Abstract
The interaction of a dimeric membrane anchor-free form of the envelope protein E (sE dimer) from tick-borne encephalitis virus with liposomes at acidic pH levels leads to its conversion into membrane-inserted sE trimers. Electron microscopy shows that these trimers have their long dimensions along the threefold molecular axis, which is oriented perpendicularly to the plane of the membrane, where the protein inserts via the internal fusion peptide. Liposomes containing sE at their surface display paracrystalline arrays of protein in a closely packing arrangement in which each trimer is surrounded by six others, suggesting cooperativity in the insertion process. sE trimers, solubilized with nonionic detergents, yielded three-dimensional crystals suitable for X-ray diffraction analysis.
Membrane fusion is a key step during entry of enveloped viruses into cells. This function is mediated by viral surface glycoproteins (fusion proteins) that undergo extensive conformational changes upon receptor binding (fusion at the plasma membrane) or exposure to low pH (fusion at the endosomal membrane) (11, 23). In many cases, these triggers have been shown to release the fusion proteins from a metastable state and allow their conversion to a lower energy state. This structural change leads to the exposure of a previously buried functional element (fusion peptide) and is believed to provide the energy required for the merger of the lipid bilayers (4, 5). So far, viral fusion proteins have been shown to fall into two different structural classes, designated class I and class II (15). Class I fusion proteins of orthomyxo-, paraymxo-, retro-, and filoviruses possess amino-terminal or amino-proximal fusion peptides and have a postfusion structure in which a triple-stranded alpha-helical coiled coil is a central feature (5, 19, 23). The atomic structures of protein fragments containing the trimeric six-helix bundle have been determined for a number of class I fusion proteins (reviewed in reference 5).
Class II viral fusion proteins, which have so far been found in flavi- and alphaviruses, have a completely different structure. They are oriented parallel to the membrane, possess an internal fusion peptide, and form part of an icosahedral network in the virion envelope. Fusion is triggered by acidic pH, which leads to structural changes that convert the metastable fusion proteins present in mature virions into stable homotrimers (reviewed in references 8 and 13).
We are studying the structural basis of class II-mediated viral membrane fusion using the flavivirus tick-borne encephalitis virus (TBEV). As shown previously, exposure of the native, homodimeric form of the fusion protein E to low pH leads to its irreversible conversion into a homotrimer. In contrast, a truncated form of the E dimer (sE dimer) lacking the carboxy-terminal membrane anchor and the consecutive 40 amino acids (referred to as “stem”) (3, 20) was shown to dissociate at low pH but not to form trimers in the absence of membranes (20). Trimerization did occur, however, when sE was acidified in the presence of liposomes, and these trimers remained bound to the target membrane (21). Similar to the lipid-induced trimerization of sE dimers of TBEV, it has been shown that a C-terminally truncated form of the alphavirus class II fusion protein is able to trimerize at low pH in the presence of liposomes, but not when membranes are absent (14).
Here we show that sE trimers from TBEV form paracrystalline arrays on the surface of liposomes. Upon solubilization with nonionic detergents, these trimers yielded crystals suitable for structure determination by X-ray diffraction analysis.
Liposome-associated sE trimers.
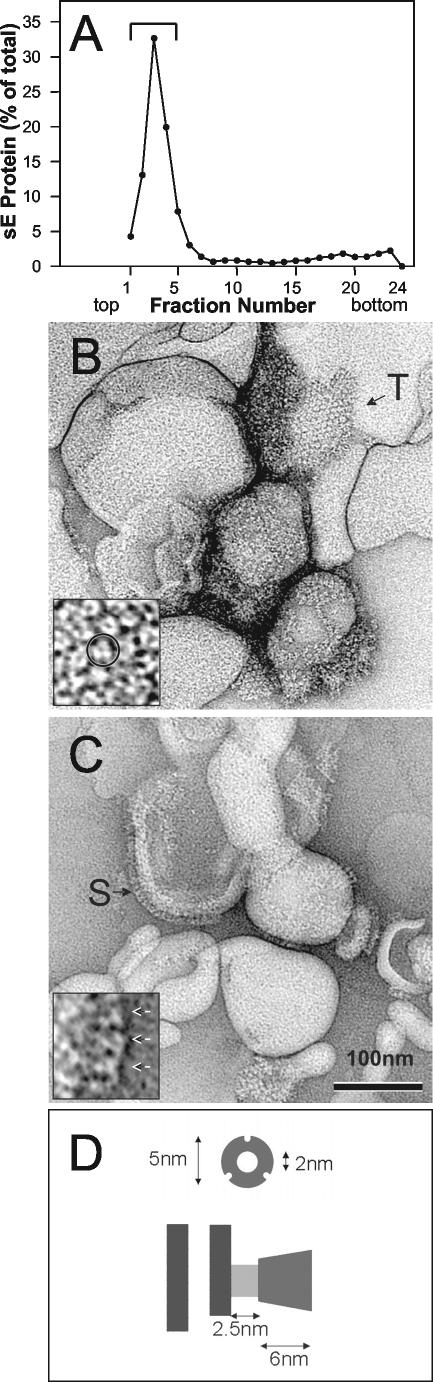
As starting material, native sE dimers were first prepared by limited trypsin digestion of purified TBEV and purification via anion-exchange chromatography (9). To convert the dimers to trimers, we then exposed them to acidic pH in the presence of large unilamellar liposomes, as described previously (21). Briefly, sE dimers were mixed with liposomes consisting of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (PC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (PE), and 1-cholesterol (CH) (molar ratio, 1:1:2) at a ratio of 1 μg of sE to 15 nmol of lipid (22). This mixture was acidified with 300 mM morpholineethanesulfonic acid (MES), incubated for 30 min at 37°C at pH 5.4, back neutralized, adjusted to 20% (wt/wt) sucrose in 20 mM triethanolamine (TEA) and 130 mM NaCl (pH 8.0) (TAN buffer), and was then applied to a 50% cushion, overlaid with 15% (wt/wt) sucrose and 5% (wt/wt) sucrose. Centrifugation was carried out for 1.5 h at 50,000 rpm at 4°C in a Beckman SW 55 rotor, and the top fractions were collected by upward displacement as described in reference 21. Under these conditions, about 60 to 80% of the sE proteins were found to bind and float with the liposomes to the top of the gradient (Fig. 1A). Electron microscopy of these fractions stained with uranyl acetate revealed that the resulting sE trimers were distributed in patches of regular lattices on the surface of the liposomes, and structures with threefold rotational symmetry could be observed in the photomicrographs (Fig. 1B and C). Top views of the E trimer display a donut appearance with inner and outer diameters of about 2 and 5 nm, respectively (Fig. 1B and D). The side views show conical shapes with mean width and height of about 5 and 9 nm, respectively (Fig. 1C and D). The donut-shaped structure displaying threefold rotational symmetry is in agreement with the biochemical data demonstrating that sE forms trimers during its insertion into target membranes (21). All recognizable trimers inserted in the liposomes are closely packed, forming a lattice in which each trimer is surrounded by six others. The arrangement of sE trimers at the liposome surface is different from that observed in the case of the Semliki Forest virus (SFV) E1* fragment, which displays a hexagonal lattice with a central hole (7) instead of a central trimer, as observed here. The neutral pH form of E is an elongated, brick-shaped dimeric molecule in which each subunit is contained within a cylinder having a diameter and height of about 3 and 11 nm, respectively (17). In the dimer, the long dimension of the molecule is parallel to the viral membrane. The fact that the long dimension of the trimer is now along the threefold molecular axis, perpendicular to the liposome membrane, indicates that the E subunit has reoriented so that it lies perpendicular to its original orientation on the virus. All these results are similar to those obtained with the class II fusion protein E1 of the alphavirus Semliki Forest virus, which also clusters in the plane of the membrane to form hexagonal lattices of E1 trimers (7). It can therefore be proposed that the strong protein interactions leading to these arrays are an important parameter for the membrane fusion event in both virus systems. Differences between flavi- and alphaviruses, however, exist in terms of the lipid dependence of membrane insertion, trimerization, and fusion (6, 14, 22, 24). In contrast to alphaviruses, these processes in flaviviruses are not absolutely dependent on cholesterol and do not require sphingomyelin (6, 22).
FIG.1.
(A) Coflotation of TBEV sE protein with liposomes at low pH. sE dimers were mixed with liposomes and acidified (pH 5.4), and sucrose was added to a final concentration of 20% (wt/wt). The mixture was applied to a 50% sucrose cushion (wt/wt), overlaid with 15% sucrose (wt/wt) and 5% sucrose (wt/wt), and subjected to ultracentrifugation as described previously (21). The amount of E protein in each fraction was determined by a quantitative four-layer ELISA after denaturation of the samples with 0.4% sodium dodecyl sulfate (10). The bracket indicates the top fractions, containing sE bound to liposomes. (B and C) Electron photomicrographs of liposome-bound sE trimers. The fraction containing sE inserted in liposome was negatively stained with a 2% uranyl acetate solution. sE forms clusters at the surface of the liposomes, displaying closely packed arrays. Two characteristic views, top and side, are shown. Panel B shows top views. The marked area (T) is enlarged in the inset. Note that the protein packing is much denser than in the case of E1 of SFV (7). Panel C shows mainly side views of the protein. The marked area (S) is enlarged in the inset. Top and side views have a donut and conical appearance, respectively. (D) Schematic of top (donut) and side (conical) views of sE trimers.
Isolated sE trimers.
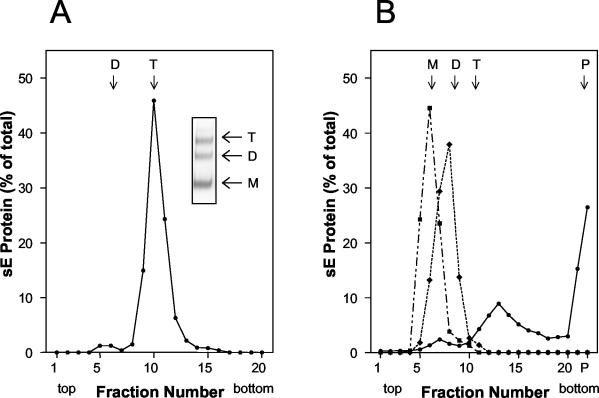
To isolate the sE trimers from liposomes and to remove the lipids, the membranes were solubilized with 1.5% n-octylglucoside (n-OG), followed by ultrafiltration using Vivaspin20 concentrators with a cutoff of 100 kDa (Vivascience AG, Hannover, Germany) and TAN buffer (pH 8.0) containing 0.8% n-OG. As revealed by rate-zonal centrifugation in sucrose density gradients containing 0.8% n-OG (22), more than 90% of the material formed a defined peak at the position corresponding to an E trimer, and only a small amount (5 to 10%) sedimented as a dimer (Fig. 2A). The trimeric state of the protein in the major peak was further confirmed by cross-linking with dimethylsuberimidate (DMS) (Fig. 2A, inset).
FIG. 2.
Sedimentation analysis of sE trimers in the presence (A) and absence (B) of detergent. The gradients were fractionated, and the amount of E protein in each fraction was determined by a quantitative four-layer ELISA after denaturation of the samples with 0.4% sodium dodecyl sulfate (10). The sedimentation direction is from left to right, and the arrows indicate the sedimentation positions of sE monomer (M), dimer (D) , and trimer (T) analyzed in the presence of detergent. P indicates sE recovered from the pellet. (A) sE trimers were subjected to sedimentation in 7 to 20% sucrose gradients in TAN buffer (pH 8.0) containing 0.8% n-OG as described in reference 22. The inset shows the Western blot analysis after chemical cross-linking of the trimer peak fraction with dimethylsuberimidate as described in reference 18. Panel B shows data for sE trimers (•) that were subjected to sedimentation in detergent-free 7 to 20% (wt/wt) sucrose gradients in TAN buffer (pH 8.0). As controls, sE dimers (▴) and sE monomers (▪) were used. The monomers were generated by incubation of sE dimers at pH 5.4, and sedimentation was carried out at pH 5.4 in MES buffer (50 mM MES, 100 mM NaCl) instead of pH 8.0.
To investigate whether the sE trimer remains soluble after removal of the detergent, we also analyzed its sedimentation behavior in detergent-free sucrose gradients. sE monomers produced by dissociation of sE dimers at low pH (20) and sE dimers were used as sedimentation standards; these behaved as expected for soluble proteins in the absence of detergent (Fig. 2B). In the case of sE trimers, however, most of the material was found as a pellet at the bottom of the gradient, indicating that the removal of the detergent had caused aggregation due to the exposure of hydrophobic sequence elements (Fig. 2B). The sE trimer thus appears to be more hydrophobic than either the sE monomer or sE dimer. Because the fusion peptide is presumably exposed in both the monomer and the trimer (21), it is probable that other still-unidentified regions of the trimer contribute to this increased hydrophobicity. All further experiments with isolated sE trimers were therefore carried out in the presence of detergents.
MAb reactivity of liposome-associated and isolated sE trimers.
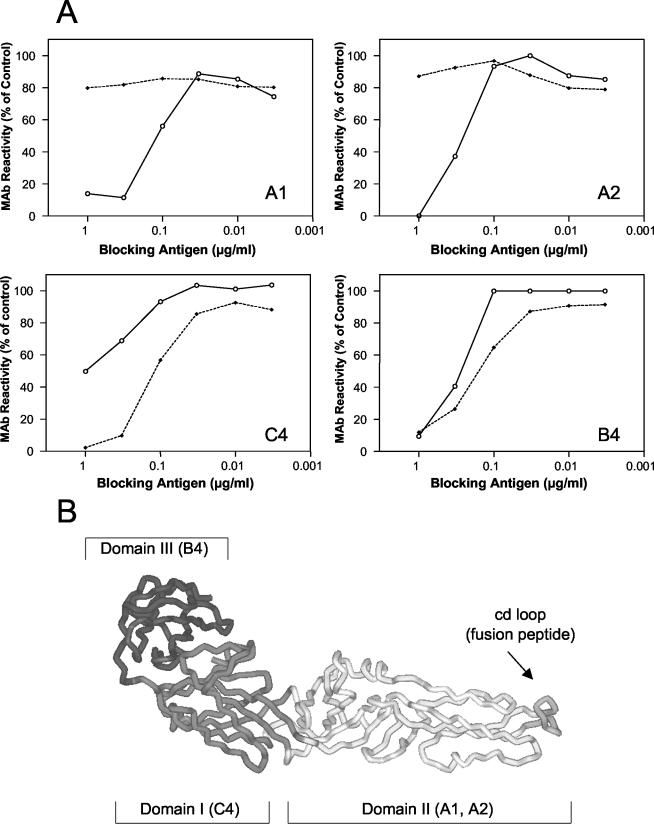
Site-directed mutagenesis experiments have provided evidence that the sequence element (designated the cd loop) at the tip of domain II of the E protein functions as an internal fusion peptide and that it forms part of the epitopes recognized by the monoclonal antibodies (MAbs) A1 and A2 (2) (Fig. 3B). To obtain information about interactions of this sequence element with the liposomal membrane, we analyzed the reactivity of these two MAbs with the membrane-bound and detergent-solubilized form of the sE trimer in a blocking enzyme-linked immunosorbent assay (ELISA) (12). In the course of this assay we investigated whether the binding of the MAbs to virions coated to the solid phase could be inhibited by the two different trimer preparations in solution. Although the fusion peptide loops and thus the epitopes recognized by A1 and A2 are normally buried in the native state of E, they become accessible due to partial denaturation of the virions adsorbed to the plastic surface (12), allowing MAbs that are not bound to trimer preparations to be detected by ELISA.
FIG. 3.
(A) Blocking ELISA with liposome-associated and solubilized sE trimers. A predetermined fixed dilution of the indicated MAb was mixed with decreasing concentrations of isolated detergent-solubilized trimers (solid lines) or liposome-bound trimers (dashed lines) as indicated. The mixture was added to microtiter plates that had been coated with purified virus at a concentration of 1 μg/ml. Free antibody that was not blocked by the antigen in solution was detected by using peroxidase-labeled rabbit anti-mouse immunoglobulin G as described in reference 12. Results are expressed as percentage of the absorbance value obtained with each MAb in the absence of a blocking antigen. (B) Top view of a single monomeric subunit of sE in its native conformation (16). The three domains of E are shown in different shades of grey. The MAbs reacting with each of the domains are indicated within the brackets.
As shown in Fig. 3A, the liposome-associated form of sE (shown in Fig. 1) did not react with MAbs A1 and A2. Their reactivity, however, was restored when the liposome-protein complex was dispersed by detergent treatment (Fig. 3B). In contrast, the control MAbs C4 (specific for domain I) and B4 (specific for domain III) bound to their epitopes on the liposome-associated form. These data provide evidence that the cd loop interacts directly with the target membrane and is inserted in the liposomal membrane after the low-pH-induced conversion to a trimer, consistent with its function as a fusion peptide (Fig. 3B). A similar result was obtained with the truncated form of SFV E1, which was shown to insert into liposomes at low pH via its fusion peptide (1). At present, however, it is still unknown how far domain II penetrates into the membrane. MAb C4 consistently reacted better with the liposome-associated form than with the solubilized form, but so far we do not have a structural explanation for this finding.
Crystallization of sE trimers.
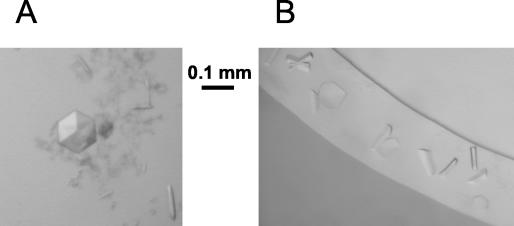
Crystallization trials were carried out with isolated sE trimers in different detergents by using the hanging-drop method (16). The initial buffer (TAN buffer containing 0.8% n-OG) was exchanged for 0.1 M ammonium acetate (pH 5.0) containing the corresponding detergent at a concentration above the critical micellar concentration (1.5 mM undecyl maltoside, 1.0 mM dodecyl maltoside, 3 mM N,N-dimethyl dodecylamine oxide [LDAO], or 15 mM N,N-dimethyl decylamine oxide [DDAO]) by using the following procedure. The solution was concentrated to 2 to 5 mg of trimer per ml by ultrafiltration with a Vivaspin500 concentrator (cutoff, 100 kDa). The trimer solution was then diluted 10-fold in the ammonium acetate buffer and reconcentrated to 2 to 5 mg/ml. The procedure was repeated four times before crystallization trays were set up. Crystals were only obtained in the presence of DDAO. Several crystal forms were characterized (Table 1). They grew either at pH 7.8 in 0.1 M Tris, 2 M ammonium sulfate, 2% polyethylene glycol (PEG) 400 or at pH 4.5 in 0.1 M sodium acetate with PEG as the precipitant (20 to 30% PEG 2000, 15 to 20% PEG 4000, 10 to 20% PEG 10000, 10 to 20% PEG 20000). At pH 7.8, the crystals belonged either to the cubic space group P213 with two molecules per asymmetric unit (yielding eight trimers to make up the unit cell) or to the tetragonal space group P4222 with one trimer per asymmetric unit. These two forms were easily distinguished by their appearance (cubic and rodlike, respectively) (Fig. 4A). At pH 4.5, several orthorhombic crystal forms were grown that could not be visually distinguished (Fig. 4B). The diffraction quality of these crystals is sufficient to allow structure determination (Table 1).
TABLE 1.
Growth conditions and characteristics of sE trimer crystals
| Growth conditions (precipitant, pH) | Space groupa | No. of molecules per asymmetric unit | Resolution (Å) |
|---|---|---|---|
| Ammonium sulfate, 7.8 | P4222 | 3 | 3.5b |
| Ammonium sulfate, 7.8c | P213 | 2d | 3.5 |
| PEG, 4.5e | P21212 | 3 | 3.2 |
| PEG, 4.5e | C222 | 3 | 3.2 |
| PEG, 4.5e | P212121 | 6 | 2.7 |
As determined from analysis of a dataset (92 to 100% complete) and extending to at least 3.5 Å.
Due to strong anisotropy, the diffraction pattern extends to 3 Å in one direction but not beyond 3.8 Å in the orthogonal plane, yielding an overall approximate resolution limit of 3.5 Å.
Crystals appeared in the same conditions (sometimes in the same drops) as those of the tetragonal form.
For this crystal form, the threefold axis of the trimer coincides with a crystallographic axis.
The three orthorhombic crystal forms appeared in similar conditions with various PEGs (2000 to 20000) and could only be distinguished by X-ray diffraction analysis.
FIG. 4.
Crystals of sE trimers grown in the presence of detergent (15 mM DDAO). (A) Cubic and tetragonal (rods) crystal forms that grow simultaneously in the same drops at pH 7.8. (B) Orthorhombic crystals grown at pH 4.5.
The described results represent a major step forward toward a more detailed understanding of the fusion process mediated by class II viral fusion proteins. The formation of paracrystalline arrays of sE trimers upon membrane insertion suggests strong trimer-trimer interactions that may be important for cooperative effects during the fusion process. Together with the already known structure of the native E dimer (17), the resolution of the low-pH structure will reveal details of the structural transitions of E that are involved in fusion and thereby provide new insights into the fusion process mediated by class II proteins at a molecular level.
Acknowledgments
We thank Steven Allison for helpful discussions and for critical reading of the manuscript, Don Gibbons for technical discussions, and Walter Holzer, Silvia Röhnke, and Armelle Vigouroux for technical assistance.
Stéphane Bressanelli has a long-term EMBO fellowship.
REFERENCES
- 1.Ahn, A., D. L. Gibbons, and M. Kielian. 2002. The fusion peptide of Semliki Forest virus associates with sterol-rich membrane domains. J. Virol. 76:3267-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, S. L., J. Schalich, K. Stiasny, C. W. Mandl, and F. X. Heinz. 2001. Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. J. Virol. 75:4268-4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allison, S. L., K. Stiasny, K. Stadler, C. W. Mandl, and F. X. Heinz. 1999. Mapping of functional elements in the stem-anchor region of tick-borne encephalitis virus envelope protein E. J. Virol. 73:5605-5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bentz, J., and A. Mittal. 2000. Deployment of membrane fusion protein domains during fusion. Cell Biol. Int. 24:819-838. [DOI] [PubMed] [Google Scholar]
- 5.Colman, P. M., and M. C. Lawrence. 2003. The structural biology of type I viral membrane fusion. Nat. Rev. Mol. Cell Biol. 4:309-319. [DOI] [PubMed] [Google Scholar]
- 6.Corver, J., A. Ortiz, S. L. Allison, J. Schalich, F. X. Heinz, and J. Wilschut. 2000. Membrane fusion activity of tick-borne encephalitis virus and recombinant subviral particles in a liposomal model system. Virology 269:37-46. [DOI] [PubMed] [Google Scholar]
- 7.Gibbons, D. L., I. Erk, B. Reilly, J. Navaza, M. Kielian, F. A. Rey, and J. Lepault. 2003. Visualization of the target-membrane-inserted fusion protein of Semliki Forest virus by combined electron microscopy and crystallography. Cell 114:573-583. [DOI] [PubMed] [Google Scholar]
- 8.Heinz, F. X., and S. L. Allison. 2001. The machinery for flavivirus fusion with host cell membranes. Curr. Opin. Microbiol. 4:450-455. [DOI] [PubMed] [Google Scholar]
- 9.Heinz, F. X., C. W. Mandl, H. Holzmann, C. Kunz, B. A. Harris, F. Rey, and S. C. Harrison. 1991. The flavivirus envelope protein E: isolation of a soluble form from tick-borne encephalitis virus and its crystallization. J. Virol. 65:5579-5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heinz, F. X., K. Stiasny, G. Puschner-Auer, H. Holzmann, S. L. Allison, C. W. Mandl, and C. Kunz. 1994. Structural changes and functional control of the tick-borne encephalitis virus glycoprotein E by the heterodimeric association with protein prM. Virology 198:109-117. [DOI] [PubMed] [Google Scholar]
- 11.Hernandez, L. D., L. R. Hoffman, T. G. Wolfsberg, and J. M. White. 1996. Virus-cell and cell-cell fusion. Annu. Rev. Cell Dev. Biol. 12:627-661. [DOI] [PubMed] [Google Scholar]
- 12.Holzmann, H., G. Utter, E. Norrby, C. W. Mandl, C. Kunz, and F. X. Heinz. 1993. Assessment of the antigenic structure of tick-borne encephalitis virus by the use of synthetic peptides. J. Gen. Virol. 74:2031-2035. [DOI] [PubMed] [Google Scholar]
- 13.Kielian, M. 2002. Structural surprises from the flaviviruses and alphaviruses. Mol. Cell 9:454-456. [DOI] [PubMed] [Google Scholar]
- 14.Klimjack, M. R., S. Jeffrey, and M. Kielian. 1994. Membrane and protein interactions of a soluble form of the Semliki Forest virus fusion protein. J. Virol. 68:6940-6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lescar, J., A. Roussel, M. W. Wien, J. Navaza, S. D. Fuller, G. Wengler, G. Wengler, and F. A. Rey. 2001. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105:137-148. [DOI] [PubMed] [Google Scholar]
- 16.McPherson, A. 1989. Preparation and analysis of protein crystals. Krieger Publishing Co., Malabar, Fla.
- 17.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2-Å resolution. Nature 375:291-298. [DOI] [PubMed] [Google Scholar]
- 18.Schalich, J., S. L. Allison, K. Stiasny, C. W. Mandl, C. Kunz, and F. X. Heinz. 1996. Recombinant subviral particles from tick-borne encephalitis virus are fusogenic and provide a model system for studying flavivirus envelope glycoprotein functions. J. Virol. 70:4549-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skehel, J. J., and D. C. Wiley. 1998. Coiled coils in both intracellular vesicle and viral membrane fusion. Cell 95:871-874. [DOI] [PubMed] [Google Scholar]
- 20.Stiasny, K., S. L. Allison, A. Marchler-Bauer, C. Kunz, and F. X. Heinz. 1996. Structural requirements for low-pH-induced rearrangements in the envelope glycoprotein of tick-borne encephalitis virus. J. Virol. 70:8142-8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stiasny, K., S. L. Allison, J. Schalich, and F. X. Heinz. 2002. Membrane interactions of the tick-borne encephalitis virus fusion protein E at low pH. J. Virol. 76:3784-3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stiasny, K., C. Koessl, and F. X. Heinz. 2003. Involvement of lipids in different steps of the flavivirus fusion mechanism. J. Virol. 77:7856-7862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weissenhorn, W., A. Dessen, L. J. Calder, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1999. Structural basis for membrane fusion by enveloped viruses. Mol. Membr. Biol. 16:3-9. [DOI] [PubMed] [Google Scholar]
- 24.Wilschut, J., J. Corver, J. L. Nieva, R. Bron, L. Moesby, K. C. Reddy, and R. Bittman. 1995. Fusion of Semliki Forest virus with cholesterol-containing liposomes at low pH: a specific requirement for sphingolipids. Mol. Membr. Biol. 12:143-149. [DOI] [PubMed] [Google Scholar]