Abstract
The core protein P3 of Rice dwarf virus constructs asymmetric dimers, one of which is inserted by the amino-terminal region of another P3 protein. The P3 proteins with serial amino-terminal deletions, expressed in a baculovirus system, formed particles with gradually decreasing stability. The capacity for self-assembly disappeared when 52 of the amino-terminal amino acids had been deleted. These results demonstrated that insertion of the amino-terminal arm of one P3 protein into another appears to play an important role in stabilizing the core particles.
Rice dwarf virus (RDV), an icosahedral double-shelled particle of approximately 70 nm in diameter, belongs to the genus Phytoreovirus in the family Reoviridae. The virus has seven structural proteins, namely, P1, P2, P3, P5, P7, P8, and P9, which are encoded by the S1, S2, S3, S5, S7, S8, and S9 segments, respectively, of the double-stranded RNA (dsRNA) genome (7, 9). The outer shell of the RDV capsid is composed of the P2, P8, and P9 proteins, while the core consists of the P1, P5, and P7 proteins enclosed by the P3 core capsid protein. The P3 core and P8 outer capsid proteins constitute the major framework of the architecture of RDV. The atomic structure of RDV determined by X-ray crystallography showed that the core particle was composed of 120 copies of the P3 protein (6). It had two icosahedrally independent P3 subunits, P3A and P3B. The amino-terminal arm of 30 residues of P3B interacted intimately with the P3A molecule, and this arm stabilized the dimer of the two P3 subunits. This dimer is considered to behave as the building blocks of a larger unit and finally conforms the core particles. The outer shell of RDV has a T=13 structure formed by 260 trimers of the P8 protein, which has positively charged patches on the inner surface that correspond to the concentration of negative charge on the outer surface of the P3 subunits that locate at icosahedral threefold axes of the core particles. Such configurations of the heterologous core capsid P3 and outer capsid P8 proteins are considered to guide the assembly of the outer shell, aided by lateral interactions between the P8 trimers, and complete the particle construction.
It has recently been reported that the major capsid protein P3, produced by using a baculovirus expression system, had the ability to form single-shelled core-like particles (CLPs) in the absence of any other structural proteins (2). To identify the role of the amino-terminal region of the P3 protein in the conformation of RDV particles, a series of deletion mutants of P3 was produced by means of the expression system; we then analyzed the stability of the core particles obtained with these mutants in the presence of high concentrations of MgCl2.
To produce P3 mutants with deletions of amino acids 2 to 10, 2 to 29, and 2 to 52 (N10del-P3, N29del-P3, and N52del-P3, respectively), we designed the following primers: 5′-AACCATG11GCGTCTGAATTCAAAAGTGTC-3′ (S3F/10-deletion), 5′-AACCATG30GAAGTATATAACATCCTCGAT-3′ (S3F/29-deletion), and 5′-AACCATG53GTGTCACGTACTCCAATTCCT-3′ (S3F/52-deletion). The numbers written as superscripts indicate the positions of the beginning of the amino acid sequence of each deletion mutant in the full-length P3 protein. Three types of deletion DNA were amplified by PCR with these primers and the reverse primer S3R-NcoI, as described previously (2). After digestion with the appropriate restriction enzyme, the PCR products were ligated into the transfer vector pBlueBacIII (Invitrogen, Carlsbad, Calif.). We determined the sequence of each resultant plasmid to confirm that the coding sequence of each deleted variant of S3 DNA was oriented appropriately with respect to the baculovirus promoter of the gene for polyhedrin.
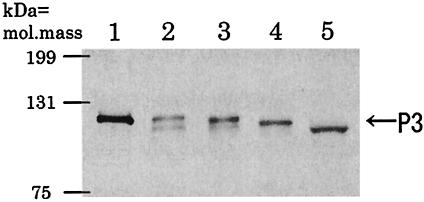
We used linearized Autographa californica multiple nucleopolyhedrovirus DNA (Invitrogen) and the various transfer plasmids that encoded the amino-terminally deleted forms of P3 to cotransfect insect cultured cells derived from Spodoptera frugiperda (Sf9) in the presence of CellFECTIN (Invitrogen) in accordance with the manufacturer's instructions. We examined the expression of the deletion mutants of P3 by Western blotting with P3-specific antibodies. On the Western blots, deleted forms of P3 were detected at the expected positions (Fig. 1), indicating that the three types of amino-terminally deleted P3 proteins had been expressed correctly in the baculovirus system. To solubilize the expressed proteins, we mixed Sf9 cells that had been infected with recombinant baculovirus with BugBuster protein extraction reagent (Novagen, Madison, Wis.). After the mixture was incubated for 30 min at 25°C, it was centrifuged for 5 min at 30,000 × g and the supernatant was collected. Then, after density gradient centrifugation of the supernatant on 10 to 40% sucrose for 70 min at 94,500 × g, the banded material was collected and pelleted by centrifugation for 60 min at 155,000 × g. CLPs were observed after the expression of N10del-P3 and N29del-P3 (Fig. 2a) but not after the expression of N52del-P3 (Table 1). These results indicated that the amino-terminal region from residue 30 to residue 52 was essential for self-assembly. A comparison of the structures of N29del-P3 and N52del-P3 based on extrapolation of the wild-type P3 dimer structure (6) revealed that N52del-P3 had lost the α-helical domain from residue 32 to residue 51 (Fig. 3), suggesting that the amino acids in this region might be important for maintenance of the overall conformation of P3 or for the specific interactions between P3A and P3B that are essential for the self-assembly of CLPs.
FIG. 1.
Western blotting analysis of amino-terminally deleted variants of the P3 protein. P3 proteins lacking the amino-terminal 10, 29, and 52 amino acids were expressed by the baculovirus system in insect cells. Samples were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and proteins were detected by Western blotting with P3-specific antibodies. Lane 1, purified RDV particles; lane 2, cell extract containing full-length P3; lanes 3, 4, and 5, cell extracts containing N10del-P3, N29del-P3, and N52del-P3, respectively. Mass markers and the mobility of P3 are indicated.
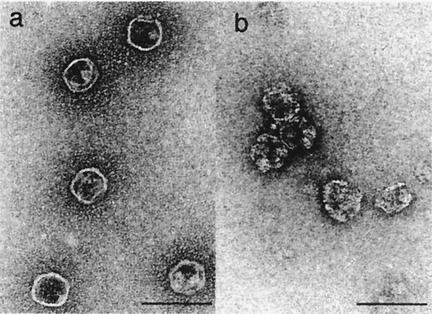
FIG. 2.
Electron micrographs of uranyl acetate-stained core particles. (a) CLPs of N29del-P3 that had been purified from Sf9 cells that had been infected with recombinant baculovirus. (b) Purified CLPs of N29del-P3 after treatment with 2.0 M MgCl2. Bars represent 100 nm.
TABLE 1.
Stabilitiy of various particles in the presence of high concentrations of MgCl2a
| Particle type | Stability in MgCl2
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 1.0 M | 2.0 M | 2.1 M | 2.2 M | 2.3 M | 2.4 M | 2.5 M | 2.6 M | |
| RDV particles | +++ | +++ | +++ | ++ | ++ | + | + | − |
| P3 CLPs | +++ | +++ | +++ | ++ | ++ | + | + | − |
| N10del-P3 CLPs | +++ | ++ | ++ | + | + | − | − | − |
| N29del-P3 CLPs | +++ | + | + | − | − | − | − | − |
| N52del-P3 CLPs | NDb | ND | ND | ND | ND | ND | ND | ND |
The symbols +++ and ++ indicate that more than 80 and 50% the particles remained intact, respectively. The symbol + indicates that more than 80% of the particles were disrupted. The symbol − indicates that the particles were completely destroyed. In each case, we examined more than 100 particles in three replicate experiments.
ND, not done; stability could not be measured because no CLPs were obtained.
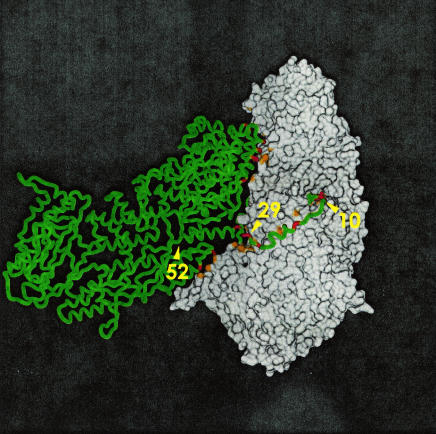
FIG. 3.
Structure of the asymmetric dimer composed of P3A and P3B, as shown by this surface representation of the P3A molecule (right, gray) and the Cα-trace of the P3B molecule (left, green), as viewed from the inside of the virus particle. Regions involved in hydrogen bonding or in a salt bridge between the P3 monomers are colored orange (P3A) and red (P3B). The extent of each of the three deletions (residues 10, 29, and 52) is indicated. The figure was produced with the DINO visualization program (http://www.dino3d.org) and rendered with the Povray graphics program (http://www.povray.org).
To compare the stabilities of the various particles obtained by the deleted form of P3 with those of intact RDV particles, we treated CLPs prepared from full-length P3, N10del-P3, and N29del-P3 with MgCl2 at various concentrations and monitored the results under the electron microscope. The stability of CLPs generated from full-length P3 and that of intact RDV particles were the same, and the particles were completely disrupted by 2.6 M MgCl2 (Table 1). By contrast, CLPs prepared from N10del-P3 were disrupted by 2.4 M MgCl2, and CLPs prepared from N29del-P3 were even more sensitive to MgCl2. Most of the CLPs prepared from N29del-P3 were disrupted by 2.0 M MgCl2 (Fig. 2b) and all were completely destroyed by treatment with 2.2 M MgCl2. These results indicated that the amino-terminal 29 amino acids of P3 were important for the stabilization of CLPs at a high concentration of MgCl2.
To visualize the interactions and deletion points of the amino-terminal arm, the structure of the P3 asymmetric dimer as viewed from inside the core particle was reconstructed (Fig. 3) based on the atomic structure of the P3 core reported previously (6). This reconstruction clearly showed the interactions of P3A (Fig. 3, right) and P3B (Fig. 3, left) and the deletion positions of the amino-terminal arm of P3B. The P3 asymmetric dimer had 11 strong hydrogen bonds and 4 strong salt bridges within this arm among 19 strong H bonds and 12 strong salt bridges, respectively (Table 2). Thus, the total energies of P3A-P3B interactions were gradually reduced by the serial deletions of amino-terminal regions. These results suggest that the reduction in stability of the CLPs obtained with the amino-terminally deleted mutants was caused by the loss of the chemical bonds described above.
TABLE 2.
Summary of the interactions between monomers in asymmetric dimers of reoviral proteinsa
| Protein | No. of strong H bonds (<3.3 Å) | No. of strong salt bridges (<3.3 Å) | No. of weak H bonds (3.3-3.6 Å) | No. of weak salt bridges (3.3-3.6 Å) | No. of van der Waals interactions | Total no. of interactions | Total energy of interactions (kcal/mol)b | Contact area (Å2) |
|---|---|---|---|---|---|---|---|---|
| RDV P3 | 19 | 12 | 5 | 1 | 341 | 378 | 207.3 | 4,100 |
| N10del-P3 | 13 | 10 | 4 | 1 | 236 | 264 | 149.8 | 3,000 |
| N29del-P3 | 10 | 10 | 2 | 1 | 165 | 188 | 115.5 | 2,100 |
| N52del-P3 | 8 | 8 | 2 | 0 | 134 | 152 | 92.2 | 1,700 |
| BTV VP3c | 10 | 3 | 3 | 0 | 297 | 313 | 134.1 | 3,100 |
| Reovirus N230-delλ1d | 14 | 3 | 0 | 1 | 165 | 183 | 99.5 | 2,500 |
The numbers of bonds and the estimated total energy for each pair are shown. Atomic pairs with distances of less than 4 Å are grouped under van der Waals interactions. An interaction between the positively charged side chain (arginine or lysine) and the negatively charged side chain (asparate or glutamate) is defined as a salt bridge.
The total energy of the interactions was estimated from the energy for each interaction as follows: strong H bonds and strong salt bridges, 3 kcal/mol; weak H bonds and weak salt bridges, 2 kcal/mol; and van der Waals interactions, 0.3 kcal/mol.
See reference 1.
The structural property shared by viruses in the family Reoviridae is the enclosure of 10 to 12 dsRNAs and certain proteins that are required for transcription within two or three concentric layers of an icosahedral capsid. Asymmetric dimers with 120 copies of an individual protein that form the thin layer of an inner core particle have been identified in crystallographic studies of bluetongue virus (BTV) (1), reovirus (8), and RDV (6), which are the only members of Reoviridae that have been analyzed at the atomic level to date. In RDV, moreover, the insertion of the amino-terminal arm of one P3 protein (P3B) into another P3 protein (P3A) seemed likely, from such analysis, to be important in the dimeric association of components of this core configuration (6). The biochemical analysis in the present study supports this hypothesis.
We calculated the total energies of interaction of asymmetric dimers in reoviruses based on their atomic structure (Table 2) and found that asymmetric P3 dimers of RDV had a higher energy of interaction (207.3 kcal/mol) than those of the BTV VP3 protein (134.1 kcal/mol), and the amino-terminal region seemed to be responsible for this difference. In the case of N52del-P3 of RDV, which had lost the capacity for self-assembly, the total energy of the P3A-P3B interaction was less than half (92.2 kcal/mol) that of native P3. The relatively high energy of interaction associated with P3 dimers in RDV, due to the amino-terminal region of P3, might allow the core of RDV to be generated in the absence of other structural proteins. The requirement for an additional protein VP7 in BTV (3, 5) for the construction of the CLPs might be due to the low energy of the interaction between monomers that correspond to P3 in this virus. In reovirus, the capability of the formation of inner core particles with the core capsid protein λ1, from which the amino-terminal 230 amino acids have been removed (energy of N230del-λ1, 99.5 kcal/mol) (Table 2), might be owing to the assistance of the σ2 protein (4).
Inside the P3 protein, RDV contains 25.7 kbp of dsRNA in 12 segments (7), the largest genome among dsRNA viruses studied by X-ray crystallography. It also contains the P1 protein, an RNA-dependent RNA polymerase, the P5 protein, a guanylyltransferase, and the P7 protein, a nonspecific nucleic acid binding protein. It is reasonable to consider that a large cavity is required to enclose the molecules involved in transcription. The sophisticated mechanism for the generation of tightly interacting dimers that allow the side-by-side binding of the very thin P3 proteins, which are only 2.5 to 4.5 nm thick (6), would be able to create a large cavity for packaging the nucleic acids and proteins described above.
Acknowledgments
This work was supported by Ministry of Agriculture Forestry and Fisheries grants 14051 and 15151 and by a grant from the Japan Society for the Promotion of Science. This project was also partly supported by grants-in-aid for scientific research (no. 14380319) from the 21st Century COE program and the national projects on protein structure and functional analysis from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
REFERENCES
- 1.Grimes, J. M., J. N. Burroughs, P. Gouet, J. M. Diprose, R. Malby, S. Zientara, P. P. C. Mertens, and D. I. Stuart. 1998. The atomic structure of the bluetongue virus core. Nature 395:470-478. [DOI] [PubMed] [Google Scholar]
- 2.Hagiwara, K., T. Higashi, K. Namba, T. Uehara-Ichiki, and T. Omura. 2003. Assembly of single-shelled cores and double-shelled virus-like particles after baculovirus expression of major structural proteins P3, P7 and P8 of Rice dwarf virus. J. Gen. Virol. 84:981-984. [DOI] [PubMed] [Google Scholar]
- 3.Hewat, E. A., T. F. Booth, P. T. Loudon, and P. Roy. 1992. Three-dimensional reconstruction of baculovirus expressed bluetongue virus core-like particles by cryo-electron microscopy. Virology 189:10-20. [DOI] [PubMed] [Google Scholar]
- 4.Kim, J., X. Zhang, V. E. Centonze, V. D. Bowman, S. Nobel, T. S. Baker, and M. L. Nibert. 2003. The hydrophilic amino-terminal arm of reovirus core shell protein λ1 is dispensable for particle assembly. J. Virol. 76:12211-12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loudon, P. T., and P. Roy. 1991. Assembly of five bluetongue virus proteins expressed by recombinant baculoviruses: inclusion of the largest protein VP1 in the core and virus-like particles. Virology 180:798-802. [DOI] [PubMed] [Google Scholar]
- 6.Nakagawa, A., N. Miyazaki, J. Taka, H. Naitow, A. Ogawa, Z. Fujimoto, H. Mizuno, T. Higashi, Y. Watanabe, T. Omura, R. H. Cheng, and T. Tsukihara. 2003. The atomic structure of Rice dwarf virus reveals the self-assembly mechanism of component proteins. Structure 11:1227-1238. [DOI] [PubMed] [Google Scholar]
- 7.Omura, T., and J. Yan. 1999. Role of outer capsid proteins in transmission of Phytoreovirus by insect vectors. Adv. Virus Res. 54:15-43. [DOI] [PubMed] [Google Scholar]
- 8.Reinisch, K. M., M. L. Nibert, and S. C. Harrison. 2000. Structure of the reovirus core at 3.6 Å resolution. Nature 404:960-967. [DOI] [PubMed] [Google Scholar]
- 9.Zhong, B., A. Kikuchi, Y. Moriyasu, T. Higashi, K. Hagiwara, and T. Omura. 2003. A minor outer capsid protein, P9, of Rice dwarf virus. Arch. Virol. 148:2275-2280. [DOI] [PubMed] [Google Scholar]