Abstract
In Huntington’s disease (HD), polyglutamine expansions in the huntingtin (Htt) protein cause subtle changes in cellular functions that, over-time, lead to neurodegeneration and death. Studies have indicated that activation of the heat shock response can reduce many of the effects of mutant Htt in disease models, suggesting that the heat shock response is impaired in the disease. To understand the basis for this impairment, we have used genome-wide chromatin immunoprecipitation followed by massively parallel sequencing (ChIP-Seq) to examine the effects of mutant Htt on the master regulator of the heat shock response, HSF1. We find that, under normal conditions, HSF1 function is highly similar in cells carrying either wild-type or mutant Htt. However, polyQ-expanded Htt severely blunts the HSF1-mediated stress response. Surprisingly, we find that the HSF1 targets most affected upon stress are not directly associated with proteostasis, but with cytoskeletal binding, focal adhesion and GTPase activity. Our data raise the intriguing hypothesis that the accumulated damage from life-long impairment in these stress responses may contribute significantly to the etiology of Huntington’s disease.
Keywords: Huntington Disease, heat shock transcription factor, Heat-Shock Response, Chromatin Immunoprecipitation, cDNA Microarrays, Deep Sequencing
INTRODUCTION
Huntington’s disease (HD) is an autosomal dominant genetic disorder that leads to neurodegeneration and death; it is caused by expansion of a repeating stretch of glutamine residues (polyQ) in the protein Htt [1, 2]. Although the effects of polyQ expanded Htt are multifaceted, there is strong evidence that this form of the protein severely disrupts pathways required for the maintenance and degradation of proteins [3]. Based on this observation, several therapeutic strategies have been proposed to restore normal function to these proteostasis pathways. For example, compounds that activate the heat shock response, a critical component of homeostasis, have been shown to be effective in reducing the toxicity of expanded Htt in diverse models of HD from flies [4] to mice [5].
Heat Shock Factor 1 (HSF1) is the master regulator of the heat shock response [6]. This DNA-binding protein may provide an important avenue for restoring proteostasis in the presence of HD [7, 8]. HSF1 activates transcription of heat shock proteins (HSPs) that protect proteins from misfolding and aggregation. Two compounds that mitigate the toxicity of polyQ-expanded Htt, celastrol and geldanamycin, are believed to function by activating HSF1 [9–11] and thereby increasing the capacity of cells to respond to proteostatic stress [12, 13]. However, HSF1 is likely to have roles beyond those in proteostasis. HSF1 is highly conserved in evolution, and its homologs in other species bind to a variety of non-heat shock related gene targets [6, 14]. These targets span a range of functional categories, including metabolism, cytoskeleton, transport, signaling, and apoptosis in yeast, flies, and humans [15–17]. The diverse roles of HSF1 are highlighted by the fact that while loss of HSF1 causes damage to the central nervous system [18], it also profoundly protects mice against tumors [19].
To understand the complex role of HSF1 in Huntington’s disease we examined murine striatal neuron-derived cells that express full-length polyQ-expanded Htt [20]. These mutant polyQ-expanded Htt cells show reduced ATP levels and mitochondrial dysregulation and represent an excellent model for the early stages of the disease. To determine whether there was any defect in the transcriptional regulation of proteostasis, we used genome wide chromatin immunoprecipitation, followed by massively parallel sequencing (ChIP-Seq) to reveal HSF1 targets across the entire genome. Our study provides the first genome-wide view of how full-length polyQ-expanded Htt alters the activity of HSF1 under conditions of cellular stress. Previous studies, which had focused on a limited number of genes, suggested that polyQ-expanded Htt had only modest effects on the binding of HSF1 [21]. Our comprehensive study reveals that this is true only under normal (33°C) conditions. By contrast, in stressed cells after heat shock, polyQ-expanded Htt severely blunts the normal HSF1 response, causing the protein to bind to only one-third of the targets it regulates in wild-type cells. These changes in HSF1 binding are associated with changes in HSF1-dependent gene expression.
Surprisingly, the HSF1 targets most affected by polyQ-expanded Htt are not directly associated with proteostasis, but rather with a wide range of functions, including cytoskeletal binding, focal adhesion, and GTPase activity. Our data raise the intriguing hypothesis that polyQ-expanded Htt disturbs a broad range of HSF1 regulated cellular functions necessary for the response to stress conditions [22]. These disruptions may have only modest effects in the short term. However, with time, the accumulated damage may become significant, as has been proposed previously in other contexts [23, 24]. Our results suggest that therapies able to increase HSF1 function in pre-manifest patients might serve to protect neurons from damage.
RESULTS
Genome-wide map of HSF1 targets in STHdhQ7/Q7 cells and ST HdhQ111/Q111 cells
To explore the role of HSF1 in wild-type STHdhQ7/Q7 (from hereon referred to as ST HdhQ7) and polyQ-expanded STHdhQ111/Q111 (from hereon referred to as ST HdhQ111) striatal cells, we employed genome-wide chromatin immunoprecipitation followed by sequencing (ChIP-seq). All the libraries we generated had more than 5 millions reads and a very similar number of reads across different conditions (Figure 1A).
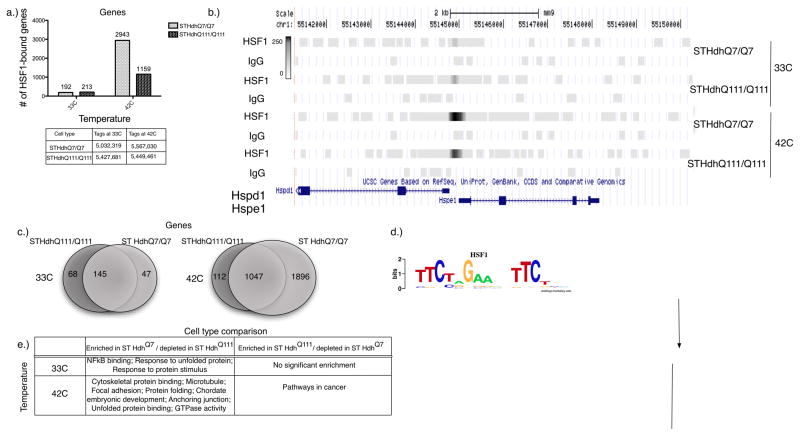
Figure 1. Mutant polyQ-expanded Htt cells show significant global reduction in HSF1 genomic occupancy.
a) Number of HSF1-bound genes under normal (33°C) and heat-shock (42°C) conditions in the wild-type and polyQ-expanded mutant Htt cells. Genes are associated with a binding event if they reside within 10,000bp upstream or downstream of a genomic region bound by HSF1. The table shows the numbers of uniquely mapping reads in each cell type under both normal and heat-shock conditions are shown. b) An example of two HSF1 targets, HSPD1 /HSPE1. The gray bars indicate the number of sequence tags in both ST HdhQ7 and ST HdhQ111 under normal (33°C) and heat-shock (42°C) conditions. IgG control binding levels are included for each condition. Genome browser bars are set on a gray-scale of 250 for all conditions. c) Number of overlapping HSF1-bound genes at normal growth and heat-shock temperatures between wild-type and mutant Htt cells. Binding events are determined by comparison to a non-specific IgG control using MACS. d) A refined HSF1 binding motif resembles the canonical HSF1 binding site. The motif was identified using 200bp sequences around the peak center of each of the top 1000 (where available) HSF1 bound regions in every condition. Motif enrichment was evaluated using the hypothesis-driven motif analysis program THEME at a p-value cutoff of 0.05. e) Functional enrichment analysis by DAVID of all genes bound significantly more (“enriched”) or less (“depleted”) in wild-type ST HdhQ7 Htt cells relative to the mutant ST HdhQ111 Htt cells at normal growth and heat-shock temperatures, as determined by MACS (p-value threshold < 1e-9). (The selected categories have a P-value< 0.0002)
HSF1 binding targets relative to an IgG control
In wild-type cells (ST HdhQ7) at normal growth temperatures (33°C), HSF1 bound a total of 260 genomic loci. These sites were strongly enriched for the expected HSF1 binding site sequence (Table S1; GEO entry GSE38002), even though the presence of a canonical binding site sequence was not required to identify a locus as bound. The bound regions were associated with 192 genes (“targets”, Table S2), consisting mostly of the expected proteostatic genes, such as CCT7, HSP90AB1, DNAJB7, CCT4, HSP90AA1, HSPE1, HSPD1 and HSPA8 (Figure 1B, Figure S1).
To examine the effect of polyQ expanded Htt on HSF1, we repeated our experiments in cells expressing full-length Htt containing a 111 glutamine repeat (ST HdhQ111). We found that the majority of genes bound by HSF1 at 33°C were the same in ST HdhQ7 and ST HdhQ111 cells (Table S1; Figure 1C). In ST HdhQ111 cells, 292 sites were bound, and these binding events mapped to 213 genes (see Table S3 for a summary of bound genes using alternative window sizes). Of the genes bound in ST HdhQ7 cells, 145 were also bound in ST HdhQ111 cells (p<3e-270). Overall, our results demonstrate that despite some minor differences, a core set of HSF1 binding remains intact in both cell types (Figure 1C).
ST HdhQ111 cells are sensitized to heat shock
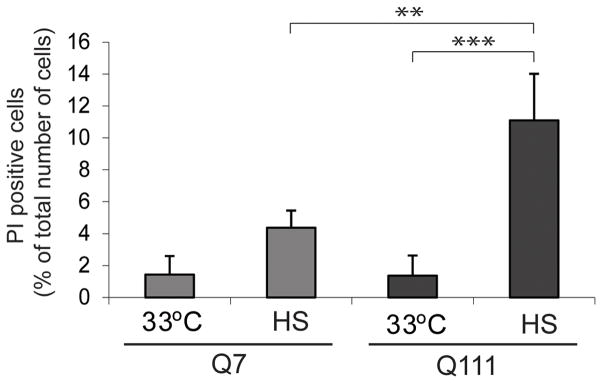
Our ChIP-Seq findings suggested that despite the presence of polyQ expanded Htt, ST HdhQ111 cells maintained mostly normal HSF1 function. To examine whether ST HdhQ111 cells had altered response under stress conditions, we tested their sensitivity to heat shock. Using FACS analysis of propidium iodide staining, which only stains the nuclei of severely damaged cells [25], we found that ST HdhQ7 cells showed a modest loss of viability upon heat shock, while ST HdhQ111 cells showed a significantly higher loss of viability (Figure 2).
Figure 2. Cells expressing polyQ-expanded Htt are sensitized to heat shock.
FACS results for ST HdhQ7 and ST HdhQ111 cells that were exposed to heat shock, and monitored for propidium iodide fluorescence for cellular injury. Bar graph presents quantification of the results pooled from three independent experiments. ST HdhQ7 and ST HdhQ111 cells were grown at 33°C, and one set of cells was exposed to heat shock for six hours at 42°C. Subsequently, all cells were stained with propidium iodide and analyzed by FACS. The error bars represent SDs. ST HdhQ111 cells show a significantly higher percentage of PI positive cells after heat shock as compared to ST HdhQ111 at 33°C and compared to ST HdhQ7 after heat shock, *** p<0.001 and **p<0.01, respectively (One-way ANOVA, followed by Tukey test).
Global reduction in HSF1 genomic occupancy in ST HdhQ111 cells
In order to understand the difference in sensitivity to heat shock between the cell lines, we reexamined HSF1 binding in both cell lines after heat shock at 42°C for 6 hours. In stark contrast to our findings at normal temperature (33°C), HSF1 had very different binding targets under heat shock conditions. In ST HdhQ7 cells 6,173 regions were bound, while only 2,161 regions were bound in ST HdhQ111 cells (Table S1). These binding events mapped to 2,943 genes in ST HdhQ7 cells and only 1,159 genes in the polyQ-expanded ST HdhQ111 cells (Figure 1C, Table S2; see Table S3 for summary of bound genes using alternative window sizes). HSF1 bound genes in ST HdhQ111 cells represented a subset of those bound in ST HdhQ7 cells: a staggering 90% (1,047; Figure 1C) of the genes bound in ST HdhQ111 cells after heat shock were also bound in ST HdhQ7 cells, but in ST HdhQ7 cells HSF1 bound an additional 1,896 genes.
The dramatic change in HSF1 binding could arise from a number of causes. We analyzed the sequence motifs associated with bound regions to determine what role DNA binding specificity might play. Using the hypothesis-driven motif discovery program THEME, we examined the 1,000 target sequences that had the highest signal. These sequences turned out to be largely the same for both ST HdhQ7 and ST HdhQ111 cells, and the most significant refined motif element for both was the canonical triple inverted repeat TTCxxGAAxxTTC (Figure 1D). The regions bound only in ST HdhQ7 were also enriched for the same motif, suggesting that binding specificity of HSF1 was unchanged in the two conditions. Interestingly, we also observed that canonical HSP targets (target genes) have a higher frequency of HSF1 motifs than novel ones, such as cytoskeletal genes, but in almost all conditions the number of motifs in HSF1-bound regions was significantly higher than expected by chance(Table S4).
HSF1 binding targets enriched in wild-type, relative to polyQ-expanded cells
To understand the functional consequences of the differences in binding that occur during heat shock, we examined the HSF1-bound genes using DAVID (Database for Annotation, Visualization and Integrated Discovery) [26, 27]. Genes bound in both cell lines were enriched for the expected functional categories of protein folding, chaperone and stress response, but also for some unexpected functional categories such as chromatin, nucleosome assembly and, to a lesser extent, cytoskeleton. Surprisingly, the genes that were bound only in STHdhQ7 cells were strongly enriched for functional categories that included cytoskeletal binding, microtubule and actin cytoskeleton, focal adhesion, anchoring junctions, and GTPase activity (Table S5). The small number of genes bound only in STHdhQ111 cells showed no significant enrichment for functional categories (Table S5).
In summary, the polyQ expanded Htt had a dramatic effect on the ability of HSF1 to respond to heat shock, but many of the genes that were no longer bound were not canonical HSF1 targets. Rather, they were associated with diverse biological processes that may have broad influences on cell viability.
Loss of HSF1 binding in ST HdhQ111 cells leads to a significant shift in mRNA expression
Since changes in the binding of regulatory regions do not necessarily lead to changes in transcription, we used microarrays to examine gene expression ST HdhQ7 cells and ST HdhQ111 cells at normal (33°C) and heat-shock conditions (42°C) (Table S6). Expression data for these cell lines under other experimental conditions have been reported by others [28, 29]. We identified 1,711 differentially upregulated and 559 differentially downregulated genes in ST HdhQ7 cells as compared to ST HdhQ111 cells after heat shock. We then classified genes based on whether they were bound in the upstream, coding, or downstream regions in one or more of the ChIP-Seq experiments. We observed that intronic HSF1 binding contributed significantly to the changes in expression between genes bound in both cell types and genes bound only in ST HdhQ7. Based on these observations, we reformulated our definition of bound genes to include binding events within the body of a gene (using a 2000bp window), which while distant from a gene’s transcription start site (TSS), can have a significant effect on the expression of the same gene (see Table S7 for summary statistics on bound genes).
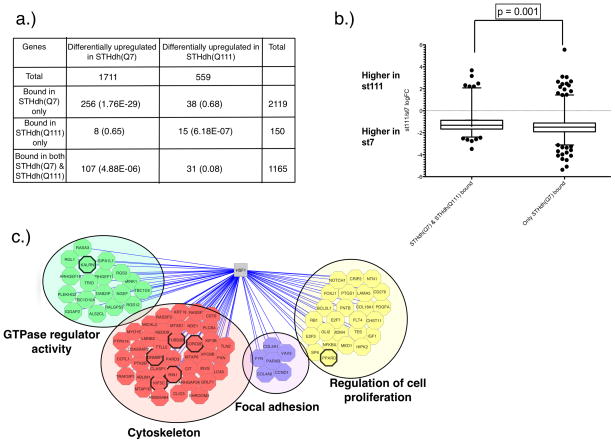
Under heat shock conditions, we find a statistically significant overlap between genes that are differentially expressed and those that are bound in only one cell type. Specifically, genes bound only in ST HdhQ7 cells overlapped significantly with those that were expressed at a higher level in these cells (p<1.76E-29). Similarly, genes bound only in STHdhQ111 were over-represented among those that had higher expression in the presence of polyQ-expanded Htt (p<6.18E-7; Figure 3A). These results are consistent with the primary role of HSF1 as a transcriptional activator.
Figure 3. Loss of HSF1 binding in the mutant Htt cells at 42°C is associated with a significant shift in mRNA expression.
a) Number of differentially upregulated and downregulated genes in the wild-type vs. mutant Htt cells under heat-shock conditions (42°C) (p-value < 0.01). For genes with more than one probe, logFC is estimated as the median of the representative significant probes. Significantly differentially expressed genes at 42°C were further subdivided into three categories: genes bound by HSF1 only in the wild-type (ST HdhQ7) cells at 42°C, those bound only in the expanded polyQ cells at 42°C, and those bound in both ST HdhQ7 and ST HdhQ111 cells at 42°C. Binding events were associated with genes, based on the distance (2000bp) relative to the body of the gene. The numbers in parentheses are p-values testing whether the overlap of bound and differentially expressed genes is greater than expected by chance using the hypergeometric distribution. b) Comparison of logFC changes in mRNA expression between wild-type and mutant Htt cells at 42°C in two categories: genes bound in both the wild-type and polyQ-expanded Htt cells vs. genes bound only in the wild-type cells. The significance of global logFC mRNA expression differences was evaluated using a Mann-Whitney test (one-sided p-value). Whiskers are set at 5th and 95th percentiles. c) Functional enrichment and representative genes in categories associated with genes that show higher levels of expression and binding in wild-type cells compared to the polyQ-expanded mutant cells at 42°C. Functional enrichment was assessed using DAVID at a corrected p-value cutoff of 0.05. In all three panels, genes were considered bound by HSF1 if they reside within 2,000bp of a significant HSF1 binding event (p-value < 1e-9). Each gene discussed in the text is indicated with a thick black line around its corresponding node.
Interestingly, we also find that the genes bound in both cell types are more highly expressed in wild-type cells. This difference can be seen by examining the overlap of genes bound in both conditions and those that are called differentially expressed (p<4.88E-6). Furthermore, we observed a small, but statistically significant drop in expression levels between genes that lost HSF1 binding in STHdhQ111 cells and genes bound in both cell types (Figure 3B, p-value = 0.001; Mann-Whitney test, one-sided p-value). To address any potential biases associated with the distance of the genes to their corresponding binding events, we also examined a larger gene mapping window around binding events (10,000bp; Table S7; Figure S2). The difference in mRNA expression between the two classes of genes remained statistically significant (p-value = 0.0005; Mann-Whitney test, one-sided p-value).
Processes that show loss of HSF1 activation in the presence of polyQ-expanded Htt
To understand whether quantitative differences in the strength of HSF1 binding between the wild-type and polyQ-expanded cells were relevant, we examined the functional enrichment of genes that showed decreased HSF1 binding and decreased expression in the presence of polyQ-expanded Htt relative to the wild-type cells (see Figure 3C and Table S8) using MACS [53]. This group of genes includes some well-established heat-shock protein targets of HSF1, such as DNAJB5, DNAJB12 and HSPB6. Interestingly, DNAJB5 has been previously found to be downregulated in the R6/2-Q150 mouse model of HD [17]. However, overall the genes that lost binding and expression were not enriched for heat shock proteins. In fact, many of the genes and functional categories that lost binding and expression did not show any obvious connection to previously described HSF1 pathways and genes (such as chaperones). Instead, these genes were enriched for GTPase regulator activity (p<4.43E-08; Figure 3C), cytoskeleton (p<2.19E-06), regulation of cell proliferation (p<8.31E-05) and focal adhesion (p<1.98E-04).
Hierarchical cluster analysis uncovers interesting relationships between HSF1 binding and expression changes upon heat shock
To understand the relationship of binding and expression in more detail, we used hierarchical clustering (Figure 4A). One of the main conclusions from our clustering analysis is that many chaperones respond in very similar ways in the two cell lines. One cluster of genes (A) contains constitutively HSF1-bound genes - canonical HSF1 binding targets that were bound under normal conditions (33°C) and remained bound upon heat shock (42°C). This cluster was enriched for molecular chaperones (Figure 4B). The majority of genes in this cluster showed no differences in expression between the two cell lines under heat shock, though some genes were downregulated in STHdhQ111 compared to STHdhQ7. A second cluster (C) largely contains genes that were bound by HSF1 exclusively upon heat shock in both cell lines, and showed increased expression in both cell lines upon heat shock. This cluster was also enriched for molecular chaperones and various genes involved in stress response. These two clusters represent a large fraction of the proteostatic machinery that we would expect to respond to HSF1, and show similar responses in both cell lines.
Figure 4. Heatmap of HSF1 binding and expression data.
a) Each column represents a gene and each row represents HSF1 binding or expression data. For each gene, binding is represented by the sum of the number of reads in all significantly bound peaks within a 2000 bp window upstream or downstream of the gene, regardless of the transcription start site location. Each expression value represents the fold change (log2) between a pair of conditions, as follows: ST HdhQ111 vs. ST HdhQ7 at 33°C, ST HdhQ111 vs ST HdhQ7 at 42°C, ST HdhQ111 at 42°C vs. 33°C, and ST HdhQ7 at 42°C vs. 33°C. b) Functional enrichment of the genes in the indicated regions of the heatmap.
Surprisingly, we find that the genes that show the biggest differences between the cell lines are not involved in proteostasis, per se. Clusters B, D, E and F all show changes in binding upon heat shock, which in some cases are associated with changes in expression. Interestingly, we find that these clusters were significantly enriched for actin binding, cytoskeletal protein binding, and transcription. Thus, the dominant effect of Htt on HSF1 appears to be altering the regulation of the least-explored aspects of HSF1 function.
Comparative analysis of the differentially expressed genes in mouse and human HD studies and differentially expressed genes between ST HdhQ7 and ST HdhQ111 under heat shock
To understand the physiological relevance of our findings, we compared the changes we observed in the ST HdhQ7 and ST HdhQ111 to previously published expression data from two mouse models (22-month CHL2 (HdhCAG150) knock-in mice [30], as well as 12- week R6/2 mice) as well as human HD patients.
We found a significant overlap between genes identified in our study and these previous studies. Considering only the expression data, we find that differentially upregulated genes in ST HdhQ111 show a significant overlap with genes that increased expression in human HD patients, as well as 22-month CHL2 mice. We observe the same trend for the differentially downregulated genes in ST HdhQ111 and their corresponding genes in the same studies. Similarly, if we examine the changes in binding, regardless of expression, we find that genes bound by HSF1 only in ST HdhQ7 as well as genes that are bound in both ST HdhQ7 and ST HdhQ111 significantly overlap with the genes that significantly change their expression levels between human HD patient samples, 12-week R6/2, or 22-month CHL2 mice and their corresponding control models (Table S9). Finally, we observe a significant overlap when we compare genes that are both bound by HSF1 and have significantly higher expression levels in ST HdhQ7 vs. ST HdhQ111 compared to the genes with higher expression in controls than in human HD patients (p-value=9.05E-3), 12-week R6/2 (p-value=1.08E-3), and 22-month CHL2 (HdhQ150; p-value=8.20E-4) mice (Table S9).
DISCUSSION
We have shown that polyglutamine-expanded full-length Htt sensitizes cells to heat shock, and we have traced the origins of this sensitivity to changes in the binding of the master heat shock regulator HSF1. Under normal conditions, there are few differences in the binding of HSF1 to its targets between cells carrying wild-type or polyQ expanded Htt. Many critical proteostatic genes, including HSP90AB1, HSPA8, HSPE1 and its co-chaperonin HSPD1, are bound by HSF1 in both cell types. Upon heat shock, much more dramatic differences emerge in the binding of HSF1. Approximately one-third of the genes bound by HSF1 in wild-type cells are not bound in the presence of mutant Htt, and loss of HSF1 binding to these genes is strongly associated with loss of expression. Unexpectedly, we find that the genes no longer regulated by HSF1 in polyQ expanded Htt cells are not the classical HSF1 targets, but are associated with a range of other important biological functions.
We note that many of these processes are also targeted by Htt through other mechanisms. Mutant Htt affects cytoskeletal protein integrity in the early stages of HD [31] and actin cytoskeleton is downregulated in the pre-symptomatic disease stages [32]. Proteins involved in cytoskeletal organization interact with Htt and are modifiers of the neurodegeneration phenotype [33]. Additionally, mutant Htt causes defective actin remodeling during stress [34] and disrupts cellular pathways involved in vesicle transport along cytoskeletons [34–36]. Furthermore, dynamin shows a striking loss by immunocytochemistry beginning in early stage HD tissue suggesting abnormal distribution of this protein [31]. Many of these effects have been shown to involve direct physical association of mutant Htt with other proteins. For example, Htt associates with microtubules in vivo and increases the efficiency of microtubule transport, facilitating dynein/dynactin-mediated vesicle transport [35] to enhance BDNF vesicular transport along microtubules [36]. Our study suggests that mutant Htt also has effects on the regulation of expression of cytoskeleton genes. Indeed, we find that some kinesin proteins (e.g. KIF3B and KIF5C) and a dynamin-binding protein that links dynamin with actin-regulating proteins (DNMBP) are differentially bound and expressed under heat-shock conditions. In addition, two highly relevant cytoskeletal genes - CORO2B and TUBB2B - show a loss of HSF1 binding and expression in the presence of mutant Htt under these conditions. CORO2B was identified as a molecular indicator of early disease mechanisms as it showed consistent gene expression changes in multiple early-stage HD mouse models [37]. TUBB2B is the major constituent of microtubules and interacts with Htt [38].
Htt has been suggested to normally function in vesicle trafficking [39, 40] and endocytosis inhibition, and our study shows disruption of HSF1 regulation in these processes. We observe a loss of binding and expression of Ston2, a component of the endocytic pathway, in the presence of polyQ-expanded Htt. LRP1, another gene involved in endocytosis, also shows decreased expression. LRP1 plays a central role in maintaining brain lipid homeostasis, which is essential for synaptic and neuronal integrity. Interestingly, this gene shows an age-dependent reduction in expression and its knock out causes neurodegeneration in a mouse model [41].
We also observe loss of binding and expression in GTPases and genes associated with small GTPase regulatory activity. Indeed, small GTPases are involved in several processes including vesicle trafficking, actin cytoskeleton remodeling and cellular growth. Dysfunction of GTPase signaling in HD has been previously reported [42]. In our study, RAB11FIP3, RAB6B, RIN1 and KALRN expression levels are reduced in the presence of mutant Htt compared to wild-type cells. RIN1 decreased expression has been reported in another study considering a HD mouse model [43].
PPARD is another important gene that we find under heat shock to be bound only in ST HdhQ7 and differentially upregulated in those cells, as compared to ST HdhQ111. The expression level of PPARD is significantly reduced in the striatum of the NLS-N171-82Q transgenic mouse model of HD [44], and has important roles in regulating cellular metabolism and suppressing inflammatory response [45]. Our findings provide a possible mechanism to explain the previous observations. Mutant Htt is also reported to disrupt cellular processes involved in protein clearance through autophagy [46–50]. Indeed, two autophagy-related genes Atg16l2, Atg3, as well as Hspb6 [51] are all HSF1 targets that show decreased expression in the presence of mutant Htt, implying that HSF1 is unable to activate autophagy in the presence of polyQ-expanded Htt.
The partially impaired ability of HSF1 to respond to stress in these cells is consistent with slow onset of Huntington’s disease. Although polyglutamine-expanded Htt, is constitutively expressed throughout the body, it primarily affects striatum and cortex relatively late in life. An intriguing hypothesis is that polyQ expanded Htt sensitizes the cells to various stresses, but is not sufficiently toxic on its own to cause cell death. On this hypothesis, striatal and cortical cells carrying mutant Htt are unable to cope with the accumulation of damage over many years. It is possible that the effects of Htt on HSF1 have an important role in this process. We have shown that polyQ Htt significantly blunts, but does not completely eliminate the HSF1 mediated stress response. Over time, the reduced HSF1 response may lead to significant damage and cell death.
Our findings raise the possibility that activating HSF1 could be an effective strategy for protecting neurons from diverse types of stress and damage. In addition to the possible increase in canonical HSF1 targets associated with proteostasis, activation could limit the impact of other types of stress. However, such a strategy will have to overcome several barriers. First, HSF1 activity is highly regulated and simply increasing the expression of HSF1 may not increase the levels of the active form of HSF1. Rather, it may be necessary to induce an increase in levels of active protein by targeting signaling pathways that regulate HSF1. A second and likely more serious concern relates to the potential risks of life-long activation of HSF1 for patients. Increased HSF1 levels may raise the risk of cancer, as tumor cells depend on HSF1 activity [19]. Further analysis of the role of HSF1 in neurodegeneration and cancer are critical to uncovering a safe and effective strategy for using HSF1 activation to treat Huntington’s disease.
MATERIAL AND METHODS
Cell culture
ST HdhQ7 and ST HdhQ111 striatal cells derived from wild type ST HdhQ7 and ST HdhQ111 knock-in embryonic mice [20] were used in this study. Cells were grown at 5% CO2 at 33°C in Dulbecco’s modified Eagle’s medium (DMEM, from CellGro, Manassas, VA) with 10% fetal bovine serum (HyClone, Logan, UT), Pen/Strep/Glutamine and 400 μg/ml G418 (both from CellGro). The cells were only used up to passage 15 and at 33°C. Heat-shocked cells were incubated at 42°C for 6 hours as heat shock.
Heat-shock and assessment of cell death by flow cytometry
ST HdhQ7 and ST HdhQ111 striatal cell lines were grown at 33°C in DMEM supplemented with 10% fetal bovine serum (FBS) to 70% confluency. The culture medium was changed to low serum/low glucose DMEM medium for 24 hours. The following day, cells were placed in 42°C incubator for 6 hours as heat shock. Subsequently, for flow cytometry assessment of cell death, adherent cells were trypsinized and combined with detached cells from the medium. All cells were spun, washed in D-PBS and filtered through a 37um mesh, followed by staining with 5ug/ml propidium iodide (PI, Sigma) in D-PBS for 5 min, short spinning and resuspension in 1 ml D-PBS. Flow cytometric analysis of the PI-stained injured or dead cells was carried out on a Becton Dickinson FACS LSR II HTS. Gates were set with unstained cells. Cells were excited with a 561 nm laser and analyzed on 695 nm detectors for PI signal. For all flow cytometry procedures, FACSdiva software programs (BD Bio-sciences) were used to analyze the data.
Expression changes induced by mutant Htt
Total RNA from triplicate cell cultures was isolated and purified using RNeasy columns (Qiagen RNeasy plus Mini Kit, Qiagen Valencia, CA) and was labeled according to Affymetrix protocol. Arrays were washed and stained in the Affymetrix Fluidics Station 450 and they were scanned using the GeneChip scanner 3000-7G controlled by Affymetrix GeneChip Operating Software (GCOS). Microarray experiments were carried out at the MIT BioMicro Center using Affymetrix Mouse Genome 430–2.0 arrays.
Gene expression data analysis
Expression data were analyzed using R and Bioconductor (http://www.bioconductor.org/). We normalized gene expression data using Robust Multichip Average (RMA) algorithm [52] and we used the LIMMA package [53], which implements a moderated t-test, to identify differentially expressed genes. We compared the expression profiles of ST HdhQ7 and ST HdhQ111 cells at each of two temperatures (33°C or 42°C), as well as the profile of each cell line between normal (33°C) and heat-shock (42°C) conditions. We selected genes with Benjamini-Hochberg adjusted p-values of 0.01 as statistically significantly differentially expressed. For genes with multiple statistically significant probes, we calculated the median logFC and used it as the representative logFC for the gene in further analyses.
Chromatin Immunoprecipitation and sequencing
Striatal cells expressing either wild-type or mutant Htt were crosslinked with 1% formaldehyde and immunoprecipitated using antibody sc-9144 (Santa Cruz Biotech) for HSF1. The specificity of the HSF1 antibody was previously described [54]. Chromatin Immunoprecipitation (ChIP) was performed as described previously [55]. Library preparation for sequencing was performed according to the standard Illumina protocol. Sequencing was performed using the Illumina Genome Analyzer II.
ChIP-Seq data analysis
Raw data were processed using the Illumina Genome Analyzer Pipeline Software. Reads were aligned to the mouse reference genome (UCSC, mm9) using ELAND extended. Significant peaks (bound regions) were identified using MACS [56] against IgG controls at different p-value thresholds (1e-5, 1e-7, 1e-9, 1e-11, 1e-13) and subsequent peak processing was done using an in-house pipeline analysis tool. Final p-value threshold used in the main text was 1e-9; results for additional p-value thresholds are included in Table S10. For direct comparisons between reads from two experimental conditions, i.e. ST HdhQ7 and ST HdhQ111 at 42°C, MACS was run at a p- value threshold of 1e-6.
Genes were associated with enriched peak regions using the mm9 (July 2007) table from the UCSC genome database [57]. We used two different distance thresholds (2,000bp or 10,000bp) between a gene and a binding event. Specifically, genes with transcription start sites (TSS) within either 2,000bp or 10,000bp of a peak were called HSF1-bound.
HSF1 binding and expression analysis integration
We identified common genes between each ChIP-seq HSF1 dataset and its matched mRNA expression profiling dataset. For the HSF1-binding data, a gene was associated with a significant binding peak if the enriched peak fell within 2,000 or 10,000bp upstream or downstream of the gene, regardless of the transcription start site location (in Table S3 we report the genes found using a window of 2,000bp). These two separate windows were selected to match either proximal promoter windows upstream of transcription start sites, or the windows of observed distant transcription factor regulation as shown before [55]. Furthermore, in calling binding events we included events within the body of a gene, because while distant from a gene’s transcription start site, they can have a significant effect on the regulation of the expression of the same gene. Significant differentially expressed probes at a p-value cutoff of 0.01 were mapped to their corresponding genes symbols using the mouse4302. db (Version 2.5.0) annotation package from Bioconductor. For genes represented by multiple significant probes, the median logFC across those probes was used as the final gene logFC.
For clustering, we assembled a matrix containing both gene expression and Chip-Seq binding data. Specifically, for each differentially expressed gene we used the fold change (log2) between each of the following pairs of conditions: ST HdhQ111 vs ST HdhQ7 at 33°C, ST HdhQ111 vs. ST HdhQ7 at 42°C, ST HdhQ111 at 42°C vs 33°C, and ST HdhQ7 at 42°C vs 33°C. For binding, we used the sum of all reads within all peaks associated with a gene. Each significant peak was associated with a gene if the peak fell within 2,000 or 10,000bp upstream or downstream of the gene, regardless of the transcription start site location. The data were normalized and clustered using centered correlation distance and complete linkage. Annotations were evaluated using DAVID [26, 27]. The ChIP-seq and cDNA microarray data have been deposited in the Gene Expression Omnibus (GEO) database with the series entry GSE38002.
Motif analysis
Motif analysis was performed on the sequences of significantly enriched peaks in all conditions. Motif enrichment was performed using the algorithm THEME [58]. All sequences from the top 1,000 peaks sorted by p-value were selected for motif enrichment analysis. A comparative background set was generated to match the size, sequence length and composition of the original (foreground) sequence set. To narrow in on relevant binding site locations, we selected only 200bp on each side of the peak center (400bp total) for motif analysis. To evaluate the significance of the identified motifs, we selected relevant parameters in THEME, i.e. beta value of 0.7, 5-fold cross-validation, and 25 randomized cross-validation trials each motif. We scanned the entire TRANSFAC database. Motifs were ranked based on cross-validation errors and the significance of the cross-validation errors (p-value cutoff = 0.05).
We scanned the sequences of HSF1-bound regions identified in ST HdhQ7 and ST HdhQ111 at 33°C and 42°C for the presence of two TRANSFAC HSF1 motifs (TRANSFAC matrices M01023 and M00146) with three different thresholds each (0.773, 0.6, and 0.5 for matrix M01023; 0.907, 0.6, and 0.5 for matrix M00146). Additionally, we scanned the sequences of known HSF1 target genes (HSPs), i.e. genes classically involved in protein folding, and of non-canonical HSF1 target genes, such as cytoskeletal genes, i.e. genes annotated as cytoskeleton-related. To evaluate the significance of the motif results, we performed a Monte Carlo simulation, generating 100 comparative background sequences with a matched sequence length and base composition to the original sequence set. We used a one-tailed binomial test to assess significance. All results are reported in detail in Table S10.
Functional enrichment analysis
To assess functional enrichment, we analyzed HSF1-bound genes in all conditions for enriched Gene Ontology categories and significant KEGG, Biocarta, or Reactome pathways using DAVID [26, 27]. Functional enrichment p-values reported in the main and supplemental text have been Benjamini-Hochberg adjusted for multiple hypothesis testing.
Comparative analysis with expression data of human and mouse models of HD
We identified differentially expressed genes between the striatum of 12-week R6/2 mice or 22-month CHL2 mice and their corresponding control models. Gene expression data for the striatum of 12-week R6/2 and control models were obtained from NCBI Gene Expression Omnibus (GEO) entries GSE9803 and GSE9804 [37], while data for the striatum of 22-month CHL2 and control models were obtained from NCBI GEO entries GSE7958 [30]. Expression levels of each probe series were normalized, using R and Bioconductor packages, by robust multi-array average (RMA) method [52], and log base2 transformed. The expression value of each gene was calculated by averaging the expression level of its corresponding probes. Differentially expressed genes between each HD model and its corresponding control model were identified using R limma package [53].
Differentially expressed genes in the caudate nucleus of normal and HD patients were identified by analyzing the transcriptional profiles of post mortem human brain tissues of 32 control and 38 HD patients obtained from NCBI GEO entry GSE3790 [59]. We identified differentially expressed genes between control and HD patients using the same procedure as outlined above. To perform comparative analysis with the results in our study, we obtained the orthologs of mouse genes in human using Bioconductor annotationTools package and the homoloGene data base [60].
The significance of the overlap between the results in our study and the differentially expressed genes in the human and mouse models of HD described above were measured by calculating the p-value of the overlap using the hypergeometric distribution.
Supplementary Material
Figure S2: Comparison of logFC changes in mRNA expression between ST HdhQ7 and ST HdhQ111 at 42°C in two categories: genes bound in both Q7 and Q111 vs. genes bound only in Q7 and within 10,000bp of a significant HSF1 binding event (p-value < 1e-9).
Bound genes have at least one significant binding peak within 10000 and 2000 bps from their transcription start site.
Table S4: Individual gene-level HSF1 motif analysis for different classes of HSF1 target genes: i) sequences of HSF1-bound regions identified in ST HdhQ7 and ST HdhQ111 at 33°C and 42°C; ii) sequences of known HSF1 target genes (HSPs); iii) sequences of non-canonical HSF1 target genes, such as cytoskeletal genes.
Bound genes have at least one significant binding peak within 2,000 or 10,000bp upstream or downstream of the gene, regardless of the transcription start site location
Acknowledgments
We would like to thank Sidhartha M. Chafekar for advice on the conditions for cell culture.
FUNDING
This work was supported by the National Institute of Health grants (R24 DK-090963 and R01-GM089903) support for sequencing (P30-ES002109) and support for computing resources from the National Science Foundation (Award No. DB1-0821391).
ABBREVIATIONS
- ChIP
chromatin immunoprecipitation
- ChIPseq
chromatin immunoprecipitation followed by sequencing
- HD
Huntington’s Disease
- Htt
huntingtin protein
- HS
heat shock
- HSR
heat shock response
- HSF1
heat shock transcription factor 1
- HSP
heat shock protein
Footnotes
Conflict of Interest statement. The authors state no conflict of interest.
References
- 1.A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell. 1993;72(6):971–83. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 2.MacDonald ME, et al. Huntington’s disease. Neuromolecular Med. 2003;4(1–2):7–20. doi: 10.1385/NMM:4:1-2:7. [DOI] [PubMed] [Google Scholar]
- 3.Voisine C, Pedersen JS, Morimoto RI. Chaperone networks: tipping the balance in protein folding diseases. Neurobiol Dis. 40(1):12–20. doi: 10.1016/j.nbd.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fujikake N, et al. Heat shock transcription factor 1-activating compounds suppress polyglutamine-induced neurodegeneration through induction of multiple molecular chaperones. J Biol Chem. 2008;283(38):26188–97. doi: 10.1074/jbc.M710521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang J, et al. Reversal of a full-length mutant huntingtin neuronal cell phenotype by chemical inhibitors of polyglutamine-mediated aggregation. BMC Neurosci. 2005;6:1. doi: 10.1186/1471-2202-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akerfelt M, et al. Heat shock factors at a crossroad between stress and development. Ann N Y Acad Sci. 2007;1113:15–27. doi: 10.1196/annals.1391.005. [DOI] [PubMed] [Google Scholar]
- 7.Fujimoto M, et al. Active HSF1 significantly suppresses polyglutamine aggregate formation in cellular and mouse models. J Biol Chem. 2005;280(41):34908–16. doi: 10.1074/jbc.M506288200. [DOI] [PubMed] [Google Scholar]
- 8.Westerheide SD, Morimoto RI. Heat shock response modulators as therapeutic tools for diseases of protein conformation. J Biol Chem. 2005;280(39):33097–100. doi: 10.1074/jbc.R500010200. [DOI] [PubMed] [Google Scholar]
- 9.Zhang YQ, Sarge KD. Celastrol inhibits polyglutamine aggregation and toxicity though induction of the heat shock response. J Mol Med (Berl) 2007;85(12):1421–8. doi: 10.1007/s00109-007-0251-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trott A, et al. Activation of heat shock and antioxidant responses by the natural product celastrol: transcriptional signatures of a thiol-targeted molecule. Mol Biol Cell. 2008;19(3):1104–12. doi: 10.1091/mbc.E07-10-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sittler A, et al. Geldanamycin activates a heat shock response and inhibits huntingtin aggregation in a cell culture model of Huntington’s disease. Hum Mol Genet. 2001;10(12):1307–15. doi: 10.1093/hmg/10.12.1307. [DOI] [PubMed] [Google Scholar]
- 12.Neef DW, Jaeger AM, Thiele DJ. Heat shock transcription factor 1 as a therapeutic target in neurodegenerative diseases. Nat Rev Drug Discov. 10(12):930–44. doi: 10.1038/nrd3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neef DW, Turski ML, Thiele DJ. Modulation of heat shock transcription factor 1 as a therapeutic target for small molecule intervention in neurodegenerative disease. PLoS Biol. 8(1):e1000291. doi: 10.1371/journal.pbio.1000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anckar J, Sistonen L. Heat shock factor 1 as a coordinator of stress and developmental pathways. Adv Exp Med Biol. 2007;594:78–88. doi: 10.1007/978-0-387-39975-1_8. [DOI] [PubMed] [Google Scholar]
- 15.Hahn JS, et al. Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol Cell Biol. 2004;24(12):5249–56. doi: 10.1128/MCB.24.12.5249-5256.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Birch-Machin I, et al. Genomic analysis of heat-shock factor targets in Drosophila. Genome Biol. 2005;6(7):R63. doi: 10.1186/gb-2005-6-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Page TJ, et al. Genome-wide analysis of human HSF1 signaling reveals a transcriptional program linked to cellular adaptation and survival. Mol Biosyst. 2006;2(12):627–39. doi: 10.1039/b606129j. [DOI] [PubMed] [Google Scholar]
- 18.Homma S, et al. Demyelination, astrogliosis, and accumulation of ubiquitinated proteins, hallmarks of CNS disease in hsf1-deficient mice. J Neurosci. 2007;27(30):7974–86. doi: 10.1523/JNEUROSCI.0006-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dai C, et al. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130(6):1005–18. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trettel F, et al. Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum Mol Genet. 2000;9(19):2799–809. doi: 10.1093/hmg/9.19.2799. [DOI] [PubMed] [Google Scholar]
- 21.Labbadia J, et al. Altered chromatin architecture underlies progressive impairment of the heat shock response in mouse models of Huntington disease. J Clin Invest. 121(8):3306–19. doi: 10.1172/JCI57413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trinklein ND, et al. The role of heat shock transcription factor 1 in the genome-wide regulation of the mammalian heat shock response. Mol Biol Cell. 2004;15(3):1254–61. doi: 10.1091/mbc.E03-10-0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bano D, et al. Neurodegenerative processes in Huntington’s disease. Cell Death Dis. 2:e228. doi: 10.1038/cddis.2011.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li XJ, Li S. Proteasomal dysfunction in aging and Huntington disease. Neurobiol Dis. 43(1):4–8. doi: 10.1016/j.nbd.2010.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore A, et al. Simultaneous measurement of cell cycle and apoptotic cell death. Methods Cell Biol. 1998;57:265–78. doi: 10.1016/s0091-679x(08)61584-8. [DOI] [PubMed] [Google Scholar]
- 26.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 27.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sadri-Vakili G, et al. Histones associated with downregulated genes are hypo-acetylated in Huntington’s disease models. Hum Mol Genet. 2007;16(11):1293–306. doi: 10.1093/hmg/ddm078. [DOI] [PubMed] [Google Scholar]
- 29.Lee JM, et al. Unbiased gene expression analysis implicates the huntingtin polyglutamine tract in extra-mitochondrial energy metabolism. PLoS Genet. 2007;3(8):e135. doi: 10.1371/journal.pgen.0030135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin CH, et al. Neurological abnormalities in a knock-in mouse model of Huntington’s disease. Hum Mol Genet. 2001;10(2):137–44. doi: 10.1093/hmg/10.2.137. [DOI] [PubMed] [Google Scholar]
- 31.DiProspero NA, et al. Early changes in Huntington’s disease patient brains involve alterations in cytoskeletal and synaptic elements. J Neurocytol. 2004;33(5):517–33. doi: 10.1007/s11068-004-0514-8. [DOI] [PubMed] [Google Scholar]
- 32.Zabel C, et al. A large number of protein expression changes occur early in life and precede phenotype onset in a mouse model for huntington disease. Mol Cell Proteomics. 2009;8(4):720–34. doi: 10.1074/mcp.M800277-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaltenbach LS, et al. Huntingtin interacting proteins are genetic modifiers of neurodegeneration. PLoS Genet. 2007;3(5):e82. doi: 10.1371/journal.pgen.0030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munsie L, et al. Mutant huntingtin causes defective actin remodeling during stress: defining a new role for transglutaminase 2 in neurodegenerative disease. Hum Mol Genet. 20(10):1937–51. doi: 10.1093/hmg/ddr075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trushina E, et al. Mutant huntingtin impairs axonal trafficking in mammalian neurons in vivo and in vitro. Mol Cell Biol. 2004;24(18):8195–209. doi: 10.1128/MCB.24.18.8195-8209.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gauthier LR, et al. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004;118(1):127–38. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 37.Kuhn A, et al. Mutant huntingtin’s effects on striatal gene expression in mice recapitulate changes observed in human Huntington’s disease brain and do not differ with mutant huntingtin length or wild-type huntingtin dosage. Hum Mol Genet. 2007;16(15):1845–61. doi: 10.1093/hmg/ddm133. [DOI] [PubMed] [Google Scholar]
- 38.Hoffner G, Kahlem P, Djian P. Perinuclear localization of huntingtin as a consequence of its binding to microtubules through an interaction with beta-tubulin: relevance to Huntington’s disease. J Cell Sci. 2002;115(Pt 5):941–8. doi: 10.1242/jcs.115.5.941. [DOI] [PubMed] [Google Scholar]
- 39.Tukamoto T, et al. Huntington’s disease gene product, huntingtin, associates with microtubules in vitro. Brain Res Mol Brain Res. 1997;51(1–2):8–14. doi: 10.1016/s0169-328x(97)00205-2. [DOI] [PubMed] [Google Scholar]
- 40.Caviston JP, et al. Huntingtin facilitates dynein/dynactin-mediated vesicle transport. Proc Natl Acad Sci U S A. 2007;104(24):10045–50. doi: 10.1073/pnas.0610628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu X, et al. Protein expression in the striatum and cortex regions of the brain for a mouse model of Huntington’s disease. J Proteome Res. 2007;6(8):3134–42. doi: 10.1021/pr070092s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X, et al. Mutant huntingtin impairs vesicle formation from recycling endosomes by interfering with Rab11 activity. Mol Cell Biol. 2009;29(22):6106–16. doi: 10.1128/MCB.00420-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas EA. Striatal specificity of gene expression dysregulation in Huntington’s disease. J Neurosci Res. 2006;84(6):1151–64. doi: 10.1002/jnr.21046. [DOI] [PubMed] [Google Scholar]
- 44.Chaturvedi RK, et al. Impaired PGC-1alpha function in muscle in Huntington’s disease. Hum Mol Genet. 2009;18(16):3048–65. doi: 10.1093/hmg/ddp243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bishop-Bailey D, Bystrom J. Emerging roles of peroxisome proliferator-activated receptor-beta/delta in inflammation. Pharmacol Ther. 2009;124(2):141–50. doi: 10.1016/j.pharmthera.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 46.Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292(5521):1552–5. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 47.Bennett EJ, et al. Global impairment of the ubiquitin-proteasome system by nuclear or cytoplasmic protein aggregates precedes inclusion body formation. Mol Cell. 2005;17(3):351–65. doi: 10.1016/j.molcel.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 48.Bennett EJ, et al. Global changes to the ubiquitin system in Huntington’s disease. Nature. 2007;448(7154):704–8. doi: 10.1038/nature06022. [DOI] [PubMed] [Google Scholar]
- 49.Diaz-Hernandez M, et al. Neuronal induction of the immunoproteasome in Huntington’s disease. J Neurosci. 2003;23(37):11653–61. doi: 10.1523/JNEUROSCI.23-37-11653.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zemskov EA, Nukina N. Impaired degradation of PKCalpha by proteasome in a cellular model of Huntington’s disease. Neuroreport. 2003;14(11):1435–8. doi: 10.1097/00001756-200308060-00006. [DOI] [PubMed] [Google Scholar]
- 51.Fuchs M, et al. Identification of the key structural motifs involved in HspB8/HspB6-Bag3 interaction. Biochem J. 425(1):245–55. doi: 10.1042/BJ20090907. [DOI] [PubMed] [Google Scholar]
- 52.Irizarry RA, et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31(4):e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 54.Trinklein ND, et al. Transcriptional regulation and binding of heat shock factor 1 and heat shock factor 2 to 32 human heat shock genes during thermal stress and differentiation. Cell Stress Chaperones. 2004;9(1):21–8. doi: 10.1379/481.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.MacIsaac KD, et al. A quantitative model of transcriptional regulation reveals the influence of binding location on expression. PLoS Comput Biol. 6(4):e1000773. doi: 10.1371/journal.pcbi.1000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuhn RM, et al. The UCSC Genome Browser Database: update 2009. Nucleic Acids Res. 2009;37(Database issue):D755–61. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Macisaac KD, et al. A hypothesis-based approach for identifying the binding specificity of regulatory proteins from chromatin immunoprecipitation data. Bioinformatics. 2006;22(4):423–9. doi: 10.1093/bioinformatics/bti815. [DOI] [PubMed] [Google Scholar]
- 59.Hodges A, et al. Regional and cellular gene expression changes in human Huntington’s disease brain. Hum Mol Genet. 2006;15(6):965–77. doi: 10.1093/hmg/ddl013. [DOI] [PubMed] [Google Scholar]
- 60.Sayers EW, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 39(Database issue):D38–51. doi: 10.1093/nar/gkq1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S2: Comparison of logFC changes in mRNA expression between ST HdhQ7 and ST HdhQ111 at 42°C in two categories: genes bound in both Q7 and Q111 vs. genes bound only in Q7 and within 10,000bp of a significant HSF1 binding event (p-value < 1e-9).
Bound genes have at least one significant binding peak within 10000 and 2000 bps from their transcription start site.
Table S4: Individual gene-level HSF1 motif analysis for different classes of HSF1 target genes: i) sequences of HSF1-bound regions identified in ST HdhQ7 and ST HdhQ111 at 33°C and 42°C; ii) sequences of known HSF1 target genes (HSPs); iii) sequences of non-canonical HSF1 target genes, such as cytoskeletal genes.
Bound genes have at least one significant binding peak within 2,000 or 10,000bp upstream or downstream of the gene, regardless of the transcription start site location