Abstract
The disappointing efficacy of blood-stage malaria vaccines may be explained in part by allele-specific immune responses that are directed against polymorphic epitopes on blood-stage antigens. FMP2.1/AS02A, a blood-stage candidate vaccine based on apical membrane antigen 1 (AMA1) from the 3D7 strain of Plasmodium falciparum, had allele-specific efficacy against clinical malaria in a phase II trial in Malian children. We assessed the cross-protective efficacy of the malaria vaccine and inferred which polymorphic amino acid positions in AMA1 were the targets of protective allele-specific immune responses. FMP2.1/AS02A had the highest efficacy against AMA1 alleles that were identical to the 3D7 vaccine-type allele at 8 highly polymorphic amino acid positions in the cluster 1 loop (c1L) but differed from 3D7 elsewhere in the molecule. Comparison of the incidence of vaccine-type alleles before and after vaccination in the malaria vaccine and control groups and examination of the patterns of allele change at polymorphic positions in consecutive malaria episodes suggest that the highly polymorphic amino acid position 197 in c1L was the most critical determinant of allele-specific efficacy. These results indicate that a multivalent AMA1 vaccine with broad efficacy could include only a limited set of key alleles of this extremely polymorphic antigen.
Keywords: Malaria, Plasmodium falciparum, apical membrane antigen 1, AMA1, vaccine trial, allele-specific efficacy, cluster 1 loop
Antigenic diversity in Plasmodium falciparum surface antigens may pose a major obstacle to the development of an effective malaria vaccine [1, 2]. P. falciparum apical membrane antigen 1 (AMA1) is considered a promising vaccine candidate [3], but in vitro studies [4, 5] and molecular epidemiological studies [2, 6, 7] suggest that extreme polymorphism in AMA1 may limit its efficacy as a vaccine antigen. The extracellular domain of AMA1 is divided into 3 subdomains on the basis of the pattern of disulfide bonds [8]. Domain 1 is divided into 3 clusters (clusters 1, 2, and 3) on the basis of amino acid spatial proximity [9], and the most polymorphic region of AMA1 is a loop within cluster 1 (the cluster 1 loop; c1L) that contains 8 highly polymorphic amino acid positions (Figure 1).
Figure 1.
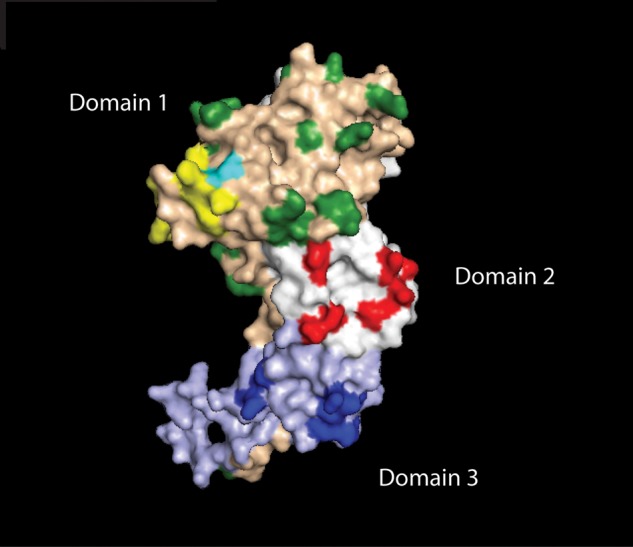
Side view of the apical membrane 1 (AMA1) crystal structure. Domain 1 amino acids are shown in tan, with polymorphic residues highlighted in green. Cluster one loop (c1L) amino acids, which are located in domain 1, are shown in yellow. Within c1L, amino acid 197 is in turquoise. Domain 2 residues are shaded in white. Polymorphic residues in this region are highlighted in red. Domain 3 residues are shown in light blue, with polymorphic residues in dark blue.
Allelic exchange experiments [5, 10], in vitro analysis of amino acid relevance [11–13], and molecular epidemiology studies [7] identified polymorphic amino acid positions in c1L (residues 196, 197, 199, 200, 201, 204, 206, and 207) as being the main targets of naturally acquired protective antibodies. It was therefore hypothesized that these amino acids could be important in determining allele-specific efficacy.
We recently demonstrated the first evidence of allele-specific efficacy against clinical malaria of an AMA1 vaccine (FMP2.1/AS02A) in a phase II trial conducted in Malian children [14]. FMP2.1/AS02A, developed by the Walter Reed Army Institute of Research, is a monovalent blood-stage vaccine that is based on an AMA1 sequence corresponding to that of the 3D7 strain of P. falciparum and is formulated with GlaxoSmithKline Biologicals' Adjuvant System AS02A. A phase II trial of a different, bivalent vaccine containing 2 versions of AMA1, which are based on sequences derived from the 3D7 and FVO strains of P. falciparum, showed neither overall nor allele-specific efficacy [15]. The leading preerythrocytic vaccine, RTS,S/AS01, which is based on the circumsporozoite protein of P. falciparum, has shown about 50% overall efficacy against clinical malaria, but thus far no allele-specific efficacy has been reported on the basis of sequencing of T-cell epitopes [16–18].
The analyses reported here were designed to understand which AMA1 alleles contributed to the observed allele-specific efficacy of the FMP2.1/AS02A vaccine. We previously found that sera from Malian adults vaccinated with FMP2.1/AS02A inhibited parasite growth in a non–allele-specific fashion [19]. Here we present the results of exploratory analyses designed to assess systematically the degree of cross-protective efficacy provided by FMP2.1/AS02A against clinical malaria with diverse AMA1 alleles, with the ultimate goal of informing development of an AMA1 malaria vaccine that offers broad cross-protective efficacy.
METHODS
Ethics Approval
Ethics approval for the vaccine trial was obtained from the institutional review boards of the School of Medicine and Pharmacy (Bamako, Mali), the University of Maryland (Baltimore, MD), the Walter Reed Army Institute of Research (Silver Spring, MD), and the US Army Surgeon General.
Overall Study Design
The FMP2.1/AS02A vaccine was evaluated in a randomized, controlled, phase II clinical trial designed to assess the efficacy, safety, and immunogenicity of the AMA1 malaria vaccine FMP2.1/AS02A versus rabies vaccine in Bandiagara, Mali [14]. The study population comprised 2 groups of children aged 1–6 years old. One group (199 children) received the malaria vaccine (50 μg of FMP2.1 adjuvanted with 0.5 mL of AS02A), and the control group (201 children) received the rabies vaccine. During participant screening, at each cross-sectional survey (days 0, 30, 60, 90, 120, 150, 180, 210, and 240) and each clinic visit with malaria symptoms, children were clinically examined, and blood was collected on filter paper for ama1 gene sequencing. A clinical malaria episode was defined as an axillary temperature of ≥37.5°C and a parasitemia of ≥2500 asexual P. falciparum parasites per microliter of blood [20]. Allele-specific analyses consisted of an intention-to-treat analysis and a per-protocol analysis (starting at day 74, 2 weeks after the last vaccination at day 60).
DNA Extraction, ama1 Gene Amplification, and Sequencing
The QIAamp manufacturer's instructions for the 96 DNA blood kit (Qiagen, Valencia, CA) were followed to extract malaria parasite DNA from filter paper blood spots. The entire 1861-bp ectodomain coding sequence of the pfama1 gene was amplified following a previously described nested polymerase chain reaction protocol [15]. Purified products were sequenced as described by Duan et al [21], using an ABI 3730XL automatic sequencer (Applied Biosystems, Foster City, CA).
Sequences were edited and aligned using 3D7 (GenBank accession number AF512508) as the reference sequence. Sequencher 4.8 software (Gene Codes, Ann Arbor, MI) was used to align and edit DNA sequences. Sequences were defined as collected from single/predominant clone infections or polyclonal infections on the basis of the peak height of the electropherogram. Multiple-allele infections were defined as those with a secondary peak height of ≥50% of the primary peak height at any polymorphic site.
Statistical Analysis
Haplotypes were defined only for single/predominant infections. To assess the cross-protective efficacy of the FMP2.1/AS02A vaccine, SAS 9.2 (Cary, NC) was used to assess the time to the first clinical malaria episode with the 3D7 (DERHFDKY) or a non-3D7 AMA1 c1L haplotype, using Kaplan-Meier survival curves. To evaluate the cross-protective efficacy of the malaria vaccine, the time to the first clinical malaria episode with AMA1 c1L haplotypes matching Fab9 (DQRHFDKY), DD2 (DRRLLDED), M5 (NGRDLNEY), and FVO (NGRDFNEY) during the first malaria transmission season (days 0–240) was also estimated using a Kaplan-Meier approach. Cox proportional hazards models were used to assess the association between vaccine group and risk of a clinical episode with parasites having an AMA1 c1L haplotype identical to 3D7 or any of the aforementioned alleles, and χ2 tests were used to compare the frequency of c1L haplotypes in the 2 vaccine groups.
To assess whether c1L was the main target of vaccine-induced immune responses, haplotypes were defined on the basis of polymorphic positions that discriminate between 3D7 and FVO in domain 2 (positions 308, 330 332, and 404) and domain 3 (positions 439 and 451). Haplotypes defined on the basis of these positions were used to estimate the time to the first clinical malaria episode with a 3D7-type haplotype. Furthermore, we categorized sequences into groups of high, medium, and low homology to 3D7 on the basis of the number of amino acid differences in c1L. These groups were used in a time to the first clinical malaria episode analysis, using a Kaplan-Meier approach and a Cox proportional hazards analysis.
Polymorphic positions relevant to vaccine escape were identified by comparing the incidence of vaccine alleles across all polymorphic sites in the 2 vaccine groups during and just after the vaccination period (days 0–74, with the last vaccination occurring on day 60) and starting 2 weeks after the last vaccination (after day 74). Data collected from the 2 vaccine groups were used to compute the relative risk ratio (RRR) of the incidence of vaccine-type amino acids at each polymorphic AMA1 amino acid position. The RRR was estimated as follows: [(incidence of amino acid in rabies group after vaccination)/(incidence of amino acid in AMA1 group after vaccination)]/[(incidence of amino acid in rabies group during vaccination)/(incidence of amino acid in AMA1 group during vaccination)].
To investigate the shift from vaccine-type to non–vaccine-type alleles at individual polymorphic amino acid positions in the 2 vaccine groups, we compared the frequency of residues that changed from a 3D7-type haplotype to a non–3D7-type haplotype in consecutive clinical malaria episodes in sequences identified prior to the final vaccine dose to the frequency of those collected after the final vaccination. Data from the 2 vaccine groups were used to estimate the RRR of having a shift from vaccine-type to non–vaccine-type amino acids at each polymorphic position in AMA1. The formula used to compute the RRR of amino acid shifts from vaccine-type to non–vaccine-type alleles (amino acid dynamics) can be written as follows: {[(change from a 3D7 type to a non–3D7 type) in AMA1 group after vaccination]/[(all changes) in AMA1 group after vaccination]}/{[(change from a 3D7 type to a non–3D7 type) in AMA1 group during vaccination]/[(all changes) in AMA1 group during vaccination]}
RESULTS
Overall, 600 ama1 gene sequences (GenBank accession numbers JQ812138–JQ812610) were obtained from both clinical malaria episodes and asymptomatic infections during the first 6 months of follow-up. Single and/or predominant clone infections represented 78.83% of these sequences (473 of 600). Among the 473 single and/or predominant clone sequences, 221 were observed in the FMP2.1/AS02A vaccine group, and 252 were observed in the rabies group.
Allele-Specific Efficacy
Cross-protective efficacy of the FMP2.1/AS02A vaccine against clinical malaria due to heterologous alleles was measured by comparing the hazard of the first clinical malaria episode with various AMA1 c1L haplotypes (3D7, Fab9, DD2, M5, and FVO strains) in the 2 vaccine groups. Table 1 shows estimates of vaccine efficacy and the incidence of clinical malaria episodes observed for each haplotype. The incidence of clinical malaria with the Fab9 haplotype, which differed from 3D7 by only 1 amino acid (at position 197), was comparable to the incidence of 3D7-type clinical episodes. However, the vaccine did not have significant efficacy against the Fab9 AMA1 c1L haplotype (vaccine efficacy, 1%; 95% confidence interval [CI], .45%–2.17%). Analyses targeting the 3 remaining haplotypes had similar results. These analyses indicate that the vaccine had no cross-protective efficacy against parasites with AMA1 c1L sequences that were not fully identical to the vaccine 3D7 haplotype.
Table 1.
Cross-Protective Efficacy of the Malaria Vaccine Against Malaria Episodes Due to Alleles Defined on the Basis of Amino Acids in Cluster One Loop (c1L)
| Variable | Amino Acid Differences From 3D7 c1L, No.a | Sequences, No. | HR (95% CI) | Estimated Vaccine Efficacy, % |
|---|---|---|---|---|
| AMA1 c1L allele (allele name) | ||||
| DERHFDKY (vaccine allele 3D7) | 0 | 22 | 0.36 (.14–.92) | 64 |
| DQRHFDKY (Fab9) | 1 | 25 | 0.99 (.45–2.17) | 1.0 |
| NGRDFNEY (FVO) | 5 | 10 | 1.28 (.39–4.92) | −38 |
| DDRLLDED (Dd2) | 5 | 24 | 0.90 (.41–2.01) | 10 |
| NGRDLNEY (M5) | 6 | 12 | 0.65 (.21–2.05) | 35 |
| Allele grouping | ||||
| High agreement with 3D7 | 1–2 | 26 | 0.88 (.36–2.16) | 12 |
| Medium agreement with 3D7 | 3–4 | 50 | 0.85 (.48–1.50) | 15 |
| Low agreement with 3D7 | 5–8 | 61 | 1.1 (.66–1.80) | −10 |
| All non-3D7 | 1–8 | 179 | 0.93 (.69–1.25) | 7 |
| 3D7 based on domain 2 | 1–4 | 5 | NA | NA |
| 3D7 based on domain 3 | 1–2 | 45 | 0.73 (.41–1.30) | 27 |
| Amino acid at position 197 (allele name) | ||||
| E (3D7) | 0 | 22 | 0.36 (.14–0.92) | 64 |
| Q (Fab9, HB3) | 1 | 35 | 0.94 (.50–1.80) | 6 |
| G (FVO, M5) | 1 | 30 | 0.80 (.38–1.67) | 20 |
| D (DD2, L32) | 1 | 40 | 0.90 (.48–1.67) | 10 |
| H (7G8, 425) | 1 | 24 | 1.09 (.49–2.43) | −9 |
Abbreviations: CI, confidence interval; HR, hazard ratio; NA, not available.
a Range, 0–8 amino acid differences.
To determine whether the malaria vaccine had activity against clinical malaria episodes with regard to the number of amino acid differences between the vaccine 3D7 c1L sequence and non-3D7 c1L sequence, we grouped non-3D7 haplotypes into those with low (5–8 amino acid differences), medium (3–4 differences), and high (1–2 differences) degrees of similarity to 3D7. Haplotypes identified using these groupings were used to assess the time to the first clinical malaria episode in the 2 vaccine groups. The time to the first clinical malaria episode with any of these haplotypes was comparable in the 2 vaccine groups, and neither vaccine was associated with a reduction of the risk of clinical malaria episodes with any of the haplotypes (Table 1).
Finally, to confirm a lack of cross-protective efficacy, we estimated vaccine efficacy against any non-3D7 c1L haplotypes. The time to a clinical malaria episode with any non-3D7 c1L haplotype was comparable in the 2 vaccine groups (P = .62), and no vaccine efficacy was observed (hazard ratio, 0.93; 95% CI, .69–1.25; Table 1).
Vaccine-Induced Selection
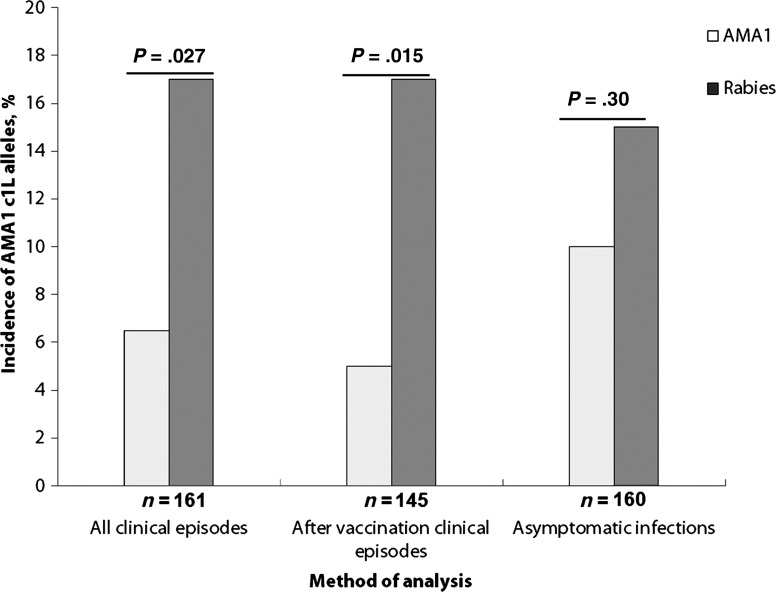
By using the amino acids in c1L to define haplotypes, we identified incident cases of clinical malaria caused by infections with 3D7-type c1L haplotypes in the malaria and rabies vaccine groups. The incidence of clinical malaria episodes with parasites having AMA1 with the 3D7 c1L haplotype was reduced in the malaria vaccine group as compared to the rabies vaccine group: 16 episodes with 3D7-type c1L haplotypes were seen in the rabies vaccine group, while only 6 were seen in the AMA1 vaccine group (intention-to–treat analysis: χ2 = 4.9 and P = .027; Figure 2). In a similar analysis using only sequences from clinical malaria episodes occurring in individuals who received all 3 doses of vaccine, the reduction of the incidence of 3D7-type c1L haplotype in the malaria vaccine group was even greater (per-protocol analysis: χ2 = 5.97 and P = .015; Figure 2). In contrast, sequences with the 3D7 c1L haplotype detected in asymptomatic infections during active surveillance (cross-sectional surveys) were similarly distributed between the 2 vaccine groups (P = .3; Figure 2). These observations suggest that selective pressure induced by the malaria vaccine significantly reduced the incidence of homologous alleles during clinical malaria episodes but not in asymptomatic infections.
Figure 2.
Incidence of 3D7-type apical membrane antigen 1 (AMA1) cluster one loop (c1L) haplotypes detected during clinical episodes and in asymptomatic infections detected by active surveillance in the malaria vaccine and rabies groups. The incidence of vaccine-type AMA1 c1L haplotypes was significantly lower in the AMA1 vaccine group in clinical episodes (but not in asymptomatic infections detected during active surveillance).
Identifying Important Amino Acids
To identify specific amino acid positions targeted by vaccine-induced immune responses, we explored the amino acid composition across the whole ectodomain of sequences identified through study day 240. This analysis showed that only 5 sequences exactly matched the 3D7 haplotype at all polymorphic positions. Six of the 22 sequences with c1L amino acids matching 3D7 corresponded to P. falciparum strain PC26 on the basis of the whole ectodomain sequence, 3 matched the sequence of the S35 strain, and 8 were of other types (Supplementary Figure 1). This variation outside of c1L suggests that polymorphic residues located outside c1L may not play as important a role in recognition by inhibitory antibodies. Moreover, we used residues that differentiate 3D7 and FVO in domain 2 (positions 308, 330, 332, and 404 [3D7: QPIT; FVO: ESNR]) and discriminate amino acids in domain 3 (positions 439 and 451 [3D7: NM; FVO: HK]) to define haplotypes. Haplotypes identified by these methods were then used to model the time to a clinical malaria episode with a 3D7 haplotype. Participants who received the malaria vaccine had survival times comparable to those who received the rabies vaccine (domain 2: long rank test χ2 = 2.67, P = .10; domain 3: log rank test χ2 = 1.16, P = .28). Moreover, we observed no difference in vaccine efficacy against clinical malaria between the 2 vaccine groups (Table 1).
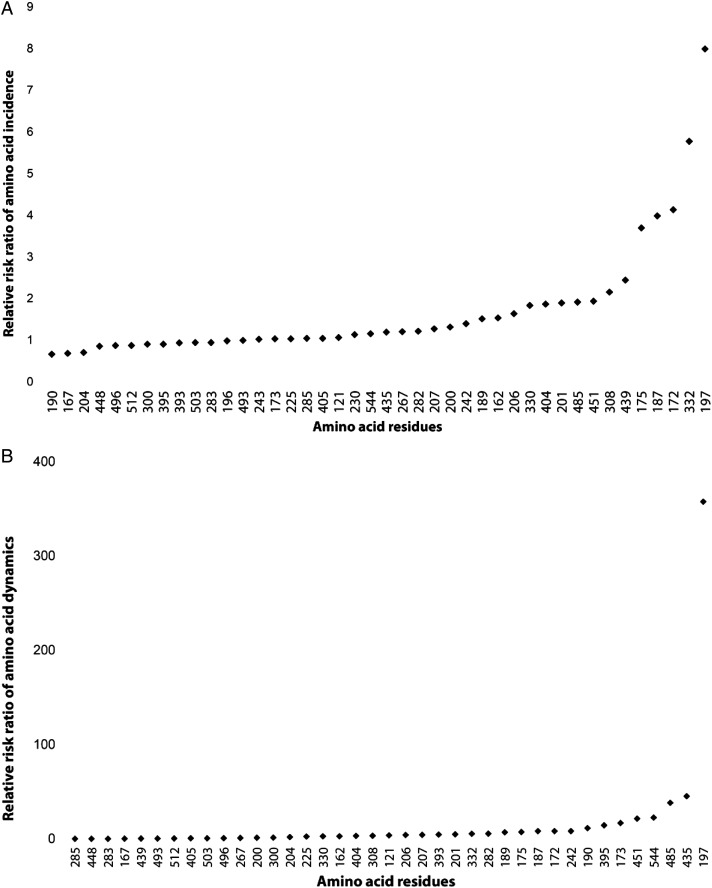
To identify specific polymorphisms driving allele-specific immunity, we assessed whether the incidence of AMA1 polymorphisms was equally distributed in the 2 vaccine groups before and after vaccination. This was done by comparing the distribution of vaccine-type alleles at each individual amino acid position in sequences collected from clinical malaria episodes occurring in the 2 treatment arms during and up to 2 weeks after the vaccination period, as well as during the period starting 2 weeks after vaccination was completed (Figure 3), accounting for multiple comparisons. The RRR of the incidence of vaccine-type amino acids showed that vaccine-type amino acids at positions 175, 187, 172, 197, and 332 were less frequent in the malaria vaccine group as compared to the rabies vaccine group (Figure 4A) following vaccination. In contrast, the incidences of vaccine-type amino acids at the rest of the polymorphic positions were similarly distributed in the 2 vaccine groups before and after vaccination.
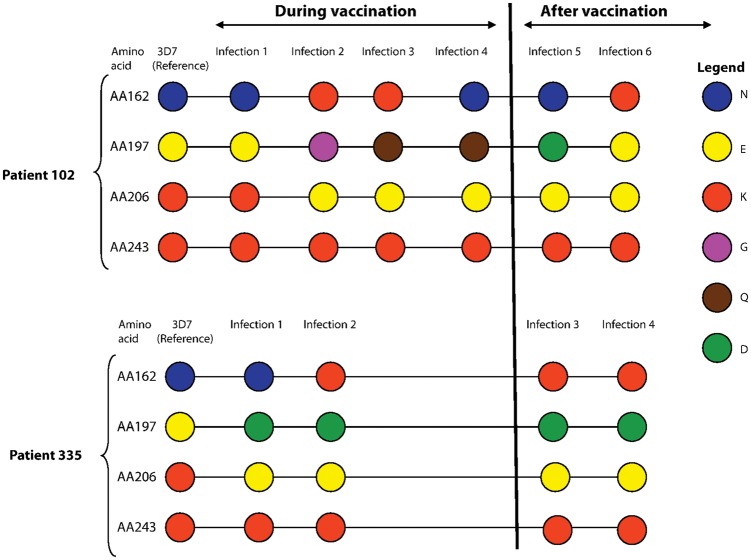
Figure 3.
Shifts from vaccine-type to non–vaccine-type alleles at 4 apical membrane antigen 1 polymorphic amino acid positions (162, 197, 206, and 243) identified in 2 representative study participants during 6 months of follow-up. The shift from vaccine-type to non–vaccine-type alleles between consecutive infections was assessed during the vaccination period (vaccinations occurred on days 0, 30, and 60), from day 0 through day 74 (ie, 2 weeks after the last vaccination, before vaccine-induced selection should be strong), and after vaccination (ie, after day 74, when vaccine-induced selection should be most evident). Malaria vaccine (3D7) amino acids are shown in the first column, with each color representing a different amino acid. Amino acid changes (or lack of change) occurring during consecutive infections are shown along the horizontal lines. The dynamics of amino acid changes observed during the vaccination period were compared to those observed starting 2 weeks after the third and last vaccination, with the hypotheses that, at amino acid positions under selection by the malaria vaccine, (1) non–vaccine-type amino acids would be observed more often in the period after vaccination in the malaria vaccine group than in the control group, and (2) in paired consecutive infections, changes from vaccine-type amino acids to non–vaccine-type amino acids would be observed more often in the period after vaccination in the malaria vaccne group than in the control group.
Figure 4.
Relative risk ratio (RRR) of the incidence of vaccine-type amino acids (A) and shifts from vaccine-type to non–vaccine-type amino acids (B) during the vaccination period (up to day 74) as compared to the period after vaccination (days 75–240) in the malaria vaccine and rabies vaccine groups. A, At positions 175, 187, 172, 332, and 197, 3D7-type amino acids were more prevalent after vaccination in the rabies vaccine group as compared to the malaria vaccine group. Polymorphic positions for which the 2 vaccine groups were significantly different have a RRR of >3 (the cutoff for a significant difference in amino acid prevalence between sequences generated before and after vaccination). B, Amino acids from consecutive clinical malaria episodes experienced by the same study participants may change from 3D7 type (ie, vaccine type) to 3D7 type, from 3D7 type to other type, from other type to 3D7 type, or from other type to other type. The incidence of changes from 3D7 type to other type, divided by the incidence of all other types of changes, should increase after vaccination in the malaria vaccine group but not in the rabies vaccine group if an amino acid position is under selection by the vaccine. The relative risk observed after vaccination, divided by relative risk during vaccination, is plotted for all polymorphic positions of the AMA1 ectodomain. This ratio is high at positions where the change from 3D7 type to other type increased after vaccination, indicating vaccine selection. A significantly increased RRR of > 38 (the cutoff for a significant difference in amino acid shift from the vaccine type between sequences generated before and after vaccination) is observed at positions 485, 435, and 197.
In further support of the relevance of specific amino acid positions, an analysis of the shift from vaccine-type to non–vaccine-type alleles in consecutive malaria episodes revealed that the incidence of alleles that changed from a 3D7 type to a non–3D7 type was higher in the malaria vaccine group as compared to the rabies control group in sequences identified from days 75 through 240 for the 3 amino acid positions 197, 435, and 485 (Figure 4B).
Interestingly, position 197 was identified as the most important polymorphic site for characterizing AMA1 allelic identity by all 3 methods of analysis used to assess the role of specific amino acid positions. We used the amino acid at this position to define alleles to assess the time to the first clinical malaria episode with a 3D7 allele. The allele-specific efficacy data calculated using only position 197 to define alleles revealed vaccine efficacy identical to that for the whole c1L haplotype against 5 known c1L haplotypes (Table 1), suggesting that amino acid residues at position 197 may be the driving force of allele-specific efficacy in c1L. This was confirmed by an analysis of the amino acid composition at position 197 in the whole sequence data set. Among the 473 single or predominant clone sequences used in this analysis, we identified 49 sequences with c1L amino acids exactly matching the 8 amino acids in 3D7 c1L. In all of these sequences, when the amino acid at position 197 is a glutamate (E), the remaining amino acids in c1L were, by definition, 3D7 type. However, the same observation did not hold when any of the other amino acid positions were used to define the 3D7 c1L allele. This observation indicates that, at least within this data set, amino acid 197 can be used as a surrogate for the amino acid identity of 3D7 c1L. Therefore, instead of defining AMA1 alleles on the basis of the entire c1L, it may be possible to use the amino acid at position 197 to reduce the number of alleles that would need to be considered in vaccine design.
DISCUSSION
Identification of the relevant amino acids driving allele-specific immunity is an important step in the design of malaria vaccines that target polymorphic antigens. The characterization of the AMA1 protein crystal structure [9] has allowed identification of immunologically important amino acid residues in this malaria vaccine antigen, using both in vitro [5] and in vivo approaches [7]. In particular, molecular epidemiological tools and approaches have the potential to inform the choice of antigenic variants to include in multivalent, broadly efficacious vaccines [1], and these tools can be used to assess the impact of malaria vaccines on the frequency of alleles observed in field trials [14].
The finding that a monovalent AMA1 vaccine was efficacious only against homologous alleles in a human vaccine trial is consistent with in vitro studies [4] and animal studies [2, 22] suggesting that AMA1 generates allele-specific immune responses. The observed patterns of natural acquisition of allele-specific immunity to AMA1 are consistent with a gradual accumulation of protective responses to a repertoire of AMA1 variants by people repeatedly exposed to malaria over years [23].
In contrast to a bivalent AMA1 vaccine that showed no overall efficacy nor any suggestion of allele-specific efficacy [15], the FMP2.1/AS02A vaccine was highly efficacious against clinical malaria with vaccine-type AMA1 c1L. The mean antibody responses to FMP2.1/AS02A vaccine were much higher and more sustained as compared to antibody responses to the bivalent AMA1 vaccine, suggesting that the latter vaccine failed simply because it was not immunogenic enough. In secondary analyses, the FMP2.1/AS02A vaccine was shown to have an efficacy of about 20% against all clinical malaria episodes [14], with varying statistical significance. This marginal overall efficacy may represent the vaccine having an effect exclusively directed against vaccine-type and closely related alleles with respect to the cluster of highly polymorphic amino acids located in domain 1 of AMA1.
The allele-specific efficacy of the FMP2.1/AS02A vaccine was confirmed by an analysis of the incidence of the vaccine and nonvaccine c1L haplotypes in clinical episodes before and after vaccination, which revealed a decreased incidence of the 3D7 AMA1 c1L haplotype in the malaria vaccine group as compared to the rabies group following vaccination. With 3D7 representing 13.5% of c1L haplotypes, an overall efficacy of approximately 20% against clinical malaria episodes suggests that there may be other AMA1 amino acid positions outside of c1L that are targeted by protective antibodies. While it is possible that FMP2.1/AS02A would offer protective efficacy against clinical malaria caused by parasites with c1L that matches 3D7 at position 197 but is not identical to 3D7 at other positions within c1L, we found no such haplotypes among the 600 samples sequenced in this study.
The finding that this malaria vaccine was efficacious only against clinical malaria with AMA1 alleles homologous to the vaccine allele in c1L points to the need for a multiple-allele vaccine. Although a multivalent vaccine including all unique AMA1 alleles would be practically impossible, by using molecular epidemiological approaches it may be possible to group sequences that are closely related and/or to identify amino acids that play a fundamental role in allele-specific efficacy. Representative alleles that cover a large proportion of the population may be selected and used as vaccine antigens, as has been done for a successful multivalent pneumococcal vaccine [24]. Focusing on representative alleles may reduce the number of alleles to a number that can feasibly be managed by vaccine developers. A multivalent vaccine that comprises 10 of the most prevalent alleles may cover >70% of alleles identified in the study area (Supplementary Figure 2). In corroboration of this finding, immunological analyses have been used to assess the allele specificity of the immune response by characterizing antibody responses against variants of AMA1 [13]. These analyses revealed that epitopes within AMA1 of the 3D7 strain of P. falciparum are representative of both D10 and S35 epitopes (which are identical in domain 1 but differ in domains 2 and 3), suggesting that 3D7 may be used to achieve cross-protection against both D10 and S35 strains in a multivalent malaria vaccine.
Even though a barrier blocking clinical episodes with vaccine-type alleles (a sieve effect) [25] was observed at several amino acid positions in domains 1 and 3, the strongest barrier induced by the malaria vaccine was seen at amino acid position 197, which is located in c1L of domain 1. Alleles defined on the basis of this position showed vaccine efficacy identical to that for the whole c1L, suggesting that this position may be the most critical amino acid in antibody binding. Therefore, instead of considering all polymorphic positions of AMA1 to define haplotypes, position 197 alone might be used to define which alleles to include in a vaccine. This would reduce the number of alleles required to design a multivalent AMA1-based vaccine to only 6, which would represent 100% coverage of all alleles, where “alleles” is defined on the basis of AMA1 position 197.
The latest findings that a mixed-antigen formulation may produce the same immunological response as sequential infections with the same antigens [26] further support the feasibility of developing a multivalent AMA1 vaccine, either as a stand-alone blood-stage malaria vaccine or, more likely, as a component of a multistage, multiple antigen vaccine [27], with higher efficacy than that achieved by the most advanced malaria vaccine, RTS,S/AS01, which is currently being assessed in a large phase III trial in Africa [28].
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank the study participants and the population of Bandiagara, for their compliance and commitment; the Bandiagara Malaria Project field staff, for their assistance; the members of A. O.'s PhD thesis committee (Samer El-Kamary, Colin Stine, Patricia Langenberg, and Xin-zhuan Su), for advice and useful suggestions; and Adrian Batchelor, for providing the AMA1 crystal structure.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases (contract N01AI85346 and cooperative agreement U19AI065683), the Fogarty International Center of the National Institutes of Health (grant D43TW001589), both the US Department of Defense and US Agency for International Development (contract W81XWH-06-1-0427), the Howard Hughes Medical Institute and the Doris Duke Charitable Foundation (support for training and genetics analysis), and the University of Maryland Multidisciplinary Clinical Research Career Development Program (NIH award K12RR023250 to S. T.-H.).
Potential conflicts of interest. S. D. holds a proprietary patent for the FMP2.1 vaccine antigen. J. V. and J. C. are or were employees of GlaxoSmithKline Biologicals, the manufacturer of the proprietary Adjuvant System AS02A, and hold shares in GlaxoSmithKline. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Takala SL, Plowe CV. Genetic diversity and malaria vaccine design, testing and efficacy: preventing and overcoming ‘vaccine resistant malaria. Parasite Immunol. 2009;31:560–73. doi: 10.1111/j.1365-3024.2009.01138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barclay VC, Chan BH, Anders RF, Read AF. Mixed allele malaria vaccines: host protection and within-host selection. Vaccine. 2008;26:6099–107. doi: 10.1016/j.vaccine.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stowers AW, Kennedy MC, Keegan BP, Saul A, Long CA, Miller LH. Vaccination of monkeys with recombinant Plasmodium falciparum apical membrane antigen 1 confers protection against blood-stage malaria. Infect Immun. 2002;70:6961–7. doi: 10.1128/IAI.70.12.6961-6967.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dutta S, Dlugosz LS, Clayton JW, et al. Alanine mutagenesis of the primary antigenic escape residue cluster, c1, of apical membrane antigen 1. Infect Immun. 2010;78:661–71. doi: 10.1128/IAI.00866-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dutta S, Lee SY, Batchelor AH, Lanar DE. Structural basis of antigenic escape of a malaria vaccine candidate. Proc Natl Acad Sci U S A. 2007;104:12488–93. doi: 10.1073/pnas.0701464104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osier FH, Weedall GD, Verra F, et al. Allelic diversity and naturally acquired allele-specific antibody responses to Plasmodium falciparum apical membrane antigen 1 in Kenya. Infect Immun. 2010;78:4625–33. doi: 10.1128/IAI.00576-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takala SL, Coulibaly D, Thera MA, et al. Extreme polymorphism in a vaccine antigen and risk of clinical malaria: implications for vaccine development. Sci Transl Med. 2009;1:2ra5. doi: 10.1126/scitranslmed.3000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hodder AN, Crewther PE, Matthew ML, et al. The disulfide bond structure of Plasmodium apical membrane antigen-1. J Biol Chem. 1996;271:29446–52. doi: 10.1074/jbc.271.46.29446. [DOI] [PubMed] [Google Scholar]
- 9.Bai T, Becker M, Gupta A, et al. Structure of AMA1 from Plasmodium falciparum reveals a clustering of polymorphisms that surround a conserved hydrophobic pocket. Proc Natl Acad Sci U S A. 2005;102:12736–41. doi: 10.1073/pnas.0501808102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Healer J, Murphy V, Hodder AN, et al. Allelic polymorphisms in apical membrane antigen-1 are responsible for evasion of antibody-mediated inhibition in Plasmodium falciparum. Mol Microbiol. 2004;52:159–68. doi: 10.1111/j.1365-2958.2003.03974.x. [DOI] [PubMed] [Google Scholar]
- 11.Dutta S, Sullivan JS, Grady KK, et al. High antibody titer against apical membrane antigen-1 is required to protect against malaria in the Aotus model. PLoS One. 2009;4:e8138. doi: 10.1371/journal.pone.0008138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Healer J, Triglia T, Hodder AN, Gemmill AW, Cowman AF. Functional analysis of Plasmodium falciparum apical membrane antigen 1 utilizing interspecies domains. Infect Immun. 2005;73:2444–51. doi: 10.1128/IAI.73.4.2444-2451.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coley AM, Parisi K, Masciantonio R, et al. The most polymorphic residue on Plasmodium falciparum apical membrane antigen 1 determines binding of an invasion-inhibitory antibody. Infect Immun. 2006;74:2628–36. doi: 10.1128/IAI.74.5.2628-2636.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thera MA, Doumbo OK, Coulibaly D, et al. A field trial to assess a blood-stage malaria vaccine. N Engl J Med. 2011;365:1004–13. doi: 10.1056/NEJMoa1008115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ouattara A, Mu J, Takala-Harrison S, et al. Lack of allele-specific efficacy of a bivalent AMA1 malaria vaccine. Malar J. 2010;9:175. doi: 10.1186/1475-2875-9-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enosse S, Dobano C, Quelhas D, et al. RTS,S/AS02A malaria vaccine does not induce parasite CSP T cell epitope selection and reduces multiplicity of infection. PLoS Clin Trials. 2006;1:e5. doi: 10.1371/journal.pctr.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alloueche A, Milligan P, Conway DJ, et al. Protective efficacy of the RTS,S/AS02 Plasmodium falciparum malaria vaccine is not strain specific. Am J Trop Med Hyg. 2003;68:97–101. [PubMed] [Google Scholar]
- 18.Waitumbi JN, Anyona SB, Hunja CW, et al. Impact of RTS,S/AS02(A) and RTS,S/AS01(B) on genotypes of P. falciparum in adults participating in a malaria vaccine clinical trial. PLoS One. 2009;4:e7849. doi: 10.1371/journal.pone.0007849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thera MA, Doumbo OK, Coulibaly D, et al. Safety and immunogenicity of an AMA-1 malaria vaccine in Malian adults: results of a phase 1 randomized controlled trial. PLoS One. 2008;3:e1465. doi: 10.1371/journal.pone.0001465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dicko A, Mantel C, Kouriba B, et al. Season, fever prevalence and pyrogenic threshold for malaria disease definition in an endemic area of Mali. Trop Med Int Health. 2005;10:550–6. doi: 10.1111/j.1365-3156.2005.01418.x. [DOI] [PubMed] [Google Scholar]
- 21.Duan J, Mu J, Thera MA, et al. Population structure of the genes encoding the polymorphic Plasmodium falciparum apical membrane antigen 1: implications for vaccine design. Proc Natl Acad Sci U S A. 2008;105:7857–62. doi: 10.1073/pnas.0802328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crewther PE, Matthew ML, Flegg RH, Anders RF. Protective immune responses to apical membrane antigen 1 of Plasmodium chabaudi involve recognition of strain-specific epitopes. Infect Immun. 1996;64:3310–7. doi: 10.1128/iai.64.8.3310-3317.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weiss GE, Traore B, Kayentao K, et al. The Plasmodium falciparum-specific human memory B cell compartment expands gradually with repeated malaria infections. PLoS Pathog. 2010;6:e1000912. doi: 10.1371/journal.ppat.1000912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Obaro SK. The new pneumococcal vaccine. Clin Microbiol Infect. 2002;8:623–33. doi: 10.1046/j.1469-0691.2002.00424.x. [DOI] [PubMed] [Google Scholar]
- 25.Gilbert P, Self S, Rao M, Naficy A, Clemens J. Sieve analysis: methods for assessing from vaccine trial data how vaccine efficacy varies with genotypic and phenotypic pathogen variation. J Clin Epidemiol. 2001;54:68–85. doi: 10.1016/s0895-4356(00)00258-4. [DOI] [PubMed] [Google Scholar]
- 26.Kusi KA, Faber BW, van der Eijk M, Thomas AW, Kocken CH, Remarque EJ. Immunization with different PfAMA1 alleles in sequence induces clonal imprint humoral responses that are similar to responses induced by the same alleles as a vaccine cocktail in rabbits. 550. Malar J. 2011;10:40. doi: 10.1186/1475-2875-10-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heppner DG, Jr., Kester KE, Ockenhouse CF, et al. Towards an RTS,S-based, multi-stage, multi-antigen vaccine against falciparum malaria: progress at the Walter Reed Army Institute of Research. Vaccine. 2005;23:2243–50. doi: 10.1016/j.vaccine.2005.01.142. [DOI] [PubMed] [Google Scholar]
- 28.First results of phase III trial of RTS,S/AS01 malaria vaccine in African children. N Engl J Med. 2011;365:1863–75. doi: 10.1056/NEJMoa1102287. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.