21). From 107 transformants, 79 colonies grew on plates lacking histidine and supplemented with 3-AT. Nine of the 79 colonies were also able to grow on plates lacking histidine and/or adenine. To test whether Rep was required for interaction with the products of the nine selected cDNAs, all of the clones were retransformed into the yeast strain PJ696 together with pBD-GAL4 or a plasmid encoding other Gal4 DB fusion (pDBREn, TYLCSV REn gene cloned in pBDGAL4). Two library clones were found to interact specifically with DB-Rep, and subsequent analyses indicated that both clones contained the same cDNA sequence. The two clones contained a single ORF of 160 amino acids (Fig. 1A) that has high homology to the AtSCE1a gene, which codes for an E2 ubiquitin-conjugating-like enzyme from Arabidopsis (accession no. Q42551), and to UBC9 from Saccharomyces cerevisiae. Like ScUBC9 and AtSCE1a, NbSCE1 belongs to a family of conjugating enzymes, all of which have a conserved UBCc domain (for Ub-conjugating enzyme catalytic) with a positionally conserved cysteine that forms the thiol ester linkage with the peptide modifier (Ub or SUMO). The predicted amino acid sequence for the N. benthamiana gene (NbSCE1) differs by seven and five amino acids from SCE1 orthologs from A. thaliana and rice (OsSCE1), respectively. It has 63 and 61% identity to S. cerevisiae (56) and human UBC9 (61), respectively (Fig. 1B). In this screen, we did not isolate any of the cellular proteins already described as RepAC1 binding proteins.
FIG. 1.
(A) Nucleotide and amino acid sequences of the isolated cDNA. The NbSCE1 ORF is underlined. (B) Comparison of the amino acid sequence of NbSCE1 to those of UBC9 from different organisms: Oriza sativa (OsSCE1), A. thaliana (AtSCE1a), Homo sapiens (HsUBC9), and S. cerevisiae (ScUBC9). Identical amino acids are shown in white letters on a black background. The conserved cysteine residue is marked by an asterisk.
NbSCE1 complements an S. cerevisiae ubc9 temperature-sensitive mutant.
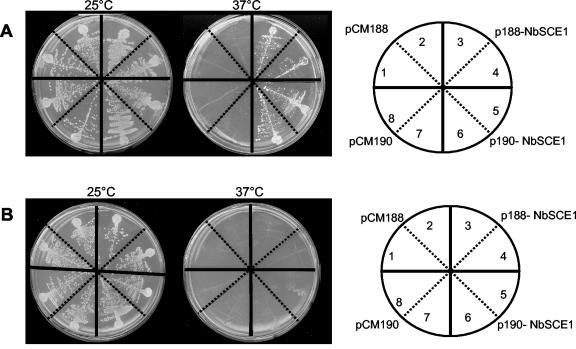
To test whether NbSCE1 can functionally replace S. cerevisiae UBC9, we used a yeast mutant strain (ubc9-2) containing a temperature-sensitive mutation in the UBC9 gene (7). Wild-type yeast and the ubc9-2 temperature-sensitive mutant (YWO2 and YWO98, respectively) were transformed with empty plasmids (a centromeric plasmid, pCM188, and a multicopy plasmid, pCM190) or with the plasmids expressing NbSCE1 (p188-NbSCE1 and p190-NbSCE1). Both vectors express NbSCE1 from a tetracycline-regulatable promoter so that expression is inhibited in the presence of doxycycline (a tetracycline analog) (20). The temperature-sensitive ubc9-2 mutant was able to grow at 37°C when the NbSCE1 gene was expressed from either the centromeric or the multicopy plasmid (Fig. 2A). When expression of NbSCE1 was repressed, the temperature-sensitive ubc9-2 mutant did not grow at 37°C (Fig. 2B). Wild-type yeast cells transformed with NbSCE1 grew at both temperatures and at any of the doxycycline concentrations assayed (data not shown). These data indicated that NbSCE1 is functionally homologous to the S. cerevisae UBC9 gene.
FIG. 2.
Functional complementation of the S. cerevisiae ubc9-2 mutant (YW098) by the NbSCE1 gene. Transformants of the temperature-sensitive mutant (YWO98) harboring pCM188 (1 and 2), p188-NbSCE1 (3 and 4), p190-NbSCE1 (5 and 6), or pCM190 (7 and 8) were streaked on selective plates and incubated at 25 and 37°C in the absence (A) or presence (B) of doxycycline (10 μg/ml). Photographs were taken after a 5-day incubation.
DNA and RNA gel blot analysis of NbSCE1.
The Arabidopsis genome contains two loci for UBC9—AtSCE-1a and AtSCE-1b. AtSCE-1a encodes a full-length E2 that is highly expressed in all tissues. In contrast, AtSCE-1b does not encode the first 53 residues, and no mRNA or protein has been detected for this pseudogene (33). To determine the number of loci for SCE1 in N. benthamiana, total leaf DNA was digested with EcoRI (which does not cut in the NbSCE1 ORF) or HindIII (which cuts once in NbSCE1 ORF) and probed with an NbSCE1-specific probe. Even though high-stringency conditions were used, bands with different intensities were observed in both digestions (data not shown). These results suggested that NbSCE1-related sequences occur at more than one locus in N. benthamiana. Further studies will be needed to confirm if there is only a single functional locus, as in the A. thaliana genome.
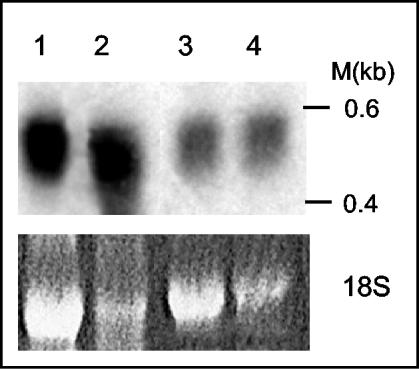
To characterize the expression profile of NbSCE1 gene, gel blots of total RNA isolated from leaves, flowers, stems, and veins were probed with NbSCE1 ORF (Fig. 3). A single 600-nucleotide band was detected in all RNA samples, indicating that, like in Arabidopsis (33) and humans (61), NbSCE1 is ubiquitously expressed in N. benthamiana. When RNA loading differences were considered, NbSCE1 mRNA levels were higher in flowers than in other tissues.
FIG. 3.
RNA gel blot analysis from N. benthamiana. (Top panel) RNA gel blot analysis of total RNA extracted from N. benthamiana leaves, flowers, stems, and veins (no. 1 to 4, respectively). The membrane was hybridized with a double-stranded NbSCE1 ORF probe. Ten micrograms of total RNA was loaded in each lane. As a loading control, the ethidium bromide-stained form of 18S rRNA is shown (bottom panel).
NbSCE1 interacts with Rep from other geminiviruses.
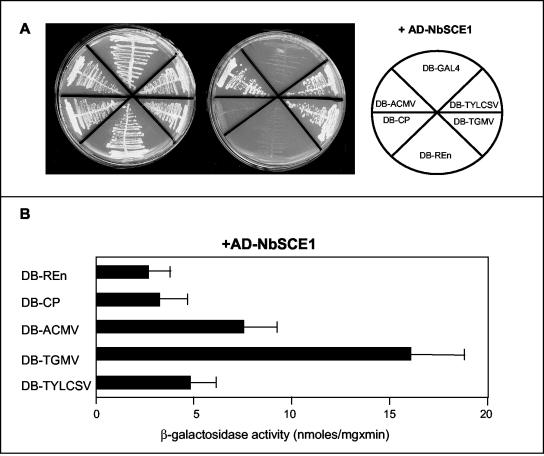
To investigate whether the NbSCE1 protein is able to interact with Rep proteins from others geminiviruses, we fused the GAL4 DB to the Rep coding sequences from the begomoviruses TGMV and ACMV-KE (African cassava mosaic virus, Kenyan isolated) and asked whether they were able to interact with AD-NbSCE1 in PJ696. Cotransformants were isolated and plated on different selective media to detect activation of the reporter genes HIS3 and ADE2. Figure 4A shows that the replication proteins from TGMV (DB-TGMV) and ACMV-KE (DB-ACMV) also interact with NbSCE1, because the cells were able to grow in a media lacking histidine and adenine. The interaction between NbSCE1 and Rep from these begomoviruses was quantified by using the two-hybrid system. The activation of a GAL4-responsive promoter driving the lacZ reporter gene in PJ696 was assayed by measuring β-galactosidase activity in total soluble protein extracts (Fig. 4B). The β-galactosidase activity for the yeast coexpressing AD-NbSCE1 with DB-TGMV or DB-ACMV was higher than that in the negative controls (DB-REn and AD-NbSCE1 or DB-CP and AD-NbSCE1). Because the highest β-galactosidase activity, as well as the expression of the other two reporter genes (HIS3 and ADE2), was obtained with the fusion protein DB-TGMV, we decided to use the RepAC1 protein from TGMV to characterize the interaction with NbSCE1 further. The variance observed in the activation of the reporter genes among begomovirus Rep proteins could reflect different strengths of interaction or different levels of protein accumulation.
FIG. 4.
Interaction between NbSCE1 and Rep/RepAC1 from different begomoviruses. The AD-NbSCE1 protein was coexpressed in yeast with plasmids corresponding to DB-Rep bait proteins from TGMV (DB-TGMV), ACMV-KE (DB-ACMV), and TYLCSV (DB-TYLCSV). As negative controls, a plasmid expressing a fusion protein between the GAL4 DB and REn or CP from TYLCSV or the empty plasmid expressing DB-Gal were used as baits. (A) Growth on nonselective (left plate) and selective media lacking histidine and adenine and supplemented with 2 mM 3-AT (right plate). Positions of the different cotransformed yeasts in the plates are indicated. (B) Measurement of β-galactosidase activity in total protein extracts (nanomoles of ONPG [o-nitrophenyl-β-d-galactopyranoside] per minute per milligram of protein). The error bars correspond to 2 standard errors.
NbSCE1 protein interacts with TGMV RepAC1 in coimmunoprecipitation assays.
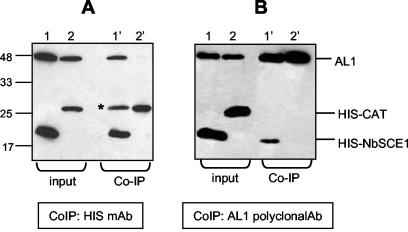
To confirm the interaction between TGMV RepAC1 and NbSCE1 in a different system, we carried out a coimmunoprecipitation assay. RepAC1 and the fusion protein HIS-NbSCE1 were expressed in insect cells by using a baculovirus expression system (36). We coimmunoprecipitated protein extracts from cells coexpressing HIS-NbSCE1 and RepAC1 (lane 1), or HIS-CAT (chloramphenicol acetyltransferase) and RepAC1 (lane 2) with either anti-HIS monoclonal antibody (Fig. 5A) or anti-RepAC1 polyclonal antibody (Fig. 5B). When RepAC1 was coexpressed with HIS-NbSCE1 and the protein extract was incubated with anti-HIS antibody, RepAC1 was detected in both the input (Fig. 5A, lane 1) and the precipitated fractions (Fig. 5A, lane 1′). In contrast, RepAC1 was not detected in the precipitated fraction when it was coexpressed with HIS-CAT (Fig. 5A, lane 2′) under the same conditions. The same results were obtained when the coimmunoprecipitation was done with anti-RepAC1 serum (Fig. 5B). Together, these results demonstrated that HIS-NbSCE1 interacts specifically with RepAC1.
FIG. 5.
Total protein extracts from insect cells coexpressing TGMV RepAC1 and either HIS-NbSCE1 (lanes 1 and 1′) or HIS-CAT proteins (lanes 2 and 2′) were incubated first with either anti-HIS monoclonal antibody (mAb) (A) or TGMV anti-RepAC1 polyclonal antibody (Ab) (B) and next with protein A-Sepharose. Total (input) and coimmunoprecipitated (Co-IP) proteins were analyzed by immunoblotting with anti-HIS monoclonal antibody and TGMV anti-RepAC1 monoclonal antibody. Input (lanes 1 and 2) and coimmunoprecipitated (lanes 1′ and 2′) fractions are shown for the interactions. A subunit of mouse immunoglobulin G that has the same size as the HIS-CAT fusion protein is marked by an asterisk in lane 1′ of panel A. This band is not present in panel B, because the coimmunoprecipitation was done with a rabbit polyclonal antibody.
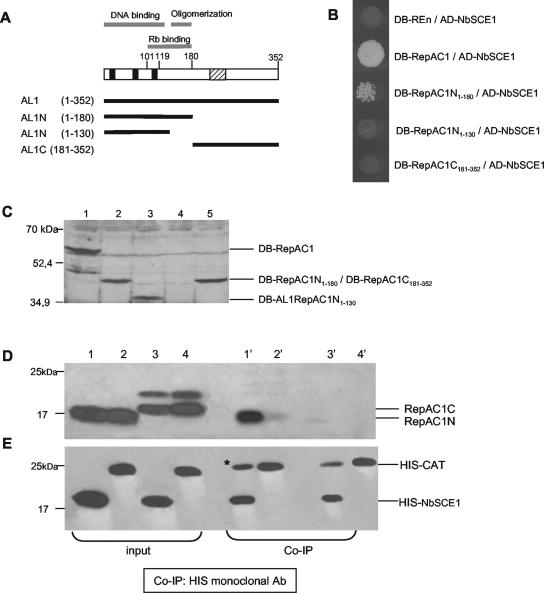
NbSCE1 protein interacts with the N-terminal half of TGMV RepAC1.
To further explore the interaction between TGMV RepAC1 and NbSCE1, we used the yeast two-hybrid system to ask whether the interaction occurs at the N terminus or C terminus of RepAC1. Figure 6A shows a diagram of TGMV RepAC1 and its functional domains. The deletion analysis revealed that the N-terminal region of RepAC1 (DB-RepAC1N1-180, amino acids 1 to 180) interacts with AD-NbSCE1, whereas there is no activation of the reporter genes in yeast coexpressing AD-NbSCE1 and the C-terminal region of RepAC1 (DB-RepAC1C181-352, amino acids 181 to 352) (Fig. 6B). Analysis of growth of the cotransformed yeast on selective medium showed that the interaction between the N-terminal RepAC1 and NbSCE1 is weaker than with full-length RepAC1 protein and that this interaction is lost when 50 amino acids from the central region of RepAC1 are removed (DB-RepAC1N1-130). We verified by immunoblot analysis that the truncated forms of RepAC1 and full-length RepAC1 accumulated to the same levels in yeast. Total protein extracts from cotransformants grown under nonselective conditions were resolved by SDS-PAGE, and the immunoblots were probed with anti-RepAC1 polyclonal antibody (Fig. 6C). There was no difference in levels of protein accumulation among the four bait proteins, so the lack of interaction of AD-NbSCE1 with DB-RepAC1C181-352 or DB-RepAC1N1-130 is not due to different expression levels. These results indicated that the domain for binding to NbSCE1 is located at the first half of RepAC1 and requires residues between amino acids 130 and 180 for interaction.
FIG. 6.
Interaction between NbSCE1 and truncated forms of TGMV RepAC1. (A) Diagram of TGMV RepAC1 and its functional domains. The shaded box indicates an ATP-binding consensus, and the solid boxes mark motifs conserved among rolling circle replication initiator proteins. The gray lines above the protein mark the locations of the overlapping DNA-binding, oligomerization, and pRb-binding domains. Complete (RepAC11-352) and truncated RepAC1 proteins (RepAC1N1-180, RepAC1N1-130, and RepAC1C182-352) used in the interaction assays are diagrammed below. (B) The AD-NbSCE1 protein was coexpressed in yeast with the full-length or the truncated forms of TGMV RepAC1 fused to the DB-Gal4. Growth on plates lacking histidine and supplemented with 2 mM 3-AT is shown. As a negative control, yeasts coexpressing AD-NbSCE1 and DB-REn from TYLSCV were used. (C) Immunoblot analysis of protein extracts from yeast cells coexpressing AD-NbSCE1 and either DB-RepAC1 (lane 1), DB-RepAC1N1-180 (lane 2), DB-RepAC1N1-130 (lane 3), DB-REn (lane 4), or DB-RepAC1C182-352 (lane 5), using TGMV anti-RepAC1 polyclonal antibody. The expected positions of the DB fusion proteins are marked on the left in kilodaltons. (D and E) Immunoblots showing in vitro interaction between NbSCE1 and the N terminus (RepAC1N1-180) or C terminus (RepAC1C181-352) of TGMV RepAC1. Total protein extracts from insect cells coexpressing RepAC1N1-180 and HIS-NbSCE1 or HIS-CAT proteins (lanes 1, 2, 1′, and 2′) or coexpressing RepAC1C181-352 and HIS-NbSCE1 or HIS-CAT proteins (lanes 3, 4, 3′, and 4′) were incubated first with anti-HIS monoclonal antibody (Ab) and afterwards with protein A-Sepharose. Total and coimmunoprecipitated (Co-IP) proteins were analyzed by immunoblotting with TGMV anti-RepAC1 polyclonal antibody (D) or anti-HIS monoclonal antibody (E). Input (lanes 1, 2, 3, and 4) and coimmunoprecipitated (lanes 1′, 2′, 3′, and 4′) fractions are shown for the interactions. A subunit of mouse immunoglobulin G that has the same size as the HIS-CAT fusion protein is marked by an asterisk in lane 1′.
We also confirmed the interaction between the N-terminal end of TGMV RepAC1 and NbSCE1 by coimmunoprecipitation. The fusion protein HIS-NbSCE1 was coexpressed in insect cells with either the N terminus (RepAC1N1-180) or the C terminus of RepAC1 (RepAC1C181-352) by using a baculovirus expression system (36). As a negative control, HIS-CAT was also coexpressed in insect cells with either RepAC1N1-180 or RepAC1C181-352. We coimmunoprecipitated protein extracts from the insect cells with anti-HIS monoclonal antibody (Fig. 6D and E). When the N-terminal end of RepAC1 was coexpressed with HIS-NbSCE1, RepAC1N1-180 was detected in the input and immunoprecipitated fractions (Fig. 6D, lanes 1 and 1′) when the immunoblot was done with anti-RepAC1 polyclonal antibody. In contrast, RepAC1N1-180 was not detected in the coimmunoprecipitated fraction when it was coexpressed with HIS-CAT (Fig. 6D, lane 2′) under the same conditions. The C-terminal half of RepAC1 was observed in the input fraction when it was coexpressed with either HIS-NbSCE1 or HIS-CAT (Fig. 6D, lanes 3 and 4), but was not detected in the coimmunoprecipitated fraction for both assays (Fig. 6D, lanes 3′ and 4′). Figure 6E corresponds to an immunoblot with anti-HIS monoclonal antibody to show that HIS-NbSCE1 and HIS-CAT were correctly expressed in all of the coimmunoprecipitations. These results confirmed those obtained by the two-hybrid assay showing that the N-terminal half of TGMV RepAC1 (residues 1 to 180) contains the domain required for the interaction with NbSCE1.
Expression of sense and antisense RNAs of tomato SUMO (LeSUMO) impairs TGMV replication.
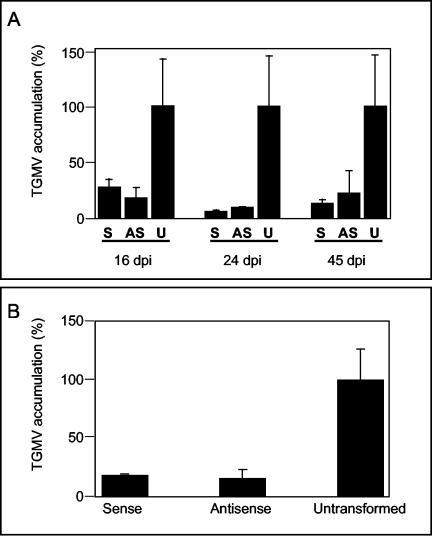
To test if sumoylation has a role in the geminivirus infection process, we used TGMV to infect transgenic tobacco cultivar Samsum expressing sense or antisense RNA of LeSUMO (SUMO from tomato, homolog to Arabidopsis SUMO1/2 and human SUMO2/3) (24). Previous experiments showed that transgenic plants harboring the sense construct expressed a higher level of SUMO RNA than wild-type plants, while transgenic plants harboring the antisense construct expressed a lower level of SUMO RNA than did wild-type plants (Adi Avni, personal communication). We agroinfected six to eight T2 kanamycin-resistant plants of the under- and overexpressing SUMO lines and a similar number of wild-type plants in two independent experiments. Leaves were collected 16, 24, and 45 dpi, and accumulation of viral DNA was quantified on DNA gel blots (Fig. 7A). Viral DNA was detected at 16 dpi in plants from all lines, and the percentages of infected plants were similar for all lines. However, the amount of viral DNA in transformed plants was significantly smaller than those in wild-type plants. These differences occurred at all infection times, peaking at 24 dpi, when the amount of viral DNA in transformed plants was <10% of that of the wild type. No significant differences were detected between the sense and antisense lines.
FIG. 7.
Altered SUMO levels affect TGMV replication. (A) Transgenic tobacco plants expressing sense (S) or antisense (AS) RNA of LeSUMO and untransformed (U) plants were agroinfected with TGMV A and B. Six to eight plants per transformant were infected in two independent assays. Total DNA was isolated from young leaves 16, 24, and 45 dpi and analyzed by DNA gel blot hybridization with a radiolabeled double-stranded TGMV A probe. The amount of viral DNA accumulation was measured in FUJIFILM Bas1500 and normalized to total genomic DNA loaded on the gel. (A) The percentage of viral DNA accumulation (single and double strands) in infected plants 16, 24, and 45 dpi was normalized to the maximum value of untransformed plants collected the same day. (B) Percentage of viral DNA accumulation in leaf disks from tobacco transgenic (sense and antisense LeSUMO RNA) or unstransformed plants agroinfected with TGMV A. Total DNA was extracted from leaf disk at 8 dpi and analyzed by DNA gel blot hybridization. The DNA probe and viral DNA quantification were done as described above. DNA accumulation of all samples was normalized to levels in leaf disks from untransformed plants. The error bars correspond to 2 standard errors.
This reduction in viral DNA accumulation could be due to impairment of viral replication or movement. Because all TGMV proteins required for replication are encoded by the A component, we asked whether the same reduction also occurs in the absence of viral movement in leaf disks inoculated with TGMV A alone. Leaf disks from tobacco plants (six plants per group of sense, antisense, and wild-type genotypes) were agroinfected with TGMV A. Accumulation of viral DNA was quantified from DNA gel blots at 8 dpi (Fig. 7B). The accumulation of viral DNA was severely impaired in leaf disks from sense and antisense LeSUMO plants compared to the wild type. These data suggested that the reduction in viral accumulation in plants expressing sense and antisense LeSUMO RNA interferes with viral replication and not with movement.
DISCUSSION
Using the yeast two-hybrid system, we identified an N. benthamiana protein (NbSCE1) that is a homolog of the E2-SUMO-conjugating enzyme UBC9 and interacts with Rep from the geminivirus TYLCSV. NbSCE1 has a high degree of amino acid identity with AtSCE1a from A. thaliana and other eukaryotic UBC9 proteins and contains all of the residues known to be involved in the human UBC9-substrate interaction (6). We confirmed the identity of NbSCE1 by genetic complementation of an S. cerevisiae ubc9 temperature-sensitive mutant. Like Arabidopsis, NbSCE1 is transcribed in all plant tissues assayed and may occur as multiple copies in the N. benthamiana genome. Together these results strongly suggested that the protein identified by interaction with Rep is the E2-conjugating enzyme of the sumoylation system of N. benthamiana. Like UBC9 from S. cerevisiae, Arabidopsis, and humans (22, 30, 33), NbSCE1 is predicted to conjugate SUMO, not ubiquitin, to target proteins. The NbSCE1 protein also interacts with Rep proteins from the bipartite begomoviruses TGMV and ACMV-KE (RepAC1), both of which infect N. benthamiana, suggesting that binding to SCE1/UBC9 is a conserved function of begomovirus replication proteins.
The consensus sequence for UBC9/SCE1 binding is not yet defined. As previously described by Rangasamy and Wilson (49), analysis of the interaction domain of several mammalian UBC9 binding proteins (c-Jun, E2A, and BPV-E1) suggested that UBC9 frequently binds to hydrophobic regions that contain LK and/or KL dipeptides. There are two conserved LK pairs in the TGMV RepAC1 protein at positions 270/271 and 327/328, but they are at the C terminus of the protein, which does not bind to UBC9. Our truncation mapping studies indicated that the interaction between Rep and NbSCE1 occurs at the N-terminal end of TGMV RepAC1, and this was confirmed by an in vitro assay. Analysis of the amino acid sequence of the RepAC1 N-terminal half identified a potential NbSCE1 interaction domain between positions 30 and 47. Although this region does not have a LK motif, as occurs in other UBC9/SCE1-interacting proteins (49), it is hydrophobic and highly conserved among geminiviruses, suggesting that it could be responsible for the interaction with NbSCE1 (Fig. 8A). The truncated form RepAC1N1-130 contains this putative UBC9 binding domain but does not interact with NbSCE1. Immunoblotting experiments verified that this lack of interaction is not due to reduced levels of the truncated protein in yeast. One possibility is that sequences outside the putative binding domain contribute stabilizing contacts needed for the interaction with NbSCE1. Alternatively, the truncated RepAC1N1-130 protein may fold incorrectly in yeast.
FIG. 8.
Potential NbSCE1-interacting domain and putative sumoylation sites of Rep/RepAC1. (A) Sequence alignment of the UBC9/SCE1-interacting region of the mammalian proteins c-Jun, E2A, and E1 from the bovine papillomavirus and Rep/RepAC1 from the geminiviruses BCTV (Beet curly top virus), TYLCSV, TGMV, and ACMV-KE. (B) Potential sumoylation sites of Rep/RepAC1 deduced by sequence comparison with the consensus sequence of sumoylation (ΨKXE). The N-terminal half of TGMV RepAC1 is underlined, the four helices are indicated (H1 to -4), and the putative NbSCE1-interacting domain is emphasized. Potential sumoylation sites with a polar, uncharged amino acid instead of a hydrophobic one before the lysine are marked with open rectangles The two putative sites in the N terminus are conserved among geminivirus Rep proteins.
Many proteins, including some from mammalian viruses, interact with UBC9/SCE1 in yeast two-hybrid assays (60, 62). Although most of these UBC9/SCE1-interacting proteins are also sumoylated, some are not sumoylation substrates. We do not know yet if Rep/RepAC1 is a substrate for sumoylation. In plants, it is not easy to prove Rep sumoylation because of the difficulty detecting Rep/RepAC1 in infected tissues on immunoblots and problems associated with its overexpression (unpublished results). In mammalian cells and yeast, SUMO is covalently attached to specific lysines in the target protein. The precise lysine residues modified by SUMO have been identified in more than a dozen known substrates (41). The majority of these modification sites conform to a consensus sequence defined by four amino acids, ΨKXE, where Ψ is a large hydrophobic residue and K serves as the acceptor for SUMO. However, there are sumoylated proteins that do not share this exact sequence, such as HIPK2, yeast Cdc3, and one of the PML sites (62). Rep/RepAC1 does not contain any protein motif that exactly matches the consensus, but it does have lysine residues surrounded by similar amino acids that are potential sumoylation sites (Fig. 8B). We identified three sites in TGMV RepAC1—two in the N terminus and one in the C terminus where the residue before lysine is a polar, uncharged amino acid instead of a hydrophobic one (Fig. 8B, open rectangles). The two N-terminal sites are conserved among geminiviruses that infect dicotyledonous plants, while the C-terminal one is not. The results obtained by Sampson et al. (52) indicated that there is a correlation between the ability of proteins to interact with UBC9 and their ability to be modified by SUMO. They demonstrated that conserved residues surrounding the SUMO modification site are critical for efficient SUMO modification and that this consensus sequence mediates UBC9 binding. There is one potential sumoylation site at the beginning of the putative binding domain for NbSCE1 (lysine 26).
To determine if sumoylation plays a role in TGMV infection, we used transgenic tobacco plants expressing sense or antisense RNA of LeSUMO (the tomato ortholog of AtSUMO1/2) (24). Although none of the tobacco SUMO genes (NtSUMO) has been isolated, sequence comparison between LeSUMO and an N. benthamiana ortholog of AtSUMO1/2 (M. Sánchez-Durán et al., unpublished observations) showed very high identity. There is central region of 267 nucleotides with 93% identity that contains 74 consecutive identical nucleotides. Moreover, RNA blot analysis of untransformed tobacco plants by using a LeSUMO probe hybridized to a band of the expected size for the SUMO transcript (24), indicating that sequence identity between NtSUMO and LeSUMO RNA is sufficient to hybridize under high-stringency conditions. RNA blots of transgenic plants expressing LeSUMO sense RNA showed a stronger hybridization signal than untransformed plants, while the signal was clearly reduced in tobacco plants expressing LeSUMO antisense RNA compared to nontransgenic plants, suggesting that the endogenous expression levels of NtSUMO orthologs are reduced. After TGMV infection, we detected a reduction in viral DNA accumulation in transgenic plants expressing either sense or antisense LeSUMO compared to wild-type plants. Leaf disk assays established that this decrease reflected a reduction in viral DNA replication and not viral movement.
These results indicated that positive as well as negative changes in SUMO levels disturb viral replication. It is difficult to provide a simple hypothesis that explains these results, because the biological effects of sumoylation are quite diverse, and the mechanisms and signal pathways involved in most of them remain unclear. Recent results show that the accumulation profiles of SUMO1/2 and SUMO3 differ in Arabidopsis. SUMO1/2 conjugates increase rapidly during stress, while SUMO3 conjugates were unaffected (33). It has been proposed that SUMO1/2 conjugation plays a regulatory role in the stress response, modifying the activity or localization of critical effectors. SUMO1/2 conjugation could impact a battery of nuclear regulatory proteins when plants are exposed to stress signals. Potential targets under negative regulation could include factors that promote cell division and other general physiological processes that are repressed while plants cope with adverse environments. Proteins under positive regulation are likely to include those whose activation promotes stress control (33). Two previous studies suggested that SUMO plays an important role in plant pathogen defense response (24, 47). In one study, transgenic tobacco plants expressing sense LeSUMO showed an altered response to EIX from the fungus T. viridiae, a strong elicitor of the rapid defense response in tomato (24). Thus, interference with viral DNA replication observed in LeSUMO-overexpressing transgenic plants could be related to changes in the levels and/or the profiles of SUMO conjugates. These changes may repress proteins required for viral replication or induce a stronger defense response.
However, we cannot rule out a direct effect on Rep as a consequence of altered SUMO levels in the plants. Viruses have evolved numerous mechanisms to overcome host defenses and to utilize host biochemical pathways to their advantage. One type of virus-host interaction that is well established and widespread is the modulation of viral protein function by posttranslational modification systems such as phosphorylation, glycosylation, ubiquitination, and sumoylation. The interaction between viral proteins and the cellular sumoylation system has been described previously for mammalian viruses, and the effects are target specific and very diverse. Interaction with UBC9 and/or sumoylation of these mammalian viral proteins determines their nuclear uptake and/or retention, the complete transactivation of their activity, or changes in the sumoylation state of specific cellular proteins (60). It is difficult to suggest a role for the interaction between Rep and NbSCE1, but interestingly, some of the putative sumoylation sites described in Fig. 8B are located in known functional domains of TGMV RepAC1. The two sites in the N terminus of RepAC1 are located in motifs that are conserved in all dicot-infecting geminiviruses (44). The first one (amino acids 25 to 28) overlaps the DNA binding and cleavage domains of RepAC1 and is required for infectivity (mutations in it inhibit DNA binding and cleavage in vitro and replication in vivo) (45). The second sumoylation site (amino acids 143 to 146) is in the helix 4 motif, which plays a role in TGMV RepAC1 oligomerization and binding to RBR, the plant homolog of the retinoblastoma protein (46). Mutation of the second putative sumoylation site severely impairs binding to RBR and alters symptomology (31). The third sequence, located in the C terminus of RepAC1 (amino acids 255 to 258), is in the helicase domain very close to two conserved aspartic residues implicated in this function. Sumoylation of a lysine residue in any of these sites could promote a particular subset of RepAC1 activities.
Acknowledgments
We thank GENTECH Co. for providing some plasmids and yeast strains. We thank L. Jongejan for providing the cDNA library from N. benthamiana, C. Mann for giving the ubc9 thermosensitive mutant, A. Avni for providing the transgenic tobacco lines, and L. Cruzado for excellent technical assistance. We thank M. A. Botella, C. Gutiérrez, R. Flores, and J. Jiménez for helpful suggestions and discussions.
This research was supported by a grant from the Spanish Ministerio de Ciencia y Tecnología (AGF98-0439-C05-05). A.G.C. was awarded a Predoctoral Fellowship from the Spanish Ministerio de Educación y Cultura and an EMBO Short Term Fellowship (ASTF no. 9886).
REFERENCES
- 1.Ach, R. A., T. Durfee, A. B. Miller, P. Taranto, L. Hanley-Bowdoin, P. C. Zambryski, and W. Gruissem. 1997. RRB1 and RRb2 encode maize retinoblastoma-related proteins that interact with a plant D-type cyclin and geminivirus replication protein. Mol. Cell. Biol. 17:5077-5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akbar Behjatnia, S. A., I. B. Dry, and M. Ali Rezaian. 1998. Identification of the replication-associated protein binding domain within the intergenic region of tomato leaf curl geminivirus. Nucleic Acids Res. 26:925-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F., R. Brent, R. Kingston, D. Moore, J. Seidman, J. Smith, and K. Struhl. 1998. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 4.Bartel, P., C. T. Chien, R. Sternglanz, and S. Fields. 1993. Elimination of false positives that arise in using the two-hybrid system. BioTechniques 14:920-924. [PubMed] [Google Scholar]
- 5.Bejarano, E. R., and C. P. Lichtenstein. 1994. Expression of TGMV antisense RNA in transgenic tobacco inhibits replication of BCTV but not ACMV geminiviruses. Plant Mol. Biol. 24:241-248. [DOI] [PubMed] [Google Scholar]
- 6.Bernier-Villamor, V., D. A. Sampson, M. J. Matunis, and C. D. Lima. 2002. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108:345-356. [DOI] [PubMed] [Google Scholar]
- 7.Betting, J., and W. Seufert. 1996. A yeast Ubc9 mutant protein with temperature-sensitive in vivo function is subject to conditional proteolysis by a ubiquitin- and proteasome-dependent pathway. J. Biol. Chem. 271:25790-25796. [DOI] [PubMed] [Google Scholar]
- 8.Castellano, M. M., A. P. Sanz-Burgos, and C. Gutierrez. 1999. Initiation of DNA replication in a eukaryotic rolling-circle replicon: identification of multiple DNA-protein complexes at the geminivirus origin. J. Mol. Biol. 290:639-652. [DOI] [PubMed] [Google Scholar]
- 9.Castillo, A., D. Collinet, S. Deret, A. Kashoggi, and E. Bejarano. 2003. Dual interaction of plant PCNA with geminivirus replication accessory protein (REn) and viral replication protein (Rep). Virology 312:381-394. [DOI] [PubMed] [Google Scholar]
- 10.Church, G. M., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81:1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dellaporta, S. L., J. Wood, and J. B. Hicks. 1983. A plant DNA minipreparation: version II. Plant Mol. Biol. Rep. 1:19-21. [Google Scholar]
- 12.Duttweiler, H. M. 1996. A highly sensitive and non-lethal beta-galactosidase plate assay for yeast. Trends Genet. 12:340-341. [DOI] [PubMed] [Google Scholar]
- 13.Elledge, S. J., J. T. Mulligan, S. W. Ramer, M. Spottswood, and R. W. Davis. 1991. Lambda YES: a multifunctional cDNA expression vector for the isolation of genes by complementation of yeast and Escherichia coli mutations. Proc. Natl. Acad. Sci. USA 88:1731-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elmer, J. S., L. Brand, G. Sunter, W. E. Gardiner, D. M. Bisaro, and S. G. Rogers. 1988. Genetic analysis of the tomato golden mosaic virus. II. The product of the AL1 coding sequence is required for replication. Nucleic Acids Res. 16:7043-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fauquet, C. M., D. P. Maxwell, B. Gronenborn, and J. Stanley. 2000. Revised proposal for naming geminiviruses. Arch. Virol. 145:1743-1761. [DOI] [PubMed] [Google Scholar]
- 16.Fauquet, C. M., D. M. Bisaro, R. W. Briddon, J. K. Brown, B. D. Harrison, E. P. Rybicki, D. C. Stenger, and J. Stanley. 2003. Revision of taxonomic criteria for species demarcation in the family Geminiviridae, and an updated list of begomovirus species. Arch. Virol. 148:405-421. [DOI] [PubMed] [Google Scholar]
- 17.Fields, S., and O. Song. 1989. A novel genetic system to detect protein-protein interactions. Nature 340:245-246. [DOI] [PubMed] [Google Scholar]
- 18.Fontes, E. P. B., V. A. Luckow, and L. Hanley-Bowdoin. 1992. A geminivirus replication protein is a sequence specific DNA binding protein. Plant Cell 4:597-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fontes, E. P. B., P. A. Eagle, P. S. Sipe, V. A. Luckow, and L. Hanley-Bowdoin. 1994. Interaction between a geminivirus replication protein and origin DNA is essential for viral replication. J. Biol. Chem. 269:8459-8465. [PubMed] [Google Scholar]
- 20.Gari, E., L. Piedrafita, M. Aldea, and E. Herrero. 1997. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast 13:837-848. [DOI] [PubMed] [Google Scholar]
- 21.Gietz, R. D., and R. H. Schiestl. 1995. Transforming yeast with DNA. Methods Mol. Cell Biol. 5:255-269. [Google Scholar]
- 22.Gong, L., T. Kamitani, K. Fujise, L. S. Caskey, and E. T. Yeh. 1997. Preferential interaction of sentrin with a ubiquitin-conjugating enzyme, Ubc9. J. Biol. Chem. 272:28198-28201. [DOI] [PubMed] [Google Scholar]
- 23.Gutierrez, C. 2000. DNA replication and cell cycle in plants: learning from geminiviruses. EMBO J. 19:792-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanania, U., N. Furman-Matarasso, M. Ron, and A. Avni. 1999. Isolation of a novel SUMO protein from tomato that suppresses EIX-induced cell death. Plant J. 19:533-541. [DOI] [PubMed] [Google Scholar]
- 25.Hanley-Bowdoin, L., S. B. Settlage, B. M. Orozco, S. Nagar, and D. Robertson. 1999. Geminiviruses: models for plant DNA replication, transcription, and cell cycle regulation. Crit. Rev. Biochem. Mol. Biol. 35:105-140. [PubMed] [Google Scholar]
- 26.Hayashi, T., M. Seki, D. Maeda, W. Wang, Y. Kawabe, T. Seki, H. Saitoh, T. Fukagawa, H. Yagi, and T. Enomoto. 2002. Ubc9 is essential for viability of higher eukaryotic cells. Exp. Cell Res. 280:212-221. [DOI] [PubMed] [Google Scholar]
- 27.Heyraud-Nitschke, F., S. Schumacher, J. Laufs, S. Schaefer, J. Schell, and B. Gronenborn. 1995. Determination of the origin cleavage and joining domain of geminivirus Rep proteins. Nucleic Acids Res. 23:910-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iraki, N., R. Bressan, P. Hasegawa, and N. Carpita. 1989. Alteration of the physical and chemical structure of the primary cell wall of growth-limited plant cells adapted to osmotic stress. Plant Physiol. 91:39-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.James, P., J. Halladay, and E. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson, E. S., and G. Blobel. 1997. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J. Biol. Chem. 272:26799-26802. [DOI] [PubMed] [Google Scholar]
- 31.Kong, L. J., B. M. Orozco, J. L. Roe, S. Nagar, S. Ou, H. S. Feiler, T. Durfee, A. B. Miller, W. Gruissem, D. Robertson, and L. Hanley-Bowdoin. 2000. A geminivirus replication protein interacts with the retinoblastoma protein through a novel domain to determine symptoms and tissue specificity of infection in plants. EMBO J. 19:3485-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kong, L. J., and L. Hanley-Bowdoin. 2002. A geminivirus replication protein interacts with a protein kinase and a motor protein that display different expression patterns during plant development and infection. Plant Cell 14:1817-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurepa, J., J. M. Walker, J. Smalle, M. M. Gosink, S. J. Davis, T. L. Durham, D. Y. Sung, and R. D. Vierstra. 2003. The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. Accumulation of SUMO1 and -2 conjugates is increased by stress. J. Biol. Chem. 278:6862-6872. [DOI] [PubMed] [Google Scholar]
- 34.Laufs, J., I. Jupin, C. David, S. Schumacher, F. Heyraud-Nitschke, and B. Gronenborn. 1995. Geminivirus replication—genetic and biochemical characterization of Rep protein function, a review. Biochimie 77:765-773. [DOI] [PubMed] [Google Scholar]
- 35.Lois, L. M., C. D. Lima, and N. H. Chua. 2003. Small ubiquitin-like modifier modulates abscisic acid signaling in Arabidopsis. Plant Cell 15:1347-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luckow, V. A., S. C. Lee, G. F. Barry, and P. O. Olins. 1993. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J. Virol. 67:4566-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luque, A., A. P. Sanz-Burgos, E. Ramirez-Parra, M. M. Castellano, and C. Gutierrez. 2002. Interaction of geminivirus Rep protein with replication factor C and its potential role during geminivirus DNA replication. Virology 302:83-94. [DOI] [PubMed] [Google Scholar]
- 38.Melchior, F. 2000. SUMO—nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. 16:591-626. [DOI] [PubMed] [Google Scholar]
- 39.Mittnacht, S. 1998. Control of pRB phosphorylation. Curr. Opin. Genet. Dev. 8:21-27. [DOI] [PubMed] [Google Scholar]
- 40.Moriones, E., and J. Navas-Castillo. 2000. Tomato yellow leaf curl virus, an emerging virus complex causing epidemics worldwide. Virus Res. 71:123-134. [DOI] [PubMed] [Google Scholar]
- 41.Müller, S., C. Hoege, G. Pyrowolakis, and S. Jentsch. 2001. SUMO, ubiquitin's mysterious cousin. Nat. Rev. Mol. Cell Biol. 2:202-210. [DOI] [PubMed] [Google Scholar]
- 42.Nagar, S., T. Pedersen, K. Carrick, L. Hanley-Bowdoin, and D. Robertson. 1995. A geminivirus induces expression of a host DNA synthesis protein in terminally differentiated plant cells. Plant Cell 7:705-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Orozco, B. M., and L. Hanley-Bowdoin. 1996. A DNA structure is required for geminivirus replication origin function. J. Virol. 70:148-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orozco, B. M., A. B. Miller, S. B. Settlage, and L. Hanley-Bowdoin. 1997. Functional domains of a geminivirus replication protein. J. Biol. Chem. 272:9840-9846. [DOI] [PubMed] [Google Scholar]
- 45.Orozco, B. M., and L. Hanley-Bowdoin. 1998. Conserved sequence and structural motifs contribute to the DNA binding and cleavage activities of a geminivirus replication protein. J. Biol. Chem. 273:24448-24456. [DOI] [PubMed] [Google Scholar]
- 46.Orozco, B. M., L. J. Kong, L. A. Batts, S. Elledge, and L. Hanley-Bowdoin. 2000. The multifunctional character of a geminivirus replication protein is reflected by its complex oligomerization properties. J. Biol. Chem. 275:6114-6122. [DOI] [PubMed] [Google Scholar]
- 47.Orth, K., Z. Xu, M. B. Mudgett, Z. Q. Bao, L. E. Palmer, J. B. Bliska, W. F. Mangel, B. Staskawicz, and J. E. Dixon. 2000. Disruption of signaling by Yersinia effector YopJ, a ubiquitin-like protein protease. Science 290:1594-1597. [DOI] [PubMed] [Google Scholar]
- 48.Pichler, A., and F. Melchior. 2002. Ubiquitin-related modifier SUMO1 and nucleocytoplasmic transport. Traffic 3:381-387. [DOI] [PubMed] [Google Scholar]
- 49.Rangasamy, D., and V. G. Wilson. 2000. Bovine papillomavirus E1 protein is sumoylated by the host cell Ubc9 protein. J. Biol. Chem. 275:30487-30495. [DOI] [PubMed] [Google Scholar]
- 50.Saitoh, H., and J. Hinchey. 2000. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 275:6252-6258. [DOI] [PubMed] [Google Scholar]
- 51.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 52.Sampson, D. A., M. Wang, and M. J. Matunis. 2001. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J. Biol. Chem. 276:21664-21669. [DOI] [PubMed] [Google Scholar]
- 53.Schwartz, D. C., and M. Hochstrasser. 2003. A superfamily of protein tags: ubiquitin, SUMO and related modifiers. Trends Biochem. Sci. 28:321-328. [DOI] [PubMed] [Google Scholar]
- 54.Settlage, S. B., A. B. Miller, and L. Hanley-Bowdoin. 1996. Interactions between geminivirus replication proteins. J. Virol. 70:6790-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seufert, W., J. P. McGrath, and S. Jentsch. 1990. UBC1 encodes a novel member of an essential subfamily of yeast ubiquitin-conjugating enzymes involved in protein degradation. EMBO J. 9:4535-4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seufert, W., B. Futcher, and S. Jentsch. 1995. Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature 373:78-81. [DOI] [PubMed] [Google Scholar]
- 57.Sherr, C. J., and J. M. Roberts. 1999. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13:1501-1512. [DOI] [PubMed] [Google Scholar]
- 58.Tatham, M. H., E. Jaffray, O. A. Vaughan, J. M. Desterro, C. H. Botting, J. H. Naismith, and R. T. Hay. 2001. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276:35368-35374. [DOI] [PubMed] [Google Scholar]
- 59.Verger, A., J. Perdomo, and M. Crossley. 2003. Modification with SUMO. EMBO Rep. 4:137-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilson, V. G., and D. Rangasamy. 2001. Viral interaction with the host cell sumoylation system. Virus Res. 81:17-27. [DOI] [PubMed] [Google Scholar]
- 61.Yasugi, T., and P. M. Howley. 1996. Identification of the structural and functional human homolog of the yeast ubiquitin conjugating enzyme UBC9. Nucleic Acids Res. 24:2005-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yeh, E. T., L. Gong, and T. Kamitani. 2000. Ubiquitin-like proteins: new wines in new bottles. Gene 248:1-14. [DOI] [PubMed] [Google Scholar]