Abstract
Chronic myeloid leukemia (CML) is characterized by a specific chromosome translocation, and its pathobiology is considered comparatively well understood. Thus, quantitative analysis of CML and its progression to blast crisis may help elucidate general mechanisms of carcinogenesis and cancer progression. Hitherto it has been widely postulated that CML blast crisis originates mainly via cell-autonomous mechanisms such as secondary mutations or genomic instability, rather than by intercellular interactions. However, recent results suggest that intercellular interactions play an important role in carcinogenesis. In this study, we analyzed alternative mechanisms, including pairwise intercellular interactions, for CML blast crisis origination. A quantitative, mechanistic cell population dynamics model was employed. This model used recent data on imatinib-treated CML; it also used earlier clinical data, not previously incorporated into current mathematical CML/imatinib models. With the pre-imatinib data, which include results on many more blast crises, we obtained evidence that the driving mechanism for blast crisis origination is intercellular interaction between specific cell types. Assuming leukemic-normal interactions resulted in a statistically significant improvement over assuming either cell-autonomous mechanisms or interactions between leukemic cells. This conclusion was robust with regard to changes in the model’s adjustable parameters. Application of the results to patients treated with imatinib suggests that imatinib may act not only on malignant blast precursors, but also, to a limited degree, on the malignant blasts themselves.
Major Findings
A comprehensive mechanistic model gives evidence that the main driving mechanisms for CML blast crisis origination are interactions between leukemic and normal cells.
Keywords: mathematical/computational model, CML cell population dynamics, cancer progression, stochastic mass-action kinetics, interactions between cancer cells and normal cells
Introduction
Chronic myeloid leukemia (CML) is regarded as one of the best understood cancers, being characterized by Ph+ cells, i.e. cells having a Philadelphia (BCR-ABL) chromosome translocation (1). In the 1990’s the tyrosine kinase inhibitor imatinib mesylate (“imatinib”), which suppresses bcr-abl oncoprotein action, was found to improve prognosis dramatically. However, imatinib does not definitively cure the disease. Many patients may need to continue taking the drug indefinitely. In some cases the treatment fails and the disease progresses (2). The use of other tyrosine kinase inhibitors has mitigated but not fully solved this problem (3).
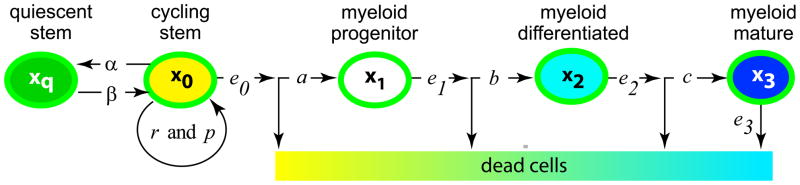
CML has a simpler etiology than most cancers (4); moreover, its time course is comparatively easy to monitor in the clinic (5–7). Consequently, despite being considerably less prevalent than major solid tumors, CML has often been regarded as a kind of “model organism” for quantitative modeling of carcinogenesis (8). Relevant to our approach is a recent cell population dynamical model of CML and its response to imatinib treatment (9). This model was grounded in the biology of hematopoietic differentiation [reviewed in (10, 11)], extended by parameter calibration using data from CML clinical trials with imatinib (12), and used to analyze treatment protocols (13). Figure 1 summarizes some of its methods. Many other mathematical/computational CML/imatinib models have been suggested in the last decade [reviewed, e.g., in (5, 8, 14)]. In the Discussion section below we will compare our present approach to some of these models.
Figure 1. Hematopoiesis as modeled in reference (9).
Hematopoietic cells form a differentiation hierarchy. Shown are cell types with various proliferation capabilities. Variables (xq,…,x3) denote the time dependent number of Ph− cells in each subpopulation; other letters denote adjustable parameters. The least differentiated cells, the stem cells, are, as has recently become common in quantitative CML modeling (14), divided into two sub-types: cycling and quiescent. Cycling stem cells can self-renew, i.e. increase in numbers by symmetric division, or they can produce progenitor cells by asymmetric division (10, 11). A similar figure is postulated for Ph+ cells.
Untreated CML usually progresses to the accelerated phase and blast crisis, whose prognosis is poor. Malignant blasts arise that apparently differ irreversibly from any of the cell types found in the chronic stage (15–17). The malignant blasts often have additional, heterogeneous karyotype alterations (7, 8, 18), and exhibit genomic instability (19, 20). Morphologically and immunophenotypically, they generally show characteristics of both leukemic stem cells and some other cell type: in most cases, granulocyte-monocyte progenitor cells (17); in a substantial minority of cases, immature members in the lymphoid, rather than myeloid, branch of the hematopoietic developmental tree (17, 21); and in a few cases cell types in the myeloid branch different from granulocyte-monocyte progenitors [e.g. (22–24)]. The mechanism of malignant blast origination remains unclear. Often, an additional point mutation or other genetic/epigenetic alteration in a Ph+ cell is postulated (15). Such alterations have been modeled quantitatively as autonomous “one-cell” processes not driven mainly by intercellular interactions [e.g. (16)].
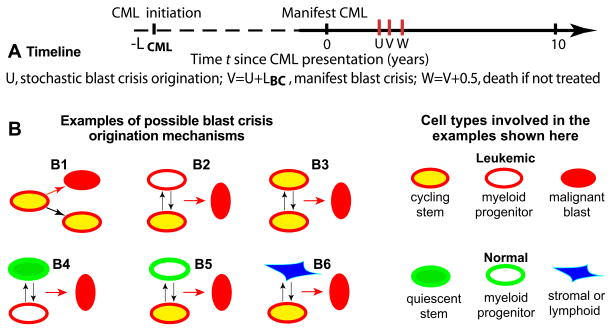
However, there is evidence [e.g. (25)] that intercellular interactions play an important role in tumor development for both leukemias and solid tumors. Among relevant growth-modulating intercellular interactions reported are the following: cell-cell signaling [e.g. (26–29)], which has been suggested as underlying a feedback mechanism responsible for CML blast crisis (30); tumor interactions with the immune system [e.g. (31, 32)]; and niche-epithelial cell contact effects [e.g. (33)], in particular for the hematopoietic system (34, 35). Therefore we here analyze quantitatively the possibility that intercellular interactions, among Ph+ cells or perhaps even between Ph+ and normal cells, play a key role in blast crisis origination. For concreteness we consider only pairwise interactions (Figure 2); our results do not exclude 3-way or higher-order intercellular interactions as blast originating mechanisms.
Figure 2. Blast crisis model.
Panel A depicts a timeline for the events we will analyze. The time origin t=0 is chosen at the time of manifest (i.e. clinically diagnosable) CML. This choice focuses the analysis on observable events, rather than on CML initiation, which occurs during times (dashed line) for which usually no data are available. LCML, U, and LBC are characterized in the text. The stochastic variable U is here shown occurring after t=0, but occurrence before t=0 (most probably shortly before) is also possible.
Panel B shows six examples of one-cell and two-cell blast crisis origination mechanisms. B1 exemplifies the usual assumption, that a one-cell alteration in a leukemic cell creates a malignant blast. B2 shows a pairwise mechanism where interaction (vertical arrows) of leukemic stem and progenitor cells (by paracrine signaling or some other interaction mechanism) induces a blast cell. B3–B6 show some additional pairwise mechanisms. Other cell types and pairs were also considered (see text). Equations for different mechanisms typically differ. In B3, for example, the mass-action-kinetics blast cell origination probability per unit time is proportional to the square of the number of Ph+ cycling stem cells; but in B1 the commonly assumed linear proportionality is more appropriate.
Current computational CML/imatinib models deemphasize clinical CML data from the pre-imatinib era. However, earlier data remain important. They not only contribute to understanding untreated CML as a cancer paradigm, they can also, as we shall show, help calibrate the parameters of recent models. A 1987 data compilation (36), not previously incorporated into the recent models, includes many more observed blast crises than do the imatinib-era data sets, and thus gives better statistics. We first use this pre-imatinib data to help establish the most likely mechanisms of blast crisis origination, then briefly compare resulting predictions to data on blast crises in imatinib-treated patients.
Quick Guide to Equations and Assumptions
Our mathematical model consists of:
coupled, nonlinear, ordinary differential equations for time-dependent cell numbers in a cell population consisting of various normal (Ph−) and leukemic (Ph+) hematopoietic cell subpopulations;
parameter values and initial conditions for the differential equations;
a stochastic formalism for determining blast crisis incidence from solutions of the differential equations.
The differential equations for Ph− and Ph+ cells are respectively:
In Equation [1], Equation [2], and Figure 1:
t is time, with the origin of time taken as the time of clinical CML presentation (Figure 2A).
Variables x with subscripts denote numbers of normal (i.e. Ph−) cells (Figure 1). For example, xq(t) is the number of normal quiescent stem cells at time t.
Leukemic (Ph+) cell numbers are denoted similarly, by y.
n0 ≡ x0 + y0 is the total number of cycling stem cells at time t, leukemic plus normal, used to model a non-linear coupling between Equations [1] and [2].
The remaining letters are adjustable parameters which characterize cellular proliferation, differentiation, density effects, apoptosis, and transitions between compartments (Figure 1). The subsection “Parameter Interpretation and Values” below gives details.
The underlying assumptions Equations [1] and [2] implement are stated and motivated in (9) and its references. In brief, with appropriate parameter values the solutions of the differential equations can explain clinically observed behavior of imatinib-treated CML in terms of different proliferation properties for different cell types in the hematopoietic hierarchy.
Initial conditions for the differential equations are obtained as follows (see Figure 2A). In the absence of Ph+ cells there is an equilibrium solution to Equation [1] that is approached after a long time and corresponds to dynamical homeostasis of a normal mature blood system. At negative time t = −LCML equilibrium with no Ph+ cells present is assumed; then one Ph+ cycling stem cell is introduced. Here LCML is the “CML latency time”, calculated as follows: the system is tracked using Equations [1] and [2] and LCML is the interval until there are 1012 mature Ph+ cells (“manifest CML” in Figure 2A). That manifest CML time is identified with the time of clinical presentation, and, for treated patients, with the start of imatinib treatment; it is chosen as the time origin t=0.
The blast crisis model compares mechanisms involving pairwise intercellular interactions for blast crisis origination (e.g. Figure 2B2) with standard one-cell mechanisms (e.g. Figure 2B1), using stochastic irreversible mass-action kinetics. Specifically, our basic assumption is:
Blast crisis origination is a time-inhomogeneous Poisson process with rate λΛ(t-LBC), where
Here the following interpretations and comments hold.
“Origination” of blast crisis is defined as the appearance of a blast clone sufficiently large that its probability of stochastic extinction is negligible. Blast crisis origination (labeled by t=U in Figure 2A) could occur probabilistically at any time t > −LCML.
The interplay between stochastic vs. deterministic calculations is based, as often (5), on small vs. large subpopulation numbers; it is summarized in Supplementary Materials section S2.
Λ(t) is the mass-action term for the incoming cell types involved. For example, if blast cells are produced by a one-cell mechanism, e.g. Figure 2B1, then zb =1 and Λ(t) is simply the appropriate cell number, e.g. Λ=y0(t) for the mechanism of Figure 2B1. If the mechanism for producing blast cells is a cell-cell interaction event, then Λ(t) in Equation [3] is bilinear or quadratic. For example, for the mechanism in Figure 2B5ya = y0. and zb = x1 in Equation [3].
λ is an adjustable parameter for each mechanism, referred to henceforth as the “intensity parameter”. The dimensions of λ are inverse time.
After blast crisis origination there is an unknown (and with present techniques not directly observable) “blast crisis latency time” LBC until “manifest” blast crisis, i.e. until accelerated phase or blast crisis can be diagnosed by standard criteria. We treat LBC as a parameter to be adjusted from the data. Manifest CML and manifest blast crisis are different events and blast crisis latency time LBC is less than CML latency time LCML (Figure 2A).
In Figure 2B6 we consider non-myeloid and non-leukemic cells, assuming their number is time-independent; this number can be absorbed into the intensity parameter λ and then the mathematical treatment of the cases in Figures 2B1 and 2B6 becomes identical, despite the biological differences, so we shall not separately consider the case of Figure 2B6 or similar cases in the further calculations. As will be seen below, this procedure leaves 45 different blast crisis mechanisms to consider.
Of note, the present mathematical treatment of blast crisis mechanisms does not distinguish between paracrine signaling and other pairwise intercellular interaction mechanisms. Rather it distinguishes between autonomous one-cell mechanisms and intercellular interaction mechanisms.
Methods
Figure 1 on hematopoietic cell population dynamics depicts our starting point. Figure 2 on blast crisis and the Quick Guide to Equations and Assumptions summarize our approach. This section gives some details.
Parameter Interpretation and Values
In Equation [1], Equation [2], and Figure 1 the following intuitive remarks apply.
The parameter p determines how much the cycling stem cell proliferation rate is decreased by crowding, with leukemic cells tolerating overcrowding better than normal ones (Table 1).
The parameter ratio α/β determines the ratio of quiescent to cycling stem cells at equilibrium. Biologically, α/β > 1 (i.e. more quiescent than cycling stem cells) is preferred (37).
With no leukemic cells and the parameter values used in our main calculation (Table 1), dynamical equilibrium for Equation [1] occurs when the number of quiescent stem, cycling stem, progenitor, differentiated, and mature cells is respectively 9×106, 106, 108, 1010, and 1012
The parameter r is a maximum rate for cycling stem cell proliferation by symmetric division, estimated to be larger for leukemic than for normal cells in the absence of treatment (Table 1). This parameter is closely related to the calculated CML mean latency time LCML (Supplementary Materials S1 gives some details).
The ej parameters are compartment outflows due to both apoptosis and (except in the case of e3) cell differentiation.
Parameters a–c are the effective input rates for formation of more differentiated by less differentiated cells, taking into account multiplication within downstream compartments. For example, the effect of divisions within the compartment of myeloid progenitors, involving an increase in numbers coupled with transition to more differentiated subcompartments, is included in the parameter a. In the model, Ph+ cells have somewhat larger values for a and b than do normal cells. However, for Ph+ cells, imatinib treatment decreases a (Table 1) putatively due to increased apoptosis of Ph+ cycling stem cells and/or less proliferation within the Ph+ progenitor cell compartment. Similarly imatinib decreases b for Ph+ cells (Table 1).
Overall, imatinib treatment is modeled as follows (Table 1): a drastic decrease in some leukemic cell compartment input parameters reflecting both apoptosis and decreased downstream proliferation; and a moderate decrease in the maximum symmetric proliferation rate for cycling leukemic stem cells.
Table 1.
Parameter values for the main calculation.‡
| parameters (days−1) | normal cells | leukemic cells* | ||
|---|---|---|---|---|
| imatinib− | imatinib+ | |||
| cycling/quiescent stem cell transitions | α | 9.0E-4 | ||
| β | 1.0E-4 | |||
| cell number outflow | e0 | 3.0E-3 | ||
| e1 | 8.0E-3 | |||
| e2 | 5.0E-2 | |||
| e3 | 1.0 | |||
| cell number inflow and proliferation | a | 0.8 | 1.6 | 1.6E-2 |
| b | 5.0 | 10 | 1/75 | |
| c | 1.0E2 | |||
| stem cell proliferation | r | 5.0E-3 | 8E-3 | 2.0E-3 |
| crowding** | p | 6.7E-7 | 1.7E-7 | |
Values shown were taken from (9). The blast crisis latency time was LBC=0. The CML latency time (LCML=11.83) years and the blast crisis intensity parameters (λ, listed in Supplementary Materials S1) were calculated from the model.
Parameter symbols have primes (for example α′, etc.). Empty cells have same values as the cells to their left.
p is dimensionless.
The Sokal Data Set
In a data set presented by Sokal in 1987 (36), the number of blast crisis cases, 1635, was markedly larger than in any recent data set. Moreover, serendipitously or by foresight, Sokal’s approach was very well suited for our present purposes. He emphasized cell population dynamics, discussed why he thought the clinical treatments involved did not substantially affect blast crisis onset kinetics, estimated the time between manifest blast crisis and death, and discussed issues related to heterogeneity in time of actual clinical diagnosis compared to the time when the disease could in principle have been diagnosed. All these are important to our use of the data.
Unfortunately the database Sokal used to prepare the paper was lost several years ago during a computer upgrade. We thus resorted to automated analysis of Sokal’s Figure 1 in his 1987 paper, a semi-logarithmic actuarial survival curve for patients diagnosed with CML but not blast crisis or accelerated phase. Sokal interpreted subsequent deaths in his data set as due to blast crises and took the time between manifest blast crisis and death to be 6 months, the same value as suggested by a more recent estimate (38).
Using CurveUNscan curve recognition software (www.curveunscan.com), we scanned Sokal’s figure to estimate empirical survival fractions. The output consisted of “survival” (defined here as being alive and not having manifest blast crisis or accelerated phase) for 1635 equi-spaced time intervals from year 0 (clinical presentation) to 15 years. Data cleansing involved the following: changing a few initial survival fractions so that survival never increased as time progressed; translating the curve by 6 months corresponding to Sokal’s estimate of the time interval between manifest blast crisis and death; and determining an over-all scale from the condition that survival (as defined above) was 100% for this cohort at the time of clinical CML presentation.
Blast Crisis Mechanisms
With y and z as in Equation [3], there are 5*11=55 products and 10 redundancies (for example y0y1 ≡ y1y0) so the number of distinct autonomous one-cell plus pairwise interaction mechanisms is 45 (examples are shown in Figure 2B). Many of these mechanisms give quite similar results computationally, reflecting close coupling among some of the cell subpopulation numbers. Our main assumption implies by Poisson statistics that the survival S(t) (i.e. the probability that manifest blast crisis has not occurred by time t) involves “blast integrals” (with integrand Λ given by Equation [3]), as the following calculation shows
For a given LBC, μ was calculated directly from the model characterized earlier (in Equation [1], Equation [2], and Table 1). Then λ was estimated by fitting to Sokal’s data, using a non-linear least squares method. Details on the statistical techniques used are described in Supplementary Materials S3.
Results
We first performed the main calculation as follows: a) we calculated the time-dependent number of cells of various types; b) given the cell numbers, we calculated the timing of blast crisis origination for 45 different candidate blast crisis mechanisms (a few of which are shown in Figure 2B), using comparatively extensive data from the pre-imatinib era to determine intensity parameters; and c) we identified preferred mechanisms. After carrying out the main calculation, we checked robustness of the results by a number of auxiliary calculations. Finally our results were applied to estimate the (small) chance of blast crisis for imatinib-treated CML, and briefly compared with current data.
Main Calculation
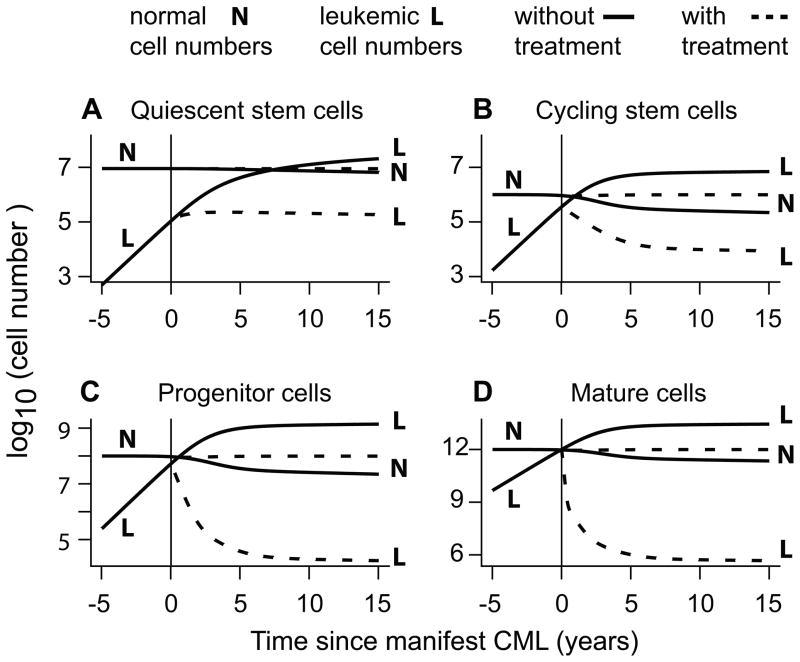
Our main calculation used the parameter values specified in Table 1, e.g. assumed that in equilibrium prior to initiation of CML there were more quiescent than cycling normal stem cells. The calculation used blast crisis latency time LBC=0, corresponding to very rapid blast expansion after blast crisis origination (Figure 2A). Equations [1] and [2] gave the cell numbers shown in Figure 3 and a CML latency time of LCML=11.83 years.
Figure 3. Cell numbers.
The figure shows results for our main calculation. A normal cell population at equilibrium numbers was perturbed by a leukemic cycling stem cell, whose clone eventually produced the time-dependent leukemic populations shown here for times in the neighborhood of the simulated diagnosis (“manifest CML”) time t=0. Numbers of differentiated but not mature cells (not shown) are intermediate between the numbers shown in panels C and D. In the absence of treatment, competition among cycling stem cells, eventually reflected in the other populations, ultimately produces a large number of leukemic cells. However, in case of imatinib treatment the model predicts mostly normal cells at close to their normal numbers (dashed lines). Equation [3] predicts a large chance of originating blast crisis in the former, but not the latter case. In reference (33) the detailed structure of the curve for mature leukemic cells under imatinib treatment (the dashed L curve in panel D) was used to calibrate parameters, and timing of imatinib response was successfully related to the duration of steps from pluripotent to mature, terminally differentiated cells in hematopoiesis.
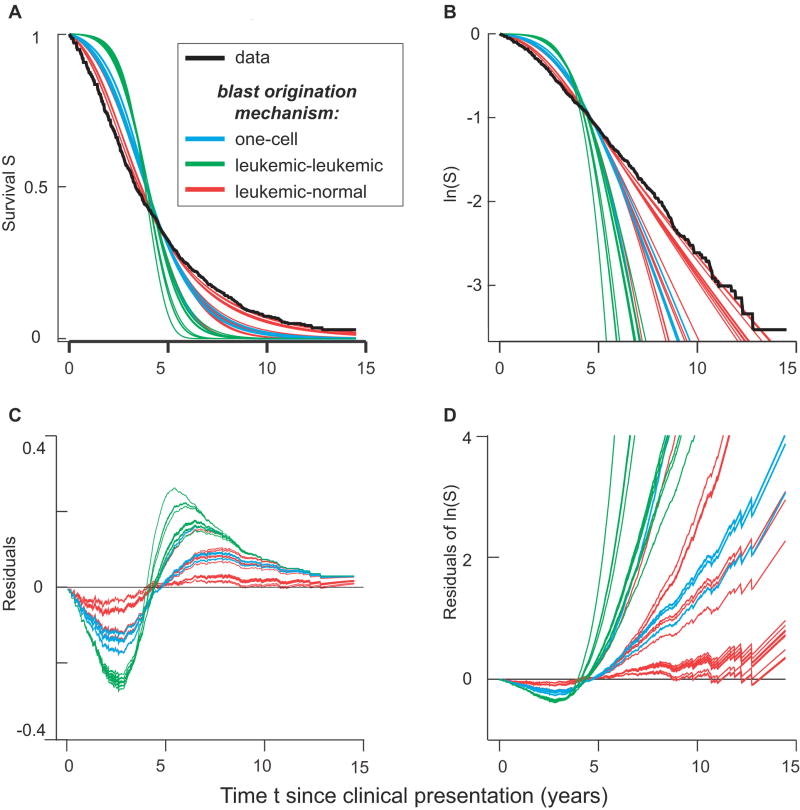
Using the cell numbers in Figure 3 we compared 45 possible blast crisis mechanisms (some of which are shown schematically in Figure 2B) to the data of (36) by using Equations [3] and [4]. The results are shown graphically in Figure 4.
Figure 4. Blast crisis mechanisms.
Clinical data for survival (i.e. fraction of patients with no manifest blast crisis yet, among patients diagnosed at time t=0 with manifest CML but without manifest accelerated phase or blast crisis) is shown in black. Predictions for three kinds of blast crisis origination mechanisms are color coded: blue for one-cell mechanisms (e.g. Figure 2B1); green for pairwise interactions between leukemic cells (e.g. Figures 2B2 and 2B3); and red for pairwise interactions between leukemic and normal myelocytic cells (e.g. Figures 2B4 and 2B5). The red curves provide the best fits, as can be seen in panels A and B, which differ only insofar as B is semilogarithmic. Residuals are shown in panels C and D, for example, log(data/theoretical) = log(data)-log(theoretical) in panel D. The correlated jags in the residuals come from corresponding jags in the data.
Predictions for all the mechanisms agree qualitatively with Sokal’s comment that, starting a few years after clinical presentation, the slope of the semilogarithmic curves is nearly constant (i.e. the time-specific incidence −d(lnS)/dt is nearly constant in time). However, for most of the mechanisms the theoretical slope is steeper than the observed slope, corresponding, intuitively speaking, to an excess predicted delay followed by excess predicted time-specific incidence.
Using non-linear regression and a log-rank test to estimate goodness of fit, as described in Supplementary Materials S3, we assigned, to each of the 45 mechanisms, p-values for the null hypothesis that the predicted survival and Sokal data do not differ. The p-values are organized in Table 1A according to the interactions; for example the (y0,x1) box in the table refers to interactions between cycling leukemic stem cells (y0) and normal myeloid progenitor cells (x1) in Equation [3]. It was apparent throughout that in such tables the entries involving y2, y3, x2, and/or x3 are always quite similar to entries involving progenitor cells instead. The intuitive reason is that changes in the differentiated cell numbers mirror changes in progenitor cell numbers with only a comparatively small time delay. We omitted the columns and rows for differentiated cells in Table 2 to focus attention on essential differences, and to take into account the fact that stem and progenitor cells, unlike more differentiated cells, are found predominantly in the bone marrow, where there are niches and more opportunities for orchestrated intercellular interactions. Tables without the omissions, from which Table 2 was condensed, are given in the Supplementary Materials S1 and S4; they do not contain important extra information.
Table 2.
Blast mechanism p-values.
| yq* | y0 | y1 | 1 | xq | x0 | x1 | |
|---|---|---|---|---|---|---|---|
| yq | <E-22 | <E-15 | <E-16 | <E-11 | <E-10 | <E-5 | <E-5 |
| y0 | <E-7 | <E-8 | 3E-3 | 0.01 | 0.76 | 0.89 | |
| y1 | <E-9 | <E-3 | 2E-3 | 0.40 | 0.54 |
Columns yq to y1 specify pairwise leukemic-leukemic cell interaction mechanisms, the column labeled “1” specifies one-cell mechanisms, columns xq to x1 specify pairwise leukemic-normal cell interaction mechanisms. Underlined values are the preferred ones for these three cases.
Results in Table 2 show statistically significant differences between observations and predictions (p-value < 0.05) for all but the four leukemic-normal cell interactions in the lower right. Of these four mechanisms, it is seen that particularly high p-values are obtained for the two mechanisms involving cycling leukemic stem (i.e. y0) cells. For leukemic-leukemic or one-cell mechanisms, the mechanism with the highest p-value also includes y0 cells, but the p-values are markedly smaller. Underlined entries In Table 2 were chosen as preferred representatives of the three different colors in Figure 4. The difference between colors is striking even allowing for Bonferroni-like corrections for multiple comparison bias (39). We estimate that at most 9 different possibilities, corresponding to y = (yq, y0) and z = (yq, y0, 1, xq, x0) in Equation [3], are effectively independent for our purposes.
Effect of Changes in the Blast Crisis Latency Time
We next considered results obtained by replacing blast crisis latency time LBC=0 with various time lags. We conjectured that large times LBC>3 years would lead to poor fits, the rationale being the following. LBC>3 years would imply that more than half of the patients who present with CML and no manifest blast crisis already have cryptic malignant blast clones, since absent treatment most manifest blast crises in such patients occur within 3 years of manifest CML (36). Given this pattern, imatinib treatment would not be able to stave off blast crisis for long since the drug is not fully effective against malignant blasts (3). But the imatinib-era data shows that, to the contrary, imatinib is very effective in preventing or long-postponing blast crises in a large majority of cases. This contradiction indicates that LBC<3 years. For small values of LBC the contradiction does not arise since the action of imatinib on blast cell precursors could explain its effectiveness.
Table 3 shows results for three non-zero values of LBC, using the same format as Table 2. Table 3 shows that leukemic-normal pairwise interactions are robustly preferred as blast crisis origination mechanisms even for LBC ≠ 0. It is also seen that, as conjectured, large blast crisis lag times are inconsistent with data on untreated CML. This confirmation of our conjecture highlights how a quantitative model can integrate disparate data sets (here pre- and post- imatinib era data) in unexpected ways to elucidate the observations.
Table 3.
Blast mechanism p-values for non zero values of blast crisis latency time LBC.
| LBC | yq* | y0 | y1 | 1 | xq | x0 | x1 | |
|---|---|---|---|---|---|---|---|---|
| 4 months | yq | <E-24 | <E-17 | <E-18 | <E-12 | <E-11 | <E-6 | <E-6 |
| y0 | <E-9 | <E-10 | <E-3 | 2E-3 | 0.51 | 0.66 | ||
| y1 | <E-11 | <E-4 | .02 | 0.20 | 0.30 | |||
|
| ||||||||
| 11 months | yq | <E-28 | <E-22 | <E-23 | <E-15 | <E-14 | <E-8 | <E-7 |
| y0 | <E-14 | <E-15 | <E-5 | <E-4 | 0.16 | 0.23 | ||
| y1 | <E-17 | <E-7 | <E-6 | 0.03 | 0.05 | |||
|
| ||||||||
| 19 months | yq | <E-32 | <E-27 | <E-29 | <E-18 | <E-18 | <E-10 | <E-10 |
| y0 | <E-21 | <E-23 | <E-9 | <E-9 | 7E-3 | 0.01 | ||
| y1 | <E-25 | <E-12 | <E-11 | <E-3 | <E-3 | |||
Other Robustness Calculations
We performed further robustness checks by varying parameters (in a “one at a time” approach), as described in Supplementary Materials. For example, analyses varying the leukemic stem cell proliferation parameter r′ showed that interaction between leukemic and normal cells remains the preferred blast crisis origination mechanism in all cases (Supplementary Materials S1). This result was particularly important because the value used for r′ in our main calculation is somewhat problematical. Originally, r′ was estimated from results on the Japanese atomic bomb survivors using older data (12), which have meanwhile been revised (40–42); moreover the estimate of r′ neglected some complications due to possible stochastic extinction of small Ph+ clones (see Supplementary Materials S2). Robustness of our main result under variations of r′ shows that these problems are technical rather than crucial.
We also examined how changing the statistical methods used affected our conclusions (see Supplementary Materials S3). We found our main results and conclusions were robust with respect to alterations of the statistical approach.
Blast Crises in Imatinib-Treated CML
Starting from a formalism originally designed for analyzing CML response to imatinib treatment, we found that data on blast crises in the pre-imatinib era can be used to characterize and calibrate a model that analyzes the cause and estimates the time of blast crisis origination. We therefore used the calibrated results for estimates of blast crisis incidence in imatinib treated patients, assuming imatinib prevents or delays blast crisis by decreasing the number of leukemic cells that can act as precursors of the blast cells, rather than by acting directly on blast cells. The estimates were compared to recent data (43), with no further adjustable parameters introduced. It was found that, despite the absence of adjustable parameters, the predictions and data have the same order of magnitude and similar shapes. Because the shapes correspond, introducing one extra adjustable parameter for each curve, i.e. allowing λ to be modified by imatinib treatment (corresponding to some imatinib action directly on the malignant blasts, not just on their precursors), can give a close match between predictions and observations (results not shown).
Discussion
Recapitulation
To asses the role of intercellular interactions in CML blast crisis formation we analyzed alternative mechanisms for blast crisis origination. Throughout, we emphasized data for the actual endpoints of interest – human hematopoiesis and CML – rather than surrogate (i.e. in silico, in vitro, animal, or loosely related human) endpoints. We used data, not previously incorporated explicitly into current quantitative CML models, from the pre-imatinib era, when many more blast crises were observed. Assuming a pairwise interaction mechanism between leukemic and normal cells for blast crisis origination robustly gave a statistically significant improvement over assuming either pairwise interactions between leukemic cells or autonomous one-cell mechanisms not driven by intercellular interactions. Applying the model with pairwise interactions between normal progenitor and leukemic cycling stem cells to blast crises for imatinib-treated patients gave predictions qualitatively consistent with the data. Thus, within the framework of our model, we found that interaction of leukemic with normal cells is the indicated mechanism for origination of blast crisis. We suggested this result could provide fundamental insights into the development of other cancers.
Other Computational Models of CML
One limitation of the present treatment is a need for more systematic calibration of the parameters in the underlying CML/imatinib model summarized above in Equations [1] and [2]; at present we have not found a convincing way to assign error bars to the values in Table 1 and thus have not developed an estimate of whether the model is overparameterized. However, a more general limitation is the fact that robustness of our basic conclusions under variations in the underlying CML/imatinib model has not been demonstrated.
In other quantitative CML/imatinib models that have been suggested in the last decade (5, 8), the mathematics involved is usually quite straightforward, but there are marked differences in the underlying assumptions and in the parameter estimates. A series of papers [e.g. (14, 44, 45)] emphasize additional modern clinical data, cell cycle effects, and the possibility of stochastic extinction of small Ph+ clones. Another series of papers incorporate additional stochastic effects and more specific biological information about hematopoiesis [reviewed in (46)]. Spatial effects, potentially important to any model involving intercellular interactions, have also been considered recently [e.g. (47)], as have immune system responses to leukemic cells [e.g. (31, 32)]. A model which considers feedback from differentiated to pluripotent cells (30) emphasizes, as we did here, intercellular interaction blast crisis mechanisms. We could not compare it in detail to our approach because its parameters have not been calibrated with clinical data.
These and other recent CML/imatinib models disagree substantially on the relevant cell population dynamics – for example on how many Ph+ cycling stem cells there are; whether these have a growth advantage over their Ph− counterparts; whether imatinib affects them; how rapidly alterations in progenitor cell numbers are reflected in the number of differentiated cells; etc. Blast crisis origination models also differ on key points. For example several [e.g. (30, 48)] regard blast crisis as a different dynamic state of the same cell types present in the chronic phase of CML rather than assuming the alternative (15–17), that malignant blasts differ irreversibly. We here assumed the later and suggest the blast phenotypic diversity reviewed in the Introduction perhaps indicates origination by interactions of Ph+ cells with diverse cell types.
Prospects
In view of the wide range of suggested formalisms, and disagreements among them, deeper quantitative analysis of blast crisis now likely depends on first choosing and calibrating a consensus mathematical/computational model for CML under imatinib treatment. Developing a consensus model will require use of large, up-to-date clinical databases and careful selection of which biological effects to deemphasize in order to avoid over-parameterization. Critically comparing various current models that emphasize cell population dynamics and intercellular interactions while using subcellular data as auxiliary rather than primary is perhaps the most promising starting point.
It is already clear from the results of the present paper that a major effort to record detailed information on the proportions of various cell subpopulations at the time of CML presentation and shortly thereafter would be an important aid to such modeling. Almost all current models consider the time-course of untreated CML. With rare exceptions, corresponding longitudinal studies are no longer carried out in the clinic. However, due to variability in presentation times, the cross sectional information available from blood and marrow samples at the time of presentation, together with modeling, could help fill the resulting gap.
The primary reason for constructing a credible, generally accepted quantitative model of the action of imatinib and tyrosine kinase inhibitors on CML would of course be finding insights that improve CML treatment. The results of the present paper suggest strongly that such a model would also improve our understanding of how blast crisis originates – whether it is basically a unicellular or, as our findings suggest, a multicellular process. This improvement in turn should help clarify analyses of how other blood and solid cancers, more complex than CML, evolve.
Supplementary Material
Acknowledgments
We are grateful to R. Hehlmann and S. Saussele for making available unpublished details on the data of reference (43) and to D. Levy and P.S. Kim for making available the source code used in reference (44).
Financial support. NASA NSCOR NNJ06HA28G (RS, JL, LH); NIH ICBP 1U54CA149233 (RS, LH); NIH grant R01-GM68423-01(AC); DOE grant DE-SC0001434 (PH).
Footnotes
Note. The following supplementary sections are available at Cancer Research online (http://cancerres.aacrjournals.org/): S1. Robustness relative to changes in the stem cell proliferation parameter. S2. Stochastic effects. S3. Statistical methods. S4. Sensitivity of main calculation to changes in the blast crisis latency time LBC. S5. Sensitivity of main calculation to changes in stem cell transition parameters. S6. Customized computer program used. S7. Supplement bibliography. In addition there is the file Sokal.csv used in the computer program S6.
Potential conflicts of interest. None.
Bibliography
- 1.Carella A, Daley G, Eaves C, Goldman J, Hehlmann R. Chronic myeloid leukaemia: biology and treatment. London: Martin Dunitz; 2001. [Google Scholar]
- 2.Cortes J, Deininger M. Chronic Myeloid Leukemia. New York: informa; 2007. [Google Scholar]
- 3.Hehlmann R, Hochhaus A, Baccarani M. Chronic myeloid leukaemia. Lancet. 2007;370:342–350. doi: 10.1016/S0140-6736(07)61165-9. [DOI] [PubMed] [Google Scholar]
- 4.Quintas-Cardama A, Cortes J. Molecular biology of bcr-abl1-positive chronic myeloid leukemia. Blood. 2009;113:1619–1630. doi: 10.1182/blood-2008-03-144790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whichard ZL, Sarkar CA, Kimmel M, Corey SJ. Hematopoiesis and its disorders: a systems biology approach. Blood. 2010;115:2339–2347. doi: 10.1182/blood-2009-08-215798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hehlmann R, Berger U, Hochhaus A. Chronic myeloid leukemia: a model for oncology. Ann Hematol. 2005;84:487–497. doi: 10.1007/s00277-005-1039-z. [DOI] [PubMed] [Google Scholar]
- 7.Hehlmann R, Saussele S. Treatment of chronic myeloid leukemia in blast crisis. Haematologica. 2008;93:1765–1769. doi: 10.3324/haematol.2008.001214. [DOI] [PubMed] [Google Scholar]
- 8.Melo JV, Barnes DJ. Chronic myeloid leukaemia as a model of disease evolution in human cancer. Nat Rev Cancer. 2007;7:441–453. doi: 10.1038/nrc2147. [DOI] [PubMed] [Google Scholar]
- 9.Foo J, Drummond MW, Clarkson B, Holyoake T, Michor F. Eradication of chronic myeloid leukemia stem cells: a novel mathematical model predicts no therapeutic benefit of adding G-CSF to imatinib. PLoS Comput Biol. 2009;5:e1000503. doi: 10.1371/journal.pcbi.1000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morrison SJ, Uchida N, Weissman IL. The biology of hematopoietic stem cells. Annu Rev Cell Dev Biol. 1995;11:35–71. doi: 10.1146/annurev.cb.11.110195.000343. [DOI] [PubMed] [Google Scholar]
- 11.Dingli D, Traulsen A, Michor F. (A)symmetric stem cell replication cancer. PLoS Comput Biol. 2007;3:e53. doi: 10.1371/journal.pcbi.0030053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michor F, Hughes TP, Iwasa Y, Branford S, Shah NP, Sawyers CL, Nowak MA. Dynamics of chronic myeloid leukaemia. Nature. 2005;435:1267–1270. doi: 10.1038/nature03669. [DOI] [PubMed] [Google Scholar]
- 13.Michor F. Quantitative approaches to analyzing imatinib-treated chronic myeloid leukemia. Trends Pharmacol Sci. 2007;28:197–199. doi: 10.1016/j.tips.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 14.Roeder I, d’Inverno M. New experimental and theoretical investigations of hematopoietic stem cells and chronic myeloid leukemia. Blood Cells Mol Dis. 2009;43:88–97. doi: 10.1016/j.bcmd.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stuart SA, Minami Y, Wang JY. The CML stem cell: evolution of the progenitor. Cell Cycle. 2009;8:1338–1343. doi: 10.4161/cc.8.9.8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michor F. Chronic myeloid leukemia blast crisis arises from progenitors. Stem Cells. 2007;25:1114–1118. doi: 10.1634/stemcells.2006-0638. [DOI] [PubMed] [Google Scholar]
- 17.Reid AG, De Melo VA, Elderfield K, Clark I, Marin D, Apperley J, Naresh KN. Phenotype of blasts in chronic myeloid leukemia in blastic phase-Analysis of bone marrow trephine biopsies and correlation with cytogenetics. Leuk Res. 2009;33:418–425. doi: 10.1016/j.leukres.2008.07.019. [DOI] [PubMed] [Google Scholar]
- 18.Radich JP. The Biology of CML blast crisis. Hematology Am Soc Hematol Educ Program. 2007:384–391. doi: 10.1182/asheducation-2007.1.384. [DOI] [PubMed] [Google Scholar]
- 19.Calabretta B, Perrotti D. The biology of CML blast crisis. Blood. 2004;103:4010–4022. doi: 10.1182/blood-2003-12-4111. [DOI] [PubMed] [Google Scholar]
- 20.Koptyra M, Cramer K, Slupianek A, Richardson C, Skorski T. BCR/ABL promotes accumulation of chromosomal aberrations induced by oxidative and genotoxic stress. Leukemia. 2008;22:1969–1972. doi: 10.1038/leu.2008.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Motoji T. Immunophenotypes on blast cells of chronic myelogenous leukemia. Nippon Rinsho. 2001;59:2342–2347. [PubMed] [Google Scholar]
- 22.Oh SH, Park TS, Kim HR, Lee JY, Kim JH, Shin JH, Lee JN. Chronic myelogenous leukemia showing biphenotypic blast crisis followed by lineage switch to B lymphoblastic leukemia. Leuk Res. 2009;33:e195–198. doi: 10.1016/j.leukres.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 23.Pullarkat ST, Vardiman JW, Slovak ML, Rao DS, Rao NP, Bedell V, Said JW. Megakaryocytic blast crisis as a presenting manifestation of chronic myeloid leukemia. Leuk Res. 2008;32:1770–1775. doi: 10.1016/j.leukres.2008.02.025. [DOI] [PubMed] [Google Scholar]
- 24.Westfall DE, Zhang L, Song S, Lee S. Concurrent megakaryocytic and erythroid chronic myelogenous leukemia blast crisis. Arch Pathol Lab Med. 2008;132:1021–1025. doi: 10.5858/2008-132-1021-CMAECM. [DOI] [PubMed] [Google Scholar]
- 25.Baker SG, Kramer BS. Paradoxes in carcinogenesis: new opportunities for research directions. BMC Cancer. 2007;7:151. doi: 10.1186/1471-2407-7-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhowmick NA, Moses HL. Tumor-stroma interactions. Curr Opin Genet Dev. 2005;15:97–101. doi: 10.1016/j.gde.2004.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sachs RK, Chan M, Hlatky L, Hahnfeldt P. Modeling intercellular interactions during carcinogenesis. Radiat Res. 2005;164:324–331. doi: 10.1667/rr3413.1. [DOI] [PubMed] [Google Scholar]
- 28.Almog N, Ma L, Raychowdhury R, Schwager C, Erber R, Short S, Hlatky L, Vajkoczy P, Huber PE, Folkman J, Abdollahi A. Transcriptional switch of dormant tumors to fast-growing angiogenic phenotype. Cancer Res. 2009;69:836–844. doi: 10.1158/0008-5472.CAN-08-2590. [DOI] [PubMed] [Google Scholar]
- 29.Trosko JE. From adult stem cells to cancer stem cells: Oct-4 Gene, cell-cell communication, and hormones during tumor promotion. Ann N Y Acad Sci. 2006;1089:36–58. doi: 10.1196/annals.1386.018. [DOI] [PubMed] [Google Scholar]
- 30.Wodarz D. Stem cell regulation and the development of blast crisis in chronic myeloid leukemia: Implications for the outcome of Imatinib treatment and discontinuation. Med Hypotheses. 2008;70:128–136. doi: 10.1016/j.mehy.2007.03.040. [DOI] [PubMed] [Google Scholar]
- 31.Moore H, Li NK. A mathematical model for chronic myelogenous leukemia (CML) and T cell interaction. J Theor Biol. 2004;227:513–523. doi: 10.1016/j.jtbi.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 32.Kim PS, Lee PP, Levy D. Dynamics and potential impact of the immune response to chronic myelogenous leukemia. PLoS Comput Biol. 2008;4:e1000095. doi: 10.1371/journal.pcbi.1000095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walker MR, Patel KK, Stappenbeck TS. The stem cell niche. J Pathol. 2009;217:169–180. doi: 10.1002/path.2474. [DOI] [PubMed] [Google Scholar]
- 34.Kirouac DC, Madlambayan GJ, Yu M, Sykes EA, Ito C, Zandstra PW. Cell-cell interaction networks regulate blood stem and progenitor cell fate. Mol Syst Biol. 2009;5:293. doi: 10.1038/msb.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carlesso N, Cardoso AA. Stem cell regulatory niches and their role in normal and malignant hematopoiesis. Curr Opin Hematol. 2010;17:281–286. doi: 10.1097/MOH.0b013e32833a25d8. [DOI] [PubMed] [Google Scholar]
- 36.Sokal JE. Prognosis in chronic myeloid leukaemia: biology of the disease vs. treatment. Baillieres Clin Haematol. 1987;1:907–929. doi: 10.1016/s0950-3536(87)80032-x. [DOI] [PubMed] [Google Scholar]
- 37.Cheshier SH, Morrison SJ, Liao X, Weissman IL. In vivo proliferation and cell cycle kinetics of long-term self-renewing hematopoietic stem cells. Proc Natl Acad Sci U S A. 1999;96:3120–3125. doi: 10.1073/pnas.96.6.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kantarjian H, O’Brien S, Cortes J, Giles F, Thomas D, Kornblau S, Shan J, Beth Rios M, Keating M, Freireich E, Talpaz M. Sudden onset of the blastic phase of chronic myelogenous leukemia: patterns and implications. Cancer. 2003;98:81–85. doi: 10.1002/cncr.11477. [DOI] [PubMed] [Google Scholar]
- 39.Hochberg Y. A sharper Bonferroni procedure for multiple tests of significance. Biometrika. 1988;75:800–802. [Google Scholar]
- 40.Little MP, Weiss HA, Boice JD, Jr, Darby SC, Day NE, Muirhead CR. Risks of leukemia in Japanese atomic bomb survivors in women treated for cervical cancer and in patients treated for ankylosing spondylitis. Radiat Res. 1999;152:280–292. [PubMed] [Google Scholar]
- 41.Little MP, Hoel DG, Molitor J, Boice JD, Wakeford R, Muirhead CR. New models for evaluation of radiation-induced lifetime cancer risk and its uncertainty employed in the UNSCEAR 2006 report. Radiat Res. 2008;169:660–676. doi: 10.1667/RR1091.1. [DOI] [PubMed] [Google Scholar]
- 42.Richardson D, Sugiyama H, Nishi N, Sakata R, Shimizu Y, Grant EJ, Soda M, Hsu WL, Suyama A, Kodama K, Kasagi F. Ionizing radiation and leukemia mortality among Japanese Atomic Bomb Survivors, 1950–2000. Radiat Res. 2009;172:368–382. doi: 10.1667/RR1801.1. [DOI] [PubMed] [Google Scholar]
- 43.Saussele S, Lauseker M, Pfirrmann M, Leitner A, Pletsch N, Stegelmann F, Ho AD, Schlimok G, Lindemann A, Beelen D, Gratwhol A, Haenel M, Haferlach C, Schlegelberger B, Mueller MC, Erbern P, Schnittger S, Jung-Munkwitz S, Shazi S, Hasford J, Hochhaus A, Hehlmann R. Evolution of Blast Crisis (BC) in Chronic Myeloid Leukemia (CML) in the Imatinib-Era: A Rare Event with High Proportions of Low Risk Patients and of Early Bc; Need for Rapid Detection. Results of the German CML Study IV. Blood. 2009:114. Abstract 3287. [Google Scholar]
- 44.Kim PS, Lee PP, Levy D. Modeling imatinib-treated chronic myelogenous leukemia: reducing the complexity of agent-based models. Bull Math Biol. 2008;70:728–744. doi: 10.1007/s11538-007-9276-z. [DOI] [PubMed] [Google Scholar]
- 45.Roeder I, Horn M, Glauche I, Hochhaus A, Mueller MC, Loeffler M. Dynamic modeling of imatinib-treated chronic myeloid leukemia: functional insights and clinical implications. Nat Med. 2006;12:1181–1184. doi: 10.1038/nm1487. [DOI] [PubMed] [Google Scholar]
- 46.Lenaerts T, Pacheco JM, Traulsen A, Dingli D. Tyrosine kinase inhibitor therapy can cure chronic myeloid leukemia without hitting leukemic stem cells. Haematologica. 2009 doi: 10.3324/haematol.2009.015271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Komarova NL, Wodarz D. Combination therapies against chronic myeloid leukemia: short-term versus long-term strategies. Cancer Res. 2009;69:4904–4910. doi: 10.1158/0008-5472.CAN-08-1959. [DOI] [PubMed] [Google Scholar]
- 48.Ainseba B, Benosman C. Optimal control for resistance and suboptimal response in CML. Math Biosci. 2010;227:81–93. doi: 10.1016/j.mbs.2010.06.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.