Abstract
The UGT1A1*28 variant has been associated with hyperbilirubinemia and atazanavir discontinuation. Protocol A5202 randomly assigned human immunodeficiency virus type 1 (HIV-1)–infected patients to receive atazanavir/ritonavir (atazanavir/r) or efavirenz, with tenofovir/emtricitabine or abacavir/lamivudine. A total of 646 atazanavir/r recipients were evaluable for UGT1A1. Homozygosity for *28/*28 was present in 8% of whites, 24% of blacks, and 18% of Hispanics and was associated with increased bilirubin concentrations. There was an association between *28/*28 and increased atazanavir/r discontinuation among Hispanic participants (P = .005) but not among white or black participants (P = .79 and P = .46, respectively). The positive predictive value of 28*/28* for atazanavir/r discontinuation among Hispanic participants was only 32% (95% confidence interval, 16%–52%).
Keywords: Gilbert syndrome, pharmacogenetics, atazanavir, UGT1A1, HIV therapy
Bilirubin elimination in bile requires glucuronidation by hepatic UDP-glucuronosyltransferase (UGT). Interindividual differences in plasma unconjugated bilirubin concentrations are due in part to a UGT1A1 promoter tandem (TA) repeat that varies from 5 to 8 repeats. The UGT1A1*28 (TA)7 variant reduces UGT1A1 transcription as compared to the UGT1A1*1 (TA)6 allele [1, 2].
Hyperbilirubinemia with the human immunodeficiency virus type 1 (HIV-1) protease inhibitor atazanavir results from inhibition of UGT1A1-mediated bilirubin glucuronidation [3]. Although such hyperbilirubinemia does not indicate hepatic injury, some patients discontinue atazanavir because of jaundice. Among patients receiving once-daily atazanavir 300 mg with ritonavir 100 mg, the reported frequency of grade ≥3 elevation in total bilirubin level at least once is approximately 40% [4–6] and the reported frequency of grade 4 elevation is approximately 5% [5, 6]. Atazanavir discontinuation due to hyperbilirubinemia, however, is infrequent [7–9].
Among atazanavir recipients, UGT1A1*28 has been associated with unconjugated hyperbilirubinemia [10] and increased likelihood of atazanavir discontinuation. In the Swiss HIV Cohort, among 121 patients (80% of whom were white) receiving atazanavir boosted with ritonavir (atazanavir/r), homozygosity for low-expresser UGT1A1 alleles (*28/*28 or *28/*37) was associated with increased risk of treatment discontinuation over 1 year, with cumulative rates of 62.5% among 18 homozygous, 23.8% among 48 heterozygous, and 14.6% among 55 noncarrier individuals [11].
We sought to replicate the association between low-expresser UGT1A1 genotypes and atazanavir/r treatment discontinuation.
MATERIALS AND METHODS
Study Participants
Protocol A5202 (ClinTrials.gov identifier NCT00118898) was a phase IIIb randomized study of 4 once-daily regimens for initial treatment of HIV-1 infection [8, 12]. Participants were randomly assigned to receive open-label 300-mg atazanavir plus 100-mg ritonavir or 600-mg efavirenz, with placebo-controlled 600-mg abacavir–300-mg lamivudine or 300-mg tenofovir DF–200-mg emtricitabine. Study drugs were provided at no cost. Study evaluations were done before entry; at entry; at weeks 4, 8, 16, and 24; and every 12 weeks until the last enrolled subject had been followed for 96 weeks. Site personnel were required to report all grade ≥2 central nervous system adverse events, nausea, diarrhea, all other grade ≥3 signs, symptoms and toxicities, and all signs or symptoms that caused a change in study treatment. Approximately 2.5 years after the study opened, in response to a planned interim data safety monitoring board review, participants with a screening HIV-1 RNA level of ≥100 000 copies/mL were unblinded, and those receiving abacavir/lamivudine could switch to tenofovir/emtricitabine or continue to receive their current regimen [12].
Decisions to discontinue atazanavir/r for bilirubin-related issues were at the discretion of study participants and site investigators, in consultation with the protocol team. Participants who discontinued atazanavir/r could switch to other antiretrovirals (eg, lopinavir/r or fosamprenavir), which were provided at no cost.
Identifying Genetic Polymorphisms
Consent for genetic testing was obtained under AIDS Clinical Trials Group protocol A5128 [13]. Genotyping of UGT1A1 rs8175347 (ie,*28, *36, and *37) by fragment analysis involved polymerase chain reaction followed by capillary electrophoresis. Laboratory personnel with no knowledge of clinical data performed genotyping. Genotype assignment was based on visual inspection for peaks representing promoter TA tandem repeats *36 (TA)5, *1 (TA)6, *28 (TA)7, and *37 (TA)8.
Statistical Analyses
The primary study end point for this analysis was time from documented first dose of atazanavir/r to permanent discontinuation of atazanavir/r. Additional end points of interest included maximum total bilirubin concentration from baseline to week 24, incident cases of reported jaundice, and time to bilirubin-associated atazanavir/r discontinuation. For the latter, attribution of association to bilirubin was based on site report. Questionable attributions were reviewed for final determination by 2 investigators (E. S. D. and D. W. H.). Since follow-up in A5202 was truncated for some individuals because of closure of clinical research sites, participants discontinuing treatment due to site closure were censored for all end points [8, 12, 14]).
Deviation from Hardy–Weinberg equilibrium expectations was evaluated using exact tests. Proportions of subjects with incident cases of hyperbilirubinemia-attributed atazanavir/r discontinuations across ordered genotypes were compared with a Cochran-Mantel Haenszel test for non-0 correlation stratified by race; comparisons within racial subgroups used the Cochran-Mantel Haenszel trend test. The distribution of time to atazanavir/r discontinuation was estimated using Kaplan-Meier methods and compared across ordered genotypes, using the exact Tarone test for trend. Cox proportional hazards models were used to estimate the relative hazards of atazanavir/r discontinuation by UGT1A1 genotype (low-risk *1/*1 as reference), with adjustment for randomized nucleoside reverse-transcriptase inhibitor (NRTI; abacavir/lamivudine vs tenofovir/emtricitabine), sex, age, and body mass index (BMI [defined as the weight in kilograms divided by the height in meters squared]; <18, 18–25, >25–30, and >30), and plasma HIV-1 RNA concentration at study entry. The discriminatory properties (sensitivity, specificity, and positive predictive value) of the UGT1A1 high-risk 28*/*28* genotype for predicting atazanavir/r discontinuation and hyperbilirubinemia-attributed atazanavir/r discontinuation within 48 weeks of antiretroviral initiation (binary outcome) were estimated with exact 95% confidence intervals (CIs).
On the basis of the low expected frequencies of UGT1A1*36 (TA)5 and *37 (TA)8 and their known effects on UGT1A1 expression, UGT1A1*36 (TA)5 and *37 (TA)8 were grouped with *1 (TA)6 and *28 (TA)7 alleles, respectively. This provided a 3-level ordered genotype based on (allele 1)/(allele 2): (*1 or *36)/(*1 or *36), (*1 or *36)/(*28* or *37), and (*28* or *37)/(*28* or *37). These are hereafter referred to as *1/*1, *1/*28*, and 28*/*28*, respectively.
Genetic association analyses involved all self-identified non-Hispanic and Hispanic white and black individuals who were randomly assigned to and initiated an atazanavir/r-containing regimen, had consented to genetic testing under A5128, and were successfully genotyped for UGT1A1 rs8175347. Self-identified race/ethnicity categories “white, non-Hispanic,” “black, non-Hispanic,” and “Hispanic” are hereafter referred to as white, black, and Hispanic, respectively. All analyses are performed separately by race/ethnicity groups and overall with stratification by race/ethnicity.
This study complied with the Helsinki Declaration and was approved by institutional review boards for each site, and subjects gave written informed consent.
RESULTS
Study Participants and UGT1A1 Genotypes
A total of 928 participants in A5202 were randomly assigned to receive atazanavir/r, of whom 646 were included in the present analyses. Derivation of these 646 study participants and baseline characteristics are provided in Supplementary Figure 1. Among these 646 participants, 544 (84%) were male, 291 (45%) were white, 202 (31%) were black, and 153 (24%) were Hispanic. The UGT1A1*1 (TA)6 allele was most frequent overall (69% in whites, 47% in blacks, and 60% in Hispanics), followed by UGT1A1*28 (TA)7 (31% in whites, 40% in blacks, and 39% in Hispanics). The UGT1A1*36 (TA)5 and UGT1A1*37 (TA)8 alleles were extremely infrequent in white and Hispanic participants (only 3 white participants and 1 Hispanic participant had at least 1 such allele). Allelic frequencies of UGT1A1*36 (TA)5 and UGT1A1*37 (TA)8 were 7% and 6%, respectively, in black participants. After grouping *36 with *1 and *37 with *28, homozygosity for UGT1A1*28/*28 was present in 100 individuals, including 24 (8%), 48 (24%), and 28 (18%) of white, black, and Hispanic participants, respectively. Genotype frequencies did not deviate significantly from Hardy-Weinberg equilibrium.
UGT1A1 Genotype, Serum Bilirubin Concentrations, and Jaundice
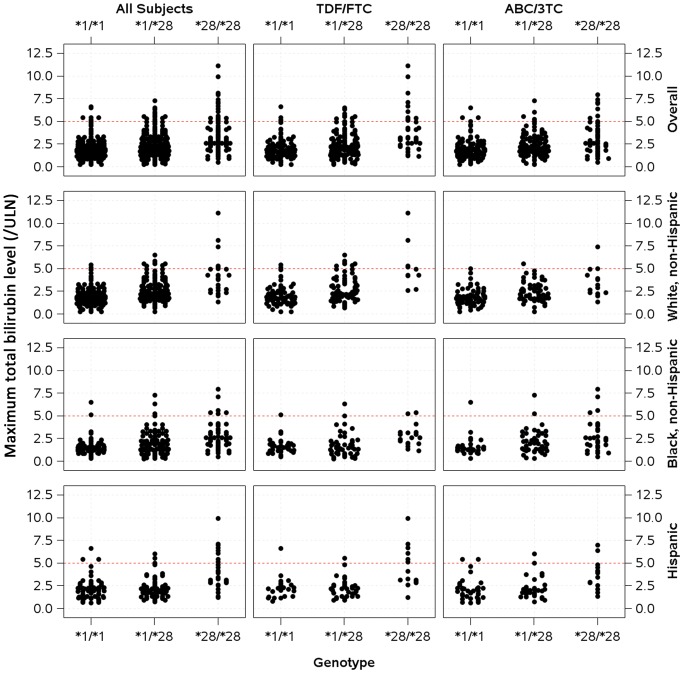
During the first 24 weeks of atazanavir/r treatment, across all race/ethnicity groups, higher proportions of individuals homozygous for UGT1A1*28/*28 experienced grade 4 elevations in bilirubin level (defined as a level ≥5 times the upper limit of normal). Relationships between maximum total bilirubin concentration and UGT1A1 genotype are shown in Figure 1. Incident reports of jaundice among individuals homozygous for UGT1A1*28/*28 were significantly higher among white (25%) and Hispanic (29%) participants, but not among black participants (10%; Supplementary Table 1).
Figure 1.
Relationships between UGT1A1 genotype and maximum total bilirubin level during the first 24 weeks of atazanavir/ritonavir therapy. Data are stratified by race/ethnicity and by nucleoside reverse-transcriptase inhibitor to which participants were randomly assigned. The dashed horizontal line indicates grade 4 total bilirubin level (defined as 5 times the upper limit of normal [ULN]). Abbreviations: TDF/FTC, tenofovir DF/emtricitabine; ABC/3TC, abacavir/lamivudine.
UGT1A1 Genotype and Atazanavir/r Discontinuation
Among the 646 qualifying participants, 27% prematurely discontinued atazanavir/r (24%, 32%, and 27% among white, black, and Hispanic participants, respectively). Among 177 participants who prematurely discontinued atazanavir/r, this was attributed to bilirubin-associated issues in 3 (4%) of 70 white participants, 7 (11%) of 65 black participants, and 9 (21%) of 42 Hispanic participants (P = .046). Most atazanavir/r discontinuations (79%) were determined to not be due to bilirubin-associated issues (Supplementary Table 2).
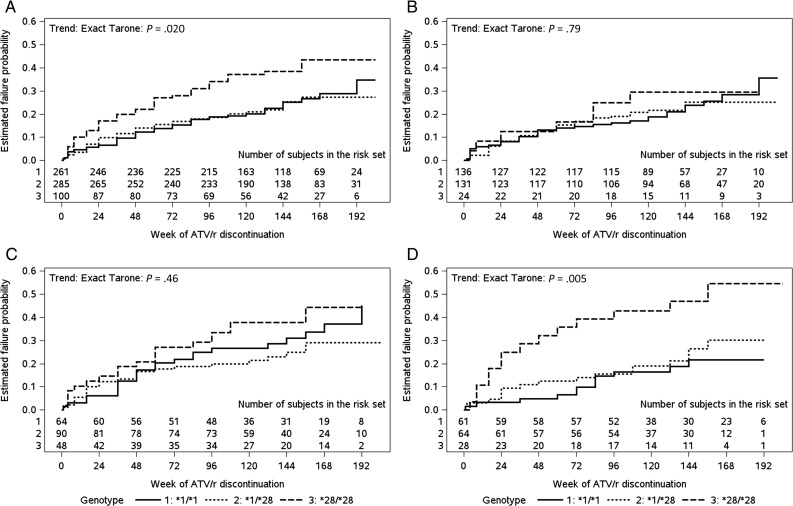
The number of UGT1A1*28 alleles was associated with time to discontinuation across the study population as a whole (P = .02), but there was no apparent association among white participants and black participants (P = .79 and P = .46 respectively). Among Hispanic participants, having a greater number of UGT1A1*28 alleles was associated with shorter time to atazanavir/r discontinuation, driven by *28/*28 homozygosity (P = .005; Figure 2). Findings were similar in Cox proportional hazards models stratified by race/ethnicity and adjusted for age, BMI, concomitant NRTI use, baseline HIV-1 RNA concentration, and sex (P = .003). Over >3 years of follow-up, the estimated cumulative incidence of atazanavir/r discontinuation attributed to bilirubin-associated issues among all participants was <3% for the combined UGT1A1 *1/*1 and *1/*28 genotypes and 8% for UGT1A1 *28/*28. The positive predictive value of the UGT1A1 *28/*28 genotype for atazanavir/r discontinuation was low among white (13%; 95% CI, 3%–32%), black (21%; 95% CI, 10%–35%), and Hispanic (32%; 95% CI, 16%–52%) participants.
Figure 2.
Time to premature atazanavir/ritonavir (ATV/r) therapy discontinuation, stratified by UGT1A1 genotype. According to UGT1A1 genotype, within each panel the lines estimate the cumulative probability of discontinuing ATV/r overall (A) and among whites (B), blacks (C), and Hispanics (D). P values are given by exact Tarone tests. Solid lines represent UGT1A1*1/*1, dotted lines represent *1/*28, and dashed lines represent *28/*28. On each panel, numbers of participants in the risk set at each time point are provided.
It was unclear why UGT1A1 *28/*28 would be associated with atazanavir/r discontinuation only in Hispanic participants. We found no suggestive trends regarding patterns of atazanavir/r discontinuations across clinical study sites and potential associations with race/ethnicity and clustering of UGT1A1 genotypes. Furthermore, results were unchanged in sensitivity analyses that adjusted for population stratification by incorporating EIGENSTRAT values generated from whole genome data [13]. To investigate whether associations in white or black participants may have been masked by discontinuations that were possibly attributable to clinical trial participation, potential clinical-trial-associated reasons (eg, inability to get to study visits, nonadherence with study visits, and withdrawal of consent) were censored in a post hoc competing risks approach. A higher proportion of clinical-trial-associated reasons for discontinuation were observed among black participants (20% vs 11% and 12% for white and Hispanic participants, respectively). After censoring these events, there was a trend toward a higher hazard of non–clinical-trial-associated atazanavir/r discontinuation with a greater number of *28 alleles among black participants (P = .1), while results for Hispanic and white participants were unchanged.
DISCUSSION
In a previous observational study involving Swiss HIV Cohort Study patients who had initiated atazanavir-containing regimens, low-expresser UGT1A1 genotypes (primarily UGT1A1*28) were reported to be associated with increased likelihood of atazanavir/r discontinuation during the first year of therapy [11]. In the present study, the low-expresser UGT1A1 genotype (primarily UGT1A1*28) was not significantly associated with an increased likelihood of atazanavir discontinuation among either white or black participants. Although an association was seen among Hispanics, the *28/*28 genotype did not reliably predict atazanavir/r discontinuation. We presently cannot explain why this association was seen only among Hispanics.
There are several possible reasons for the discrepant findings of the present study with the Swiss HIV Cohort Study [11]. The present analyses involved participants in a prospective clinical trial in the United States, rather than patients in clinical care in Switzerland. It is possible that clinical trial participants were for some reason more inclined to continue assigned study medications despite similar degrees of elevated bilirubin levels, and despite availability of alternative antiretrovirals at no cost. It is well appreciated that unrecognized population stratification can generate spurious associations, which could have contributed to discrepant findings between the 2 studies. A post hoc sensitivity analysis also suggested that a genotype association may have been masked because of discontinuations that were possibly attributable to clinical trial participation rather than to tolerability.
The present study had several limitations. While we focused on the UGT1A1 promoter tandem TA repeat, a more comprehensive, high-throughput genotyping approach might identify novel predictors of atazanavir/r discontinuation. However, as our goal was to replicate the Swiss HIV Cohort Study findings, the present genotyping strategy was appropriate. In addition, while UGT1A1 is the dominant genetic determinant of bilirubin concentrations, we cannot exclude the possibility that additional polymorphisms could improve prediction of bilirubin-driven discontinuations. Nongenetic causes of atazanavir/r discontinuation (eg, nonadherence) could obscure genetic associations.
In summary, the UGT1A1*28 genotype was associated with an increased likelihood of hyperbilirubinemia in a large, prospective clinical trial and was associated with an increased likelihood of atazanavir/r discontinuation among Hispanic participants but not among white or black participants. The association among Hispanic participants did not appear to be driven by decisions related to bilirubin level. The low positive predictive value from these data does not support routine testing for UGT1A1*28 before starting atazanavir/r-based regimens.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We are grateful to the many persons with HIV infection who volunteered for protocols A5202 and A5128; the study teams and site staff involved in protocols A5202 and A5128; and Kasey D. Lawrence of the Vanderbilt DNA Sequencing Core and Paul Leger and Melissa Potter of the Vanderbilt Center for Human Genetics Research, for UGT1A1 genotyping support.
Disclaimer. The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases (NIAID) or the National Institutes of Health (NIH).
Financial support. This work was supported by the NIAID (U01AI068636), the National Institute of Mental Health, and the National Institute of Dental and Craniofacial Research. The ACTG is funded by the NIAID (AI-068636, AI-038858, AI-068634, and AI-038855). Grant support included AI-069439, RR-024975, AI-054999 (to D. W. H.), AI-077505 (to D. W. H. and H. J. R.), BRS-ACURE-06-00140-T001 (to G. D. M.), and AI-069434 (to A. C. C.). Clinical research sites that participated in ACTG protocol A5202 and collected DNA under protocol A5128 were supported by the NIAID (AI-069477, AI-027675, AI-073961, AI-069474, AI-069432, AI-069513, AI-069423, AI-050410, AI-069452, AI-69450, AI-054907, AI-069428, AI-069439, AI-069467, AI-045008, AI-069495, AI-069415, AI-069556, AI-069484, AI-069424, AI-069532, AI-069419, AI-069471, AI-025859, AI-069418, AI-050409, AI-069423, AI-069501, AI-069502, AI-069511, AI-069434, AI-069465, AI-069471, AI-069494, AI-069424, AI-069472, AI-069501, AI-069470, AI-046376, AI-069511, AI-072626, AI-069472, AI-038858, AI-069472, AI-069472, AI-069471, AI-027661, AI-027661, AI-034853, AI-069472, AI-069447, AI-032782, AI-027658, AI-27666, AI-027661, AI-058740, AI-046370, AI-069423, AI-069470, AI-069511, AI-069470, and AI-069494) and the National Center for Research Resources (RR-00051, RR-00046, RR-025747, RR-025777, RR-024160, RR-024996, RR-024156, RR024160, and RR-024160). Study drugs were provided by Bristol-Myers Squibb and GlaxoSmithKline.
Potential conflicts of interest. E. D. is a consultant for Merck, Gilead, Bristol Myers Squibb, and ViiV and receives grant support from Abbott, Merck, Pfizer, Gilead, and ViiV. C. T. is a member of a data monitoring committee for a hepatitis drug manufactured by Tibotec. P. S. is as a consultant for Abbott, BMS, Gilead, GSK, Merck, Tibotec, and ViiV and receives grant support from BMS, Gilead, Merck, Tibotec, and ViiV. A. C. C. has received grant support from Boehringer-Ingelheim, Gilead Sciences, Merck, Schering-Plough, and Tibotec-Virco; is a former member of a data safety and monitoring board for a Merck-sponsored study; and owns stock in Abbott Laboratories, Bristol Myers Squibb, Johnson and Johnson, and Pfizer. D. W. H. receives grant support from Bristol Myers Squibb, Boehringer Ingelheim, Merck, and Gilead Sciences. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Bosma PJ, Chowdhury JR, Bakker C, et al. The genetic basis of the reduced expression of bilirubin UDP-glucuronosyltransferase 1 in Gilbert's syndrome. New Engl J Med. 1995;333:1171–5. doi: 10.1056/NEJM199511023331802. [DOI] [PubMed] [Google Scholar]
- 2.Monaghan G, Ryan M, Seddon R, Hume R, Burchell B. Genetic variation in bilirubin UPD-glucuronosyltransferase gene promoter and Gilbert's syndrome. Lancet. 1996;347:578–81. doi: 10.1016/s0140-6736(96)91273-8. [DOI] [PubMed] [Google Scholar]
- 3.Zhang D, Chando TJ, Everett DW, Patten CJ, Dehal SS, Humphreys WG. In vitro inhibition of UDP glucuronosyltransferases by atazanavir and other HIV protease inhibitors and the relationship of this property to in vivo bilirubin glucuronidation. Drug Metab Dispos. 2005;33:1729–39. doi: 10.1124/dmd.105.005447. [DOI] [PubMed] [Google Scholar]
- 4.Cleijsen RM, van de Ende ME, Kroon FP, et al. Therapeutic drug monitoring of the HIV protease inhibitor atazanavir in clinical practice. J Antimicrob Chemo. 2007;60:897–900. doi: 10.1093/jac/dkm298. [DOI] [PubMed] [Google Scholar]
- 5.Molina JM, Andrade-Villanueva J, Echevarria J, et al. Once-daily atazanavir/ritonavir versus twice-daily lopinavir/ritonavir, each in combination with tenofovir and emtricitabine, for management of antiretroviral-naive HIV-1-infected patients: 48 week efficacy and safety results of the CASTLE study. Lancet. 2008;372:646–55. doi: 10.1016/S0140-6736(08)61081-8. [DOI] [PubMed] [Google Scholar]
- 6.Torti C, Lapadula G, Antinori A, et al. Hyperbilirubinemia during atazanavir treatment in 2404 patients in the Italian atazanavir expanded access program and MASTER Cohorts. Infection. 2009;37:244–9. doi: 10.1007/s15010-008-8010-6. [DOI] [PubMed] [Google Scholar]
- 7.Puls RL, Srasuebkul P, Petoumenos K, et al. Efavirenz versus boosted atazanavir or zidovudine and abacavir in antiretroviral treatment-naive, HIV-infected subjects: week 48 data from the Altair study. Clin Infect Dis. 2010;51:855–64. doi: 10.1086/656363. [DOI] [PubMed] [Google Scholar]
- 8.Daar ES, Tierney C, Fischl MA, et al. Atazanavir plus ritonavir or efavirenz as part of a 3-drug regimen for initial treatment of HIV-1. Ann Intern Med. 2011;154:445–56. doi: 10.1059/0003-4819-154-7-201104050-00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McDonald C, Uy J, Hu W, et al. Clinical significance of hyperbilirubinemia among HIV-1-infected patients treated with atazanavir/ritonavir through 96 weeks in the CASTLE Study. AIDS Patient Care STDs. 2012;25:259–64. doi: 10.1089/apc.2011.0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rotger M, Taffe P, Bleiber G, et al. Gilbert syndrome and the development of antiretroviral therapy-associated hyperbilirubinemia. J Infect Dis. 2005;192:1381–6. doi: 10.1086/466531. [DOI] [PubMed] [Google Scholar]
- 11.Lubomirov R, Colombo S, di Iulio J, et al. Association of pharmacogenetic markers with premature discontinuation of first-line anti-HIV therapy: an observational cohort study. J Infect Dis. 2011;203:246–57. doi: 10.1093/infdis/jiq043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sax PE, Tierney C, Collier AC, et al. Abacavir-lamivudine versus tenofovir-emtricitabine for initial HIV-1 therapy. N Engl J Med. 2009;361:2230–40. doi: 10.1056/NEJMoa0906768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haas DW, Wilkinson GR, Kuritzkes DR, et al. A multi-investigator/institutional DNA bank for AIDS-related human genetic studies: AACTG Protocol A5128. HIV Clin Trials. 2003;4:287–300. doi: 10.1310/MUQC-QXBC-8118-BPM5. [DOI] [PubMed] [Google Scholar]
- 14.Sax PE, Tierney C, Collier AC, et al. Abacavir/lamivudine versus tenofovir DF/emtricitabine as part of combination regimens for initial treatment of HIV: final results. J Infect Dis. 2011;204:1191–201. doi: 10.1093/infdis/jir505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.