Abstract
We have described previously the generation of an escape variant of human immunodeficiency virus type 1 (HIV-1), under the selection pressure of AD101, a small molecule inhibitor that binds the CCR5 coreceptor (A. Trkola, S. E. Kuhmann, J. M. Strizki, E. Maxwell, T. Ketas, T. Morgan, P. Pugach, S. X. L. Wojcik, J. Tagat, A. Palani, S. Shapiro, J. W. Clader, S. McCombie, G. R. Reyes, B. M. Baroudy, and J. P. Moore, Proc. Natl. Acad. Sci. USA 99:395-400, 2002). The escape mutant, CC101.19, continued to use CCR5 for entry, but it was at least 20,000-fold more resistant to AD101 than the parental virus, CC1/85. We have now cloned the env genes from the the parental and escape mutant isolates and made chimeric infectious molecular clones that fully recapitulate the phenotypes of the corresponding isolates. Sequence analysis of the evolution of the escape mutants suggested that the most relevant changes were likely to be in the V3 loop of the gp120 glycoprotein. We therefore made a series of mutant viruses and found that full AD101 resistance was conferred by four amino acid changes in V3. Each change individually caused partial resistance when they were introduced into the V3 loop of a CC1/85 clone, but their impact was dependent on the gp120 context in which they were made. We assume that these amino acid changes alter how the HIV-1 Env complex interacts with CCR5. Perhaps unexpectedly, given the complete dependence of the escape mutant on CCR5 for entry, monomeric gp120 proteins expressed from clones of the fully resistant isolate failed to bind to CCR5 on the surface of L1.2-CCR5 cells under conditions where gp120 proteins from the parental virus and a partially AD101-resistant virus bound strongly. Hence, the full impact of the V3 substitutions may only be apparent at the level of the native Env complex.
Several members of a new class of inhibitors based on blocking human immunodeficiency virus type 1 (HIV-1) entry into target cells are now in, or approaching, human clinical trials (8, 52, 77, 80, 85, 90, 98). These various compounds antagonize different stages in the multistep pathway by which HIV-1 fuses with susceptible cells. For example, the CD4-immunoglobulin G2 protein (CD4-IgG2; PRO 542) attaches to the viral envelope glycoprotein gp120 to prevent it from interacting with the CD4 receptor (3, 109). The T-20 and T-1249 peptides act later, by inhibiting conformational changes in the viral gp41 glycoprotein that are necessary for membrane fusion to be initiated (8, 40, 72, 115). All these inhibitors cause plasma viremia reductions in HIV-1-infected people (8, 49, 53, 62, 66, 71, 80).
A step in the entry process intermediate between gp120-CD4 attachment and gp41 conformational changes involves a coreceptor for gp120 (21, 31, 37, 85, 98). Thus, after gp120 has bound to CD4, it changes conformation to enable it to bind to a coreceptor from the G-protein-coupled receptor superfamily (21, 31, 37, 85, 98, 107, 116). The most physiologically relevant coreceptors are the chemokine receptors CCR5 or CXCR4, the former used by HIV-1 strains that usually dominate early in infection and the latter used by viruses that sometimes emerge several years later or that are detectable only transiently (21, 31, 85, 99, 122). The presence of viruses able to use CXCR4 (X4 strains) is associated with an accelerated disease course, due in part to the loss of naive CD4+ T cells that express CXCR4 but not CCR5 (32, 44, 69). Viruses using CCR5 (R5 strains) target memory CD4+ CCR5+ T cells and are lethal in their own right (32, 44, 57, 69).
Both CCR5 and CXCR4 are important targets for pharmacological intervention, and several inhibitors have been identified that are specific for each receptor (8, 51, 52, 77, 80, 85, 90, 98). Reductions in the amount of plasma X4 viruses were observed during trials of the CXCR4 inhibitor AMD3100, but the clinical development of this particular compound has been discontinued because of pharmacological and toxicology concerns (26). A small-molecule CCR5 inhibitor, SCH-C (SCH 351125), has caused significant viral load reductions in ongoing phase I clinical trials (51, 71, 101).
It is inevitable that HIV-1 will escape from the selective pressure exerted by any single replication inhibitor (65). Hence, it is prudent to study escape pathways in vitro in order to learn what might happen in clinical use. A particular concern with CCR5 inhibitors is that they might drive the evolution of X4 variants in vivo (35, 51, 73, 77). We have previously reported on the in vitro escape of an R5 HIV-1 isolate, CC1/85, from AD101 (SCH 350581), a CCR5 inhibitor structurally related to SCH-C (51, 101, 108). The escape variant continued to use CCR5 rather than acquiring the ability to enter cells via CXCR4 or any alternative coreceptor present in human peripheral blood mononuclear cells (PBMC) (108). In contrast, X4 viruses later evolved naturally in the individual from whom CC1/85 was isolated (23, 24).
We have now analyzed the sequence changes in the env gene of HIV-1 CC1/85 that correlate temporally with the evolution of AD101 escape mutants and have contrasted them with those occurring during the evolution of X4 variants of the same virus in vivo. The most relevant amino acid changes, i.e., those that arose contemporaneously with increasing levels of resistance in vitro, seemed likely to be four charges within the V3 loop of gp120. We confirmed their relevance by reconstituting the AD101-resistant phenotype in an NL4-3/env chimeric provirus and then making a series of point substitutions. Individually, each V3 change conferred partial resistance when introduced into an AD101-sensitive clone of CC1/85; collectively, the four changes were both necessary and sufficient for high-level resistance. The use of a gp120-CCR5 binding assay suggests that these amino acid substitutions alter the way in which HIV-1 interacts with CCR5, albeit in a manner that remains to be identified.
MATERIALS AND METHODS
Reagents.
AD101 and SCH-C were synthesized by Schering-Plough Research Institute (Kenilworth, N.J.) (101, 108). RANTES was from Peprotech Inc. (Rocky Hill, N.J.). The extracellular portion of CD4 encompassing domains D1 to D4 (sCD4) (27), the CD4-IgG2 (PRO 542) molecule (3), the murine anti-CCR5 monoclonal antibody (MAb) PA14 (81), and recombinant JR-FL gp120 (107) were gifts from Bill Olson (Progenics Pharmaceuticals Inc., Tarrytown, N.Y.). The human anti-gp120 MAb 17b (102) was provided by James Robinson (Tulane University).
Cloning and nomenclature of env genes.
The R5 HIV-1 isolate CC1/85 was derived in January 1985 from individual “Case C” (23, 24). The escape of CC1/85 from AD101, the cloning of full-length env genes from infected PBMC, and a preliminary analysis of their sequences have all been described (108). env clones were named according to our previous nomenclature (108). Thus, CC1/85 is the parental isolate and CC101.XX refers to the isolate made after XX passages under selection pressure from AD101. Similarly, CCcon.XX refers to the control virus that was cultured for XX passages in the same PBMC but without AD101. The clones generated and used here extend this nomenclature, such that CC1/85 cl.XX refers to clone XX from the CC1/85 isolate. The nomenclature used to designate other clones was also derived from that used elsewhere (24, 108).
Additional env genes were PCR amplified and cloned into the vector pAMP1 by using the CloneAmp system as specified by the manufacturer (Invitrogen, Carlsbad, Calif.). The nested PCR products were generated from genomic DNA from infected PBMC, as described previously (108). The outer primers were 5958F (5′-GGCTTAGGCATCTCCTATGGCAGGAGG AA-3′) and 9100R (5′-TAGCCCTTCCAGTCCCCCCTTTTCTTTTA-3′), and the inner primers were 6205F (5′-AGAAAGAGCAGAAGACAGTGGCAATGA-3′) and 8821R (5′-TTTTGACCACTTGCCACCCAT-3′).
CC101.22 was isolated after 22 passages with AD101 in vitro and then passaged 9 more times without AD101 (108). Isolates made during this secondary culture were designated CC101.22RX, where X is the number of passages made in the absence of AD101. The corresponding clones were named using the system outlined above.
To facilitate comparisons with previous reports on structure-function relationships in HIV-1 Env and to follow convention in the field, amino acid numbering for all Env proteins was based on the numbering of the prototypic HXBc2 Env (http://hiv-web.lanl.gov/content/hiv-db/LOCATE_SEQ/locate.html) (56).
Phylogenetic sequence analysis.
Phlyogenetic analysis and bootstraps were performed using the neighbor-joining method available in PHYLIP (Phylogeny Inference Package, written by J. Felsenstein in 1993, version 3.6, distributed by the author [http://evolution.genetics.washington.edu/phylip.html]), with the F84 model for base changes and a transition/transversion ratio of 2. In the final tree, there were 243 sequences, with 2,416 bases left in the alignment after gap stripping. The sequence from the initial sample most closely resembling those that emerged in culture after AD101 selection (CC1/85 cl.7) was selected as an outgroup for the tree; the use of this sequence from the initial sample gave an orientation of the tree that generally radiated outward from the time of sampling when drawn as a phenogram. Because of the reasonable outcome of the initial neighbor-joining phylogenetic analysis and because the large number of sequences in this study make likelihood methods computationally very expensive, the neighbor-joining method was deemed adequate for the purposes of this study.
To exclude problematic sequences, representative sequences from each sample and a few sequences that gave peculiar outcomes in an initial phylogenetic analysis (i.e., strikingly long branch lengths relative to the rest of the sample) were carefully screened to explore the integrity of the original set of 249 sequences. A BLAST search of the HIV database (http://hiv-web.lanl.gov/content/hiv-db/BASIC_BLAST/basic_blast.html) (4) revealed that one clone was nearly identical to the common reference strain NL4-3; it was therefore excluded as a probable contaminant. That NL4-3 clone was then used to probe the rest of the sequence set, identifying three clones containing short stretches with regional identity to NL4-3, including distinctive insertion and deletion patterns. These three clones were also excluded because they were probably PCR recombinants with the NL4-3 contaminant. Screening for hypermutation was performed using the HYPERMUT program (http://hlv-web.lanl.gov/content/hiv-db/HYPERMUT/hypermut.html) (95), revealing one clone hypermutated in its C′-terminal half, beyond position 2000; it, too, was excluded. Finally, one clone with an extensive deletion near V3 was excluded. The other 243 sequences were deemed to be valid after rigorous quality control testing.
Construction of chimeric NL4-3/env proviruses.
Chimeric proviruses were constructed from the pNL4-3 proviral plasmid (2) (AIDS Research and Reference Reagent Program, National Institute of Allergy and Infectious Diseases; contributed by Malcolm Martin) by overlapping PCR. The gp160 coding sequences were amplified from the cloning vectors by using primers EnvF (5′-AGCAGAAGACAGTGGCAATGAGAGTGAAG-3′) and EnvR (5′-TTTTGACCACTTGCCACCCATCTTATAGC-3′). A portion of the NL4-3 provirus from nucleotides 5284 to 6232 was amplified with primers NL(5284)F (5′-GGTCAGGGAGTCTCCATAGAATGGAGG-3′) and NL(6232)R (5′-CTTCACTCTCATTGCCACTGTCTTCTGCT-3′). This fragment encompasses the unique EcoRI restriction site in pNL4-3. Another fragment from the NL4-3 provirus spanning nucleotides 8779 to 9045 was amplified using primers NL(8779)F (5′-GCTATAAGATGGGTGGCAAGTGGTCAAAA-3′) and NL(9045)R (5′-GATCTACAGCTGCCTTGTAAGTCATTGGTC-3′). This fragment includes the unique XhoI restriction site in pNL4-3. Overlapping PCR was used to join the gp160-coding sequence from the desired clone to the fragment encompassing bases 8779 to 9045 that had been amplified from pNL4-3. The resulting fragment was then similarly joined to the amplified fragment encompassing bases 5284 to 6232 from pNL4-3. The product was digested with EcoRI and XhoI and subcloned into the corresponding sites in pBluescript KS(+) (Stratagene) for sequencing and subsequent manipulation. The EcoRI-XhoI fragment was sequenced after this and all subsequent manipulation steps to confirm the presence of the desired env gene and the absence of other changes. The EcoRI-XhoI fragment for each env gene was then subcloned back into pNL4-3. The end results were proviral plasmids that differ from each other only in the env gene.
Construction of chimeric and mutant env genes.
We selected one clone from the parental CC1/85 isolate (CC1/85 cl.7) and one from the AD101-resistant CC101.19 isolate (CC101.19 cl.7) for more detailed studies of genotype-phenotype associations. To generate constructs with chimeric env genes, pBluescript KS(+) plasmids containing these env fragments were cut with StuI and BsaBI. The StuI-BsaBI fragment from CC101.19 cl.7 was then ligated into the vector fragment from CC1/85 cl.7 that had been cut with the same enzymes. This chimeric construct is referred to as CC1/85 cl.7(8), the number 8 signifying that it differs from CC1/85 cl.7 by 8 amino acid changes. The reverse chimeric env gene, with the StuI-BsaBI fragment from CC1/85 cl.7 inserted into CC101.19 cl.7, was also constructed and is referred to as CC101.19 cl.7(8).
Site-directed mutagenesis was performed with the QuickChange mutagenesis kit as specified by the manufacturer (Stratagene), using the pBluescript KS(+) plasmids containing the EcoRI-XhoI fragments. EcoRI-XhoI fragments were then subcloned into pNL4-3.
Cells, viruses, and viral replication assays.
PBMC were pooled from the blood of four healthy volunteers. CD8+ T cells were removed using the RosetteSep CD8+ depletion cocktail as specified by the manufacturer (StemCell Technologies, Vancouver, Canada). The CD8+ T-cell-depleted PBMC (referred to hereafter as primary CD4+ T cells) were then activated and used for HIV-1 replication assays 3 days later (108). Primary CD4+ T cells were used to limit the interassay variability of PBMC replication assays, but similar results were obtained without CD8+ T-cell depletion (data not shown).
Clonal, replication-competent, chimeric NL4-3/env viruses were prepared by transfecting the full-length proviral plasmid into 293T cells by using Lipofectamine 2000, as specified by the manufacturer (Invitrogen). Supernatants were collected at 48 h posttransfection, filtered, and stored at −80°C. Replication assays were performed with PBMC as described for standard HIV-1 isolates (108), except that 100 50% tissue culture infective doses (TCID50) of virus were used per well of a 96-well plate.
Expression and purification of recombinant gp120.
Recombinant gp120 proteins from clones CC1/85 cl.6 and cl.7, CC101.19 cl.3, CC101.19 cl.7, and CC101.6 cl.10 were transiently expressed from pPPI4-gp120 expression vectors. These were constructed by subcloning the KpnI-BbvCI fragments from the desired env gene into the pPPI4-JR-FL gp140 vector (13). Two consecutive in-frame stop codons were then introduced by QuickChange mutagenesis (Stratagene) immediately following the lysine in the sequence REKR, the natural cleavage site between gp120 and gp41. Other than in the gp120 coding sequences, the resulting pPPI4 vectors were identical to that used to express JR-FL gp120 (107). The gp120 proteins were expressed and purified as previously described (13, 107). Their concentrations were measured by the DC modified Lowry protein assay (Bio-Rad, Hercules, Calif.). Their purity was >90% as assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Binding of gp120 to CCR5.
Capture enzyme-linked immunosorbent assays for gp120 binding to sCD4 and MAb 17b were performed as described elsewhere (76). To detect gp120 binding to cell surface CCR5, L1.2-CCR5 cells were incubated with both purified gp120 (100 μg/ml) and biotinylated CD4-IgG2 (100 μg/ml) for 1 h at 37°C (34, 111). Bound CD4-IgG2 was then detected by staining for 30 min at 25°C with phycoerythrin-conjugated streptavidin (BD BioSciences, San Jose, Calif.). After being washed the cells were fixed in paraformaldehyde and analyzed by flow cytometry.
Nucleotide sequence accession numbers.
The 243 sequences that were deemed valid have been submitted to GenBank (accession numbers AY357338 through AY357580).
RESULTS
Changes in Env are responsible for CCR5 inhibitor resistance.
We reported previously that a primary R5 isolate, CC1/85, became increasingly resistant to the CCR5 inhibitor AD101 on passaging in PBMC cultures in the presence of increasing AD101 concentrations. After 19 passages, AD101 resistance was substantial (>20,000-fold) (108). To confirm that resistance was conferred by changes in the viral envelope glycoproteins, we cloned env genes from the resistant virus (CC101.19) and used them to replace the env gene of the infectious molecular clone, HIV-1 NL4-3. Similarly, we made chimeric NL4-3 viruses by using env genes cloned from the parental virus, CC1/85, to serve as controls. Seven clonal, chimeric infectious viruses were chosen for further study: CC101.19 cl.3, CC101.19 cl.7, and CC101.19 cl.15 from the passage 19 escape mutant isolate, CC101.19; CC1/85 cl.6, CC1/85 cl.7, and CC1/85 cl.8 from the parental isolate, CC1/85; and CCcon.20 cl.11 from the passage 20 control isolate, CCcon.20.
The clonal viruses were confirmed to be replication competent in both PBMC and primary CD4+ T cells. Their titer in primary CD4+ T cells on day 12 postinfection ranged from 104 to 105 TCID50/ml, when p24 production was typically 5 to 10 ng/ml. There was no major difference in the replication rates of the CC1/85 or CC101.19 clones in primary CD4+ T cells, and the CC101.19 clones did not require AD101 for replication (data not shown). Although we have not yet performed formal fitness studies, the AD101 escape mutant isolates and clones appear to be fully replication competent in primary CD4+ T cells.
The parental and escape mutant clones faithfully recapitulated the phenotypes of the corresponding isolates. Thus, in a representative experiment, all three clones derived from CC101.19 were not inhibited by AD101 even at concentrations as high as 20 μM whereas all three clones from the parental isolate CC1/85 were sensitive to AD101, with an average 50% inhibitory concentration (IC50) of ∼2 pM (Fig. 1a). Hence in this experiment, the extent of resistance for the CC101.19-derived clones, compared to clones from CC1/85, was >107-fold. For all practical purposes, the CC101.19 clones are therefore completely resistant to the selecting compound. Clone CCcon.20 cl.11 from the passage 20 control isolate also retained the AD101-sensitive phenotype (data not shown).
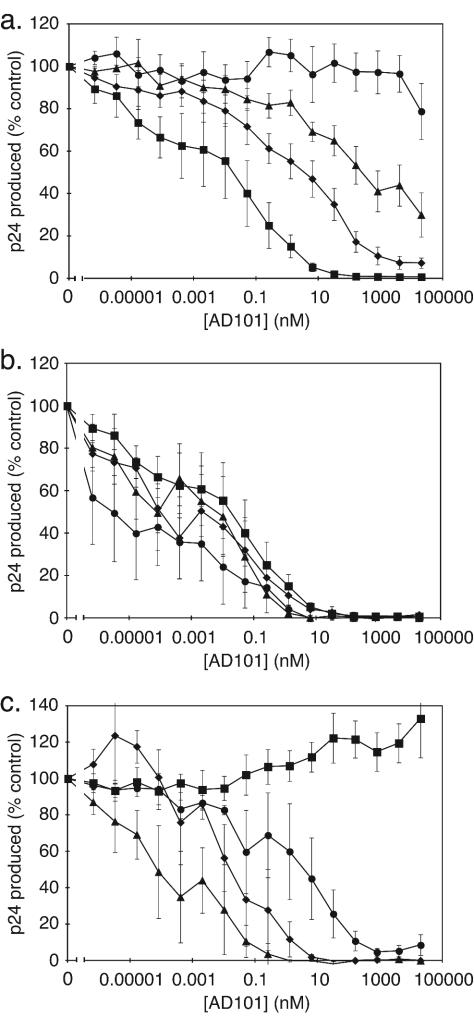
FIG. 1.
Infectious molecular clones recapitulate the phenotype of the AD101-resistant escape mutant CC101.19. (a) Chimeric molecular clones containing env genes derived from the parental CC1/85 isolate (open symbols) or the passage 19 escape mutant (solid symbols) were cultured in primary CD4+ T cells in the presence of the AD101 concentrations indicated, and the extent of HIV-1 replication was determined. The individual clones are designated CC1/85 cl.6 (open squares), CC1/85 cl.7 (open circles), CC1/85 cl.8 (open triangles), CC101.19 cl.3 (solid squares), CC101.19 cl.7 (solid circles), and CC101.19 cl.15 (solid triangles). (b to d) Four of the above clones (same symbols) were also tested for sensitivity to SCH-C (b), RANTES (c), and PA14 (d). Panel a displays data from a single representative experiment. Panels b to d show data averaged from three independent experiments, with the error bars indicating the standard error of the mean (SEM).
The CC101.19 escape mutant isolate is completely cross-resistant to SCH-C (108), a small-molecule CCR5 inhibitor chemically related to AD101 (82, 83, 101, 103, 104, 108). We therefore determined if the derived clones CC101.19 cl.3 and cl.7 were also cross-resistant. Neither clone was inhibited by SCH-C at the highest concentration tested (20 μM), whereas both parental clones, CC1/85 cl.6 and cl.7, were sensitive to SCH-C, with IC50s in the range 30 to 100 nM (Fig. 1b). The escape mutant clones were therefore completely cross-resistant to SCH-C, whereas the parental isolate clones retained the sensitivity of the input CC1/85 virus.
To see if the resistance of the CC101.19 clones to the small-molecule inhibitors reflected a more general resistance to CCR5-targeted agents, we evaluated their sensitivity to the CC chemokine RANTES and to a murine anti-CCR5 MAb, PA14 (81). We previously showed that the CC101.19 isolate was ∼10-fold more resistant to RANTES than was the parental CC1/85 isolate (108). However, the CC101.19 clones were at least as sensitive as the CC1/85 clones to RANTES (Fig. 1c). The CCR5 epitope for MAb PA14 has been mapped to extracellular loop (ECL) 2 and the N terminus (NT) of CCR5 (81). The CC101.19 isolate was ca. fivefold less sensitive than CC1/85 to PA14 (108). The pattern of sensitivities of the corresponding CC1/85 and CC101.19 clones toward PA14 was similar to what was observed with RANTES, in that all the clones were inhibited by similar PA14 concentrations (Fig. 1d). Hence, escape from the small-molecule inhibitor AD101 does not involve the acquisition of strong resistance to other classes of entry inhibitors that also target CCR5. This conclusion applies to both the CC101.19 isolate (108) and individual clones derived from it (Fig. 1). Any phenotypic differences between the CC101.19 isolate and its derivative clones are minor; presumably some CC101.19 clones will be modestly resistant to RANTES and PA14, just as the isolate is (108). It is also possible that minor differences in phenotype observed only with the isolates, and not with the clones, map to determinants outside the env gene, such as Gag regions that might influence Env incorporation into the chimeric NL4-3-based viruses.
The sensitivities of the clonal escape mutant viruses to RANTES and PA14 implies that, like the escape mutant isolate and the parental CC1/85 virus, they have an absolute dependence on CCR5 for entry (108). We confirmed this by showing that none of the NL4-3/env clones could replicate to a detectable extent in PBMC from a CCR5-Δ32 homozygous individual whereas all the clones replicated efficiently in PBMC from CCR5 wild-type donors (data not shown).
Env sequence evolution associated with the generation of AD101 resistance.
Multiple passages in PBMC culture were required to generate an escape mutant with full resistance to AD101 (108). Thus, the CC101.19 isolate acquired over 20,000-fold resistance to AD101 after 19 weekly passages. Partial resistance was detected sooner; after six passages, the CC101.6 isolate was approximately fourfold less sensitive than CC1/85 to AD101 (108). The full escape mutant had a stable phenotype, since AD101 resistance survived nine further passages in PBMC culture in the absence of AD101 (108). These observations, taken together, suggest that multiple, sequential genetic changes had conspired to create full resistance to AD101 (108). To learn how escape had occurred, we studied Env sequence evolution by using samples of virus-infected cells frozen from each stage of the long-term PBMC culture and its extension in the absence of AD101.
Multiple sequence changes had accumulated in CC1/85 gp120 over time, but gp41 changes were rare and inconsistent (see below). In a phylogenetic analysis, sequences from the 4-, 10-, and 20-week passage control viruses (CCcon.4, CCcon.10, and CCcon.20, respectively) clustered with the parental CC1/85 sequences and had a similar degree of diversity (Fig. 2). In contrast, the viruses isolated under the AD101 selection pressure branched off to form a distinct lineage with very little internal sequence diversity (Fig. 2). Sequences from escape mutant viruses cultured for nine further passages without AD101 were clustered within the same lineage, suggesting that they had changed little during the extension to the culture (Fig. 2).
FIG. 2.
Phylogenetic analysis of gp120 sequences from CC1/85 and related viruses. Sequence diversity is represented by horizontal distance. CC1/85 sequences are represented by black bars. Sequences from the CCcon.4 (passage 4), CCcon.10 (passage 10), and CCcon.20 (passage 20) isolates are shown by blue bars, with the darker bars denoting the later isolates. The sequences from viruses isolated under AD101 selection pressure are represented in yellow, orange, and red. The sequences in green represent in vivo diversification that generates the CC2/86, CC7/86, and CC12/86 isolates. The CC101.22R2, CC101.22R5, and CC101.22R9 sequences in different shades of purple are derived from escape mutant viruses cultured in the absence of AD101 for a further nine passages.
For comparison, we also analyzed gp160 sequences from viruses that had diversified in vivo from the same genetic lineage. The parental virus for the escape mutant study, CC1/85, was isolated in January 1985 from an individual who, at that time, harbored only R5 strains (23, 24). Three isolates obtained 13, 18, and 23 months later had the R5X4 phenotype and are designated CC2/86, CC7/86, and CC12/86 (23, 24). A precipitous decline in CD4+ T-cell counts occurred during this 23-month period (23, 24). Sequences from these R5X4 isolates showed a gradual progression to new forms but on a lineage distinct from those of CC1/85 and the escape mutants selected in vitro (Fig. 2).
Sequence changes in the V3 loop during the development of AD101 resistance.
Inspection of the predicted gp160 sequences from throughout the course of AD101 selection revealed a notable pattern of evolution within the gp120 V3 region (Fig. 3). One clone from the eight sequenced from CC1/85 contained a single amino acid polymorphism; a proline residue was present instead of the predominant histidine at position 308 (H308P) in the N-terminal side of the V3 loop. By passage 2 in the presence of AD101 (the CC101.2 clones), only a proline was found at position 308; the originally dominant histidine was completely absent (Fig. 3). With the exception of a few minor polymorphisms, the V3 sequence of the passage 2 clones then remained unchanged until passage 8, when an additional polymorphism (K305R) emerged nearby (Fig. 3). This change, observed in 5 of the 19 sequenced CC101.8 clones, presumably arose by de novo mutation, since arginine was not present at position 305 in any of the 41 clones from earlier time points. By passage 10, 12 of 13 clones contained arginine at position 305, and after pass as 12, arginine was the only amino acid found at this position (Fig. 3). Two more changes were observed in passage 14 clones, A316V and G321E, again presumably arising by mutation. V3 sequences carrying all four substitutions dominated from passage 16 onward and were the only ones present by passage 18. Although other scattered substitutions occurred in the V3 loop and throughout gp160, none of them showed this pattern of stabilization under selection (Fig. 3 and 4 and data not shown).
FIG.3.
V3 sequence alignments of Env clones. The first line shows the consensus V3 sequence of the CC1/85 isolate based on eight clones. The locations of specific amino acids and their corresponding amino acid numbers, based on the HXBc2 sequence, are shown above the consensus line. There are five blocks of sequence data below the consensus line. Each block includes the following information (from left to right): N, the number of clones from a given isolate having the indicated V3 sequence; the source isolate, i.e., the isolate, based on the nomenclature described in the text, from which the clones were generated; and the sequence. The sequence is shown in reference to the consensus, with a dash representing identity to the consensus. Where one or more clones differ from the consensus, the amino acid found in that clone or clones is shown. Block 1 contains the eight clones from the CC1/85 isolate, seven of which are identical to the consensus and one of which contains the H308P polymorphism. This polymorphism and others discussed in the main text are highlighted in red. Block 2 contains select isolates generated from the cultures treated with AD101; the clones are shown first in order of the isolate from which they were derived and then in order of the frequency at which each clone is present in that isolate. Block 3 contains clones derived from isolates cultured for 22 passages before AD101 was removed for an additional 9 passages. Block 4 contains clones from three control isolates passaged in the absence of AD101. Block 5 contains clones from isolates derived from patient Case C at 13 (CC2/86), 18 (CC7/86), and 23 (CC12/86) months after the CC1/85 isolate. The $ symbol in one CC101.14 clone indicates the position of a premature stop codon.
FIG. 4.
Alignments of the consensus gp160 sequences from critical time points in the AD101 escape process. The consensus amino acid sequence from the parental isolate, CC1/85, is given on the top line. Capital letters indicate that there was identity among all eight of the aligned clones. Lowercase letters indicate the amino acid sequence of most but not all of the clones. The question mark indicates either that there was a tie between amino acids at a given position or that the majority of clones had a gap at this position. A gap found in all clones is indicated by a dot. Aligned with the CC1/85 consensus are the consensus sequences from isolates from passages 4, 10, and 20 (CC101.4, CC101.10, and CC101.20, respectively), derived under AD101 selection pressure. Also depicted are consensus sequences from the CC101.22R9 isolate, which was cultured in the absence of AD101 for 9 passages after 22 passages in the presence of AD101; the passage control CCcon.20 isolate, which was cultured for 20 passages in the same PBMC as the AD101-treated cultures, but in the absence of drug; and CC12/86, which was isolated from individual Case C 23 months after the CC1/85 isolate was obtained. If the aligned sequence is identical to that of the CC1/85 consensus line, this is indicated by a dash; otherwise, the symbols described above are used to depict differences from the consensus sequence. Amino acids highlighted in red are those that were selected for in the AD101-treated cultures but not in the control cultures or in vivo. Amino acids highlighted in green are those that were selected for in both the AD101-treated and control cultures. Amino acids highlighted in blue are those that were selected for in the AD101-treated cultures and in vivo but not in the control cultures. The numbering system is based on that of HXBc2. The amino acid under the "0” of each number is the one intended to be indicated by the label. The variable and constant regions of gp120, and the gp120-gp41 cleavage site, are also indicated. The outlined amino acids are those which have been implicated in the binding of, or neutralization by, MAb 17b (59, 94, 105, 118). Amino acids on a gray background are those implicated in forming part of a conserved coreceptor binding site that partially overlaps the epitope for 17b (93, 94).
The H308P polymorphism was present in three of eight sequences from the CC2/86 R5X4 isolate (Fig. 3). The A316V substitution was absent from CC1/85 sequences but was evident in sequences from the CC2/86, CC7/86, and CC12/86 R5X4 isolates (Fig. 3). There, it appears in the context of a very different V3 loop sequence and in a different lineage in the phylogenetic tree with strong bootstrap support (Fig. 2 and 3). Hence, it most probably arose independently both in vivo and in the in vitro escape lineage.
None of the above four V3 changes was ever seen in clones of the control isolates from passages 4, 10, and 20, except for the H308P polymorphism, which was present in 1 of 11 clones from the CCcon.10 control isolate (Fig. 3). The latter observation is consistent with the hypothesis that H308P is a minor, preexisting CC1/85 variant that was rapidly selected for in the presence of AD101 but was not under selection pressure in the control cultures. The V3 sequences from the passage 20 control clones also lacked the four V3 changes, although another V3 polymorphism, F317L, was present in 9 of 12 CCcon.20 clones (Fig. 3). This particular polymorphism was absent from clones from CC1/85 or the other passage control. isolates (Fig. 3). Its significance, if any, is unclear.
The four amino acid changes in V3 remained the dominant form in the escape mutant viruses that had been cultured for 22 passages in the presence of AD101 and then for up to 9 additional passages without AD101 (designated CC101.22RX), although some variants were evident as minor forms (Fig. 3). Clearly, as with the AD101 escape phenotype itself (108), the genetic changes associated with it are stable.
Sequence evolution elsewhere in gp160 during the evolution of AD101 resistance.
Other sequence changes occurred elsewhere in Env as AD101 resistance developed (Fig. 4). These changes arose with a different time course from the V3 substitutions; they typically became fixed in the AD101-treated cultures at or before passage 4, so their occurrence did not correlate with the development of complete resistance to AD101. It is, however, possible that some non-V3 changes may play a role in the fourfold resistance to AD101 possessed by the CC101.6 isolate (108). Most of the non-V3 changes are likely to have been rapidly selected for from the input virus population; many, but not all, of them were present as minor variants in CC1/85 clones (Fig. 4 and data not shown).
Several distinct patterns of selection can be seen by inspection of the alignments of the consensus sequences from critical time points (Fig. 4). Three such patterns are as follows. First, some sequences were selected for in the AD101-treated cultures but not in the control culture (CCcon.20) or the in vivo isolates (CC12/86). These sequences are shown in red in Fig. 4. Twelve of these changes are in gp120, and seven in gp41. Of those in gp120, two were found in the V1 loop, three were found in the V3 loop, three were found in the C3 region, and four were found in the V4 loop. All the amino acid changes, including H308P, that arose in the AD101-treated cultures but not in the control cultures or in vivo, were selected for by passage 4 or earlier, with the exception of the K305R and G321E substitutions in the V3 loop (see above). Another of these variants is part of a 4-residue insertion, VTNN, in V1 between amino acids 136 and 137. This insertion adds a new N-linked glycosylation site at position 136. In contrast, an N-to-K change at the first amino acid of V4 eliminates an N-linked glycosylation site. The seven changes in gp41 are scattered throughout the protein and are mostly conservative substitutions (Fig. 4).
The second pattern observed, depicted in green in Fig. 4, involves amino acid changes that were selected for in both the AD101-treated and control cultures. Some of these are also present in the in vivo sequences. Of the changes that meet this criterion, one each can be found in C2, C3, and V4 while four are present in V5 and four more are present in gp41 (Fig. 4). The only substitution in this group that was not stabilized in the AD101-treated cultures by passage 4 was D167N (Fig. 4). This amino acid is polymorphic in the CC1/85 isolate and remains so throughout the AD101 selection process. An Asn residue gradually becomes more prevalent at this position but does not become dominant until passage 20. Given the slowness with which it emerged and given that it is also selected for in the control cultures, the D167N change is not likely to be involved in the development of AD101- resistance (Fig. 4 and data not shown).
The final pattern seen in the consensus alignments, shown in blue in Fig. 4, involves amino acids that were selected for both in the AD101-treated culture and in vivo but not in the control culture. All these changes also become fixed in the AD101-treated cultures by passage 4, with the exception of the A316V substitution in V3 (see above). Four of them occur in V1, three as part of the 4-amino-acid insertion described above, one occurs in C3, four occur in V4, and one occurs in gp41 (Fig. 4).
Overall, all the amino acid changes that occurred under AD101 selection pressure, regardless of the pattern into which they fall, remained the dominant sequences in the CC101.22R9 culture (Fig. 4). Hence they were stable after the AD101 selection pressure was withdrawn. Unlike many of the gp120 substitutions, those within gp41 were conservative and did not involve charge changes or alterations in glycosylation sites. The V4 and V5 regions were highly polymorphic in the CC1/85 isolate but became fixed to essentially invariance by passage 4. In contrast, while sequences from the control culture also lost some variability in these regions, they did so to a much lesser extent. The in vivo sequences also remained highly polymorphic in V4 and less so in V5 (Fig. 4 and data not shown).
Finally, and of particular significance, the selection pressure of AD101 caused no detectable sequence variation in the amino acids from the C4 region and the stem of the V1/V2 loops that together comprise the bridging sheet of the gp120 core, a structure which represents the most highly conserved element of the CCR5 binding site (59, 93, 94) (Fig. 4). Mutagenesis studies and the crystal structure of the complex formed between the 17b Fab fragment, the conserved core of gp120, and the first two domains of CD4 together indicate that the CD4-induced epitope for MAb 17b is formed from gp120 elements that overlap the conserved CCR5 binding site (59, 94, 105, 118). The 17b epitope was also unchanged in the AD101 escape mutant clones (Fig. 4).
Changes in the V3 loop of CC1/85 confer AD101 resistance.
The availability of NL4-3/env chimeras that reproduced the AD101-resistant and -sensitive phenotypes allowed us to investigate the genetic basis for AD101 resistance. We noted, however, that individual CC1/85 env clones were extremely sensitive to AD101 inhibition, significantly more so than the corresponding isolates. The average IC50 for a single clone, CC1/85 cl.7, over four independent experiments, was ∼4 pM (Table 1). In contrast, the average IC50 for the CC1/85 isolate was ∼1 nM, 250-fold greater (108). The difference was not a general property of the clonal virus stocks, because they were no more sensitive than the CC1/85 isolate to inhibition by SCH-C, RANTES, or PA14 (Fig. 1) (108). Instead, it presumably reflects the method and other differences involved in working with isolates and NL4-3/env chimeric clones (see below). Hence, absolute values for the fold resistance of clones to AD101 should not be directly compared with values for the corresponding isolates. Fold resistance values for clones may, however, be compared with values for other clones, and those for isolates may be compared with values for other isolates.
TABLE 1.
IC50s for AD101 inhibition of NL4-3/env chimeric virus replication in primary CD4+ T cells
| env clone, chimera, or mutant | AD101 IC50 (nM)a | Relative fold differenceb |
|---|---|---|
| CC1/85 cl.7 | 0.004 | 1 |
| CC101.19 cl.7 | >20,000 | >5 × 106, > |
| CC1/85 cl. 7(8) | >20,000 | >5 × 106, > |
| CC101.19 cl.7(8) | 0.6 | 150, > |
| CC1/85 cl.7(H308P) | 2 | 500, > |
| CC1/85 cl. 7(K305R, H308P) | 400 | 100,000, > |
| CC1/85 cl.7(K305R, H308P, A316V, G321E) | >20,000 | >5 × 106, > |
| CC1/85 cl.7(K305R) | 0.0004 | 10, < |
| CC1/85 cl.7(A316V, G321E) | 0.0006 | 7, < |
| CC1/85 cl.7(K305R, A316V, G321E) | 0.000006 | 700, < |
| CC101.19 cl.7(R305K, P308H, V316A, E321G) | 0.02 | 5, > |
| CC101.19 cl.7(P308H) | 0.0002 | 20, < |
| CC101.19 cl.7(R305K, P308H) | 1 | 250, > |
Approximate IC50s for AD101 inhibition curves were calculated by fitting to a sigmoidal dose-response curve by nonlinear regression using the program Prism (GraphPad Software) for the average data sets shown in Fig. 4 and 5.
Relative fold difference in IC50 for AD101 compared to CC1/85 cl.7, which is used as a reference. > and < indicate that IC50 is respectively greater or less than that for CC1/85 cl.7.
The pattern of Env sequence evolution described above suggested that genetic analyses should focus on the V3 loop. Although amino acid changes in other regions of gp120 and gp41 do become fixed early in the AD101 selection process (Fig. 4), only V3 amino acids were under active selection at times that correlated with the acquisition of complete AD101 resistance. Moreover, there is ample precedent for the V3 loop influencing the HIV-1 phenotype, including coreceptor choice (20-22, 31, 33, 39, 47, 48, 100, 114).
We therefore made chimeric env genes in which the coding region for amino acids 271 to 386 of the parental CC1/85 virus (clone CC1/85 cl.7) was replaced with the corresponding segment of the fully resistant AD101 escape mutant clone; CC101.19 cl.7. This chimera is referred to as CC1/85 cl.7(8). The exchanged fragment encodes part of the C2 region, the V3 and C3 regions, and the first amino acid of V4. The designation “8” in the chimera identifier signifies that there are eight amino acid changes between the clones in the region swapped. Listing the CC1/85 cl.7 amino acid first, the changes were I271V in C2; K305R, H308P, A316V, and G321E in V3; N337Q and E351K in C3; and N386K, the first amino acid of the V4 loop (Fig. 4; Table 2). The other three C3 polymorphisms (R343K, Q344H, and G379R) that were selected for in the AD101-treated cultures were already present in CC1/85 cl.7. Hence, they are invariant among the various chimeras and clones now under study (Fig. 4, Table 2). A reciprocal chimera in which the corresponding region from CC1/85 cl.7 was inserted into CC101.19 cl.7 is referred to as CC101.19 cl.7(8). The amino acid changes in the parental and chimeric env clones are summarized in Table 2.
TABLE 2.
Comparison of amino acids exchanged in the CC1/85 cl.7(8) and CC101.19 cl.7(8) Env chimeras
| Env | Amino acid identitya at following position and location
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 271, C2 | 305, V3 | 308, V3 | 316, V3 | 321, V3 | 337, C3 | 343, C3 | 344, C3 | 351, C3 | 379, C3 | 386, V4 | |
| CC1/85 consensus | I | K | H | A | G | N | R | Q | E | G | N |
| CC101.19 consensus | V | R | P | V | E | Q | K | H | K | R | K |
| CC1/85 cl.7(8) | I | K | H | A | G | N | K | H | E | R | N |
| CC101.19 cl.7(8) | V | R | P | V | E | Q | K | H | K | R | K |
The amino acids which differ between CC1/85 cl.7(8) and CC101.19 cl.7(8) are highlighted in bold type.
NL4-3-based viruses containing the above env genes were used to generate clonal virus stocks, which were then tested for AD101 sensitivity in replication assays using primary CD4+ T cells (Fig. 5). The CC101.19 cl.7(8) chimeric virus was highly sensitive to AD101 (IC50, ∼0.6 nM), whereas CC1/85 cl.7(8) resisted the highest AD101 concentration tested (20 μM) (Fig. 5; Table 1). In the context of these two env clones, complete resistance to AD101 therefore maps to the interchanged gp120 fragment spanning amino acids 271 to 386. The CC101.19 cl.7(8) chimera was ∼150-fold more resistant than CC1/85 cl.7 to AD101, with the average IC50s being ∼600 and ∼4 pM, respectively (Fig. 5; Table 1). Hence, changes in gp120 outside of residues 271 to 386 do make some contribution to AD101 resistance, but they cannot account for the complete resistance of the CC101.19 clones (Fig. 5; Table 1).
FIG. 5.
AD101 sensitivity of NL4-3/env chimeric viruses with interchanged C2, V3, and C3 regions. The AD101-sensitive clones CC1/85 cl.7 (squares) and AD101-resistant CC101.19 cl.7 (diamonds) and the chimeric clones CC1/85 cl.7(8) (triangles) and CC101.19 cl.7(8) (circles) were all tested for AD101 sensitivity in an assay of HIV-1 replication in primary CD4+ T cells. The results depicted are the average of four or five independent experiments, with the error bars indicating the SEM.
Site-directed mutagenesis of specific residues in the V3 loop.
To define which specific amino acids within residues 271 to 386 can confer complete resistance to AD101, we used the AD101-sensitive clone CC1/85 cl.7 as a basis for site-directed mutagenesis. This clone was chosen because, while it lacks many of the Env substitutions that were apparently selected for during the early passages with AD101 (i.e., prior to passage 4), including the H308P polymorphism in the V3 loop, it is the most closely related of the eight CC1/85 clones to the ones selected for under pressure from AD101 (data not shown). We first changed one or more of the four amino acids that differed between the V3 regions of CC1/85 cl.7 and the AD101-resistant clone, CC101.19 cl.7. One such mutant, CC1/85 cl.7(K305R, H308P, A316V, G321E), differs from CC1/85 by possessing all four of the substitutions in the V3 loop highlighted by the sequence analysis (Fig. 3). The CC1/85 cl.7(K305R), CC1/85 cl.7(H308P), CC1/85 cl.7(K305R, H308P), CC1/85 cl.7(A316V, G321E), and CC1/85 cl.7(K305R, A316V, G321E) env genes were also made; they differ from CC1/85 cl.7 at one, two, or three of the above V3 residues.
The mutated coding sequences were reconstituted into replication-competent viruses, and replication assays were performed with primary CD4+ T cells in the presence of increasing AD101 concentrations (Fig. 6a and b). As noted above, the IC50 for AD101 inhibition of CC1/85 cl.7 was, on average, 4 pM. Consistent with our earlier studies of the viruses bearing chimeric env genes (Fig. 5; Table 1) and the sequence evolution analysis (Fig. 3), the quadruple mutant CC1/85 cl.7(K305R, H308P, A316V, G321E) was completely resistant to AD101, even at 20 μM (Fig. 6a; Table 1). However, the single H308P change was alone sufficient to confer ∼500-fold resistance upon CC1/85 cl.7, with the IC50 being ∼2 nM (Fig. 6a; Table 1). A further ∼200-fold resistance to AD101 was created by adding the K305R change to the CC1/85 cl.7(H308P) virus to make the CC1/85 cl.7(K305R, H308P) double mutant (Fig. 6a; Table 1). Clearly, the two further changes (A316V and G321E) in the quadruple mutant generate yet further resistance (Fig. 6a; Table 1). Four amino acid changes in the V3 loop were therefore sufficient to convert the AD101-sensitive CC1/85 virus into one that was completely resistant to AD101.
FIG. 6.
Effect of specific amino acid substitutions in the V3 loop on sensitivity to AD101. (a) The parental CC1/85 cl.7 virus (squares) and mutants CC1/85 cl.7(H308P) (diamonds), CC1/85 cl.7(K305R, H308P) (triangles), and CC1/85 cl.7(K305R, H308P, A316V, G321E) (circles) were all tested for AD101 sensitivity in an assay of HIV-1 replication in primary CD4+ T cells. (b) Three additional mutants derived from CC1/85 cl.7 (squares) were also tested for AD101 sensitivity: CC1/85 cl.7(K305R) (diamonds), CC1/85 cl.7(A316V, G321E) (triangles), and CC1/85 cl.7(K305R, A316V, G321E) (circles). (c) Reverse mutants made in the context of the AD101-resistant parental clone CC101.19 cl.7 (squares) were evaluated: CC101.19 cl.7(R305K, P308H, V316A, E321G) (diamonds), CC101.19 cl.7(P308H) (triangles), and CC101.19 cl.7(R305K, P308H) (circles). The results depicted in all panels are the average of four or five independent experiments, with the error bars indicating the SEM.
The sequence analysis revealed that the K305R change and the later A316V and G321E changes arose in the context of a V3 loop in which the H308P substitution was already present (Fig. 3). We therefore determined whether the H308P substitution was important for the action of the later changes. Thus, the K305R, A316V, and G321E changes were introduced into CC1/85 cl.7 in the absence of H308P (Fig. 6b; Table 1). The mutant clones were all strongly inhibited by AD101 (Fig. 6b; Table 1). Indeed, the CC1/85 cl.7(K305R) and CC1/85 cl.7(A316V, G321E) viruses were slightly (∼10-fold) more sensitive than was CC1/85 cl.7 and the triple mutant CC1/85 cl.7(K305R, A316V, G321E) was significantly more sensitive (∼700-fold). Hence, the phenotypic influence of the K305R, A316V and G321E changes was strictly context dependent, requiring the presence of proline at residue 308. The effect of the three later-arising changes clearly differed from that of H308P. They did not themselves impart any AD101 resistance to the parental CC1/85 cl.7 clone, but they did markedly potentiate the effect of H308P by conspiring with it to impart even greater resistance to AD101.
Reverse mutagenesis confirms the role of the V3 loop in conferring AD101 resistance.
The above experiments clearly showed that four amino acid changes in V3 were sufficient to confer complete AD101 resistance on CC1/85 cl.7. We next performed reverse mutagenesis to determine whether these same changes were necessary for complete resistance. The four critical V3 residues in the AD101-resistant clone, CC101.19 cl.7, were therefore replaced with the cognate amino acids from the AD101-sensitive virus, CC1/85 cl.7 (Fig. 6c; Table 1). The quadruple mutant CC101.19 cl.7(R305K, P308H, V316A, E321G) was fully sensitive to AD101, with an IC50 (∼20 pM) only ca. fivefold greater than that of CC1/85 cl.7. Two additional substitutions introduced into the CC101.19 cl.7 background, CC101.19 cl.7(P308H) and CC101.19 cl.7(R305K, P308H), also caused an increase in AD101 sensitivity relative to the parental, AD101-resistant clone (Fig. 6c; Table 1). Hence, the four V3 loop changes in CC1/85 that were highlighted by the sequence analysis (Fig. 3) were, in fact, critical for generating AD101 resistance (Fig. 6c; Table 1).
Sequences outside the V3 loop influence the extent of AD101 resistance.
Sequences outside the V3 loop can modify the way in which sensitivity to AD101 is affected by changes within the loop. For example, the CC101.19 cl.7(P308H) mutant was more sensitive to AD101 than were the CC101.19 quadruple mutant and the parental clone, CC1/85 cl.7 (Fig. 6c; Table 1). Hence, in the context of CC101.19 cl.7, the single change P308H was alone sufficient to create a highly AD101-sensitive virus. Of note is the observation that the CC101.19 cl.7(P308H) and CC1/85 cl.7(K305R, A316V, G321E) viruses have identical V3 loops in different Env backgrounds. Both clones were very sensitive to AD101, more so than the parental clones containing the CC1/85 consensus amino acids at all four V3 positions. The difference in sensitivity is most apparent when comparing the IC50 for CC1/85 cl.7 with that for CC1/85 cl.7(K305R, A316V, G321E) and when comparing the IC50 for CC101.19 cl.7(R305K, P308H, V316A, E321G) with that for CC101.19 cl.7(P308H) (Fig. 6b and c; Table 1). Hence, the V3 sequence containing R305, H308, V316, and E321 seems able to confer the phenotype of increased sensitivity to AD101, irrespective of its context, whereas the H308P change yields either a fully or a partially resistant phenotype depending on the context into which it is introduced.
The other mutant made in the background of CC101.19 cl.7, CC101.19 cl.7(R305K, P308H), underscores the influence of regions outside the V3 loop in determining the AD101 resistance phenotypes of clonal viruses bearing mutant env genes, as follows. The CC1/85 cl.7 virus and CC101.19 cl.7 quadruple-mutant virus have identical V3 loops that contain the CC1/85 consensus amino acids at all four V3 positions: K305, H308, A316, and G321. As in the above example, these clones serve as the reference viruses for the two mutants, CC1/85 cl.7(A316V, G321E) and CC101.19 cl.7(R305K, P308H). The two mutants have identical V3 loops, each containing the V316 and E321 substitutions. However, these mutants have very different phenotypes from the reference clones from which they were derived. Thus, the CC101.19 cl.7(R305K, P308H) mutant was ∼50-fold less sensitive to AD101 than was the CC101.19 cl.7 quadruple mutant (Fig. 6c; Table 1), whereas CC1/85 cl.7(A316V, G321E) was slightly (∼7-fold) more sensitive than CC1/85 cl.7 to AD101 (Fig. 6c; Table 1).
Clearly, then, while the V3 loop appears to be the critical determinant of complete AD101 resistance in these two env clones, the V3 sequences do not act alone when influencing the relative sensitivity of specific variants to AD101; the way in which the V3 sequences interact with other regions of gp120 is also important.
The above results demonstrate that, taken together, the V3 loop changes at positions 305, 308, 316, and 321 are both necessary and sufficient for complete AD101 resistance in the backgrounds of both CC1/85 cl.7 and CC101.19 cl.7. Differences in how specific amino acid substitutions act in different mutant viruses, alone and in combination, probably reflect differences in the genetic backgrounds of CC101.19 cl.7 and CC1/85 cl.7 and the consequent influence of residues in regions outside V3 that vary between the two Env proteins.
Receptor binding properties of gp120 monomers derived from AD101-sensitive and -resistant viruses.
To gain initial insights into which alterations in the properties of Env were created by the genetic changes associated with AD101 resistance, we expressed and purified monomeric gp120 proteins from AD101-sensitive and -resistant clones. In a standard gp120 capture ELISA (76), the 50% effective concentrations (EC50s) for half-maximal CD4-IgG2 binding to gp120 were as follows: JR-FL, 60 ng/ml; CC1/85 cl.6, 69 ng/ml; CC1/85 cl.7, 40 ng/ml; CC101.19 cl.3, 56 ng/ml; CC101.19 cl.7, 94 ng/ml; CC101.6 cl.10, 75 ng/ml. The EC50s for 17b binding ranged from 33 to 190 ng/ml in the absence of sCD4 and from 6.1 to 9.3 ng/ml in the presence of 1 μg/ml sCD4, the induction of 17b binding by sCD4 being from 4- to 20-fold (data not shown). Thus in the presence of sCD4, all the CC1/85 and CC101.19 gp120s bind 17b in a similar manner. Hence all the expressed gp120 proteins were folded properly, in that they could bind with high affinity to CD4 and a MAb, 17b, which is often used as surrogate for CCR5.
We therefore tested the ability of the gp120 proteins to bind CCR5 directly, by using flow cytometry to measure the extent of binding of gp120-CD4-IgG2 complexes to CCR5-expressing L1.2 cells (34, 111). No significant binding of any gp120-CD4-IgG2 complex to CCR5-negative parental L1.2 cells could be detected (data not shown). In the absence of CD4-IgG2 or gp120, no gp120 binding to L1.2-CCR5 cells could be detected, as expected (Fig. 7 and data not shown). However, when added as CD4-IgG2 complexes, gp120 proteins from two different, AD101-sensitive clones of CC1/85 bound efficiently to L1.2-CCR5 cells, and their binding was completely inhibited by AD101 (Fig. 7). In contrast, CD4-IgG2 complexes of gp120 proteins from two different clones of the fully AD101-resistant virus, CC101.19, failed to bind to L1.2-CCR5 cells, even at the very high input concentration of 100 μg/ml and whether or not AD101 was present. We therefore tested gp120 from the partially AD101-resistant virus CC101.6. This gp120 bound efficiently to L1.2-CCR5 cells, and its binding was inhibited by AD101 (Fig. 7).
FIG. 7.
Binding of monomeric gp120s from AD101-sensitive and -resistant viruses to L1.2-CCR5 cells in the presence and absence of AD101. The monomeric gp120 proteins indicated (100 μg/ml) with or without biotinylated CD4-lgG2 (100 μg/ml) were added to L1.2-CCR5 cells with and without AD101 (1 μM). The extent of binding of the gp120-CD4-IgG2 complex was measured as the mean fluorescence intensity (MFI) and is shown in arbitrary units (A.U.). Uncorrected mean fluorescence intensity values from a single, representative experiment are presented. The intrinsic fluorescence in the absence of gp120 is indicated in the “no gp120” column.
Both the CC101.6 clone and the two CC101.19 clones contain the amino acid changes selected for during the first four passages in the presence of AD101, including the H308P change in V3, whereas only the two CC101.19 clones have all four V3 substitutions (K305R, H308P, A316V, and G321E) (see above) (Fig. 3 and 4). Indeed, CC101.6 cl.10 gp120 differs from CC101.19 cl.3 gp120 at only four positions: G78D, K305R, A316V, and G321E (the CC101.6 cl.10 amino acid is listed first, and the CC101.19 cl.3 residue is listed last). The presence of G at position 78 of CC101.6 cl.10 probably represents a sporadic polymorphism, since this substitution was not found in any other clone. Moreover, like the two CC101.19 gp120s, the two CC1/85 gp120s both contain D78. Hence, the glycine residue present at position 78 in CC101.6 cl.10 is highly unlikely to be relevant to why the CC101.19-derived gp120s fail to bind cell surface CCR5. In addition to the G78D, K305R, A316V, and G321E substitutions, the CC101.6 cl.10 gp120 protein differs from the CC101.19 cl.7 protein by only one additional substitution, D167N, a polymorphic residue discussed above. Again, an asparagine residue at position 167 is unlikely to have any impact on CCR5 binding; CC1/85 cl.7 possesses N at this position and binds to CCR5, whereas CC101.19 cl.7 also has an N but fails to bind. Finally, neither amino acid 78 nor 167 is known to be in a region associated with CCR5 binding (59, 93, 94).
The most likely explanation of why gp120 from CC101.19 fails to bind CCR5 on L1.2 cells is that one or more of the K305R, A316V, and G321E substitutions in V3 has a decisive influence on a critical aspect of Env conformation. We prepared a model of the V3 loop crown regions of CC1/85 and CC101.19 gp120s, derived from the structures of complexes between V3 loop peptides and Fab fragments from the HIV-1 neutralizing antibodies 50.1 (92) and 59.1 (41, 42). The model indicates that the replacement of H308 by proline does not increase the probability of a tight turn but that the normal β-strand observed in this region for all V3 peptide-Fab complexes would probably be disrupted (data not shown). Hence, it is clearly possible that in the context of a native Env structure, the H308P change could have a marked impact on the geometry of the V3 loop. Additional studies are required to investigate how this change could influence the resistance to CCR5 inhibitors.
DISCUSSION
Genetic correlates of escape from a small-molecule CCR5 inhibitor.
Clearly, HIV-1 can escape from the pressure exerted in vitro by a CCR5-specific, small-molecule inhibitor of virus entry (108). This is not surprising, given the propensity of HIV-1 to mutate and the ability of its Env complex to accumulate sequence changes without functional impairment (86, 119). What was less predictable was the finding that an R5 virus evolved to continue to use CCR5 rather than switching its coreceptor preference to CXCR4, in an experimental system where CXCR4 was clearly available (35, 108). We are now trying to understand why HIV-1 did not follow what was intuitively the line of least resistance but instead, adopted an apparently more complex escape route. An explanation of this point might enable us to better understand what might happen if CCR5 inhibitors are used clinically for sustained periods as front-line antiviral drugs.
We previously studied two isolates (CC101.6 and CC101.19) that were derived from the parental R5 isolate CC1/85 after passaging for 6 and 19 passages, respectively, in the presence of AD101 (108). We showed that evolution of AD101 resistance was at least a two-step process; a partial (∼4-fold) loss of sensitivity to the selecting agent was acquired after 6 passages, and essentially complete (>20,000-fold) resistance was acquired by 19 passages (108). There was no overt loss of replication competence in PBMC associated with the development of full AD101 resistance. The genetic data we have now obtained are consistent with, and help explain, the phenotypic changes.
By passage 4 of the AD101 selection process, the Env sequence had become highly homogeneous. Multiple, previously minor, amino acid sequences became dominant among the sequenced clones, to the extent that they were essentially fixed. We propose that the partial AD101 resistance of the CC101.6 isolate is explained by selection of relatively resistant clones from among the preexisting CC1/85 population. A likely mechanism is the increased ability of viruses bearing the variant Env proteins to better exploit lower levels of free CCR5; such viruses would be relatively well suited to replicate during the early stages, when AD101 concentrations were only moderately elevated (108). Because the clones derived from CC101.6 contain all the amino acid changes selected for early (by passage 4) but none of those that arose and were selected for later (after passage 8), some or all of the initially selected sequences must play a role in partial resistance to AD101. Among them is the H308P substitution in the V3 loop, and we have confirmed by mutagenesis that this change does confer partial resistance. Hence, we believe that the H308P substitution was the most critical difference selected for from the CC1/85 population during the first four passages with increasing AD101 concentrations. Other changes occurring or selected for over the same time may represent adaptations to long-term PBMC culture or may be mere hitchhikers on the galaxy of Env proteins that happened to carry the critical H308P change.
Sequence analysis of the clones obtained from isolates after passage 4 indicates that the complete AD101 escape phenotype of the passage 19 isolate is associated with three additional, sequential V3 substitutions: K305R, A316V, and G321E. These changes arise and are selected for at a time that correlates with the acquisition of full resistance (108). The four V3 loop changes associated with AD101 resistance remained stable, with only a few exceptions, when the resistant isolate CC101.22 was cultured for an additional nine passages without AD101. This is consistent with the stability of the resistance phenotype over the same time (108). We are now studying whether a more prolonged culture without AD101 allows the V3 changes to revert and whether this is associated with reacquisition of AD101 sensitivity.
In contrast to the four changes in the V3 loop, there was no detectable selection pressure on amino acids in the C4 region or the stem of the V1/V2 loop that make up the bridging sheet of the gp120 core, the most highly conserved element of the CCR5 binding site (59, 93, 94). Nor was there any change in the footprint of MAb 17b to the CD4-induced epitope that overlaps the CCR5 binding site (59, 94, 105, 118).
In vivo, X4 viruses evolved in the individual from whom CC1/85 was obtained, such that an R5X4 virus, CC2/86, was isolated 13 months later (23, 24). Additional studies are now in progress, using clonal viruses of defined phenotype derived from CC2/86, to try to define which sequence changes in a CC1/85 R5 clone are sufficient to confer CXCR4 usage. It will be instructive to compare such changes with those that create AD101 resistance while preserving the R5 phenotype.
Sequence changes in the V3 loop confer resistance to AD101.
To better define the genetics of AD101 resistance, we inserted env clones from CC1/85 and CC101.19 into the pNL4-3 backbone, in the absence of other changes. The clonal viruses qualitatively reconstitute the AD101-sensitive and -resistant phenotypes of the CC1/85 and CC101.19 isolates, respectively. There are, however, quantitative differences in the degree of resistance shown by the clonal viruses and the corresponding isolates. Much of the difference is probably methodology based. NL4-3-based Env chimeric viruses may differ from natural viruses in the extent of Env incorporatation, which could affect fusion efficiency and its inhibition (55, 87). Viruses produced by transfection of 293T cells and by natural infection of PBMC may also vary in how they attach to target cells because of differences in their complement of host-derived proteins (106). There are also more subtle issues; for example, it is not obvious why AD101 inhibits the CC1/85 cl.7 clone 250-fold more potently than it inhibits the CC1/85 isolate when no differential sensitivity was observed using the related inhibitor, SCH-C.
Mutagenesis studies using the chimeric Env clones refined our understanding of the resistance mechanism. Thus, the H308P change was sufficient to cause a 500-fold increase in the AD101 resistance of a CC1/85 clone, and the addition of both the H308P and R305K substitutions caused a further 200-fold resistance increase. The introduction of all four amino acid changes highlighted by the sequence analysis (K305R, H308P, A316V, and G321E) created a virus completely resistant to AD101 concentrations as high as 20 μM. Conversely, when the reverse changes were made at these four V3 positions, the AD101-resistant virus CC101.19 cl.7 was converted into one that was highly sensitive to AD101. Taking the mutagenesis and the evolutionary genetics data together, the >20,000-fold resistance of the CC101.19 isolate to AD101 is due primarily to the four substitutions within the V3 loop.
It is no surprise that the V3 loop plays a dominant role in altering the coreceptor interactions of an HIV-1 isolate. There is ample precedent for a major influence of the V3 region on viral tropism, which we now understand to be largely a consequence of coreceptor choice (20-22, 31, 33, 39, 47, 48, 100, 114). Moreover, the V3 loop is a component of the gp120 binding sites for both CCR5 and CXCR4 (12, 22, 25, 33, 61, 93, 94). Its ability to vary in sequence while preserving its functions renders it an obvious place for amino acid substitutions to accumulate in response to AD101 selection pressure. Other elements of the CCR5 binding site on gp120 are more highly conserved, and so sequence changes there are inherently less likely (33, 93, 94, 119).
There have been previous studies of escape from coreceptor inhibitors. The development of partial (four- to sixfold) resistance of the R5 isolate JR-FL to macrophage inflammatory protein 1α was also associated with a V3 sequence change, acting in concert with another substitution in the V2 loop (68). The generation of ∼100-fold resistance to the anti-CCR5 MAb 2D7 when using the JR-CSF isolate was not associated with V3 sequence changes, although the genetic correlates of resistance were not fully identified (1). A significant proportion of the amino acid changes that accumulated in gp120 from the X4 virus NL4-3 during prolonged passage in vitro with the CXCR4 antagonists AMD3100, SDF-1, and T134 were also located in V3 (30, 50, 97). In each case, the resistant virus continued to use CXCR4, but in an inhibitor-insensitive manner (30, 50, 97). It was not determined whether the V3 loop changes were responsible for the generation of the resistant viruses, but it is a reasonable assumption that they were, at least, relevant (30, 50, 97).
A clone of HIV-1 JR-CSF can adapt to growth on cells that express CCR5 extracellular domain mutants with changes in the defined binding sites for HIV-1 (84). The virus adapted to each CCR5 substitution by selecting a specific pattern of changes in the V3 loop. Engineered mutants of the parental virus containing the V3 substitutions replicated efficiently, in a strictly CCR5-dependent manner, in cells expressing either wild-type or mutated CCR5, whereas the parental virus could use only wild-type CCR5. Hence, as we have found in the present study, the interaction of gp120 with CCR5 can be highly plastic, with changes in the V3 loop being sufficient to significantly influence it (84).
Functional aspects of AD101 resistance.
Although the CC101.19 clones are fully resistant to AD101 and to other small-molecule CCR5 inhibitors such as SCH-C, they retained their sensitivity to CCR5-specific MAbs and to the CC chemokine RANTES. Hence, just like the CC101.19 isolate from which they were cloned, they still use CCR5; indeed, their inability to replicate in PBMC from CCR5-Δ32 homozygous individuals shows they absolutely require CCR5 for entry. Resistance to CCR5-targeted inhibitors is therefore not global; instead, resistance to the different classes of inhibitor is likely to depend on the nature of their binding sites on CCR5. The small molecules AD101 and SCH-C bind within a pocket formed between CCR5 transmembrane helices 1, 2, 3, and 7, without involvement of the NT or ECLs (111). Their binding sites are broadly similar, albeit not identical, so that cross-resistance among small molecule CCR5 inhibitors is to be anticipated (111). The CCR5 MAbs and RANTES, in contrast, have binding sites that involve, wholly or in part, the NT and ECLs (14, 15, 67, 81, 96, 117). It is perhaps not surprising, then, that the AD101-resistant virus is still sensitive to inhibitors that bind elsewhere on CCR5.
It was, however, unexpected that monomeric gp120 proteins expressed from fully AD101-resistant clones (CC101.19 cl.3 and CC101.19 cl.7) failed to bind to CCR5 on L1.2-CCR5 cells. Considering that the corresponding infectious viruses absolutely require CCR5 to enter PBMC, the inability of their gp120 proteins to bind CCR5 appears paradoxical. The gp120 proteins are properly folded; they have appropriate affinities for CD4 and MAb 17b, so that their lack of binding to CCR5 is unlikely to have a trivial explanation. Moreover, gp120 from a partially AD101-resistant virus, CC101.6 cl.10, bound CCR5 perfectly well. The three, later-arising de novo substitutions in V3, K305R, A316V and G321E, are therefore responsible for the inability of CC101.19 gp120 to bind CCR5.
We can imagine two plausible explanations for why gp120 from the fully AD101-resistant virus does not bind to L1.2-CCR5 cells. The use of monomeric gp120 proteins may not recapitulate what happens when trimeric Env on the virion surface interacts with CCR5, a concept for which there is ample precedent from studies of the CD4 and MAb interactions of gp120 and virions (58, 74, 75, 86, 119). If so, the three changes in V3 render the gp120 monomers incapable of CCR5 binding while still permitting the trimeric Env complex to do so, albeit differently from the binding of Env complexes from AD101-sensitive viruses. Alternatively, the Env complex on the fully AD101-resistant virus may have evolved to recognize a specific CCR5 isoform or conformation present only on human PBMC (or on primary cells in general) and absent from L1.2-CCR5 cells. The latter are CCR5 transfectants of a murine pre-B-cell lymphoma line (116), so that post-translational modifications of CCR5 may be subtly different from what happens in primary human cells. Again, there is precedent for chemokine receptors, including CCR5, existing in multiple isoforms and conformations (6, 10, 11, 18, 36, 63, 64, 67, 70). If this is the explanation, then full AD101 resistance involves HIV-1 acquiring the ability to use a distinctive configuration of CCR5 that, somehow, is not affected by AD101 and other small-molecule inhibitors while still being vulnerable to CCR5-specific MAbs and RANTES. Clearly, additional studies are required to reveal what, exactly, is the explanation of AD101 resistance and how the critical sequence changes in the V3 loop affect the CCR5 binding of resistant viruses.
Effects of Env context on the V3 loop changes.
Several observations suggested that introducing sequence changes into the V3 loop of chimeric env clones can have context-dependent effects. Hence, the same genetic substitutions do not always have the same effect on phenotype; their precise impact instead depends on residues elsewhere in gp120. The determining factor is presumably how the different V3 loops interact with other gp120 domains, before or after CD4 binding. Quaternary interactions within the native Env complex may also be relevant. We do not yet know how any V3 loop is oriented and functions in its native context; there is no gp120 structure that includes the V3 loop. Moreover, we have no knowledge of how sequence changes in the V3 loop might directly or indirectly affect its orientation in a native Env trimer, for which we have only models, not a structure (60). It is suspected, however, that the C-terminal flank of the V3 loop abuts elements of the C4 region proximal to the CD4 binding site (17, 78, 79, 120, 121). There are probably also associations between V3 and other variable regions (16, 19, 123). Hence, interdomain interactions, either physical or functional, that are important in CD4 and coreceptor binding might differ in viral variants with V3 sequence changes (7, 16, 17, 19, 46, 78, 79, 120, 121, 123).
The H308P change is sufficiently radical to perhaps have a significant impact on the geometry of the V3 loop, the way in which it interacts with other regions of gp120, and hence the way in which CCR5 recognition is affected, particularly in the context of a native Env trimer. However, the effect of the H308P substitution must be reinforced by the three later-arising V3 substitutions. Thus, CC101.6 contains the H308P substitution and is only partially AD101 resistant, and gp120 expressed from an H308P-containing CC101.6 clone binds CCR5 on L1.2-CCR5 cells.
Implications of in vitro resistance pathways for the clinical use of CCR5 inhibitors.
Resistance to small-molecule CCR5 inhibitors will no doubt also occur in vivo if these compounds are used for sustained periods in the clinic. However, our studies suggest that escape will probably not be a simple, one-step process. Multiple amino acid changes must accumulate in gp120 to generate first partial and then full resistance to the selecting compound. In the case of complete resistance to AD101, as many as four amino acid substitutions in V3 are needed. The particular combination of the four residues that were present at positions 305, 308, 316, and 321 in the fully AD101-resistant virus, CC101.19, is very rare in naturally occurring V3 sequences. Hence, among 6,265 sequences from North American subtype B viruses in the Los Alamos Sequence Database, none had V3 loops that contained all four, or even three, of the residues associated with AD101 resistance. Two of the residues were naturally present in 188 of the sequences (3%), while 30% of the sequences contained one of the four.
Partial resistance may be easier to generate, however, as exemplified by the selection during the first four passages of preexisting variants with natural, albeit modest resistance to AD101. It is unclear how many amino acid changes are required for the ca. fourfold resistance of CC101.6; the H308P change in V3 may or may not be sufficient, and it is not uncommon among naturally occurring V3 sequences (H was found in 57% of B clade sequences, an P was found in 16%). A more dogmatic assertion is precluded by difficulties in making direct quantitative comparisons between the IC50s for inhibition of the NL4-3/env clones and the corresponding isolates. Resistance of this magnitude, a fewfold, could be significant in vivo. However, given the diversity of Env (and gp120 in particular), inherent variability in the response to entry inhibitors is to be expected. For example, in a standard PBMC inhibition assay using 13 subtype B isolates, IC50s for SCH-C spanned a 22-fold range from 0.4 to 8.9 nM, with a mean of 2.3 ± 0.8 nM (101). It will be important to establish the normal sensitivity range for patient isolates both in vitro and in vivo before resistance can be properly defined and identified (9, 43, 45, 65). It will also be instructive to compare the rate and genetic complexity of escape from CCR5 inhibitors in vitro and in vivo with the development of resistance to reverse transcriptase and protease inhibitors under comparable conditions (9, 43, 45, 65).
Deriving patient isolates for in vitro testing is a laborious and costly process, and so the use of more practical methods for gauging the potency of, and resistance to, entry inhibitors should be considered. Clonal virus systems, based on infectious chimeras or Env-pseudotyped viruses, are becoming an increasingly common way to study entry inhibitors, including neutralizing antibodies (9, 29, 38, 43, 45, 65, 91, 110, 112, 113). We noticed, however, that quantitative, but not qualitative, estimates of AD101 resistance differed when they were derived by using clonal, infectious chimeric viruses and uncloned isolates, even with the same target cells. Thus, the infectious chimera CC1/85 cl.7(H308P) has 500-fold higher resistance to AD101 than does CC1/85 (Fig. 6a; Table 1). However, when present in the PBMC-derived isolate CC101.6, the H308P substitution confers at most a fourfold increase in resistance to AD101 over that of CC1/85 (108). As outlined above, several factors could contribute to such differences, and some probably also apply to other assay systems that use clonal or Env-pseudotyped viruses. Hence, although these types of assay can be very valuable for monitoring the relative sensitivity or resistance of different viruses and entry inhibitors, care must be taken not to overinterpret the results by comparing numerical values with those obtained using the more biologically relevant uncloned patient isolates.
Genetic methods are an alternative to phenotypic assays and are very valuable when, for example, a single, dominant amino acid change confers reverse transcriptase inhibitor resistance (9, 43, 45, 65). Our study suggests, however, that such a scenario is unlikely to apply to CCR5 inhibitors. The V3 region is a likely site for resistance mutations to accumulate, both in vitro and in vivo, but the precise pattern of sequence changes we have observed may be unique to the particular virus, CCR5 inhibitor, and PBMC donor combination that we used. It may never be seen again, even in vitro. Assays that can identify V3 loop genetic variants irrespective of the precise sequence changes that occur may therefore be useful (5, 28, 54, 88, 89). However, the influence of Env context on V3 loop function may be a significant complication.
In clinical use, CCR5 inhibitors will be combined with other entry inhibitors or with more conventional suppressors of viral replication (8, 52, 77, 80, 85, 90, 98). The use of a drug combination may mean that HIV-1 will not accumulate sufficient mutations to escape from the CCR5 inhibitor before resistance to the other components of the combination develops. Of course, when escape does occur, it will be critical to determine whether HIV-1 has switched to use CXCR4 or has instead taken a pathway similar to the one we describe here.
Acknowledgments
We are grateful to Ruth Connor for providing HIV-1 isolates CC1/85, CC2/86, CC7/86, and CC12/86; Bill Olson for JR-FL gp120 protein, sCD4, MAb PA14, and CD4-IgG2; and James Robinson for MAb 17b. We thank Mika Vesanen and Maciej Paluch for their assistance in the purification of recombinant gp120 proteins. We thank Fred Lee, Tom Morgan, and Tom Ketas for technical assistance.
This work was funded by NIH grants AI41420 (J.P.M.), GM46192 (I.A.W. and R.L.S.), HD37356 (S.M.W.), and immunology training grant T32 AI07621 (S.E.K. and P.P.); by IAVI (J.P.M. and I.A.W.); and by Schering Plough Research Institute (J.P.M.). B.T.K. is an Elizabeth Glaser Scientist of the Pediatric AIDS Foundation. J.P.M. is a Stavros S. Niarchos Scholar. The Department of Microbiology and Immunology at the Weill Medical College gratefully acknowledges the support of the William Randolph Hearst Foundation.
Footnotes
Contribution la-ur-03-5893 from the Los Alamos National Laboratory.
REFERENCES
- 1.Aarons, E. J., S. Beddows, T. Willingham, L. Wu, and R. A. Koup. 2001. Adaptation to blockade of human immunodeficiency virus type 1 entry imposed by the anti-CCR5 monoclonal antibody 2D7. Virology 287:382-390. [DOI] [PubMed] [Google Scholar]
- 2.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allaway, G. P., K. L. Davis-Bruno, G. A. Beaudry, E. B. Garcia, E. L. Wong, A. M. Ryder, K. W. Hasel, M. C. Gauduin, R. A. Koup, J. S. McDougal, and P. J. Maddon. 1995. Expression and characterization of CD4-IgG2, a novel heterotetramer that neutralizes primary HIV type 1 isolates. AIDS Res. Hum. Retrovir. 11:533-539. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 5.Alves, K., M. Canzian, and E. L. Delwart. 2002. HIV type 1 envelope quasispecies in the thymus and lymph Nodes of AIDS patients. AIDS Res. Hum. Retrovir. 18:161-165. [DOI] [PubMed] [Google Scholar]
- 6.Babcock, G. J., M. Farzan, and J. Sodroski. 2003. Ligand-independent dimerization of CXCR4, a principal HIV-1 coreceptor. J. Biol. Chem. 278:3378-3385. [DOI] [PubMed] [Google Scholar]
- 7.Bagnarelli, P., L. Fiorelli, M. Vecchi, A. Monachetti, S. Menzo, and M. Clementi. 2003. Analysis of the functional relationship between V3 loop and gp120 context with regard to human immunodeficiency virus coreceptor usage using naturally selected sequences and different viral backbones. Virology 307:328-340. [DOI] [PubMed] [Google Scholar]
- 8.Baldwin, C. E., R. W. Sanders, and B. Berkhout. 2003. Inhibiting HIV-1 entry with fusion inhibitors. Curr. Med. Chem. 10:1773-1782. [DOI] [PubMed] [Google Scholar]
- 9.Balzarini, J. 1999. Suppression of resistance to drugs targeted to human immunodeficiency virus reverse transcriptase by combination therapy. Biochem. Pharmacol. 58:1-27. [DOI] [PubMed] [Google Scholar]
- 10.Bannert, N., S. Craig, M. Farzan, D. Sogah, N. V. Santo, H. Choe, and J. Sodroski. 2001. Sialylated O-glycans and sulfated tyrosines in the NH2-terminal domain of CC chemokine receptor 5 contribute to high affinity binding of chemokines. J. Exp. Med. 194:1661-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baribaud, F., T. G. Edwards, M. Sharron, A. Brelot, N. Heveker, K. Price, F. Mortari, M. Alizon, M. Tsang, and R. W. Doms. 2001. Antigenically distinct conformations of CXCR4. J. Virol. 75:8957-8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Basmaciogullari, S., G. J. Babcock, D. Van Ryk, W. Wojtowicz, and J. Sodroski. 2002. Identification of conserved and variable structures in the human immunodeficiency virus gp120 glycoprotein of importance for CXCR4 binding. J. Virol. 76:10791-10800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Binley, J. M., R. W. Sanders, B. Clas, N. Schuelke, A. Master, Y: Guo, F. Kajumo, D. J. Anselma, P. J. Maddon, W. C. Olson, and J. P. Moore. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74:627-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blanpain, C., B. J. Doranz, A. Bondue, C. Govaerts, A. De Leener, G. Vassart, R. W. Doms, A. Proudfoot, and M. Parmentier. 2003. The core domain of chemokines binds CCR5 extracellular domains while their amino terminus interacts with the transmembrane helix bundle. J. Biol. Chem. 278:5179-5187. [DOI] [PubMed] [Google Scholar]
- 15.Blanpain, C., B. J. Doranz, J. Vakili, J. Rucker, C. Govaerts, S. S. Baik, O. Lorthioir, I. Migeotte, F. Libert, F. Baleux, G. Vassart, R. W. Doms, and M. Parmentier. 1999. Multiple charged and aromatic residues in CCR5 amino-terminal domain are involved in high affinity binding of both chemokines and HIV-1 Env protein. J. Biol. Chem. 274:34719-34727. [DOI] [PubMed] [Google Scholar]
- 16.Carrillo, A., and L. Ratner. 1996. Cooperative effects of the human immunodeficiency virus type 1 envelope variable loops V1 and V3 in mediating infectivity for T cells. J. Virol. 70:1310-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carrillo, A., and L. Ratner. 1996. Human immunodeficiency virus type 1 tropism for T-lymphoid cell lines: role of the V3 loop and C4 envelope determinants. J. Virol. 70:1301-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chelli, M., and M. Alizon. 2002. Rescue of HIV-1 receptor function through cooperation between different forms of the CCR5 chemokine receptor. J. Biol. Chem. 277:39388-39396. [DOI] [PubMed] [Google Scholar]
- 19.Cho, M. W., M. K. Lee, C. H. Chen, T. Matthews, and M. A. Martin. 2000. Identification of gp120 regions targeted by a highly potent neutralizing antiserum elicited in a chimpanzee inoculated with a primary human immunodeficiency virus type 1 isolate. J. Virol. 74:9749-9754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choe, H., M. Farzan, Y. Sun, N. Sullivan, B. Rollins, P. D. Ponath, L. Wu, C. R. Mackay, G. LaRosa, W. Newman, N. Gerard, C. Gerard, and J. Sodroski. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135-1148. [DOI] [PubMed] [Google Scholar]
- 21.Clapham, P. R., and A. McKnight. 2002. Cell surface receptors, virus entry and tropism of primate lentiviruses. J. Gen. Virol. 83:1809-1829. [DOI] [PubMed] [Google Scholar]
- 22.Cocchi, F., A. L. DeVico, A. Garzino-Demo, A. Cara, R. C. Gallo, and P. Lusso. 1996. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat. Med. 2:1244-1247. [DOI] [PubMed] [Google Scholar]
- 23.Connor, R. I., H. Mohri, Y. Cao, and D. D. Ho. 1993. Increased viral burden and cytopathicity correlate temporally with CD4+ T-lymphocyte decline and clinical progression in human immunodeficiency virus type 1-infected individuals. J. Virol. 67:1772-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Connor, R. I., K. E. Sheridan, D. Ceradini, S. Choe, and N. R. Landau. 1997. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J. Exp. Med. 185:621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cormier, E. G., and T. Dragic. 2002. The crown and stem of the V3 loop play distinct roles in human immunodeficiency virus type 1 envelope glycoprotein interactions with the CCR5 coreceptor. J. Virol. 76:8953-8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Clercq, E. 2003. The bicyclam AMD3100 story. Nat. Rev. Drug Discov. 2:581-587. [DOI] [PubMed] [Google Scholar]
- 27.Deen, K. C., J. S. McDougal, R. Inacker, G. Folena-Wasserman, J. Arthos, J. Rosenberg, P. J. Maddon, R. Axel, and R. W. Sweet. 1988. A soluble form of CD4 (T4) protein inhibits AIDS virus infection. Nature 331:82-84. [DOI] [PubMed] [Google Scholar]
- 28.Delwart, E., M. Magierowska, M. Royz, B. Foley, L. Peddada, R. Smith, C. Heldebrant, A. Conrad, and M. Busch. 2002. Homogeneous quasispecies in 16 out of 17 individuals during very early HIV-1 primary infection. AIDS 16:189-195. [DOI] [PubMed] [Google Scholar]
- 29.Derdeyn, C. A., J. M. Decker, J. N. Sfakianos, X. Wu, W. A. O'Brien, L. Ratner, J. C. Kappes, G. M. Shaw, and E. Hunter. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 74:8358-8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Vreese, K., V. Kofler-Mongold, C. Leutgeb, V. Weber, K. Vermeire, S. Schacht, J. Anne, E. de Clercq, R. Datema, and G. Werner. 1996. The molecular target of bicyclams, potent inhibitors of human immunodeficiency virus replication. J. Virol. 70:689-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doms, R. W. 2000. Beyond receptor expression: the influence of receptor conformation, density, and affinity in HIV-1 infection. Virology 276:229-237. [DOI] [PubMed] [Google Scholar]
- 32.Douek, D. C., L. J. Picker, and R. A. Koup. 2003. T cell dynamics in HIV-1 infection. Annu. Rev. Immunol. 21:265-304. [DOI] [PubMed] [Google Scholar]
- 33.Dragic, T. 2001. An overview of the determinants of CCR5 and CXCR4 co-receptor function. J. Gen. Virol. 82:1807-1814. [DOI] [PubMed] [Google Scholar]
- 34.Dragic, T., A. Trkola, D. A. Thompson, E. G. Cormier, F. A. Kajumo, E. Maxwell, S. W. Lin, W. Ying, S. O. Smith, T. P. Sakmar, and J. P. Moore. 2000. A binding pocket for a small molecule inhibitor of HIV-1 entry within the transmembrane helices of CCR5. Proc. Natl. Acad. Sci. USA 97:5639-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farber, J. M., and E. A. Berger. 2002. HIV's response to a CCR5 inhibitor: I'd rather tighten than switch! Proc. Natl. Acad. Sci. USA 99:1749-1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farzan, M., G. J. Babcock, N. Vasilieva, P. L. Wright, E. Kiprilov, T. Mirzabekov, and H. Choe. 2002. The role of post-translational modifications of the CXCR4 amino terminus in stromal-derived factor 1 alpha association and HIV-1 entry. J. Biol. Chem. 277:29484-29489. [DOI] [PubMed] [Google Scholar]
- 37.Feng, Y., C. C. Broder, P. E. Kennedy, and E. A. Berger. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872-877. [DOI] [PubMed] [Google Scholar]
- 38.Fikkert, V., P. Cherepanov, K. Van Laethem, A. Hantson, B. Van Remoortel, C. Pannecouque, E. De Clercq, Z. Debyser, A. M. Vandamme, and M. Witvrouw. 2002. env chimeric virus technology for evaluating human immunodeficiency virus susceptibility to entry inhibitors. Antimicrob. Agents Chemother. 46:3954-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fouchier, R. A., M. Groenink, N. A. Kootstra, M. Tersmette, H. G. Huisman, F. Miedema, and H. Schuitemaker. 1992. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J. Virol. 66:3183-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gallo, S. A., A. Puri, and R. Blumenthal. 2001. HIV-1 gp41 six-helix bundle formation occurs rapidly after the engagement of gp120 by CXCR4 in the HIV-1 Env-mediated fusion process. Biochemistry 40:12231-12236. [DOI] [PubMed] [Google Scholar]
- 41.Ghiara, J. B., D. C. Ferguson, A. C. Satterthwait, H. J. Dyson, and I. A. Wilson. 1997. Structure-based design of a constrained peptide mimic of the HIV-1 V3 loop neutralization site. J. Mol. Biol. 266:31-39. [DOI] [PubMed] [Google Scholar]
- 42.Ghiara, J. B., E. A. Stura, R. L. Stanfield, A. T. Profy, and I. A. Wilson. 1994. Crystal structure of the principal neutralization site of HIV-1. Science 264:82-85. [DOI] [PubMed] [Google Scholar]
- 43.Hammer, S. M. 2002. HIV drug resistance: implications for management. Top. HIV Med. 10:10-15. [PubMed] [Google Scholar]
- 44.Hazenberg, M. D., D. Hamann, H. Schuitemaker, and F. Miedema. 2000. T cell depletion in HIV-1 infection: how CD4+ T cells go out of stock. Nat. Immunol. 1:285-289. [DOI] [PubMed] [Google Scholar]
- 45.Hirsch, M. S., F. Brun-Vezinet, R. T. D'Aquila, S. M. Hammer, V. A. Johnson, D. R. Kuritzkes, C. Loveday, J. W. Mellors, B. Clotet, B. Conway, L. M. Demeter, S. Vella, D. M. Jacobsen, and D. D. Richman. 2000. Antiretroviral drug resistance testing in adult HIV-1 infection: recommendations of an International AIDS Society—USA Panel. JAMA 283:2417-2426. [DOI] [PubMed] [Google Scholar]
- 46.Hoffman, N. G., F. Seillier-Moiseiwitsch, J. Ahn, J. M. Walker, and R. Swanstrom. 2002. Variability in the human immunodeficiency virus type 1 gp120 Env protein linked to phenotype-associated changes in the V3 loop. J. Virol. 76:3852-3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoffman, T. L., and R. W. Doms. 1999. HIV-1 envelope determinants for cell tropism and chemokine receptor use. Mol. Membr. Biol. 16:57-65. [DOI] [PubMed] [Google Scholar]
- 48.Hwang, S. S., T. J. Boyle, H. K. Lyerly, and B. R. Cullen. 1991. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science 253:71-74. [DOI] [PubMed] [Google Scholar]
- 49.Jacobson, J. M., I. Lowy, C. V. Fletcher, T. J. O'Neill, D. N. Tran, T. J. Ketas, A. Trkola, M. E. Klotman, P. J. Maddon, W. C. Olson, and R. J. Israel. 2000. Single-dose safety, pharmacology, and antiviral activity of the human immunodeficiency virus (HIV) type 1 entry inhibitor PRO 542 in HIV-infected adults. J. Infect. Dis. 182:326-329. [DOI] [PubMed] [Google Scholar]
- 50.Kanbara, K., S. Sato, J. Tanuma, H. Tamamura, K. Gotoh, M. Yoshimori, T. Kanamoto, M. Kitano, N. Fujii, and H. Nakashima. 2001. Biological and genetic characterization of a human immunodeficiency virus strain resistant to CXCR4 antagonist T134. AIDS Res. Hum. Retrovir. 17:615-622. [DOI] [PubMed] [Google Scholar]
- 51.Kazmierski, W., N. Bifulco, H. Yang, L. Boone, F. DeAnda, C. Watson, and T. Kenakin. 2003. Recent progress in discovery of small-molecule CCR5 chemokine receptor ligands as HIV-1 inhibitors. Bioorg. Med. Chem. 11:2663-2676. [DOI] [PubMed] [Google Scholar]
- 52.Kilby, J. M., and J. J. Eron. 2003. Novel therapies based on mechanisms of HIV-1 cell entry. N. Engl. J. Med. 348:2228-2238. [DOI] [PubMed] [Google Scholar]
- 53.Kilby, J. M., S. Hopkins, T. M. Venetta, B. DiMassimo, G. A. Cloud, J. Y. Lee, L. Alldredge, E. Hunter, D. Lambert, D. Bolognesi, T. Matthews, M. R. Johnson, M. A. Nowak, G. M. Shaw, and M. S. Saag. 1998. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 4:1302-1307. [DOI] [PubMed] [Google Scholar]
- 54.Kitrinos, K. M., N. G. Hoffman, J. A. Nelson, and R. Swanstrom. 2003. Turnover of Env variable region 1 and 2 genotypes in subjects with late-stage human immunodeficiency virus type 1 infection. J. Virol. 77:6811-6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klasse, P. J., and J. P. Moore. 1996. Quantitative model of antibody- and soluble CD4-mediated neutralization of primary isolates and T-cell line-adapted strains of human immunodeficiency virus type 1. J. Virol. 70:3668-3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Korber, B., B. T. Foley, C. L. Kuiken, S. K. Pillai, and J. Sodroski. 1998. Numbering positions in HIV relative to HXB2CG, p. 102-111. In B. Korber, C. L. Kuiken, B. T. Foley, B. H. Hahn, F. McCutchan, J. W. Mellors, and J. Sodroski (ed.), Human retroviruses and AIDS. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, NM.
- 57.Kwa, D., J. Vingerhoed, B. Boeser, and H. Schuitemaker. 2003. Increased in vitro cytopathicity of CC-chemokine receptor 5-restricted human immunodeficiency virus type 1 primary isolates correlates with a progressive clinical course of infection. J. Infect. Dis. 187:1397-1403. [DOI] [PubMed] [Google Scholar]
- 58.Kwong, P. D., M. L. Doyle, D. J. Casper, C. Cicala, S. A. Leavitt, S. Majeed, T. D. Steenbeke, M. Venturi, I. Chaiken, M. Fung, H. Katinger, P. W. Parren, J. Robinson, D. Van Ryk, L. Wang, D. R. Burton, E. Freire, R. Wyatt, J. Sodroski, W. A. Hendrickson, and J. Arthos. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678-682. [DOI] [PubMed] [Google Scholar]
- 59.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kwong, P. D., R. Wyatt, Q. J. Sattentau, J. Sodroski, and W. A. Hendrickson. 2000. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J. Virol. 74:1961-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Labrosse, B., C. Treboute, A. Brelot, and M. Alizon. 2001. Cooperation of the V1/V2 and V3 domains of human immunodeficiency virus type 1 gp120 for interaction with the CXCR4 receptor. J. Virol. 75:5457-5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lalezari, J. P., K. Henry, M. O'Hearn, J. S. Montaner, P. J. Piliero, B. Trottier, S. Walmsley, C. Cohen, D. R. Kuritzkes, J. J. Eron, Jr., J. Chung, R. DeMasi, L. Donatacci, C. Drobnes, J. Delehanty, and M. Salgo. 2003. Enfuvirtide, an HIV-1 fusion inhibitor, for drug-resistant HIV infection in North and South America. N. Engl. J. Med. 348:2175-2185. [DOI] [PubMed] [Google Scholar]
- 63.Lapham, C. K., T. Romantseva, E. Petricoin, L. R. King, J. Manischewitz, M. B. Zaitseva, and H. Golding. 2002. CXCR4 heterogeneity in primary cells: possible role of ubiquitination. J. Leukoc. Biol. 72:1206-1214. [PubMed] [Google Scholar]
- 64.Lapham, C. K., M. B. Zaitseva, S. Lee, T. Romanstseva, and H. Golding. 1999. Fusion of monocytes and macrophages with HIV-1 correlates with biochemical properties of CXCR4 and CCR5. Nat. Med. 5:303-308. [DOI] [PubMed] [Google Scholar]
- 65.Larder, B. 2001. Mechanisms of HIV-1 drug resistance. AIDS 15(Suppl. 5):S27-S34. [DOI] [PubMed] [Google Scholar]
- 66.Lazzarin, A., B. Clotet, D. Cooper, J. Reynes, K. Arasteh, M. Nelson, C. Katlama, H. J. Stellbrink, J. F. Delfraissy, J. Lange, L. Huson, R. DeMasi, C. Wat, J. Delehanty, C. Drobnes, and M. Salgo. 2003. Efficacy of enfuvirtide in patients infected with drug-resistant HIV-1 in Europe and Australia. N. Engl. J. Med. 348:2186-2195. [DOI] [PubMed] [Google Scholar]
- 67.Lee, B., M. Sharron, C. Blanpain, B. J. Doranz, J. Vakili, P. Setoh, E. Berg, G. Liu, H. R. Guy, S. R. Durell, M. Parmentier, C. N. Chang, K. Price, M. Tsang, and R. W. Doms. 1999. Epitope mapping of CCR5 reveals multiple conformational states and distinct but overlapping structures involved in chemokine and coreceptor function. J. Biol. Chem. 274:9617-9626. [DOI] [PubMed] [Google Scholar]
- 68.Maeda, Y., M. Foda, S. Matsushita, and S. Harada. 2000. Involvement of both the V2 and V3 regions of the CCR5-tropic human immunodeficiency virus type 1 envelope in reduced sensitivity to macrophage inflammatory protein 1α. J. Virol. 74:1787-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McCune, J. M. 2001. The dynamics of CD4+ T-cell depletion in HIV disease. Nature 410:974-979. [DOI] [PubMed] [Google Scholar]
- 70.McKnight, A., D. Wilkinson, G. Simmons, S. Talbot, L. Picard, M. Ahuja, M. Marsh, J. A. Hoxie, and P. R. Clapham. 1997. Inhibition of human immunodeficiency virus fusion by a monoclonal antibody to a coreceptor (CXCR4) is both cell type and virus strain dependent. J. Virol. 71:1692-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Meanwell, N. A., and J. F. Kadow. 2003. Inhibitors of the entry of HIV into host cells. Curr. Opin. Drug Discov. Dev. 6:451-461. [PubMed] [Google Scholar]
- 72.Melikyan, G. B., R. M. Markosyan, H. Hemmati, M. K. Delmedico, D. M. Lambert, and F. S. Cohen. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell. Biol. 151:413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Michael, N. L., and J. P. Moore. 1999. HIV-1 entry inhibitors: evading the issue. Nat. Med. 5:740-742. [DOI] [PubMed] [Google Scholar]
- 74.Moore, J. P., Y. Cao, L. Qing, Q. J. Sattentau, J. Pyati, R. Koduri, J. Robinson, C. F. Barbas III, D. R. Burton, and D. D. Ho. 1995. Primary isolates of human immunodeficiency virus type 1 are relatively resistant to neutralization by monoclonal antibodies to gp120, and their neutralization is not predicted by studies with monomeric gp120. J. Virol. 69:101-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moore, J. P., J. A. McKeating, Y. X. Huang, A. Ashkenazi, and D. D. Ho. 1992. Virions of primary human immunodeficiency virus type 1 isolates resistant to soluble CD4 (sCD4) neutralization differ in sCD4 binding and glycoprotein gp120 retention from sCD4-sensitive isolates. J. Virol. 66:235-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moore, J. P., and J. Sodroski. 1996. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J. Virol. 70:1863-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moore, J. P., and M. Stevenson. 2000. New targets for inhibitors of HIV-1 replication. Nat. Rev. Mol. Cell. Biol. 1:40-49. [DOI] [PubMed] [Google Scholar]
- 78.Moore, J. P., M. Thali, B. A. Jameson, F. Vignaux, G. K. Lewis, S. W. Poon, M. Charles, M. S. Fung, B. Sun, P. J. Durda, L. Akerblom, B. Wahren, D. D. Ho, Q. J. Sattentau, and J. Sodroski. 1993. Immunochemical analysis of the gp120 surface glycoprotein of human immunodeficiency virus type 1: probing the structure of the C4 and V4 domains and the interaction of the C4 domain with the V3 loop. J. Virol. 67:4785-4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Morrison, H. G., F. Kirchhoff, and R. C. Desrosiers. 1993. Evidence for the cooperation of gp120 amino acids 322 and 448 in SIVmac entry. Virology 195:167-174. [DOI] [PubMed] [Google Scholar]
- 80.O'Hara, B. M., and W. C. Olson. 2002. HIV entry inhibitors in clinical development. Curr. Opin. Pharmacol. 2:523-528. [DOI] [PubMed] [Google Scholar]
- 81.Olson, W. C., G. E. Rabut, K. A. Nagashima, D. N. Tran, D. J. Anselma, S. P. Monard, J. P. Segal, D. A. Thompson, F. Kajumo, Y. Guo, J. P. Moore, P. J. Maddon, and T. Dragic. 1999. Differential inhibition of human immunodeficiency virus type 1 fusion, gp120 binding, and CC-chemokine activity by monoclonal antibodies to CCR5. J. Virol. 73:4145-4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Palani, A., S. Shapiro, J. W. Clader, W. J. Greenlee, K. Cox, J. Strizki, M. Endres, and B. M. Baroudy. 2001. Discovery of 4-[(Z)-(4-bromophenyl)-(ethoxyimino)methyl]-1′-[(2,4-dimethyl-3-pyridinyl)carbonyl]-4′-methyl-1,4′-bipiperidine N-oxide (SCH 351125): an orally bioavailable human CCR5 antagonist for the treatment of HIV infection. J. Med. Chem. 44:3339-3342. [DOI] [PubMed] [Google Scholar]
- 83.Palani, A., S. Shapiro, H. Josien, T. Bara, J. W. Clader, W. J. Greenlee, K. Cox, J. M. Strizki, and B. M. Baroudy. 2002. Synthesis, SAR, and biological evaluation of oximino-piperidino-piperidine amides. 1. Orally bioavailable CCR5 receptor antagonists with potent anti-HIV activity. J. Med. Chem. 45:3143-3160. [DOI] [PubMed] [Google Scholar]
- 84.Platt, E. J., S. E. Kuhmann, P. P. Rose, and D. Kabat. 2001. Adaptive mutations in the V3 loop of gp120 enhance fusogenicity of human immunodeficiency virus type 1 and enable use of a CCR5 coreceptor that lacks the amino-terminal sulfated region. J. Virol. 75:12266-12278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pöhlmann, S., and R. W. Doms. 2002. Evaluation of current approaches to inhibit HIV entry. Curr. Drug Targ. Infect. Dis. 2:9-16. [DOI] [PubMed] [Google Scholar]
- 86.Poignard, P., E. O. Saphire, P. W. Parren, and D. R. Burton. 2001. gp120: Biologic aspects of structural features. Annu. Rev. Immunol. 19:253-274. [DOI] [PubMed] [Google Scholar]
- 87.Reeves, J. D., S. A. Gallo, N. Ahmad, J. L. Miamidian, P. E. Harvey, M. Sharron, S. Pohlmann, J. N. Sfakianos, C. A. Derdeyn, R. Blumenthal, E. Hunter, and R. W. Doms. 2002. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc. Natl. Acad. Sci. USA 99:16249-16254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Resch, W., N. Hoffman, and R. Swanstrom. 2001. Improved success of phenotype prediction of the human immunodeficiency virus type 1 from envelope variable loop 3 sequence using neural networks. Virology 288:51-62. [DOI] [PubMed] [Google Scholar]
- 89.Resch, W., N. Parkin, E. L. Stuelke, T. Watkins, and R. Swanstrom. 2001. A multiple-site-specific heteroduplex tracking assay as a tool for the study of viral population dynamics. Proc. Natl. Acad. Sci. USA 98:176-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Richman, D. D. 2001. HIV chemotherapy. Nature 410:995-1001. [DOI] [PubMed] [Google Scholar]
- 91.Richman, D. D., T. Wrin, S. J. Little, and C. J. Petropoulos. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. USA 100:4144-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rini, J. M., R. L. Stanfield, E. A. Stura, P. A. Salinas, A. T. Profy, and I. A. Wilson. 1993. Crystal structure of a human immunodeficiency virus type 1 neutralizing antibody, 50.1, in complex with its V3 loop peptide antigen. Proc. Natl. Acad. Sci. USA 90:6325-6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rizzuto, C., and J. Sodroski. 2000. Fine definition of a conserved CCR5-binding region on the human immunodeficiency virus type 1 glycoprotein 120. AIDS Res. Hum. Retroviruses 16:741-749. [DOI] [PubMed] [Google Scholar]
- 94.Rizzuto, C. D., R. Wyatt, N. Hernandez-Ramos, Y. Sun, P. D. Kwong, W. A. Hendrickson, and J. Sodroski. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949-1953. [DOI] [PubMed] [Google Scholar]
- 95.Rose, P. P., and B. T. Korber. 2000. Detecting hypermutations in viral sequences with an emphasis on G → A hypermutation. Bioinformatics 16:400-401. [DOI] [PubMed] [Google Scholar]
- 96.Samson, M., G. LaRosa, F. Libert, P. Paindavoine, M. Detheux, G. Vassart, and M. Parmentier. 1997. The second extracellular loop of CCR5 is the major determinant of ligand specificity. J. Biol. Chem. 272:24934-24941. [DOI] [PubMed] [Google Scholar]
- 97.Schols, D., J. A. Este, C. Cabrera, and E. De Clercq. 1998. T-cell-line-tropic human immunodeficiency virus type 1 that is made resistant to stromal cell-derived factor 1α contains mutations in the envelope gp120 but does not show a switch in coreceptor use. J. Virol. 72:4032-4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schwarz, M. K., and T. N. Wells. 2002. New therapeutics that modulate chemokine networks. Nat. Rev. Drug Discov. 1:347-358. [DOI] [PubMed] [Google Scholar]
- 99.Shankarappa, R., J. B. Margolick, S. J. Gange, A. G. Rodrigo, D. Upchurch, H. Farzadegan, P. Gupta, C. R. Rinaldo, G. H. Learn, X. He, X. L. Huang, and J. I. Mullins. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489-10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shioda, T., J. A. Levy, and C. Cheng-Mayer. 1991. Macrophage and T cell-line tropisms of HIV-1 are determined by specific regions of the envelope gp120 gene. Nature 349:167-169. [DOI] [PubMed] [Google Scholar]
- 101.Strizki, J. M., S. Xu, N. E. Wagner, L. Wojcik, J. Liu, Y. Hou, M. Endres, A. Palani, S. Shapiro, J. W. Clader, W. J. Greenlee, J. R. Tagat, S. McCombie, K. Cox, A. B. Fawzi, C. C. Chou, C. Pugliese-Sivo, L. Davies, M. E. Moreno, D. D. Ho, A. Trkola, C. A. Stoddart, J. P. Moore, G. R. Reyes, and B. M. Baroudy. 2001. SCH-C (SCH 351125), an orally bioavailable, small molecule antagonist of the chemokine receptor CCR5, is a potent inhibitor of HIV-1 infection in vitro and in vivo. Proc. Natl. Acad. Sci. USA 98:12718-12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sullivan, N., Y. Sun, Q. Sattentau, M. Thali, D. Wu, G. Denisova, J. Gershoni, J. Robinson, J. Moore, and J. Sodroski. 1998. CD4-Induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J. Virol. 72:4694-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tagat, J. R., S. W. McCombie, R. W. Steensma, S. Lin, D. V. Nazareno, B. Baroudy, N. Vantuno, S. Xu, and J. Liu. 2001. Piperazine-based CCR5 antagonists as HIV-1 inhibitors. I: 2(S)-methyl piperazine as a key pharmacophore element. Bioorg. Med. Chem. Lett. 11:2143-2146. [DOI] [PubMed] [Google Scholar]
- 104.Tagat, J. R., R. W. Steensma, S. W. McCombie, D. V. Nazareno, S. I. Lin, B. R. Neustadt, K. Cox, S. Xu, L. Wojcik, M. G. Murray, N. Vantuno, B. M. Baroudy, and J. M. Strizki. 2001. Piperazine-based CCR5 antagonists as HIV-1 inhibitors. II. Discovery of 1-[(2,4-dimethyl-3-pyridinyl)carbonyl]-4-methyl-4-[3(S)-methyl-4-[1(S)-[4-(trifluoromethyl)phenyl]ethyl]-1-piperazinyl]-piperidine N1-oxide (Sch-350634), an orally bioavailable, potent CCR5 antagonist. J. Med. Chem. 44:3343-3346. [DOI] [PubMed] [Google Scholar]
- 105.Thali, M., J. P. Moore, C. Furman, M. Charles, D. D. Ho, J. Robinson, and J. Sodroski. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67:3978-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tremblay, M. J., J. F. Fortin, and R. Cantin. 1998. The acquisition of host-encoded proteins by nascent HIV-1. Immunol. Today 19:346-351. [DOI] [PubMed] [Google Scholar]
- 107.Trkola, A., T. Dragic, J. Arthos, J. M. Binley, W. C. Olson, G. P. Allaway, C. Cheng-Mayer, J. Robinson, P. J. Maddon, and J. P. Moore. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184-187. [DOI] [PubMed] [Google Scholar]
- 108.Trkola, A., S. E. Kuhmann, J. M. Strizki, E. Maxwell, T. Ketas, T. Morgan, P. Pugach, S. Xu, L. Wojcik, J. Tagat, A. Palani, S. Shapiro, J. W. Clader, S. McCombie, G. R. Reyes, B. M. Baroudy, and J. P. Moore. 2002. HIV-1 escape from a small molecule, CCR5-specific entry inhibitor does not involve CXCR4 use. Proc. Natl. Acad. Sci. USA 99:395-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Trkola, A., A. B. Pomales, H. Yuan, B. Korber, P. J. Maddon, G. P. Allaway, H. Katinger, C. F. Barbas, 3rd, D. R. Burton, D. D. Ho, and J. P. Moore. 1995. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-immunoglobulin G. J. Virol. 69:6609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Trouplin, V., F. Salvatori, F. Cappello, V. Obry, A. Brelot, N. Heveker, M. Alizon, G. Scarlatti, F. Clavel, and F. Mammano. 2001. Determination of coreceptor usage of human immunodeficiency virus type 1 from patient plasma samples by using a recombinant phenotypic assay. J. Virol. 75:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tsamis, F., S. Gavrilov, F. Kajumo, C. Seibert, S. Kuhmann, T. Ketas, A. Trkola, A. Palani, J. W. Clader, J. R. Tagat, S. McCombie, B. Baroudy, J. P. Moore, T. P. Sakmar, and T. Dragic. 2003. Analysis of the mechanism by which the small-molecule CCR5 antagonists SCH-351125 and SCH-350581 inhibit human immunodeficiency virus type 1 entry. J. Virol. 77:5201-5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 114.Westervelt, P., H. E. Gendelman, and L. Ratner. 1991. Identification of a determinant within the human immunodeficiency virus 1 surface envelope glycoprotein critical for productive infection of primary monocytes. Proc. Natl. Acad. Sci. USA 88:3097-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wild, C., T. Oas, C. McDanal, D. Bolognesi, and T. Matthews. 1992. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc. Natl. Acad. Sci. USA 89:10537-10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wu, L., N. P. Gerard, R. Wyatt, H. Choe, C. Parolin, N. Ruffing, A. Borsetti, A. A. Cardoso, E. Desjardin, W. Newman, C. Gerard, and J. Sodroski. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179-183. [DOI] [PubMed] [Google Scholar]
- 117.Wu, L., G. LaRosa, N. Kassam, C. J. Gordon, H. Heath, N. Ruffing, H. Chen, J. Humblias, M. Samson, M. Parmentier, J. P. Moore, and C. R. Mackay. 1997. Interaction of chemokine receptor CCR5 with its ligands: multiple domains for HIV-1 gp120 binding and a single domain for chemokine binding. J. Exp. Med. 186:1373-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705-711. [DOI] [PubMed] [Google Scholar]
- 119.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 120.Wyatt, R., M. Thali, S. Tilley, A. Pinter, M. Posner, D. Ho, J. Robinson, and J. Sodroski. 1992. Relationship of the human immunodeficiency virus type 1 gp120 third variable loop to a component of the CD4 binding site in the fourth conserved region. J. Virol. 66:6997-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ye, Y., Z. H. Si, J. P. Moore, and J. Sodroski. 2000. Association of structural changes in the V2 and V3 loops of the gp120 envelope glycoprotein with acquisition of neutralization resistance in a simian-human immunodeficiency virus passaged in vivo. J. Virol. 74:11955-11962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang, Y. J., and J. P. Moore. 1999. Will multiple coreceptors need to be targeted by inhibitors of human immunodeficiency virus type 1 entry? J. Virol. 73:3443-3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zwick, M. B., R. Kelleher, R. Jensen, A. F. Labrijn, M. Wang, G. V. Quinnan, Jr., P. W. H. I. Parren, and D. R. Burton. 2003. A novel human antibody against human immunodeficiency virus type 1 gp120 is V1, V2, and V3 loop dependent and helps delimit the epitope of the broadly neutralizing antibody immunoglobulin G1 b12. J. Virol. 77:6965-6978. [DOI] [PMC free article] [PubMed] [Google Scholar]