Abstract
A Turnip crinkle virus (TCV)-based system was devised to discriminate cell-to-cell and systemic long-distance spread of RNA silencing in plants. Modified TCV-GFPΔCP, constructed by replacing the coat protein (CP) gene with the green fluorescent protein (GFP) gene, replicated in single epidermal cells but failed to move from cell to cell in Nicotiana benthamiana. Mechanical inoculation of TCV-GFPΔCP induced effective RNA silencing in single epidermal cells which spread from cell to cell to form silenced foci on inoculated leaves, but no long-distance systemic spread of RNA silencing occurred. Agroinfiltration of TCV-GFPΔCP was, however, able to induce both local and systemic RNA silencing. TCV coinfection arrested TCV-GFPΔCP-mediated local induction of RNA silencing. Possible mechanisms involved in cell-to-cell and long-distance spread of RNA silencing are discussed.
RNA silencing, including gene quelling, RNA interference, and posttranscriptional gene silencing are sequence-specific RNA degradation mechanisms that operate in fungi, animals, and plants (6, 7, 30). RNA silencing is triggered by double-stranded RNA and requires a conserved set of gene products (1, 13, 14). The double-stranded RNA is processed into small interfering RNAs (siRNAs) of 21 to 25 nucleotides (nt), and these siRNAs become associated with an RNA-induced silencing complex that degrades specific target RNA sequences (3, 11, 12). RNA silencing plays a natural role in protecting fungi, plants, and animals against viral infection. To withstand the RNA-silencing defense, viruses across kingdoms have evolved diverse mechanisms either to avoid or actively suppress RNA silencing (22, 35, 40).
One intriguing feature of RNA silencing is that it is not cell autonomous. In plants, RNA silencing can be induced locally and then spread to distal parts (16, 36-38). While RNA-silencing induction and RNA degradation have been elucidated in detail, much less is known about how RNA silencing moves from cell to cell and spreads systemically in plants. No mobile silencing signal has been characterized, although the sequence specificity of RNA silencing implies that nucleic acids, possibly siRNAs, may be a component of such an RNA-silencing signal (26). We have used Turnip crinkle virus (TCV) to explore the requirements for cell-to-cell and long-distance spread of RNA silencing in plants.
TCV, a member of the Carmovirus genus, has a positive single-strand genomic RNA (4,053 nt), packaged in icosahedral capsids, which contains five major open reading frames (5). The p28 and p88 proteins are translated from genomic RNA, by readthrough of the p28 terminator, and are involved in viral RNA replication. Two overlapping proteins, p8 and p9, are expressed from subgenomic RNA1 and are required for cell-to-cell movement and systemic spread of the virus (10, 23). The 3′-proximal open reading frame encodes the 38,000-molecular-weight coat protein (CP), which also plays an essential role in cell-to-cell movement of TCV in Nicotiana benthamiana (8) and acts as an effective suppressor of RNA silencing (27, 31).
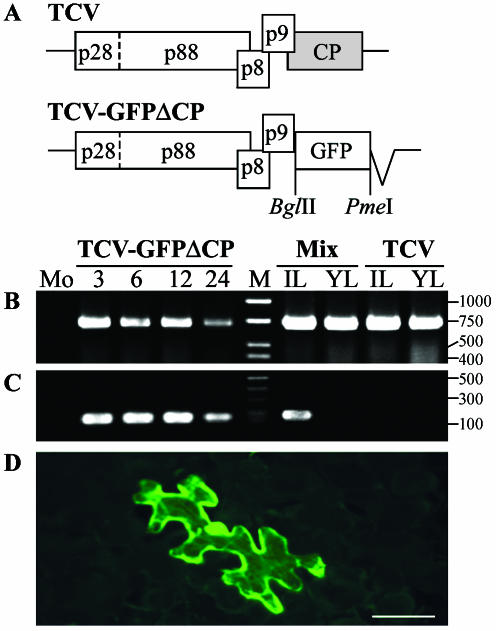
A full-length infectious cDNA of TCV was generated by reverse transcription (RT)-PCR with a genomic RNA template extracted from purified viral particles of a United Kingdom isolate derived from wild brassicas (GenBank accession no. AY312063) (Fig. 1A). Two primers specific to TCV genomic RNA contained the 5′-terminal 19 nt following a T7 RNA polymerase promoter sequence and a sequence complementary to the 3′-terminal 24 nt following a PacI site. The resulting RT-PCR fragment was cloned into pCR-BluntII-TOPO (Invitrogen) to produce pT7.TCV. Plasmid pT7.TCV was used to construct pT7.TCV-GFPΔCP, in which the CP gene was replaced with the coding sequence for green fluorescent protein (GFP). Briefly, nt 2753 to 3388, encoding the R domain and the majority of the S domain of TCV CP (18), were deleted by overlap extension PCR (15) with a pair of primers (5′-GAAAcGGAAAATGagatctggaccggtgggtttaaacCACCTACGGCCAAGGAGC-3′ and 5′-GGCCGTAGGTGgtttaaacccaccggtccagatctCATTTTCCgTTTCCAGTGTTG-3′). Modified nucleotides are shown in lowercase, and introduced restriction endonuclease sites for BglII and PmeI are underlined. The CP initiation codon (boldface) was also mutated. Unique BglII and PmeI sites were inserted downstream of the stop codon for the p9 movement protein in the modified TCV genome. The GFP coding sequence was PCR amplified with a pair of primers(5′-ggaaagatctATGGCTAGCAAAGGAGAAGAAC-3′ and5′-ggaagtttaaacTTATTTGTAGAGCTCATCCATG-3′), di-gested with BglII and PmeI, and cloned into the BglII and PmeI sites of the modified TCV cDNA to produce pT7.TCV-GFPΔCP (Fig. 1A).
FIG. 1.
Expression of GFP from TCV-GFPΔCP in single N. benthamiana epidermal cells. Construction of TCV and TCV-GFPΔCP is shown in panel A. TCV (B) and GFP (C) RNAs were detected by RT-PCR with RNA samples extracted from mock-inoculated (Mo) or TCV-GFPΔCP RNA-inoculated N. benthamiana leaves harvested at 3, 6, 12, and 24 dpi. In the case of TCV infection (TCV) or coinoculation with TCV-GFPΔCP and TCV RNAs (Mix), total RNAs were extracted from inoculated (IL) or systemically infected young (YL) leaves at 12 dpi. The sizes (nucleotides) and positions of a double-stranded DNA ladder (M) are indicated. Epifluorescence microscopic examination of GFP expression in a single epidermal cell in a TCV-GFPΔCP-inoculated N. benthamiana leaf at 6 dpi with a Zeiss Axiophot microscope through a green filter is shown in panel D. Bar = 100 μm.
To test whether the cloned TCV and recombinant TCV-GFPΔCP were infectious, RNA transcripts were produced in vitro from pT7.TCV or pT7.TCV-GFPΔCP after linearization with PacI with the mMESSAGE mMACHINE T7 kit (Ambion). Viral RNA transcripts were mechanically inoculated separately or as a mixture onto young N. benthamiana plants. Plants were maintained in an insect-free growth room at 25°C with a continuous 12-h photoperiod. Infection with transcripts of pT7.TCV alone or with pT7.TCV-GFPΔCP produced local and systemic symptoms indistinguishable from those induced by sap transmission of virions onto N. benthamiana plants. With RT-PCR assays done as previously described (34), a TCV genome-specific product (nt 3388 to 4053) was readily detectable in inoculated and systemically infected young leaves at 12 days postinoculation (dpi) (Fig. 1B). A gfp gene-specific fragment was detected in leaves inoculated with mixed viral RNA transcripts (Fig. 1C). N. benthamiana plants inoculated with the pT7.TCV-GFPΔCP transcript alone produced no obvious lesions on inoculated leaves and developed no systemic symptoms at 24 dpi, although TCV-GFPΔCP RNA replicated efficiently in the initially infected cells. Indeed, both viral and gfp-specific sequences were detected by RT-PCR in inoculated leaves at 3, 6, 12, and 24 dpi (Fig. 1C).
Fluorescence microscopy of N. benthamiana plants mechanically inoculated with TCV-GFPΔCP RNA at 3, 6, and 12 dpi revealed that GFP fluorescence was clearly visible in individual epidermal cells in inoculated leaves (Fig. 1D). Green fluorescence was confined to single epidermal cells even at 24 dpi. No GFP fluorescence was observed in young, newly emerging noninoculated leaves, consistent with the RT-PCR results for TCV and gfp RNA. Taken together, our data demonstrate that cloned TCV and recombinant TCV-GFPΔCP accumulated efficiently in initially infected single cells and that CP deletion resulted in the inability of TCV-GFPΔCP RNA to move from cell to cell in N. benthamiana. Moreover, CP expressed from TCV was unable to complement long-distance transport of TCV-GFPΔCP in plants. These results suggested that TCV-GFPΔCP may be used as a functional tool to explore the single-cell induction and cell-to-cell and long-distance movement of RNA silencing in plants.
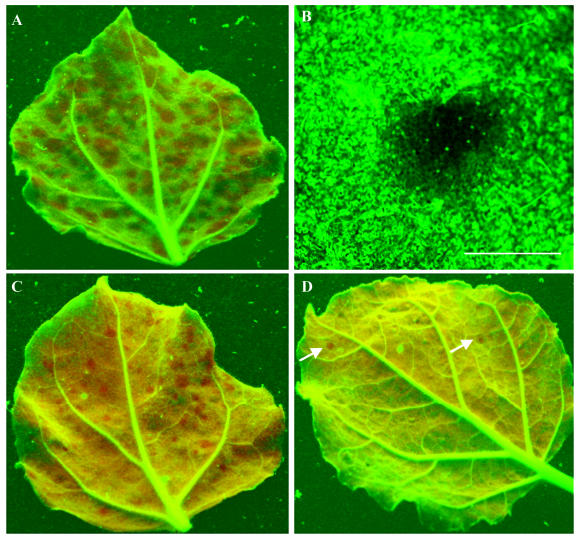
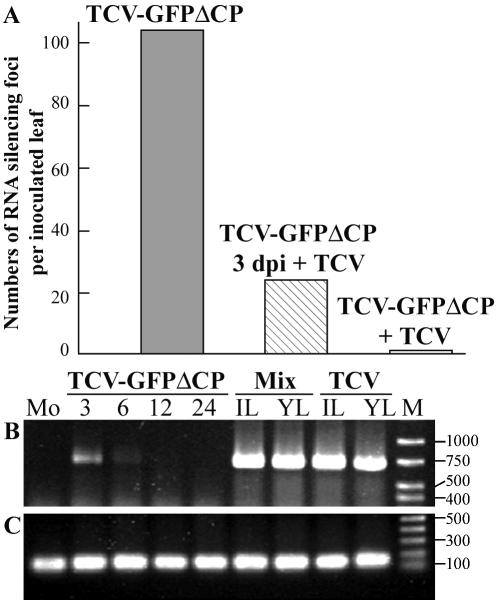
To test the potential utility of TCV-GFPΔCP, N. benthamiana plants (line 16c) carrying a highly expressed GFP transgene (4) were inoculated with pT7.TCV-GFPΔCP transcripts. Local RNA silencing was induced rapidly in individual epidermal cells on inoculated leaves, and tiny gfp RNA-silencing foci, each with dozens of cells, showing red chlorophyll fluorescence under long-wavelength UV light illumination (9), were detected at 48 h postinoculation. Expansion of these localized gfp RNA-silencing foci continued, reaching up to 6 mm in diameter at 24 dpi (Fig. 2A). Leaves of line 16c plants inoculated with wild-type TCV transcripts showed no development of gfp RNA silencing. Fluorescence microscopy of TCV-GFPΔCP RNA-inoculated leaves showed that cell-to-cell spread of gfp RNA silencing was established efficiently in upper and lower epidermal cells and in palisade and spongy mesophyll cells (Fig. 2B). It is likely that the effectiveness of TCV-GFPΔCP RNA at inducting silencing may be due to the lack of CP, an RNA-silencing suppressor (27, 31). However, the RNA-silencing suppressor activity of TCV CP had been shown only in transient agroinfiltration assays and not in natural TCV infections. To clarify this inconsistent result, leaves of line 16c plants were mechanically inoculated with TCV-GFPΔCP RNA and then 3 days later inoculated with TCV RNA. The number of gfp RNA-silencing foci was reduced approximately fourfold (Fig. 2C and 3A) compared with line 16c plant leaves mock inoculated 3 days after inoculation with TCV-GFPΔCP RNA. At 24 dpi with TCV-GFPΔCP, the sizes of the gfp RNA-silencing foci in TCV-reinoculated leaves ranged from 0.1 to 2 mm, smaller than the 1- to 6-mm foci in mock-reinoculated plants. More dramatic suppression of local gfp RNA-silencing induction was observed when the 16c plants were coinoculated with TCV and TCV-GFPΔCP RNAs. The number of gfp RNA-silencing foci decreased about 70-fold compared with TCV-GFPΔCP RNA inoculation alone, and foci were reduced in size to between 0.1 and 1 mm (Fig. 2D and 3A). Our results demonstrate that TCV infection arrested gfp RNA-silencing in plants, probably owing to expression of TCV CP, which acted as an RNA-silencing suppressor. It is likely that CP produced during TCV infection interfered with the induction of RNA silencing rather than turned off RNA silencing already established. This view is supported by an earlier finding that TCV CP suppressed RNA silencing at the initiation stage by preventing siRNA production (27). Moreover, our data also imply that TCV CP could block cell-to-cell movement of RNA silencing.
FIG. 2.
Local induction, cell-to-cell spread, and suppression of RNA silencing. N. benthamiana (line 16c) plants were mechanically inoculated with TCV-GFPΔCP RNA (A and B), inoculated with TCV-GFPΔCP RNA and then reinoculated 3 days later with TCV RNA (C), or coinoculated with TCV-GFPΔCP and TCV RNAs (D). Inoculated leaves (A, C, and D) were photographed at 24 dpi with a Nikon Coolpix990 digital camera under long-wavelength UV illumination through a yellow filter. gfp RNA-silencing foci show red chlorophyll fluorescence, and gfp RNA-expressing tissues show green fluorescence. Only two gfp RNA-silencing foci (arrow) are seen in the leaf coinoculated with TCV-GFPΔCP and TCV RNAs (D). One gfp RNA-silencing focus, examined by epifluorescence microscopy of a TCV-GFPΔCP RNA-inoculated leaf through a green filter (B), appears dark with some green fluorescent cells. Bar = 6 mm.
FIG. 3.
Effects of induction and suppression of local RNA silencing on accumulation of TCV and gfp RNAs. Average numbers of silencing foci per inoculated leaf are shown (A). Numbers were counted at 24 dpi in line 16c plants inoculated with TCV-GFPΔCP RNA, inoculated with TCV-GFPΔCP and then reinoculated 3 days later with TCV (TCV-GFPΔCP 3dpi + TCV), or coinoculated with both transcripts on day 0 (TCV-GFPΔCP + TCV). TCV (B) and GFP (C) RNAs were detected by RT-PCR with total RNA samples extracted from mock-inoculated (Mo) or TCV-GFPΔCP RNA-inoculated line 16c plant leaves harvested at 3, 6, 12, and 24 dpi. In the case of TCV infection (TCV) or coinoculation with TCV-GFPΔCP + TCV (Mix), total RNAs were extracted from inoculated (IL) and systemically infected young (YL) leaves at 12 dpi. The sizes (nucleotides) and positions of a double-stranded DNA ladder (M) are indicated.
To further demonstrate the local induction of RNA silencing by TCV-GFPΔCP RNA, and suppression of silencing by coinfection with TCV, levels of viral and gfp RNA accumulation in line 16c plants inoculated with TCV-GFPΔCP RNA alone or with TCV RNA were analyzed by RT-PCR (Fig. 3B and C). TCV-GFPΔCP RNA was detected at 3 dpi. The level of TCV-GFPΔCP RNA declined significantly at 6 dpi and was below detectable levels at 12 dpi. However, TCV RNA was easily detectable in inoculated and systemically infected young leaves of line 16c plants treated with both RNAs or with TCV transcripts alone (Fig. 3B). Consistent with limited local induction and suppression of RNA silencing, gfp RNA was detected in all of the leaf samples tested (Fig. 3C). These results suggest that the gfp sequence in TCV-GFPΔCP RNA acted as an inducer of RNA silencing that targeted both transgene-derived gfp RNA and recombinant TCV-GFPΔCP RNA. Infection by TCV-GFPΔCP RNA in line 16c plants, as in nontransformed N. benthamiana, was confined to primarily inoculated, individual epidermal cells. Thus, TCV-GFPΔCP RNA effectively initiated and induced RNA silencing in single epidermal cells, which then spread from cell to cell to form gfp silenced foci containing hundreds to thousands of cells within the inoculated leaves.
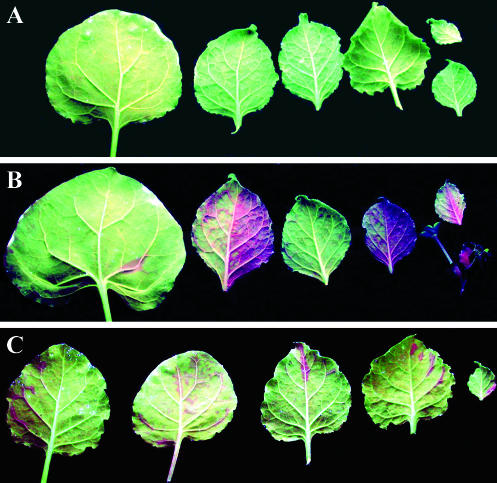
Although TCV-GFPΔCP RNA-mediated induction of gfp RNA silencing in inoculated leaves was effective, with up to 50% of the lamina exhibiting silenced gfp transgene expression, no systemic long-distance spread of RNA silencing was observed up to 6 weeks after inoculation (Fig. 4A). This phenomenon was observed consistently in 12 individual plants in three separate experiments. Thus, although gfp RNA silencing induced by TCV-GFPΔCP RNA in individual epidermal cells was able to move from cell to cell in the inoculated leaf, gfp silencing did not spread systemically to distal parts of the plant through the phloem. These results contrast with the effective induction of both local and systemic gfp RNA silencing in line 16c plants by infiltration with Agrobacterium carrying a 35S promoter-GFP binary vector (Fig. 4B) (4). It is notable the total area of local silencing in agroinfiltrated line 16c leaves did not exceed the area covered by all of the RNA-silencing foci induced by mechanical inoculation with TCV-GFPΔCP. One of the differences between the initial induction of RNA silencing by mechanical inoculation with TCV-GFPΔCP RNA and agroinfiltration of the 35S-GFP cassette is related to cell types. Mechanical inoculation of line 16c plants with TCV-GFPΔCP RNA induced gfp silencing only in individual epidermal cells, while agroinfiltration may lead to RNA silencing in a range of cells including epidermal and mesophyll cells and cells of vascular tissues in which the 35S promoter is frequently active (2).
FIG. 4.
Spread of RNA silencing induced by TCV-GFPΔCP via mechanical inoculation and agroinfiltration. No RNA silencing occurred in noninoculated older and newly emerging young leaves of line 16c plants mechanically inoculated with TCV-GFPΔCP RNA (A). Infiltration of line 16c plants with Agrobacterium cultures carrying a 35S-GFP expression cassette (B) or a TCV-GFPΔCP cassette (C) induced systemic RNA silencing in noninfiltrated young leaves. Leaves were photographed at 42 days post-RNA inoculation or -agroinfiltration. gfp RNA-silenced tissues show red fluorescence.
To establish if the initial induction of RNA silencing by TCV-GFPΔCP RNA in a range of cell types apart from epidermal cells would have an effect on the long-distance spread of RNA silencing, we constructed p35S.TCV-GFPΔCP, with the TCV-GFPΔCP cDNA inserted between the Cauliflower mosaic virus 35S promoter and terminator sequences of a binary vector in A. tumefaciens LBA4404 (17). In three independent experiments, 12 line 16c plants were infiltrated with an Agrobacterium culture carrying p35S-TCV-GFPΔCP. Six line 16c plants were also infiltrated independently with Agrobacterium carrying 35S-GFP. In the leaves of line 16c plants agroinfiltrated with 35S-TCV-GFPΔCP or 35S-GFP, local silencing of transgenic gfp expression was first visible 7 days postinfiltration; dark red rings without GFP fluorescence encircled the infiltrated areas and were clearly visible under UV light. Approximately 2 weeks after agroinfiltration with either construct, systemic gfp RNA-silencing foci appeared in upper, noninfiltrated leaves along the minor veins and in growing “carbon sink” areas. All experimental plants developed this form of systemic gfp RNA silencing (Fig. 4B and C). Our data demonstrate that TCV-GFPΔCP RNA was able to induce both local and systemic RNA silencing when introduced by agroinfiltration to different types of cells. In contrast, gfp RNA silencing induced exclusively in individual epidermal cells by mechanical inoculation with TCV-GFPΔCP transcripts only moved from cell to cell within the inoculated leaf and failed to spread long distance.
The mechanism(s) of spread of RNA silencing in plants is poorly understood, and a mobile silencing signal remains to be elucidated (16, 25, 26). The sequence specificity of RNA silencing suggests that the mobile silencing signal contains an RNA component. siRNAs are an attractive candidate as they are short enough to move easily through plasmodesmata. Other candidates for the long-distance silencing signal include aberrant RNAs that could be transported by a host RNA-trafficking system(s). Plasmodesmal (local) and phloem (systemic long-distance) trafficking of endogenous mRNAs has been reported in plants (19, 20, 24, 28). Regardless of the nature of the RNA-silencing signal, we have provided direct evidence that the spread of RNA silencing is a two-step process involving both cell-to-cell and long-distance movement of the silencing signal. The spatial or multidirectional spread of RNA silencing from TCV-GFPΔCP RNA-infected epidermal cells to adjacent cells indicates that the silencing signal(s) moves between cells by a passive diffusion mechanism. However, we cannot rule out the possibility that cell-to-cell movement of the RNA-silencing signal may require host proteins or hijack viral cell-to-cell movement proteins. For example, the two small TCV proteins, p8 and p9, may act in this regard as both proteins are essential for viral RNA cell-to-cell movement in plants (10, 23). It is obvious that TCV CP is not involved in facilitating cell-to-cell movement of the RNA-silencing signal. More likely, CP expressed during TCV infection may impede RNA-silencing signal cell-to-cell movement in addition to its role in inhibiting the production of siRNA. A more recent finding demonstrated that limited and extensive cell-to-cell movements of RNA silencing occur in plants (16). These two types of RNA-silencing cell-to-cell movement could be operated by different mechanisms involving 21- and/or 25-nt siRNA molecules. Interestingly, limited cell-to-cell movement spreads RNA silencing over only 10 to 15 cells that is thought to involve only the primary siRNAs moving from cells where RNA silencing is initially established and therefore is independent of homologous transcripts (16). In contrast, extensive cell-to-cell movement of RNA silencing requires homologous transcripts and occurs via relay amplification (16). Although we cannot distinguish these two types of cell-to-cell movement of RNA silencing in the TCV-based system, our data indeed indicate that silencing movement can be effectively promoted from one single epidermal cell to immediately neighboring cells in a three-dimensional manner and then spread to adjacent cells. This may involve both limited and extensive cell-to-cell movement of RNA silencing in TCV-GFPΔCP-inoculated leaves of line 16c plants.
Furthermore, since both local and systemic induction of RNA silencing occurs after agroinfiltration of TCV-GFPΔCP RNA into line 16c plants, it suggests that RNA silencing induced in different cell types, including phloem cells, may be a prerequisite for long-distance spread of the RNA-silencing signal. Remarkably, the patterns of spread of RNA silencing suggest that the signal moves both from cell to cell and through the phloem, mimicking viral RNA movement in plants (37). Plant viruses encode different proteins that are required for cell-to-cell and long-distance movement of genomic RNAs, respectively (21). Indeed, our data indicate that cell-to-cell RNA-silencing signal movement can be independent of long-distance spread. A recent finding reveals that systemic, but not local, spread of RNA silencing induced in plants by agroinfiltration can be blocked in the presence of low levels of cadmium (33). Intriguingly, the same concentrations of cadmium inhibited systemic movement of tobamoviruses, suggesting that the systemic spread of RNA silencing and that of viral RNA may have steps in their transport pathways in common (33). Moreover, RNA-silencing suppressors such as P1 and AC2 have contrasting effects on cell-to-cell movement and long-distance transport of RNA silencing in the vasculature (16).
Understanding the mechanisms of systemic viral movement restriction may provide a valuable insight into systemic movement of RNA silencing. It has been shown that the spread of viruses in hosts which allow cell-to-cell but not systemic movement is often restricted to the interface between bundle sheath cells and phloem parenchyma cells and the companion cell-sieve element complex (29, 32, 39). These observations suggest that the plasmodesmata which connect mesophyll cells, as well as mesophyll and bundle sheath cells, are structurally and functionally different from the plasmodesmata connecting bundle sheath cells and phloem. It is possible that the local silencing signal could move through one type of channel but would be unable to go through the other owing to an as yet unknown mechanism. In the case of TCV-GFPΔCP RNA-mediated single-cell induction of RNA silencing, the silencing signal moves between epidermal and mesophyll cells and subsequently induces RNA-silencing foci within inoculated leaves but probably was not able to enter the phloem and spread long distance to noninoculated leaves. Thus, it would be likely that lack of long-distance spread of RNA silencing initially induced in single epidermal cells by mechanical inoculation of TCV-GFPΔCP occurred at the stage of entry into the vasculature. This view is consistent with the idea that RNA silencing which is established in phloem by phloem-limited virus spreads readily into various types of cells of lamina of newly emerging young leaves (16). Nevertheless, the system devised in this study may be exploited to dissect how RNA silencing spreads in plants.
Acknowledgments
We thank D. C. Baulcombe for providing transgenic N. benthamiana line 16c seeds, J. I. Cooper for the TCV isolate, and T. M. A. Wilson for encouragement throughout this work and for critical reading of the manuscript.
This project was supported by HRI-core BBSRC funding to E.V.R. and Y.H.
REFERENCES
- 1.Bass, B. L. 2000. Double-stranded RNA as a template for gene silencing. Cell 101:235-238. [DOI] [PubMed] [Google Scholar]
- 2.Benfey, P. N., and N. H. Chua. 1990. The cauliflower mosaic virus 35S promoter: combinatorial regulation of transcription in plants. Science 250:959-966. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein, E., A. A. Caudy, S. M. Hammond, and G. J. Hannon. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363-366. [DOI] [PubMed] [Google Scholar]
- 4.Brigneti, G., O. Voinnet, W.-X. Li, L.-H. Ji, S.-W. Ding, and D. C. Baulcombe. 1998. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17:6739-6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Carrington, J. C., L. A. Heaton, D. Zudema, B. I. Hillman, and T. J. Morris. 1989. The genome structure of turnip crinkle virus. Virology 281:219-226. [DOI] [PubMed] [Google Scholar]
- 6.Carrington, J. C., K. D. Kasschau, and L. K. Johansen. 2001. Activation and suppression of RNA silencing by plant viruses. Virology 281:1-5. [DOI] [PubMed] [Google Scholar]
- 7.Cogoni, C., and G. Macino. 2000. Post-transcriptional gene silencing across kingdoms. Curr. Opin. Genet. Dev. 10:638-643. [DOI] [PubMed] [Google Scholar]
- 8.Cohen, Y., A. Gisel, and P. C. Zambrysky. 2000. Cell-to-cell and systemic movement of recombinant green fluorescent protein-tagged Turnip crinkle virus. Virology 273:258-266. [DOI] [PubMed] [Google Scholar]
- 9.Dong, X., R. van Wezel, J. Stanley, and Y. Hong. 2003. Functional characterization of the nuclear localization signal for a suppressor of posttranscriptional gene silencing. J. Virol. 77:7026-7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hacker, D. L., I. T. Petty, N. Wei, and T. J. Morris. 1992. Turnip crinkle virus genes required for RNA replication and virus movement. Virology 186:1-8. [DOI] [PubMed] [Google Scholar]
- 11.Hamilton, A. J., and D. C. Baulcombe. 1999. A novel species of small antisense RNA in post-transcriptional gene silencing. Science 286:950-952. [DOI] [PubMed] [Google Scholar]
- 12.Hamilton, A. J., O. Voinnet, L. Chappell, and D. C. Baulcombe. 2002. Two classes of short interfering RNA in RNA silencing. EMBO J. 21:4671-4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammond, S. M., A. A. Caudy, and G. J. Hannon. 2001. Post-transcriptional gene silencing by double-stranded RNA. Nat. Rev. Genet. 2:110-119. [DOI] [PubMed] [Google Scholar]
- 14.Hannon, G. J. 2002. RNA interference. Nature 418:244-251. [DOI] [PubMed] [Google Scholar]
- 15.Higuchi, R., B. Krummel, and R. K. Saaki. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16:7351-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Himber, C., P, Dunoyer, G. Moissiard, C. Ritzenthaler, and O. Voinnet. 2003. Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J. 22:4523-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoekema, A., P. R. Hirsch, P. J. J. Hooykaas, and R. A. Schilperoort. 1983. A binary plant vector based on separation of vir and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature 303:179-180. [Google Scholar]
- 18.Hogle, J. M., A. Maeda, and S. C. Harrison. 1986. Structure and assembly of turnip crinkle virus. I. X-ray crystallographic structure analysis at 3.2 Å resolution. J. Mol. Biol. 191:625-638. [DOI] [PubMed] [Google Scholar]
- 19.Kim, M., W. Canio, S. Kessler, and N. Sinha. 2001. Developmental changes due to long-distance movement of homeobox fusion transcript in tomato. Science 293:287-289. [DOI] [PubMed] [Google Scholar]
- 20.Kuhn, C., V. R. Franceschi, A. Schulz, R. Lemione, and W. B. Frommer. 1997. Macromolecular trafficking indicated by localisation and turnover of sucrose transporters in enucleate sieve elements. Science 275:1298-1300. [DOI] [PubMed] [Google Scholar]
- 21.Lazarowitz, S. G., and R. N. Beachy. 1999. Viral movement proteins as probes for intracellular and intercellular trafficking in plants. Plant Cell 11:535-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, H., W. X. Li, and S. W. Ding. 2002. Induction and suppression of RNA silencing by an animal virus. Science 296:1319-1321. [DOI] [PubMed] [Google Scholar]
- 23.Li, W.-Z., F. Qu, and T. J. Morris. 1998. Cell-to-cell movement of turnip crinkle virus is controlled by two small open reading frames that function in trans. Virology 244:405-416. [DOI] [PubMed] [Google Scholar]
- 24.Lucas, W. J., S. Bouche-Pillon, D. P. Jackeson. L. Nguyen, L. Baker, B. Ding, and S. Hake. 1995. Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science 270:1980-1983. [DOI] [PubMed] [Google Scholar]
- 25.Mallory, A. C., L. Ely, T. H. Smith, R. Marathe, R. Anandalakshmi, M. Fagard, H. Vaucheret, G. Pruss, L. Bowman, and V. B. Vance. 2001. HC-Pro suppression of transgene silencing eliminates the small RNAs but not transgene methylation or the mobile signal. Plant Cell 13:571-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mlotshwa, S., O. Voinnet, M. F. Mette, M. Matzke, H., Vaucheret, S. W. Ding, and V. B. Vance. 2002. RNA silencing and the mobile silencing signal. Plant Cell 14:S289-S301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qu, F., T. Ren, and T. J. Morris. 2003. The coat protein of turnip crinkle virus suppresses posttranscriptional gene silencing at an early initiation step. J. Virol. 77:511-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruiz-Medrano, R., B. Xoconostle-Cazares, and W. J. Lucas. 1999. Phloem long-distance transport of CmNACP mRNA: implications for supracellular regulation in plants. Development 126:4405-4419. [DOI] [PubMed] [Google Scholar]
- 29.Ryabov, E. V., I. M. Roberts, P. Palukaitis, and M. Taliansky. 1999. Host-specific cell-to-cell and long-distance movements of cucumber mosaic virus are facilitated by the movement protein of groundnut rosette virus. Virology 260:98-108. [DOI] [PubMed] [Google Scholar]
- 30.Sharp, P. A. 2001. RNA interference 2001. Genes Dev. 15:485-490. [DOI] [PubMed] [Google Scholar]
- 31.Thomas, C. L., V. Leh, C. Lederer, and A. J. Maule. 2003. Turnip crinkle virus coat protein mediates suppression of RNA silencing in Nicotiana benthamiana. Virology 306:33-41. [DOI] [PubMed] [Google Scholar]
- 32.Thompson, J. R., and F. Garcia-Arenal. 1998. The bundle sheath-phloem interface of Cucumis sativus is a boundary to systemic infection by Tomato aspermy virus. Mol. Plant-Microbe Interact. 11:109-114. [Google Scholar]
- 33.Ueki, S., and V. Citovsky. 2000. Inhibition of systemic onset of post-transcriptional gene silencing by non-toxic concentrations of cadmium. Plant J. 28:283-291. [DOI] [PubMed] [Google Scholar]
- 34.Van Wezel, R., X. Dong, H. Liu, P. Tien, J. Stanley, and Y. Hong. 2002. Mutation of three cysteine residues in Tomato yellow leaf curl virus-China C2 protein causes dysfunction in pathogenesis and post-transcriptional gene silencing suppression. Mol. Plant-Microbe Interact. 15:203-208. [DOI] [PubMed] [Google Scholar]
- 35.Voinnet, O. 2001. RNA silencing as a plant immune system against viruses. Trends Genet. 17:449-459. [DOI] [PubMed] [Google Scholar]
- 36.Voinnet, O., and D. C. Baulcombe. 1997. Systemic signalling in gene silencing. Nature 389:553. [DOI] [PubMed] [Google Scholar]
- 37.Voinnet, O., C. Lederer, and D. C. Baulcombe. 2000. A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103:157-167. [DOI] [PubMed] [Google Scholar]
- 38.Voinnet, O., P. Vain, S. Angell, and D. C. Baulcombe. 1998. Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localised induction of ectopic promoterless DNA. Cell 95:177-187. [DOI] [PubMed] [Google Scholar]
- 39.Wang, H.-L., Y. Wang, D. Giesman-Cookmeyer, S. A. Lommel, and W. J. Lucas. 1998. Mutations in viral movement protein alter systemic infection and identify an intercellular barrier to entry into the phloem long-distance transport system. Virology 245:75-89. [DOI] [PubMed] [Google Scholar]
- 40.Waterhouse, P. M., M. B. Wang, and T. Lough. 2001. Gene silencing as an adaptive defence against viruses. Nature 411:834-842. [DOI] [PubMed] [Google Scholar]