Abstract
RNA recombination has been shown to occur during circulation of enteroviruses, but most studies have focused on poliovirus. To examine the role of recombination in the evolution of the coxsackie B viruses (CVB), we determined the partial sequences of four genomic intervals for multiple clinical isolates of each of the six CVB serotypes isolated from 1970 to 1996. The regions sequenced were the 5′-nontranslated region (5′-NTR) (350 nucleotides [nt]), capsid (VP4-VP2, 416 nt, and VP1, ∼320 nt), and polymerase (3D, 491 nt). Phylogenetic trees were constructed for each genome region, using the clinical isolate sequences and those of the prototype strains of all 65 enterovirus serotypes. The partial VP1 sequences of each CVB serotype were monophyletic with respect to serotype, as were the VP4-VP2 sequences, in agreement with previously published studies. In some cases, however, incongruent tree topologies suggested that intraserotypic recombination had occurred between the sequenced portions of VP2 and VP1. Outside the capsid region, however, isolates of the same serotype were not monophyletic, indicating that recombination had occurred between the 5′-NTR and capsid, the capsid and 3D, or both. Almost all clinical isolates were recombinant relative to the prototype strain of the same serotype. All of the recombination partners appear to be members of human enterovirus species B. These results suggest that recombination is a frequent event during enterovirus evolution but that there are genetic restrictions that may influence recombinational compatibility.
Recombination is a common feature among positive-stranded RNA viruses (30). For single-stranded RNA viruses, the mechanism is probably copy choice (template switching during RNA replication) rather than true recombination; that is, the mechanism is analogous to gene conversion (14). Recombination has been shown to occur among enteroviruses, but only poliovirus has been studied in detail. Interserotypic exchange between Sabin vaccine strains is common in primary vaccinees (3, 5) and in cases of vaccine-associated paralytic poliomyelitis (8, 9, 16). Evidence for recombination among non-polio enteroviruses (NPEV) has come mainly from comparison of a small number of complete genome sequences (28) and from specific comparison of individual isolates (1, 15). Analysis of NPEV prototype strains has suggested that interserotypic recombination is a frequent event during natural transmission and that it may play a significant role in enterovirus evolution (1, 2, 22, 28). The most-detailed studies of enterovirus recombination during natural infection and circulation have been with wild or vaccine-derived polioviruses (4, 5, 9, 12, 17, 18).
Coxsackie B virus 1 (CVB1) to CBV6 constitute six of the 37 serotypes in the species Human enterovirus B (HEV-B). The other 31 HEV-B serotypes are CVA9, echovirus 1 (E1) to E7, E9, E11 to E21, E24 to E27, E29 to E33, enterovirus 69 (EV69), and EV73. The remaining HEVs are divided among four other species: (i) Poliovirus (poliovirus 1 to 3), (ii) HEV-A (CVA2 to CVA 8, CVA10, CVA12, CVA14, CVA16, and EV71), (iii) HEV-C (CVA1, CVA11, CVA13, CVA15, CVA17 to CVA22, and CVA24), and (iv) HEV-D (EV68 and EV70) (13). Recent studies suggest that polioviruses should be reclassified as members of HEV-C (2). During their initial characterization, the CVBs were shown to possess several properties unique to the group, including the ability to cause spastic paralysis in intracranially inoculated mice—hence their classification into a single group (25). It was shown subsequently that the CVBs shared many physical and genetic characteristics with other enterovirus serotypes, leading to the current classification scheme.
Our previous analysis of HEV-B prototype strains suggested that differences in pairwise sequence relationships in different genomic regions, revealed by phylogenetic reconstruction and similarity plotting, were due to interserotypic recombination (22). This analysis was limited to only prototype strains, and therefore it was not possible to compare recombination within isolates of a single serotype with recombination between serotypes. To examine recombination within serotypes in greater detail, we sequenced four intervals across the genomes of 55 CVB clinical isolates from all six serotypes and compared them with the homologous sequences from the prototype strains of CVB and other enteroviruses. The sequenced intervals include portions of the 5′-nontranslated region (5′-NTR) and the VP4-VP2, VP1, and 3D regions. Phylogenetic analysis indicates that recombination is a frequent event during NPEV transmission and that recombination both within and between serotypes plays a significant role in the evolution of the HEVs.
MATERIALS AND METHODS
Viruses.
A total of 55 CVB clinical isolates were selected (Table 1), representing each of the six CVB serotypes (at least 10 isolates each for CVB1 to CVB5; only one CVB6 isolate was available). These viruses were isolated from 1970 to 1996, and for each serotype except CVB6, the time period of isolation spanned 12 to 22 years. The complete genome sequence of each CVB prototype strain and of other representative prototype strains was obtained from GenBank.
TABLE 1.
Viruses analyzed in this study
| Type | Geographic origin
|
Yr | Strain | |
|---|---|---|---|---|
| Countrya | Stateb | |||
| CVB1 | USA | CT | 1948 | Conn-5 |
| CVB1 | USA | GA | 1970 | 10158 |
| CVB1 | USA | ME | 1971 | 10159 |
| CVB1 | USA | NH | 1977 | 10160 |
| CVB1 | USA | RI | 1980 | 10161 |
| CVB1 | USA | SD | 1981 | 10162 |
| CVB1 | USA | GA | 1982 | 10163 |
| CVB1 | ARG | 1983 | 10164 | |
| CVB1 | USA | CT | 1984 | 10165 |
| CVB1 | USA | GA | 1985 | 10166 |
| CVB1 | SOA | 1992 | 10167 | |
| CVB2 | USA | OH | 1947 | Ohio-1 |
| CVB2 | NEA | 1975 | 10168 | |
| CVB2 | USA | ND | 1976 | 10169 |
| CVB2 | USA | NE | 1977 | 10170 |
| CVB2 | USA | MS | 1979 | 10171 |
| CVB2 | USA | NC | 1979 | 10172 |
| CVB2 | USA | AL | 1980 | 10173 |
| CVB2 | USA | NH | 1981 | 10174 |
| CVB2 | USA | MA | 1982 | 10175 |
| CVB2 | USA | MA | 1983 | 10176 |
| CVB2 | USA | WA | 1991 | 10177 |
| CVB2 | SOA | 1992 | 10178 | |
| CVB2 | USA | FL | 1992 | 10179 |
| CVB2 | ARG | 1996 | 10180 | |
| CVB3 | USA | CT | 1949 | Nancy |
| CVB3 | USA | ID | 1980 | 10181 |
| CVB3 | COR | 1982 | 10182 | |
| CVB3 | USA | NH | 1983 | 10183 |
| CVB3 | USA | SD | 1985 | 10184 |
| CVB3 | USA | OH | 1983 | 10185 |
| CVB3 | USA | AL | 1986 | 10186 |
| CVB3 | USA | VA | 1986 | 10187 |
| CVB3 | USA | CT | 1987 | 10188 |
| CVB3 | USA | NM | 1992 | 10189 |
| CVB4 | USA | NY | 1951 | JVB |
| CVB4 | USA | RI | 1975 | 10190 |
| CVB4 | USA | ND | 1976 | 10191 |
| CVB4 | USA | MS | 1979 | 10192 |
| CVB4 | USA | MS | 1981 | 10193 |
| CVB4 | USA | PA | 1982 | 10194 |
| CVB4 | CAN | 1983 | 10195 | |
| CVB4 | HON | 1987 | 10196 | |
| CVB4 | USA | MA | 1987 | 10197 |
| CVB4 | USA | GA | 1991 | 10198 |
| CVB4 | SOA | 1992 | 10199 | |
| CVB5 | USA | KY | 1952 | Faulkner |
| CVB5 | DOR | 1971 | 10200 | |
| CVB5 | USA | CO | 1973 | 10201 |
| CVB5 | ECU | 1974 | 10202 | |
| CVB5 | USA | ID | 1977 | 10203 |
| CVB5 | USA | SD | 1978 | 10204 |
| CVB5 | USA | ID | 1980 | 10205 |
| CVB5 | USA | ME | 1982 | 10206 |
| CVB5 | USA | AL | 1983 | 10207 |
| CVB5 | USA | CO | 1987 | 10208 |
| CVB5 | HON | 1987 | 10209 | |
| CVB5 | HON | 1989 | 10210 | |
| CVB5 | SOA | 1992 | 10211 | |
| CVB6 | PHL | 1953 | Schmidt | |
| CVB6 | HON | 1985 | 10212 | |
Abbreviations: ARG, Argentina; CAN, Canada; COR, Costa Rica; DOR, Dominican Republic; ECU, Ecuador; HON, Honduras; NEA, Netherlands Antilles; PHL, Philippines; SOA, South Africa; USA, United States.
Abbreviations: CT, Connecticut; GA, Georgia; ME, Maine; NH, New Hampshire; RI, Rhode Island; SD, South Dakota; OH, Ohio; ND, North Dakota; NE, Nebraska; MS, Mississippi; NC, North Carolina; AL, Alabama; MA, Massachusetts; WA, Washington; FL, Florida; ID, Idaho; VA, Virginia; NM, New Mexico; NY, New York; PA, Pennsylvania; KY, Kentucky; CO, Colorido.
RT-PCR and sequencing.
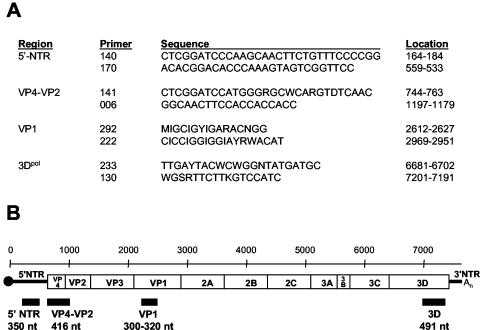
RNA was extracted from infected cell culture supernatants, using the QIAamp Viral RNA Mini Kit (Qiagen, Inc., Valencia, Calif.). Portions of the 5′-NTR, VP4-VP2, VP1, and 3D genes were amplified by reverse transcription (RT)-PCR, using standard methods and the primer pairs listed in Fig. 1A. The locations and sizes of the PCR products are depicted in Fig. 1B. PCR products were purified with the High Pure PCR Product Purification Kit (Roche Molecular Biochemicals, Indianapolis, Ind.), and both strands were sequenced by automated methods, using fluorescent dideoxy-chain terminators (Applied Biosystems, Foster City, Calif.).
FIG. 1.
Schematic representation of the enterovirus genome and primers used for RT-PCR amplification. (A) Oligonucleotide primer pairs used for amplification of four genomic intervals of each of the coxsackie B virus isolates. Nucleotide sequence coordinates are relative to the genome of poliovirus 1, Mahoney strain. Primer 006 is a modification of primer 5− (11, 23); the use of primers 292 and 222 for molecular serotyping was described previously (21, 24); primers 233 and 130 are the same as primers 3D+ and 3D−, respectively (27). (B) The approximate location and size of PCR products are shown under the enterovirus genetic map. The scale, in nucleotides, is indicated above the map.
Sequence analyses.
Sequences for each region were aligned with Pileup (Wisconsin Sequence Analysis Package [version 10.2]; Accelrys, San Diego, Calif.). Pairwise sequence differences were calculated for each of the four genome regions using Distances (Wisconsin Package). Phylogenetic trees were constructed by the neighbor-joining method using the PHYLIP programs DNADist and Neighbor (7), with 1,000 bootstrap pseudoreplicates and a transition/transversion ratio of 10. Trees were visualized using Treeview, version 1.6.6 (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html).
Nucleotide sequence accession numbers.
The sequences reported here were deposited in the GenBank sequence database, accession no. AY373036 to AY373255.
RESULTS
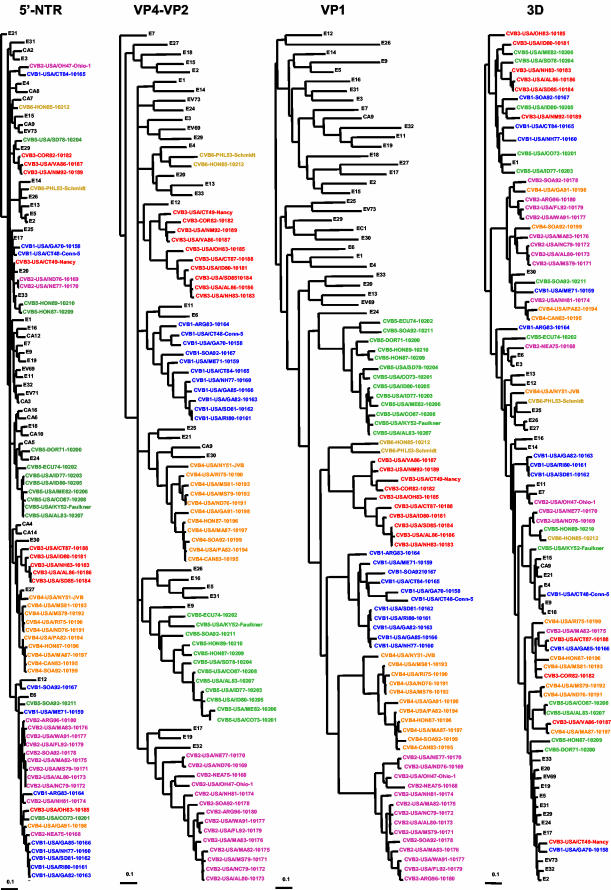
For each of the four genome intervals studied, the 55 CVB clinical isolate sequences were compared with one another and with the homologous sequences from all other HEV serotypes by calculating pairwise sequence distances and by phylogenetic reconstruction. All of the isolates analyzed are monophyletic with respect to species (HEV-B) in all regions of the genome except the 5′-NTR, where species A and B cluster together in 5′-NTR cluster II (Fig. 2 and data not shown), in accord with previous observations (26). For ease in visualization, the phylogenies were then regenerated using only the sequences of HEV-B viruses (or HEV-A and -B for the 5′-NTR) (Fig. 2). Species C and D comprise 5′-NTR cluster I (26).
FIG.2.
Phylogenetic relationships of CVB clinical isolates and other members of HEV-B. Trees were constructed by the neighbor-joining method using DNADist and Neighbor programs (PHYLIP) (7). Trees for the different genomic intervals were plotted to the same scale, indicated by the scale bar below and to the left of each tree. The CVB isolates are color coded by serotype; isolates of other serotypes are shown in black. Because the major clusters radiate from a multifurcating node at the center of the tree, their order in the tree is arbitrary.
Capsid region (VP4-VP2 and VP1).
In the VP4-VP2 interval, the CVBs vary by 0.5 to 23.1% within a serotype, by 18.3 to 28.9% between serotypes, by 19.0 to 32.5% from other HEV-B serotypes, and by 31.7 to 46.4% from serotypes in other HEV species (Table 2). The interserotypic differences are even more pronounced in the VP1 region. CVB isolates vary by 1.0 to 25.7% within a serotype, but they are at least 27.7% different from isolates of all other HEV-B serotypes and at least 42.7% different from those of HEV-A, -C, and -D (Table 2). The CVB viruses as a group are monophyletic with respect to all other members of HEV-B in VP1, except that E24 is placed near the root of the CVB group (Fig. 2). Bootstrap support for the position of E24, however, was only 11% (data not shown). By contrast, the six CVB serotypes are not monophyletic as a group with respect to other members of HEV-B in the VP4-VP2 region (Fig. 2). Despite their variability within a serotype, CVB isolates are monophyletic with respect to serotype in both VP4-VP2 and VP1 (Fig. 2). Within the individual serotype clusters, there are small differences in topology between the VP4-VP2 tree and the VP1 tree. For example, CVB1 isolate ARG83-10164 is near the root of the CVB1 cluster in both VP4-VP2 and VP1. ARG83-10164 clusters with USA/CT48-Conn-5 and USA/GA70-10158 in VP4-VP2, whereas these two isolates cluster together considerably “deeper” in the VP1 tree. Similarly, ECU74-10202 clusters with USA/KY52-Faulkner near the root of the CVB5 VP4-VP2 tree, but it clusters with SOA92-10211, also near the root of the CVB5 cluster, in VP1. On the other hand, the CVB3 and CVB4 trees are nearly identical in both parts of the capsid, with only slight differences in the relative positions of taxa within specific subbranches (e.g., the position of USA/CT-49-Nancy within the upper branch of the two CVB3 trees).
TABLE 2.
Comparison of CVB nucleotide sequences with one another, with those of other serotypes of HEV-B, and with those of other HEV species
| Sequence | Group | % Difference
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CVB1 | CVB2 | CVB3 | CVB4 | CVB5 | CVB6 | HEV species
|
|||||
| A | B | C | D | ||||||||
| 5′-NTR | CVB1 | 1.1-14.6 | 3.1-13.7 | 4.3-15.7 | 4.3-15.7 | 5.7-15.7 | 6.6-14 | 6.6-15.8 | 2.6-16.9 | 26.4-33.6 | 27.0-32.2 |
| CVB2 | 0.3-12.3 | 3.7-14 | 4.0-14.9 | 4.0-15.1 | 6.3-13.7 | 5.7-14.6 | 3.7-16.0 | 26.4-32.7 | 27.6-31.9 | ||
| CVB3 | 0.9-14 | 5.4-14.9 | 5.4-14.9 | 5.7-14.6 | 5.8-14.6 | 1.7-16.6 | 24.9-34.1 | 27.0-32.8 | |||
| CVB4 | 1.4-13.1 | 4.9-14.3 | 8.6-14.6 | 6.3-14.0 | 3.7-16.9 | 26.7-35.0 | 28.5-31.9 | ||||
| CVB5 | 1.1-14 | 8.0-15.1 | 6.3-14.9 | 4.6-17.2 | 24.9-33.3 | 27.6-32.8 | |||||
| CVB6 | 11.7 | 7.5-15.8 | 4.6-15.7 | 25.2-31.1 | 28.5-30.1 | ||||||
| Other HEV-B | 4.9-17.2 | 2.9-18.3 | 24.6-34.3 | 26.2-33.6 | |||||||
| HEV-A | 4.9-13.5 | 25.4-34.5 | 27.4-33.4 | ||||||||
| HEV-C | 8.3-18.1 | 9.8-19.8 | |||||||||
| HEV-D | 16.1 | ||||||||||
| VP4-VP2 | CVB1 | 0.5-21.4 | 20.7-27.2 | 18.3-26.2 | 22.1-28.6 | 20.4-26.0 | 22.8-27.2 | 37.3-44.2 | 20.2-32.5 | 34.6-40.1 | 40.6-44.7 |
| CVB2 | 1.7-22.1 | 21.4-26.9 | 20.9-27.4 | 20.2-26.4 | 25.0-28.9 | 37.5-45.4 | 20.7-31.7 | 33.4-41.4 | 40.1-45.9 | ||
| CVB3 | 2.2-22.8 | 20.0-26.2 | 18.5-26.4 | 20.9-27.6 | 35.8-44.0 | 19.0-30.5 | 33.2-40.4 | 40.6-45.2 | |||
| CVB4 | 1.7-18.3 | 22.1-28.1 | 20.2-25.7 | 36.3-45.2 | 20.2-31.3 | 31.7-41.1 | 38.7-44.7 | ||||
| CVB5 | 1.9-23.1 | 22.8-26.9 | 37.3-44.5 | 20.4-30.5 | 34.6-41.6 | 41.6-45.4 | |||||
| CVB6 | 21.4 | 38.5-43.5 | 21.9-30.5 | 32.9-39.9 | 42.3-44.7 | ||||||
| Other HEV-B | 35.3-44.7 | 20.4-32.7 | 32.9-44.5 | 40.1-46.4 | |||||||
| HEV-A | 25.0-34.9 | 35.6-43.5 | 36.1-43.3 | ||||||||
| HEV-C | 22.1-35.1 | 37.3-43.8 | |||||||||
| HEV-D | 24 | ||||||||||
| VP1 | CVB1 | 1.3-23.0 | 31.3-39.6 | 29.4-37.7 | 29.1-36.1 | 29.4-36.7 | 27.7-33.6 | 51.6-61.7 | 30.9-44.5 | 45.1-54.9 | 48.2-54.6 |
| CVB2 | 1.9-20.6 | 34.5-39.9 | 29.2-38.8 | 32.9-41.1 | 31.0-35.1 | 52.1-61.5 | 32.6-45.7 | 45.7-54.1 | 49.2-53.5 | ||
| CVB3 | 1.0-24.3 | 30.7-38.0 | 27.8-35.5 | 30.3-35.8 | 52.4-59.7 | 30.9-44.7 | 42.8-52.6 | 51.1-55.9 | |||
| CVB4 | 2.5-20.2 | 31.7-37.9 | 34.5-37.7 | 51.6-61.8 | 34.6-45.3 | 45.5-55.5 | 49.7-55.3 | ||||
| CVB5 | 3.1-25.7 | 28.2-35.1 | 51.0-60.8 | 29.5-45.1 | 42.7-54.4 | 47.7-53.0 | |||||
| CVB6 | 23.7 | 48.1-60.1 | 32.2-44.3 | 47.3-55.7 | 50.0-55.1 | ||||||
| Other HEV-B | 47.0-62.2 | 27.1-46.0 | 45.4-56.9 | 46.3-56.8 | |||||||
| HEV-A | 26.4-45.3 | 51.8-63.4 | 50.5-59.8 | ||||||||
| HEV-C | 26.5-48.8 | 49.5-56.8 | |||||||||
| HEV-D | 38.5 | ||||||||||
| 3D | CVB1 | 0.4-25.1 | 9.2-23.4 | 3.3-24.0 | 9.6-25.9 | 10.0-23.8 | 16.1-20.6 | 31.6-41.6 | 4.7-23.8 | 28.9-36.1 | 27.7-34.2 |
| CVB2 | 2.2-23.4 | 8.4-24.9 | 4.5-25.3 | 9.0-24.0 | 10.6-22.2 | 32.8-40.9 | 6.1-23.6 | 29.3-36.1 | 29.3-32.4 | ||
| CVB3 | 1.6-22.8 | 3.9-23.8 | 9.4-23.4 | 15.3-20.4 | 33.8-40.7 | 7.7-24.0 | 29.3-35.5 | 28.7-32.8 | |||
| CVB4 | 3.1-26.3 | 12.2-24.2 | 11.6-23.4 | 33.0-41.8 | 8.6-26.9 | 29.3-36.1 | 29.1-34.2 | ||||
| CVB5 | 2.0-23.6 | 4.5-20.8 | 33.8-40.5 | 7.3-22.6 | 29.1-35.5 | 27.7-34.4 | |||||
| CVB6 | 15.7 | 34.0-37.9 | 12.2-19.8 | 30.1-34.6 | 29.5-34.0 | ||||||
| Other HEV-B | 32.8-40.5 | 2.4-22.0 | 29.1-36.5 | 27.7-34.4 | |||||||
| HEV-A | 6.1-24.0 | 32.6-40.4 | 36.1-40.0 | ||||||||
| HEV-C | 8.0-26.0 | 29.7-36.7 | |||||||||
| HEV-D | 22.5 | ||||||||||
5′-NTR.
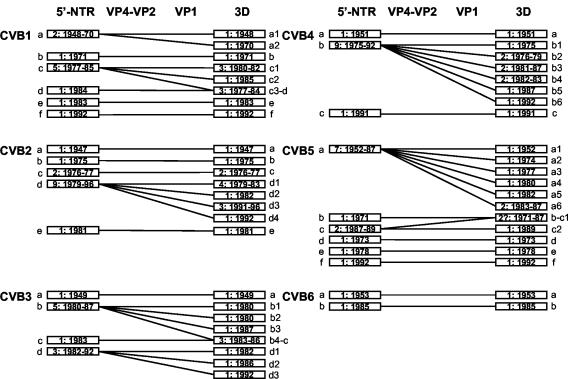
The CVB 5′-NTR sequences differ from one another by 0.3 to 15.7% and from those of the prototype strains of other HEV-B serotypes and the HEV-A prototypes by 1.7 to 17.2%. They are 24.9 to 35.0% different from the prototype strains of serotypes in HEV-C and -D (Table 2). Therefore, there is no evidence of recombination between 5′-NTR clusters. The most closely related sequences are those of CVB isolates that were temporally and geographically related to one another (Fig. 2 and data not shown). Conservation of structural features that are important for 5′-NTR function is consistent with that observed among the prototype strains of HEV-B (22). Members of HEV-B other than the CVBs differ from one another by 2.9 to 18.3%, from HEV-A by 4.9 to 17.2%, and from HEV-C and -D by 24.6 to 34.3% (Table 2). Isolates of a single serotype are not monophyletic in the 5′-NTR (Fig. 2). For each serotype, at least three branches are present (except for CVB6, for which only two isolates were available, including the prototype). For example, the CVB1 isolates are divided among six lineages, each separated from the others by one or more isolates of a different serotype. The two oldest CVB1 isolates, USA/CT48-Conn-5 and USA/GA70-10158, cluster together (lineage a), but they are distinct from all other CVB1 isolates (Fig. 3). The largest CVB1 cluster, lineage c, is composed of five isolates, which were isolated from 1977 to 1985. The remaining four CVB1 isolates cluster independently (lineages b, d, e, and f). Similarly, the other serotypes each form a major 5′-NTR cluster comprising five to nine isolates, with the remaining isolates being independent or clustering in groups of two or three (Fig. 2). There are no alternative trees in which serotype monophyly outside the capsid is supported by bootstrapping (Fig. 2 and data not shown). The CVB2 isolates form five clusters, lineages a to e, of one to nine isolates each (Fig. 2 and 3). Lineages a, b, and d each contained a single isolate. Two of these isolates, including the CVB2 prototype, are from the United States; the third isolate is from The Netherlands Antilles. Lineage c contains two U.S. isolates, which were isolated in 1976 and 1977. Lineage e is composed of seven U.S. isolates from 1979 to 1992 plus a 1992 South Africa isolate and a 1996 Argentina isolate. CVB3 is composed of four lineages—of one, one, three, and five isolates each—with the prototype strain, USA/CT49-Nancy, being independent from the other CVB3 strains (Fig. 2). The CVB4 prototype strain, USA/NY51-JVB, appears to cluster with E27, but the proximal branch is extremely short and the bootstrap value is very low (20%), making it difficult to determine whether USA/NY51-JVB truly clusters with E27 (lineage a) or whether it clusters with the nine other CVB4 isolates on the adjacent branch (lineage b). USA/GA91-10198 clusters independently from other CVB4 isolates, forming lineage c. With the possible exception of CVB4, CVB5 is the only serotype in which the largest cluster contains the prototype strain. Seven CVB5 isolates cluster together to form lineage a. The two Honduras isolates cluster in lineage c, whereas the remaining four isolates form independent lineages b, d, and f. The two CVB6 isolates cluster independently from one another.
FIG. 3.
Summary of the different recombination patterns identified for each CVB serotype. Bars represent phylogenetically distinct sequences (i.e., a monophyletic lineage of homologous serotype), defined by intervening isolates of a heterologous serotype in the trees in Fig. 2. Lowercase letters indicate the 5′-NTR lineage within each serotype. Numbers after the lowercase letters indicate the 3D sublineage of that 5′-NTR lineage. The lineages are labeled chronologically, except when their relationship in 3D dictates a different order to maintain clarity. Numbers in the bars are the number of isolates of that lineage and the range of years in which the lineage was isolated.
3D.
The CVB 3D sequences differ from each other within serotype by 0.4 to 26.3% and between serotypes by 3.3 to 25.9% (Table 2). They differ from those of other HEV-B serotypes by 4.7 to 26.9%, a range comparable to that observed among the non-CVB members of HEV-B (2.4 to 22.0%). The 3D sequences of members of HEV-A, -C, and -D are at least 27.7% different from those of all HEV-B viruses, including the CVB clinical isolates (Table 2). As in the 5′-NTR, isolates of the same serotype are not monophyletic in the 3D interval, but there are many more lineages per serotype in the 3D tree, with no more than four isolates per lineage (Fig. 2). In most cases, the 3D lineages appear to be derived by subdivision of the 5′-NTR lineages. In CVB1, for example, 5′-NTR lineages b, e, and f remain distinct in 3D, while 5′-NTR lineages a and c diversified into a1, a2, c1, and c2 in 3D (Fig. 3). The isolate in CVB1 5′-NTR lineage d (USA/CT84-10165), however, clusters with one of the c lineage isolates (USA/NH77-10160) to form the complex 3D lineage c3-d (Fig. 3). In CVB2, 5′-NTR lineage d is split among four 3D lineages (d1 to d4), and the other lineages (a, b, c, and e) are the same in both 5′-NTR and 3D. Among all isolates examined, the largest monoserotypic 3D cluster contains four CVB2 isolates (lineage d1), all isolated from 1979 to 1983. CVB3 is similar to CVB1, in that the 5′-NTR lineage c isolate (USA/OH83-10185) clusters with two 5′-NTR lineage b isolates to form 3D lineage b4-c, while the other 5′-NTR lineages either remain the same in 3D (lineage a) or are split among multiple 3D lineages (lineages b and d). CVB4 is similar to CVB2, with 5′-NTR lineage b splitting into six separate 3D lineages. In CVB5, one isolate in lineage c (USA/CO87-10208) clusters with the lineage b isolate (DOR71-10200) in 3D (lineage b-c1), leaving the other lineage c isolate (HON89-10210) alone in 3D lineage c2. The two CVB6 isolates are distinct from one another in both 5′-NTR and 3D.
DISCUSSION
Our previous analysis of HEV-B prototype strains suggested that differences in pairwise sequence relationships in different genomic regions, revealed by phylogenetic reconstruction and similarity plotting, are due to interserotypic recombination (22). The complexity of the observed pattern of relationships suggests that intertypic recombination has occurred repeatedly within HEV-B and that it may be important as a general mechanism of enterovirus evolution. We have extended these observations in the present study by analyzing multiple isolates within several HEV-B serotypes to detect phylogenetic incongruencies that are characteristic of genetic recombination.
There was no conclusive evidence for interserotypic recombination within the capsid, but intraserotypic recombination appeared to occur in several cases. For example, CVB1-USA/CT48-Conn-5 shifted from a position near the base of the CVB1 cluster in VP4-VP2 (and related to ARG83-10164) to a distal position in VP1, while ARG83-10164 remained in a basal position. Interserotypic recombination within the capsid is probably highly constrained by structural requirements of the virion shell or receptor binding, but viruses of the same serotype would be expected to be more similar in structure and, hence, more compatible. There was no evidence for interspecies recombination for any region except the 5′-NTR, consistent with the previous reports of clustering of this region between species A and B (26). The factors constraining recombination outside the capsid remain unknown. One could speculate that compatibility is required between distal elements (e.g., between the 3C protease and the proteolytic cleavage sites in the capsid), the sequences and structures of which appear to be conserved within species. The observation that the 3C proteinases of Human rhinovirus 14 (Rhinovirus genus) and CVB3 (HEV-B) can correctly process the poliovirus 1 (HEV-C) nonstructural protein precursor but not the poliovirus 1 capsid protein precursor lends support to the incompatibility hypothesis (6).
One may speculate that CVB monophyly in VP1 may be at least partly driven by the use of the same receptor, the coxsackievirus-adenovirus receptor (CAR), by all six CVB serotypes (20). CAR has been shown to bind to CVB3 in the “canyon” on the virion surface, largely through interaction with residues in VP1 (10). Unfortunately, only 3 of the ∼20 residues that comprise the canyon floor (the region that specifically interacts with CAR) are within the portion of VP1 that was sequenced for this study; therefore, we could not determine whether the canyon floor residues are more highly conserved among CVB isolates than among HEV-B viruses as a whole.
Outside the capsid region, the CVB isolates are no more closely related to one another within a serotype than they are to viruses of other HEV-B serotypes, except for those isolates that are likely to be epidemiologically related (closely related both temporally and geographically). For example, CVB2 isolates USA/ND76-10169 and USA/NE77-10170 were isolated in the upper midwestern United States in 1976 and 1977, respectively, suggesting that they may have been part of the same multiyear outbreak. CVB2 was the most frequently isolated enterovirus in the United States in 1976, accounting for 15.8% of all reported enterovirus isolates (29), so the same strain probably circulated throughout the country and carried over into the next year. This relationship between epidemiologically related viruses of the same serotype suggests that noncapsid sequences may provide a large nonrevertible marker to track virus transmission. That is, monophyly in regions outside the capsid may indicate a potential epidemiologic linkage between isolates. Such relationships, however, must be interpreted in the context of capsid sequence, as cocirculating strains of different serotypes may acquire similar noncapsid sequences (e.g., the CB3-USA/VA86-10187 and CB4-USA/MA87-10197 3D sequences in Fig. 2).
Forty-one of 55 isolates show evidence for recombination between the 5′-NTR and capsid, as indicated by incongruity between the 5′-NTR and VP4/VP2 trees (Fig. 2). All 55 isolates show evidence for recombination between the capsid and 3D, as indicated by incongruity between the VP1 and 3D trees and their positions relative to the most closely related prototype strains (Fig. 2). In both the 5′-NTR and 3D, the clusters are largely temporal within a serotype, but isolates appear to remain clustered in 3D for a shorter period, relative to clusters in the 5′-NTR. (Fig. 2). For example, nine CVB2s isolated from 1979 to 1996 cluster together in the 5′-NTR, but in 3D, these viruses are divided among four clusters of one (1992), three (1991 to 1996), four (1979 to 1983), and one (1982) isolates. The fact that isolates having the same 3D lineage were isolated no more than six years apart suggests that interserotypic recombination between the VP1 and 3D intervals is a relatively frequent event, and more frequent than that between the capsid region and the 5′-NTR, as illustrated by the larger number of 3D genetic clusters summarized in Fig. 3. That is, recombination appears to have occurred at least once every 6 years for the isolates analyzed. Because the viruses analyzed represent only a tiny fraction of those circulating during a given time period (i.e., many intermediates are missing), the true rate of recombination is probably much higher. Recombination is more likely to occur between the capsid and 3D than between the capsid and 5′-NTR, probably because of the difference in distance between markers (3.7 versus 0.2 kb, respectively), but the presence of recombination hot spots may also play a role. A higher frequency of recombination between the capsid and 3D has also been observed in circulating polioviruses which exchange noncapsid sequences with viruses of other HEV-C serotypes (12, 17, 31).
From this and previous studies, data have consistently indicated a pattern for describing the major features of recombination among HEVs. First, recombination appears to occur only among members of a given species (2, 17, 18, 22). Second, the recombination events between serotypes occur almost entirely outside of the capsid region. Finally, recombination within a serotype appears to be frequent. Therefore, it is not conceptually or practically useful to think of serotype beyond the domain of the capsid protein coding region. In a complementary manner, however, the current genetic basis for the definition of enterovirus species appears to be remarkably stable over a large number of isolates and long time period, in almost all genomic regions.
After this work was submitted, a similar study was published, comparing the sequences of 40 HEV-B clinical isolates in multiple genome regions (19). While this study was limited by the number of HEV-B prototype sequences that were available for comparison and included serotypes that are not monophyletic in VP1 as are the CVB, the conclusions were substantially similar to those of our study, namely, that isolates of a given serotype are monophyletic only in the capsid region, that recombination is a frequent event in enterovirus evolution, and that recombination appears to be restricted to members of the same species.
Acknowledgments
We appreciate the contributions of the public health laboratory virologists, in the United States and abroad, who have submitted specimens and isolates for reference testing. We also thank Olen Kew for insightful discussions of enterovirus recombination.
REFERENCES
- 1.Andersson, P., K. Edman, and A. M. Lindberg. 2002. Molecular analysis of the echovirus 18 prototype. Evidence of interserotypic recombination with echovirus 9. Virus Res. 85:71-83. [DOI] [PubMed] [Google Scholar]
- 2.Brown, B. A., K. Maher, M. S. Oberste, and M. A. Pallansch. 2003. Complete genomic sequencing shows that polioviruses and members of human enterovirus species C are closely related in the non-capsid coding region. J. Virol. 77:8973-8984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cammack, N., A. Phillips, G. Dunn, V. Patel, and P. D. Minor. 1988. Intertypic genomic rearrangements of poliovirus strains in vaccinees. Virology 167:507-514. [PubMed] [Google Scholar]
- 4.Cherkasova, E. A., E. A. Korotkova, M. L. Yakovenko, O. E. Ivanova, T. P. Eremeeva, K. M. Chumakov, and V. I. Agol. 2002. Long-term circulation of vaccine-derived poliovirus that causes paralytic disease. J. Virol. 76:6791-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cuervo, N. S., S. Guillot, N. Romanenkova, M. Combiescu, A. Aubert-Combiescu, M. Seghier, V. Caro, R. Crainic, and F. Delpeyroux. 2001. Genomic features of intertypic recombinant Sabin poliovirus strains excreted by primary vaccinees. J. Virol. 75:5740-5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dewalt, P. G., M. A. Lawson, R. J. Colonno, and B. L. Semler. 1989. Chimeric picornavirus polyproteins demonstrate a common 3C proteinase substrate specificity. J. Virol. 63:3444-3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Felsenstein, J. 1993. PHYLIP: phylogeny inference package, 3.5c ed. University of Washington, Seattle.
- 8.Furione, M., S. Guillot, D. Otelea, J. Balanant, A. Candrea, and R. Crainic. 1993. Polioviruses with natural recombinant genomes isolated from vaccine-associated paralytic poliomyelitis. Virology 196:199-208. [DOI] [PubMed] [Google Scholar]
- 9.Georgescu, M.-M., F. Delpeyroux, and R. Crainic. 1995. Tripartite genome organization of a natural type 2 vaccine/nonvaccine recombinant poliovirus. J. Gen. Virol. 76:2343-2348. [DOI] [PubMed] [Google Scholar]
- 10.He, Y., P. R. Chipman, J. Howitt, C. M. Bator, M. A. Whitt, T. S. Baker, R. J. Kuhn, C. W. Anderson, P. Freimuth, and M. G. Rossmann. 2001. Interaction of coxsackievirus B3 with the full length coxsackievirus-adenovirus receptor. Nat. Struct. Biol. 8:874-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huttunen, P., J. Santti, T. Pulli, and T. Hyypia. 1996. The major echovirus group is genetically coherent and related to coxsackie B viruses. J. Gen. Virol. 77:715-725. [DOI] [PubMed] [Google Scholar]
- 12.Kew, O. M., V. Morris-Glasgow, M. Landaverde, C. Burns, J. Shaw, Z. Garib, J. André, E. Blackburn, C. J. Freeman, J. Jorba, R. Sutter, G. Tambini, L. Venczel, C. Pedreira, F. Laender, H. Shimizu, T. Yoneyama, T. Miyamura, H. G. van der Avoort, M. S. Oberste, D. R. Kilpatrick, S. Cochi, M. Pallansch, and C. de Quadros. 2002. Outbreak of poliomyelitis in Hispanola associated with circulating type 1 vaccine-derived poliovirus. Science 296:356-359. [DOI] [PubMed] [Google Scholar]
- 13.King, A. M. Q., F. Brown, P. Christian, T. Hovi, T. Hyypiä, N. J. Knowles, S. M. Lemon, P. D. Minor, A. C. Palmenberg, T. Skern, and G. Stanway. 2000. Picornaviridae, p. 657-678. In M. H. V. Van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 14.Kirkegaard, K., and D. Baltimore. 1986. The mechamism of RNA recombination in poliovirus. Cell 47:433-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindberg, A. M., P. Andersson, C. Savolainen, M. N. Mulders, and T. Hovi. 2003. Evolution of the genome of Human enterovirus B: incongruence between phylogenies of the VP1 and 3CD regions indicates frequent recombination within the species. J. Gen. Virol. 84:1223-1235. [DOI] [PubMed] [Google Scholar]
- 16.Lipskaya, G. Y., A. R. Muzychenko, O. K. Kutitova, S. V. Maslova, M. Equestre, S. G. Drozdov, R. Perez Bercoff, and V. I. Agol. 1991. Frequent isolation of intertypic poliovirus recombinants with serotype 2 specificity from vaccine-associated polio cases. J. Med. Virol. 35:290-296. [DOI] [PubMed] [Google Scholar]
- 17.Liu, H.-M., D.-P. Zheng, L.-B. Zhang, M. S. Oberste, O. M. Kew, and M. A. Pallansch. 2003. Serial recombination during circulation of type 1 wild-vaccine recombinant polioviruses in China. J. Virol. 77:10994-11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, H.-M., D.-P. Zheng, L.-B. Zhang, M. S. Oberste, M. A. Pallansch, and O. M. Kew. 2000. Molecular evolution of a type 1 wild-vaccine poliovirus recombinant during widespread circulation in China. J. Virol. 74:11153-11161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lukashev, A. N., V. A. Lashkevich, O. E. Ivanova, G. A. Koroleva, A. E. Hinkkanen, and J. Ilonen. 2003. Recombination in circulating enteroviruses. J. Virol. 77:10423-10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martino, T. A., M. Petric, H. Weingartl, J. M. Bergelson, M. A. Opavsky, C. D. Richardson, J. F. Modlin, R. W. Finberg, K. C. Kain, N. Willis, C. J. Gauntt, and P. P. Liu. 2000. The coxsackie-adenovirus receptor (CAR) is used by reference strains and clinical isolates representing all six serotypes of coxsackie virus group B and by swine vesicular disease virus. Virology 271:99-108. [DOI] [PubMed] [Google Scholar]
- 21.Oberste, M. S., K. Maher, M. R. Flemister, G. Marchetti, D. R. Kilpatrick, and M. A. Pallansch. 2000. Comparison of classic and molecular approaches for the identification of untypeable enteroviruses. J. Clin. Microbiol. 38:1170-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oberste, M. S., K. Maher, and M. Pallansch. 2004. Evidence for frequent recombination within human enterovirus B based on complete genomic sequences of all thirty-seven serotypes. J. Virol. 78:855-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oberste, M. S., K. Maher, and M. A. Pallansch. 1998. Molecular phylogeny of all human enterovirus serotypes based on comparison of sequences at the 5′ end of the region encoding VP2. Virus Res. 58:35-43. [DOI] [PubMed] [Google Scholar]
- 24.Oberste, M. S., W. A. Nix, K. Maher, and M. A. Pallansch. 2003. Improved molecular identification of enteroviruses by RT-PCR and amplicon sequencing. J. Clin. Virol. 26:375-377. [DOI] [PubMed] [Google Scholar]
- 25.Pallansch, M. A., and R. P. Roos. 2001. Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses, p. 723-775. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 26.Pöyry, T., L. Kinnunen, T. Hyypia, B. Brown, C. Horsnell, T. Hovi, and G. Stanway. 1996. Genetic and phylogenetic clustering of enteroviruses. J. Gen. Virol. 77:1699-1717. [DOI] [PubMed] [Google Scholar]
- 27.Pulli, T., P. Koskimies, and T. Hyypia. 1995. Molecular comparison of coxsackie A virus serotypes. Virology 212:30-38. [DOI] [PubMed] [Google Scholar]
- 28.Santti, J., T. Hyypia, L. Kinnunen, and M. Salminen. 1999. Evidence of recombination among enteroviruses. J. Virol. 73:8741-8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strikas, R. A., L. J. Anderson, and R. A. Parker. 1986. Temporal and geographic patterns of isolates of nonpolio enterovirus in the United States, 1970-1983. J. Infect. Dis. 153:346-351. [DOI] [PubMed] [Google Scholar]
- 30.Worobey, M., and E. Holmes. 1999. Evolutionary aspects of recombination in RNA viruses. J. Gen. Virol. 80:2535-2543. [DOI] [PubMed] [Google Scholar]
- 31.Yang, C.-F., T. Naguib, S.-Y. Yang, E. Nasr, J. Jorba, N. Ahmed, R. Campagnoli, H. van der Avoort, H. Shimizu, T. Yoneyama, T. Miyamura, M. Pallansch, and O. Kew. 2003. Circulation of endemic type 2 vaccine-derived poliovirus in Egypt from 1983 to 1993. J. Virol. 77:8366-8377. [DOI] [PMC free article] [PubMed] [Google Scholar]