Abstract
CXCR4-using (X4) human immunodeficiency virus type 1 (HIV-1) variants evolve from CCR5-restricted (R5) HIV-1 variants. Early after their first appearance in vivo, X4 HIV-1 variants additionally use CCR5. The ability to use CCR5 in addition to CXCR4 is generally lost late in infection. Here we studied whether this evolution of the coreceptor repertoire is also reflected in a changing sensitivity of X4 variants to CXCR4 antagonists such as peptide T22 and the synthetic compound AMD3100. We observed differences in the concentrations of CXCR4 antagonists needed to suppress replication of X4 HIV variants from different patients. In general, late X4 HIV variants were less sensitive to AMD3100 than were early R5X4 HIV variants. The differences between early R5X4 HIV variants and late X4 variants were less pronounced for T22-mediated inhibition. These results suggest an ongoing evolution of X4 virus variants toward more efficient usage of the cellular entry complex.
Entry of human immunodeficiency virus type 1 (HIV-1) into CD4+ T cells is a dynamic process. Binding of envelope glycoprotein gp120 to CD4 induces a conformational change resulting in exposure and binding of the conserved binding region of gp120 to a coreceptor (33), in general, β-chemokine receptor 5 (CCR5) or α-chemokine receptor 4 (CXCR4) (5, 35).
Early in HIV-1 infection, a homogeneous population of predominantly macrophage-tropic, non-syncytium-inducing virus variants that use CCR5 for cellular entry (R5 variants) (1, 15) is present (40, 41, 46, 47). Syncytium-inducing virus variants mainly use CXCR4 as a coreceptor (X4 variants) (6, 16, 36, 42) and can be distinguished from R5 virus variants by their tendency for higher replication kinetics and a broader target cell range (7, 23, 45). Their presence in vivo has been associated with an accelerated CD4 cell decline and more rapid disease progression (11, 21). This can be explained by the fact that more CD4 T cells express CXCR4, providing X4 variants with a much larger target cell population (18, 24). More importantly, naive CD4 T cells express CXCR4 but not CCR5, which makes them selective targets for X4 HIV infection in vivo (7, 29, 30). Infection and death of these naive CD4 T cells may directly interfere with T-cell renewal (7).
In the natural course of infection, X4 HIV-1 variants evolve from R5 variants via an R5X4 phenotype, as determined by transfected U87 indicator cell lines. The ability to use CCR5 in addition to CXCR4 is generally lost late in infection (44). Whether this loss is associated with more efficient usage of CXCR4 is unknown.
Cellular entry and fusion of HIV-1 are promising new targets for the development of antiviral drugs and may have an additive effect along with the currently available drugs that interfere with reverse transcriptase and protein processing (10, 14, 26, 31, 32, 43). CXCR4-specific antagonists such as AMD3100 and T22 have been found to be highly effective at blocking entry of X4 HIV-1 variants (10, 26, 31, 34, 43).
Here we studied whether the ongoing evolution of X4 HIV-1 variants correlates with a changing sensitivity to CXCR4-specific antagonists AMD3100 and T22 and a panel of CXCR4-directed monoclonal antibodies (MAbs).
MATERIALS AND METHODS
HIV-1 variants and cells.
Clonal virus isolation was performed from peripheral blood mononuclear cells (PBMC) of five homosexual male participants of the Amsterdam Cohort studies on HIV-1 and AIDS (patients ACH208, ACH039, ACH171, ACH1120, and ACH6052), who all developed X4 variants during a progressive disease course. None of these participants ever received multidrug antiviral therapy. In the Amsterdam cohort, the presence of X4 HIV-1 variants in peripheral blood is prospectively determined at every visit (in general, every 3 months) by cocultivation of 106 patient PBMC with 106 MT2 cells. Virus replication in this coculture is considered evidence of the presence of X4 virus variants in the patient. The moment of first appearance of X4 virus was calculated as the midpoint between the last MT2-negative visit and the first MT2-positive visit.
Biological virus clones were available from previous studies (22, 44, 45) and obtained by cocultivation of patient PBMC with phytohemagglutinin (PHA)-stimulated healthy blood donor PBMC (donor PHA-PBMC) under limiting-dilution conditions as previously described (40). Briefly, patient PBMC (0.5 × 104 to 4 × 104 cells/well, 48 or 96 wells per patient cell number) were cocultivated with donor PHA-PBMC (105/well) in 96-well plates. Every week, culture supernatants were tested for the presence of p24 in an in-house antigen capture enzyme-linked immunosorbent assay (ELISA). At the same time, one-third of the cell culture was transferred to new 96-well plates and 105 fresh donor PHA-PBMC were added to propagate the culture. If fewer than one-third of the microcultures were positive at a given patient cell number, viruses were considered to be clonal. Furthermore, no evidence of mixed viral populations was obtained by sequence analyses of the viral isolates used in this study (data not shown). PBMC from cultures that tested positive in our p24 antigen capture ELISA were transferred to 25-ml culture flasks containing 5 × 106 fresh PHA-PBMC in 5 ml of medium to grow virus stocks. Cell-free supernatants with virus were stored at −70°C until use.
From each patient, three to six X4 virus variants were available from time points early after the first detection of X4 variants in vivo. In addition, five virus variants per patient, obtained after AIDS diagnosis from patients ACH208, ACH039, ACH1120, and ACH6052 or 2 years before AIDS diagnosis from patient ACH171, were used for analyses (Table 1).
TABLE 1.
Characteristics of X4 HIV-1 variants
| Patient | Serologic statuse | Early virus variants
|
Late virus variants
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | Time to seroconversion or entry (mo)a | Time to X4 (mo)a | Coreceptor usageb | CD4 T-cell count (cells/μl) | RNA load (log no. of copies/ml) | n | Time to seroconversion or entry (mo) | Time to X4 (mo) | Coreceptor usageb | CD4 T-cell count (cells/μl) | RNA load (log no. of copies/ml) | ||
| ACH208 | Sc | 3 | 18.5 | 4.5 | R5X4 (2) | 440 | 4.4 | 5 | 64.1 | 50.1 | X4 (5) | 30 | NAf |
| X4 (1) | |||||||||||||
| ACH039 | Sc | 6 | 18.0 | 1.9 | R3R5X4 (3) | 570 | 4.7 | 5 | 48.5 | 32.4 | R3X4 (5) | 10 | NA |
| X4d (3) | |||||||||||||
| ACH0171 | Sc | 4 | 66.8 | 5.2 | R3R5X4 (3) | 380 | 4.9 | 4 | 89.3 | 27.0 | R3R5X4 (1) | 130 | 5.4 |
| X4 (1) | X4 (3) | ||||||||||||
| ACH1120 | Sc | 5 | 53.1 | 7.3 | X4d | 270 | 6.4 | 4 | 66.2 | 20.4 | X4d | 10 | NA |
| ACH6052 | Sp | 5 | 0c | —c | X4d | 500 | NA | 5 | 32.0 | —c | X4d | 20 | NA |
When virus variants from multiple time points were analyzed, the average time to seroconversion or first detection of X4 HIV-1 variants is given.
The absolute number of virus variants with the indicated coreceptor preference in transfected U87 cells is given in parentheses. Data are available from a previous study (45).
Patient ACH6052 was seropositive and carried X4 variants at the time of entry into the cohort. No estimated time of first detection of X4 HIV-1 variants can therefore be given.
CXCR4 usage for entry as determined in MT2 cell line and replication in CCR5Δ/Δ PBMC. Other coreceptor preferences in transfected U87 cells were not tested.
Sc, seroconverter; Sp, seroprevalent.
NA, not available.
For all of the HIV variants studied here, the ability to replicate in the MT2 cell line was considered evidence of CXCR4 usage. In addition, CXCR4 usage was confirmed in PBMC from a healthy donor homozygous for the 32-bp deletion in the CCR5 gene (CCR5Δ/Δ). For three subjects, expanded coreceptor usage was tested in transfected U87 indicator cell lines expressing CD4 and either CCR5, CXCR4, or CCR3 (Table 1).
Sensitivity for chemokine receptor antagonists of two early virus variants and two late virus variants from patients ACH208 and ACH039 was tested on the MT2 T-cell line, on CCR5Δ/Δ PBMC from a healthy donor, and on pooled PBMC from at least two healthy donors homozygous for the CCR5 wild-type allele (CCR5+/+). All experiments, including titration of virus stocks, were performed with the same pool of cryopreserved PBMC from healthy donors to eliminate possible variation caused by differences in the susceptibility of PBMC to HIV infection. PBMC were stimulated for 2 to 3 days with 1 μg of PHA per ml in Iscove's modified Dulbecco's medium (IMDM) supplemented with 10% fetal calf serum (FCS) and were subsequently cultured in IMDM supplemented with 10% FCS and 20 U of interleukin-2 (IL-2; Chiron Benelux, Amsterdam, The Netherlands) per ml. MT2 cells were maintained in IMDM supplemented with 10% FCS, 100 U of penicillin per ml, and 100 μg of streptomycin per ml.
Sensitivity to coreceptor antagonists.
To study the sensitivity to coreceptor antagonists of two early and two late virus variants obtained from patients ACH208 and ACH039, 105 PBMC or 2.5 × 104 MT2 cells were incubated with fivefold serial dilutions of the specific antagonists in a volume of 50 μl for 2 h at 37°C in a flat-bottom 96-well plate. Ten 50% tissue culture infective doses of each virus clone were added, and medium was added to a total volume of 100 μl. Every 3 to 4 days, one-third of each MT2 culture was replaced with fresh medium. PBMC cultures were maintained for 14 days and transferred to fresh medium at day 7 after inoculation. Production of p24 in the culture supernatant was measured by ELISA at days 7 and 14 after inoculation. Each dilution of CXCR4 antagonist was tested in triplicate, and each experiment was performed at least twice. MAbs directed against CXCR4 (44708.111, 44716.111, 44717.111 [R&D Systems, Minneapolis, Minn.], and 12G5) were tested at a maximum concentration of 50 μg/ml, CXCR4 antagonist T22 (Bachem AG, Bubendorf, Switzerland) (27) was tested at a maximum concentration of 3 μM, and the bicyclam AMD3100 (9, 13, 38) was tested at a maximum concentration of 2.4 μM (concentrations apply to incubation of cells with coreceptor antagonists before addition of virus).
An expanded panel of biological virus clones from patients ACH208, ACH039, ACH171, ACH1120, and ACH6052 was tested for sensitivity to AMD3100 and T22 on PHA-PBMC by an adapted approach in which AMD3100 and T22 were added after cells were inoculated with virus. PHA-PBMC (6 × 106) were inoculated with 600 50% tissue culture infective doses in a volume of 3 ml in 15-ml tubes. After incubation for 2 h at 37°C in a shaking water bath, cultures were washed twice with IMDM and resuspended in 3 ml of IL-2-supplemented medium. A cell suspension of 105 PBMC in a volume of 50 μl was mixed with 50 μl of fivefold serial dilutions of AMD3100 (final maximum concentration of 6 μM) or twofold serial dilutions of T22 (final maximum concentration of 6 μM) in flat-bottom 96-well plates. Cultures were maintained for 14 days and transferred to fresh medium at day 7. Culture supernatants were analyzed for p24 production at days 7 and 14. All dilutions of AMD3100 and T22 were tested in triplicate, and experiments were performed at least twice. Percent inhibition relative to control infections was calculated. Differences between the 50% inhibitory concentrations (IC50s) of early and late virus variants were evaluated by the Mann-Whitney U test with SPSS software (version 10.0; SPSS Inc., Chicago, Ill.).
RESULTS
Sensitivity of early and late X4 virus variants to AMD3100 in the MT2 cell line.
X4 HIV-1 variants obtained either early or late after their first appearance in vivo were compared for sensitivity to a panel of CXCR4 antagonists. First, we tested two early and two late X4 variants from two patients. Early X4 variants from patients ACH208 (12.F4 and 13.B1) and ACH039 (20.B10 and 20.C6), which were obtained 4.5 and 1.9 months after their emergence, respectively, additionally used CCR5 (patient ACH208; R5X4) or CCR5 and CCR3 (patient ACH039; R3R5X4), as determined in transfected U87 indicator cell lines. The late X4 variants from patients ACH208 (X1.A1 and X1.B1) and ACH039 (X1.C4 and X1.H4), which were obtained, respectively, 46 and 30 months later had lost the ability to use CCR5 and used CXCR4 alone (patient ACH208; X4) or CXCR4 in combination with CCR3 (patient ACH039; R3X4) (Table 1).
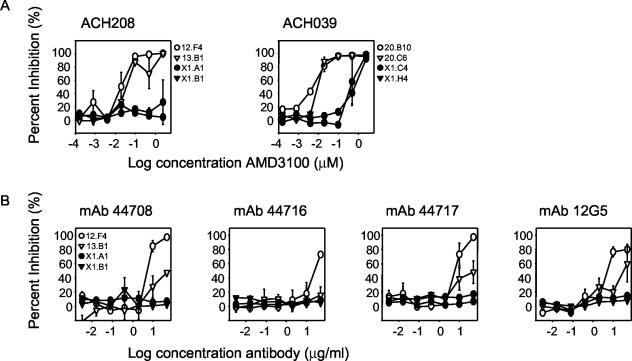
The sensitivity of these virus variants to the CXCR4-specific antagonist AMD3100 was determined in the MT2 T-cell line. For early variants from patient ACH208, the IC50s were 0.02 and 0.04 μM, whereas even the highest concentration of AMD3100 did not interfere with replication of the late X4 variants from patient ACH208 (IC50, >2.40 μM). For patient ACH039, the average IC50 of early R3R5X4 variants was 0.005 μM, which is approximately 80-fold lower than IC50s of late-stage R3X4 variants (Fig. 1A).
FIG. 1.
Sensitivity of early R3R5X4/R5X4 and late R3X4/X4 HIV-1 variants to CXCR4 antagonists in the MT2 T-cell line. (A) Sensitivity to the CXCR4-specific bicyclam AMD3100. (B) Sensitivity to CXCR4-specific MAbs. The virus variants from patient ACH208 that were used (left side) were early R5X4 (12.F4 and 13.B1) and late X4 (X1.A1 and X1.B1); those from patient ACH039 (right side) were early R3R5X4 (20.B10 and 20.C6) and late R3X4 (X1.C4 and X1.H4). Percent inhibition relative to control infections was calculated. Experiments were performed in triplicate, and average values are shown. A representative graph of at least two independent experiments is shown. Open symbols indicate early R5X4 or R3R5X4 variants; filled symbols indicate late R3X4 or X4 variants.
Sensitivity of early and late X4 virus variants to CXCR4-directed MAbs in the MT2 cell line.
To determine whether the differences in sensitivity between early and late X4 variants were specific for AMD3100, we analyzed whether these virus variants could also be inhibited by MAbs 12G5, 44708.111, 44716.111, and 44717.111, which were previously shown to recognize different conformational epitopes on CXCR4 (4, 8). Virus variant 12.F4, obtained early from patient ACH208, was inhibited by these MAbs, with IC50s of the different MAbs ranging from 5 to 26 μg/ml (Fig. 1B). Inhibition of replication of early virus variant 13.B1 from patient ACH208 was observed, albeit to a lesser extent than that of replication of variant 12.F4. The two late-stage X4 variants from patient ACH208 were not inhibited by these MAbs, not even at the highest MAb concentrations tested (IC50, >50 μg/ml). Thus, similar to our observation with AMD3100, early R5X4 variants from patient ACH208 were more sensitive to inhibition by CXCR4-directed antibodies. Replication of both the virus variants obtained early and late from patient ACH039 was not affected by the CXCR4-directed antibodies, not even at the highest MAb concentration used (50 μg/ml).
Sensitivity of early and late X4 virus variants to CXCR4 antagonists on primary cells.
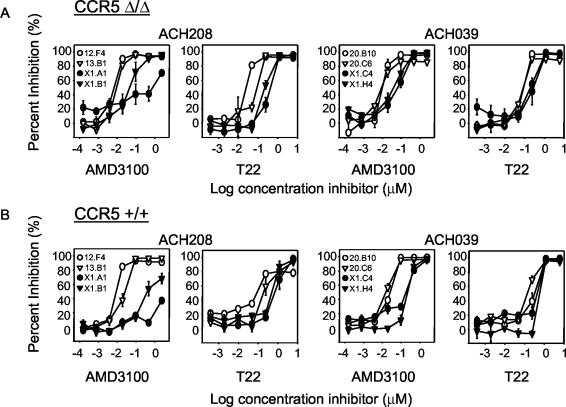
The main target cells for HIV-1 in vivo are CD4+ T lymphocytes. Since primary T cells can express CCR5 and CXCR4, CXCR4 antagonists might be unable to inhibit replication of R5X4 HIV-1 variants, as the ability to use CCR5 might provide these variants with an opportunity to circumvent inhibition by CXCR4 antagonists. To exclude this escape mechanism, we tested the sensitivity of early and late X4 HIV-1 variants to two CXCR4-specific antagonists, AMD3100 and synthetic peptide T22, on PHA-stimulated PBMC from a healthy blood donor. This blood donor is homozygous for a 32-bp deletion in CCR5 (CCR5Δ/Δ) and thus completely lacks CCR5 expression on the cell surface. On average, early R3R5X4 and R5X4 variants were 6- to 50-fold more sensitive to inhibition with AMD3100 than the late X4 variants from the same individuals. The difference in sensitivity to T22 on primary CCR5Δ/Δ cells between early and late X4 HIV-1 variants was less pronounced (Fig. 2A).
FIG. 2.
Sensitivity of early and late HIV-1 variants to CXCR4 antagonists AMD3100 and T22 in PBMC of a donor who is homozygous for a 32-bp deletion in CCR5 (CCR5 Δ/Δ) (A) and in pooled PHA-PBMC from two healthy donors with a wild-type CCR5 genotype (CCR5 +/+) (B). The same virus variants from patients ACH208 and ACH039 were used as described in the legend to Fig. 1. Percent inhibition relative to control infections was calculated. Experiments were performed in triplicate, and average values are shown. A representative graph of at least two independent experiments is shown.
Subsequent experiments with pooled PBMC from two healthy blood donors with a CCR5 wild-type genotype (CCR5+/+) confirmed that early R3R5X4 and R5X4 variants were more sensitive to inhibition by AMD3100 and T22 than were X4 variants obtained late from the same individuals (Fig. 2B). Replication of a CCR5-using HIV-1 variant was not affected by AMD3100 and T22, confirming that inhibition was specific for viral entry via CXCR4 and excludes toxicity of the compounds as an explanation for their activity (data not shown).
The maximum and dose-dependent inhibition that was achieved with AMD3100 and T22 for R3R5X4 and R5X4 HIV-1 variants on CCR5-expressing PBMC was similar to that obtained on CCR5Δ/Δ PBMC (data not shown). None of the R3R5X4, R5X4, R3X4, or X4 variants were inhibited by a combination of MIP-1α, MIP-1β, and RANTES, the natural ligands of CCR5 (IC50, >2,000 ng/ml), whereas infection of R5 HIV-1 variant 09F1 from patient ACH208 was inhibited efficiently (IC50 of 30 ng/ml; data not shown). Thus, R5X4 virus variants that were able to use CCR5 in U87 cells transfected with CD4 and CCR5 were unable to efficiently use CCR5 for infection of primary lymphocytes.
Sensitivity of a larger panel of X4 virus variants to AMD3100 and T22.
To confirm the observed differences in sensitivity to inhibition by CXCR4 antagonists between early and late X4 variants, we expanded the number of virus variants from patients ACH039 and ACH208 and added early and late X4 virus variants from three additional patients (ACH171, ACH1120, and ACH6052; Table 1). Three to six X4 virus variants obtained at a time point early after the first emergence of X4 HIV-1 variants in vivo, in addition to five late X4 virus variants obtained 2 years before AIDS diagnosis (patient ACH171) or after AIDS diagnosis (patients ACH208, ACH039, ACH1120, and ACH6052), were studied from each patient (Table 1). As described in Materials and Methods, for the extended panel, we used a modified protocol including higher maximum antagonist concentrations (final concentration, 6 μM) and twofold instead of fivefold serial dilution steps for the testing of T22-mediated inhibition. Moreover, cells were first inoculated with virus and subsequently incubated with either AMD3100 or T22. Virus variants from patients ACH208 and ACH039 that were used in the first set of experiments were included again and provided similar results, indicating that the protocol modification did not lead to different results (data not shown).
Early and late X4 HIV-1 variants were compared for sensitivity to AMD3100 and T22 in pooled PHA-PBMC. Early X4 HIV-1 variants from patients ACH208, ACH039, ACH1120, and ACH6052 were more sensitive to AMD3100 than were late X4 HIV-1 variants (P values of 0.01, <0.01, 0.02, and 0.01, respectively), whereas no difference in sensitivity to AMD3100 could be seen between early and late X4 variants from patient ACH171 (Fig. 3A). The average IC50s of early X4 variants ranged from 0.004 μM (patient ACH208) to 0.2 μM (patient ACH1120), compared to average IC50s of late X4 variants, which ranged from 2.6 μM (patient ACH208) to 1.8 μM (patient ACH1120) (Table 2). The ratio of the IC50s of early and late X4 variants was 264 for patient ACH208 (P = 0.01) and only 4 for patient ACH039 (P < 0.01) (Table 2). In addition, a large interpatient variation was observed in the IC50s of CXCR4 antagonists (Table 2).
FIG. 3.
Sensitivity of early and late X4 HIV-1 variants from the five patients to CXCR4 antagonists. (A) Sensitivities of early and late virus variants to AMD3100 in pooled PHA-PBMC from eight different healthy blood donors. (B) Sensitivities of early and late virus variants to T22 in pooled PHA-PBMC from eight different healthy blood donors. Average values and standard errors of three to six virus variants obtained early after the emergence of X4 HIV-1 variants in vivo (open circles) and of four or five variants obtained 2 to 3 years thereafter (filled circles) are indicated. Percent inhibition relative to control infections was calculated. A negative value represents an increase in viral replication compared to the control infections. Experiments were performed in triplicate, and a representative graph of at least two independent experiments is shown.
TABLE 2.
Average IC50s of early and late X4 virus variants
| Patient | IC50 (μM) of:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| AMD3100
|
T22
|
|||||||
| Early virus variantsa | Late virus variantsa | Ratio of IC50sb | P valuec | Early virus variantsa | Late virus variantsa | Ratio of IC50sb | P valuec | |
| ACH208 | <0.01 | 2.64 | >264 | 0.01 | 0.34 | 0.18 | 0.53 | 0.86 |
| ACH039 | <0.01 | 0.04 | >4 | <0.01 | 0.07 | 0.42 | 6 | 0.04 |
| ACH171 | 1.53 | 0.63 | 0.41 | 0.69 | 0.20 | 0.09 | 0.45 | 0.06 |
| ACH1120 | 0.19 | 1.76 | 9.3 | 0.02 | 0.37 | 0.55 | 1.5 | 0.20 |
| ACH6052 | 0.02 | 0.11 | 5.5 | 0.01 | 0.41 | 0.76 | 1.9 | 0.01 |
For each virus variant, the average IC50 from two independent experiments is given.
Ratio of the average IC50 for late X4 variants to the average IC50 for early X4 variants.
Per virus variant, average IC50s from two independent experiments were evaluated with the Mann-Whitney U test.
Statistically significant differences in T22-mediated inhibition between early and late X4 variants were only observed for patients ACH039 (P = 0.04) and ACH6052 (P = 0.01) and were less pronounced than differences in sensitivity observed with AMD3100 (Fig. 3B). The average IC50s of early virus variants were 0.07 μM for patient ACH039 and 0.40 μM for patient ACH6052, whereas the average IC50s for late virus variants were 0.40 μM for patient ACH039 and 0.80 μM for patient ACH6052. In contrast, for the early and late X4 variants from patients ACH208, ACH171, and ACH1120, which were differentially susceptible to AMD3100, no differences in sensitivity to T22-mediated inhibition were observed (Table 2).
DISCUSSION
In this study, we compared the sensitivity to CXCR4 antagonists of CXCR4-using virus variants obtained early and late after their appearance in vivo. We observed a decreased sensitivity to AMD3100 in late X4 virus variants compared to early X4 virus variants from the same individual in four out of five cases. This decreasing sensitivity was also observed with a panel of CXCR4-directed MAbs on early and late X4 virus variants from one out of two patients tested. The differential susceptibility to inhibition by the CXCR4 antagonist T22 was less pronounced and only observed for early and late X4 virus variants from patients ACH039 and ACH6052. Overall, these data show that the in vivo evolution of X4 HIV-1 variants tends to coincide with a decreasing sensitivity to CXCR4 antagonists. We consider differences in the interaction between gp120 envelope protein and the cellular receptor complex of CD4 and CXCR4 the most likely explanation for the observed differences between early and late X4 variants. Compared to early X4 HIV variants, late-stage X4 variants may interact with other domains or another conformation of the CXCR4 molecule or may bind the cellular entry complex with higher efficiency.
Early and late X4 variants that differed in sensitivity to AMD3100 did not necessarily differ in sensitivity to inhibition by T22, pointing to different interactions of AMD3100 and T22 with CXCR4. It is known that AMD3100 interacts with extracellular loop 2 and transmembrane region 4 of CXCR4 (25). The decreased AMD3100 sensitivity of late X4 variants could thus be due to a change toward the usage of a CXCR4 loop other than extracellular loop 2 or transmembrane region 4. T22 is thought to interact with the N-terminal domain and all extracellular domains of CXCR4, which may explain the more general inhibitory effect of T22 on the early and late X4 variants of all five of the patients in this study (28). Differences between the interactions of AMD3100 and T22 with CXCR4 were indeed supported by the observation that the T22 analog T134 was still able to inhibit replication of AMD3100- and SDF1-α-resistant clones (2, 12, 19, 37).
Various selection pressures, such as HIV-specific neutralizing antibodies and cytotoxic T cells, may drive the evolution of X4 variants during the course of infection. Furthermore, low CD4 cell numbers late in infection may select for virus variants that make the most efficient use of CD4 and/or CXCR4 for cellular entry. In line with this hypothesis, we observed a reduced sensitivity for AMD3100 in late X4 variants from patients ACH208, ACH039, ACH6052, and ACH1120, which were obtained 2 years after AIDS diagnosis, when the CD4 T-cell count had dropped below 50/μl. In contrast, reduced sensitivity for AMD3100 was not observed in late X4 variants from patient ACH171 that were obtained approximately 2 years before AIDS diagnosis, when the CD4 T-cell count was still 130/μl. Alternatively, low target cell availability may also select for the X4 variants with the highest efficiency of CD4 and/or CXCR4 usage, as these variants would have an advantage relative to coexisting X4 variants.
Another mechanism for reduced sensitivity to CXCR4 antagonists could be that late-stage virus variants use unidentified coreceptors in addition to CXCR4. As cross-reactivity of AMD3100-, T22-, and CXCR4-directed antibodies with other unknown coreceptors seems highly unlikely, the complete inhibition of replication of late X4 variants by the highest concentrations of either AMD3100 or T22 excludes the use of other coreceptors in addition to CXCR4 by late X4 variants.
Our data confirm previous findings indicating that the ability to use CCR5 and CXCR4 in transfected U87 indicator cell lines does not necessarily reflect coreceptor usage in primary T cells (17, 20, 39). R5X4 and R3R5X4 virus variants, which were able to use CCR5 (and CCR3) in addition to CXCR4 in transfected U87 indicator cell lines, were unable to infect PBMC from a CCR5+/+ donor in the presence of high concentrations of CXCR4 antagonist T22. In addition, MIP-1α, MIP-1β, and RANTES, the natural ligands of CCR5, did not affect the replication of these viruses. Thus, despite efficient usage of CCR5 in transfected cell lines, these variants were unable to use CCR5 in primary CD4 T cells.
We show here that the in vivo evolution of X4 variants in the absence of exogenous inhibitors coincides with a decreased sensitivity to CXCR4 antagonists. This natural selection in vivo is seemingly in contrast with the finding that in vitro generation of AMD3100- and T22-resistant variants is very difficult and requires extensive passaging (2, 12, 19, 37). Remarkably, IC50s of late R3X4 and X4 variants from patients ACH208 and ACH039, as determined on the MT2 T-cell line, were of the same order of magnitude as the IC50 on MT2 cells of an in vitro-generated AMD3100-resistant derivative of NL4-3 (37). Amino acid substitutions in the gp120 V1-through-C4 region that converted resistance to AMD3100 in NL4-3 (12) were not observed in the late X4 variants in our study (unpublished data), suggesting distinct mechanisms of resistance development in vitro and in vivo. In addition, in vitro-generated AMD3100-resistant strains had diminished fitness in vitro (3) whereas the late X4 variants with reduced AMD3100 sensitivity in our study were naturally selected and therefore are expected not to have diminished fitness. Earlier studies in our laboratory have indeed shown similar or enhanced replication of late compared to early X4 virus variants (45).
We observed inter- and intrapatient differences in the ratio of IC50s and the absolute IC50s of AMD3100 and T22 for early and late X4 variants, implying that the interaction of gp120 with CXCR4 can vary between different virus variants. Although our in vitro results do not necessarily translate to the in vivo situation, our findings may have implications for the putative implementation of new CXCR4 antagonists as therapeutic agents. Indeed, the large interpatient variation in X4 virus sensitivity to CXCR4 antagonists may plead for CXCR4 antagonist sensitivity screening of patients' X4 HIV-1 variants before including a CXCR4 antagonist in the therapeutic regimen.
Acknowledgments
This study was performed as part of the Amsterdam Cohort Studies on HIV Infection and AIDS, a collaboration of the Municipal Health Service, the Academic Medical Center, and CLB Sanquin, Amsterdam, The Netherlands. Recombinant human IL-2 was kindly provided by Chiron, Benelux BV. The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: MAbs to CXCR4 (clones 44708.111, 44716.111, and 44717.111, contributed by R&D Systems), MAb 12G5 (contributed by James Hoxie), and the chemokines RANTES, MIP-1α, and MIP-1β (contributed by Reprotech Inc.). We thank Michael Zwick, Frank Miedema, Fransje Koning, and Neeltje Kootstra for critical reading of the manuscript and helpful discussions.
This study was financially supported by the Dutch AIDS Funds (grants 1305 and 6006).
REFERENCES
- 1.Alkhatib, G., C. Combadiere, C. C. Broder, Y. Feng, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1996. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955-1958. [DOI] [PubMed] [Google Scholar]
- 2.Arakaki, R., H. Tamamura, M. Premanathan, K. Kanbara, S. Ramanan, K. Mochizuki, M. Baba, N. Fujii, and H. Nakashima. 1999. T134, a small-molecule CXCR4 inhibitor, has no cross-drug resistance with AMD3100, a CXCR4 antagonist with a different structure. J. Virol. 73:1719-1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armand-Ugon, M., M. E. Quinones-Mateu, A. Gutierez, J. Barretina, J. Blanco, D. Schols, E. De Clercq, B. Clotet, and J. A. Este. 2003. Reduced fitness of HIV-1 resistant to CXCR4 antagonists. Antiviral Ther. 8:1-8. [PubMed] [Google Scholar]
- 4.Baribaud, F., T. G. Edwards, M. Sharron, A. Brelot, N. Heveker, K. Price, F. Mortari, M. Alizon, M. Tsang, and R. W. Doms. 2001. Antigenically distinct conformations of CXCR4. J. Virol. 75:8957-8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger, E. A., P. M. Murphy, and J. M. Farber. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17:657-700. [DOI] [PubMed] [Google Scholar]
- 6.Bjorndal, A., H. Deng, M. Jansson, J. R. Fiore, C. Colognesi, A. Karlsson, J. Albert, G. Scarlatti, D. R. Littman, and E. M. Fenyo. 1997. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J. Virol. 71:7478-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blaak, H., A. B. Van 't Wout, M. Brouwer, B. Hooibrink, E. Hovenkamp, and H. Schuitemaker. 2000. In vivo HIV-1 infection of CD45RA+ CD4+ T cells is established primarily by syncytium-inducing variants and correlates with the rate of CD4+ T cell decline. Proc. Natl. Acad. Sci. USA 97:1269-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brelot, A., N. Heveker, O. Pleskoff, N. Sol, and M. Alizon. 1997. Role of the first and third extracellular domains of CXCR-4 in human immunodeficiency virus coreceptor activity. J. Virol. 71:4744-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bridger, G. J., R. T. Skerlj, D. Thorton, S. Padmanabhan, S. A. Martelucci, G. W. Henson, M. J. Abrams, N. Yamamoto, K. De Vreese, R. Pauwels, and E. De Clercq. 1995. Synthesis and structure-activity relationships of phenylenebis(methylene)-linked bis-tetraazamacrocycles that inhibit HIV replication. Effects of macrocyclic ring size and substituents on the aromatic linker. J. Med. Chem. 38:366-378. [DOI] [PubMed] [Google Scholar]
- 10.Chan, D. C., and P. S. Kim. 1998. HIV entry and its inhibition. Cell 93:681-684. [DOI] [PubMed] [Google Scholar]
- 11.Connor, R. I., K. E. Sheridan, D. Ceradini, S. Choe, and N. R. Landau. 1997. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J. Exp. Med. 185:621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Vreese, K., V. Kofler-Mongold, C. Leutgeb, V. Weber, K. Vermeire, S. Schacht, J. Anne, E. De Clercq, R. Datema, and G. Werner. 1996. The molecular target of bicyclams, potent inhibitors of human immunodeficiency virus replication. J. Virol. 70:689-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donzella, G., D. Schols, S. W. Lin, J. A. Este, K. A. Nagashima, P. J. Maddon, G. P. Allaway, T. P. Sakmar, G. Henson, E. De Clercq, and J. P. Moore. 1998. AMD3100, a small molecule inhibitor of HIV-1 entry via the CXCR4 co-receptor. Nat. Med. 4:72-77. [DOI] [PubMed] [Google Scholar]
- 14.Doranz, B. J., K. Grovit-Ferbas, M. P. Sharron, S. H. Mao, E. S. Daar, R. W. Doms, and W. A. O'Brien. 1997. A small-molecule inhibitor directed against the chemokine receptor CXCR4 prevents its use as an HIV-1 coreceptor. J. Exp. Med. 186:1395-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dragic, T., V. Litwin, G. P. Allaway, S. R. Martin, Y. Huang, K. A. Nagashima, C. Cayanan, P. J. Maddon, R. A. Koup, J. P. Moore, and W. A. Paxton. 1996. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381:667-673. [DOI] [PubMed] [Google Scholar]
- 16.Feng, Y., C. C. Broder, P. E. Kennedy, and E. A. Berger. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872-877. [DOI] [PubMed] [Google Scholar]
- 17.Glushakova, S., Y. Yi, J.-C. Grivel, A. Singh, D. Schols, E. De Clercq, R. G. Collman, and L. Margolis. 2000. Preferential coreceptor utilization and cytopathicity by dual-tropic HIV-1 in human lymphoid tissue ex vivo. J. Clin. Investig. 104:R7-R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grivel, J.-C., and D. B. Margolis. 1999. CCR5- and CXCR4-tropic HIV-1 are equally cytopathic for their T-cell targets in human lymphoid tissue. Nat. Med. 5:344-346. [DOI] [PubMed] [Google Scholar]
- 19.Kanbara, K., S. Sato, J.-I. Tanuma, H. Tamamura, K. Gotoh, M. Yoshimori, T. Kanamoto, M. Kitano, N. Fujii, and H. Nakashima. 2001. Biological and genetic characterization of a human immunodeficiency virus strain resistant to CXCR4 antagonist T134. AIDS Res. Hum. Retrovir. 17:615-622. [DOI] [PubMed] [Google Scholar]
- 20.Koning, F. A., D. Schols, and H. Schuitemaker. 2001. No selection for CCR5 coreceptor usage during parenteral transmission of macrophagetropic syncytium inducing human immunodeficiency virus type 1. J. Virol. 75:8848-8853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koot, M., I. P. M. Keet, A. H. V. Vos, R. E. Y. De Goede, M. T. L. Roos, R. A. Coutinho, F. Miedema, P. T. A. Schellekens, and M. Tersmette. 1993. Prognostic value of human immunodeficiency virus type 1 biological phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann. Intern. Med. 118:681-688. [DOI] [PubMed] [Google Scholar]
- 22.Koot, M., A. B. Van 't Wout, N. A. Kootstra, R. E. Y. De Goede, M. Tersmette, and H. Schuitemaker. 1996. Relation between changes in cellular load, evolution of viral phenotype, and the clonal composition of virus populations in the course of human immunodeficiency virus type 1 infection. J. Infect. Dis. 173:349-354. [DOI] [PubMed] [Google Scholar]
- 23.Koot, M., A. H. V. Vos, R. P. M. Keet, R. E. Y. De Goede, W. Dercksen, F. G. Terpstra, R. A. Coutinho, F. Miedema, and M. Tersmette. 1992. HIV-1 biological phenotype in long term infected individuals, evaluated with an MT-2 cocultivation assay. AIDS 6:49-54. [DOI] [PubMed] [Google Scholar]
- 24.Kwa, D., J. Vingerhoed, B. Boeser-Nunnink, S. Broersen, and H. Schuitemaker. 2001. Cytopathic effects of non-syncytium-inducing and syncytium-inducing human immunodeficiency virus type 1 variants on different CD4+ T-cell subsets are determined only by coreceptor expression. J. Virol. 75:10455-10459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Labrosse, B., A. Brelot, N. Heveker, N. Sol, D. Schols, E. De Clercq, and M. Alizon. 1998. Determinants for sensitivity of human immunodeficiency virus coreceptor CXCR4 to the bicylam AMD3100. J. Virol. 72:6381-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michael, N. L., and J. P. Moore. 1999. HIV-1 entry inhibitors: evading the issue. Nat. Med. 5:740-741. [DOI] [PubMed] [Google Scholar]
- 27.Murakami, T., T. Nakajima, Y. Koyanagi, K. Tachibana, N. Fujii, H. Tamamura, N. Yoshida, M. Waki, A. Matsumoto, O. Yoshie, N. Yamamoto, and T. Nagasawa. 1997. A small molecule CXCR4 inhibitor that blocks T cell line-tropic HIV-1 infection. J. Exp. Med. 186:1389-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murakami, T., T. Y. Zhang, Y. Koyanagi, Y. Tanaka, J. Kim, Y. Suzuki, S. Minoguchi, H. Tamamura, M. Waki, A. Matsumoto, N. Fujii, H. Shida, J. Hoxie, Peiper, S. C., and N. Yamamoto. 1999. Inhibitory mechanism of the CXCR4 antagonist T22 against human immunodeficiency virus type 1 infection. J. Virol. 73:7489-7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ostrowski, M. A., T.-W. Chun, S. J. Justement, I. Motola, M. A. Spinelli, J. Adelsberger, L. A. Ehler, S. B. Mizell, C. W. Hallahan, and A. S. Fauci. 1999. Both memory and CD45RA+/CD62L+ naive CD4+ T cells are infected in human immunodeficiency virus type 1-infected individuals. J. Virol. 73:6430-6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ostrowski, M. A., S. J. Justement, A. Cantanzaro, C. A. Hallahan, L. A. Ehler, S. B. Mizell, P. N. Kumar, J. Mican, T.-W. Chun, and A. S. Fauci. 1998. Expression of chemokine receptors CXCR4 and CCR5 in HIV-1-infected and uninfected individuals. J. Immunol. 161:3195-3201. [PubMed] [Google Scholar]
- 31.Owen, S. M., D. L. Rudolph, D. Schols, N. Fuyii, N. Yamamoto, and R. B. Lal. 2002. Susceptibility of diverse primary HIV isolates with varying co-receptor specificity's to CXCR4 antagonistic compounds. J. Med. Virol. 68:147-155. [DOI] [PubMed] [Google Scholar]
- 32.Pierson, T. C., and R. W. Doms. 2003. HIV-1 entry inhibitors: new targets, novel therapies. Immunol. Lett. 85:113-118. [DOI] [PubMed] [Google Scholar]
- 33.Poignard, P., E. O. Saphire, P. W. Parren, and D. R. Burton. 2001. gp120: Biologic aspects of structural features. Annu. Rev. Immunol. 19:253-274. [DOI] [PubMed] [Google Scholar]
- 34.Reeves, J. D., S. A. Gallo, N. Ahmad, J. L. Miamidian, P. E. Harvey, M. Sharron, S. Pöhlmann, J. N. Sfakianos, C. A. Derdeyn, R. Blumenthal, E. Hunter, and R. W. Doms. 2002. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density and fusion kinetics. Proc. Natl. Acad. Sci. USA 99:16249-16254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rizzuto, C. D., R. Wyatt, N. Hernández-Ramos, Y. Sun, P. D. Kwong, W. A. Hendrickson, and J. Sodroski. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949-1953. [DOI] [PubMed] [Google Scholar]
- 36.Scarlatti, G., E. Tresoldi, Å. Björndal, R. Fredriksson, C. Colognesi, H. K. Deng, M. S. Malnati, A. Plebani, A. G. Siccardi, D. R. Littman, E. M. Fenyö, and P. Lusso. 1997. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine mediated suppression. Nat. Med. 3:1259-1265. [DOI] [PubMed] [Google Scholar]
- 37.Schols, D., J. A. Esté, C. Cabrera, and E. De Clercq. 1998. T-cell-line-tropic human immunodeficiency virus type 1 that is made resistant to stromal cell-derived factor 1α contains mutations in the envelope gp120 but does not show a switch in coreceptor use. J. Virol. 72:4032-4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schols, D., S. Struyf, J. Van Damme, J. A. Este, G. Henson, and E. De Clercq. 1997. Inhibition of T-tropic HIV strains by selective antagonization of the chemokine receptor CXCR4. J. Exp. Med. 186:1383-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schramm, B., M. L. Penn, R. F. Speck, S. Y. Chan, E. De Clercq, D. Schols, R. I. Connor, and M. A. Goldsmith. 2000. Viral entry through CXCR4 is a pathogenic factor and therapeutic target in human immunodeficiency virus type 1 disease. J. Virol. 74:184-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schuitemaker, H., M. Koot, N. A. Kootstra, M. W. Dercksen, R. E. Y. De Goede, R. P. Van Steenwijk, J. M. A. Lange, J. K. M. Eeftink Schattenkerk, F. Miedema, and M. Tersmette. 1992. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus populations. J. Virol. 66:1354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuitemaker, H., N. A. Kootstra, R. E. Y. de Goede, F. de Wolf, F. Miedema, and M. Tersmette. 1991. Monocytotropic human immunodeficiency virus 1 (HIV-1) variants detectable in all stages of HIV-1 infection lack T-cell line tropism and syncytium-inducing ability in primary T-cell culture. J. Virol. 65:356-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simmons, G., D. Wilkinson, J. D. Reeves, M. T. Dittmar, S. Beddows, J. Weber, G. Carnegie, U. Desselberger, P. W. Gray, R. A. Weiss, and P. R. Clapham. 1996. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic and most can use either Lestr or CCR5 as coreceptors for virus entry. J. Virol. 70:8355-8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Starr-Spires, L. D., and R. Collman. 2002. HIV-1 entry and entry inhibitors as therapeutic agents. Clin. Lab. Med. 22:681-701. [DOI] [PubMed] [Google Scholar]
- 44.van Rij, R. P., H. Blaak, J. A. Visser, M. Brouwer, R. Rientsma, S. Broersen, A. M. De Roda Husman, and H. Schuitemaker. 2000. Differential coreceptor expression allows for independent evolution of non-syncytium-inducing and syncytium-inducing HIV-1. J. Clin. Investig. 106:1039-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van 't Wout, A. B., H. Blaak, L. J. Ran, M. Brouwer, C. Kuiken, and H. Schuitemaker. 1998. Evolution of syncytium-inducing and non-syncytium-inducing biological virus clones in relation to replication kinetics during the course of human immunodeficiency virus type 1 infection. J. Virol. 72:5099-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van 't Wout, A. B., N. A. Kootstra, G. A. Mulder-Kampinga, N. Albrecht-van Lent, H. J. Scherpbier, J. Veenstra, K. Boer, R. A. Coutinho, F. Miedema, and H. Schuitemaker. 1994. Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral and vertical transmission. J. Clin. Investig. 94:2060-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu, T., H. Mo, N. Wang, D. S. Nam, Y. Cao, R. A. Koup, and D. D. Ho. 1993. Genotypic and phenotypic characterization of HIV-1 in patients with primary infection. Science 261:1179-1181. [DOI] [PubMed] [Google Scholar]