Abstract
Although PPARγ has anti-inflammatory actions in macrophages, which macrophage populations express PPARγ in vivo and how it regulates tissue homeostasis in the steady-state and during inflammation remains unclear. We now show that lung and spleen macrophages selectively expressed PPARγ among resting tissue macrophages. In addition, Ly-6Chi monocytes recruited to an inflammatory site induced PPARγ as they differentiated to macrophages. When PPARγ was absent in Ly-6Chi-derived inflammatory macrophages, initiation of the inflammatory response was unaffected but full resolution of inflammation failed, leading to chronic leukocyte recruitment. Conversely, PPARγ activation favors resolution of inflammation in a macrophage PPARγ-dependent manner. In the steady state, PPARγ deficiency in red pulp macrophages did not induce overt inflammation in the spleen. By contrast, PPARγ deletion in lung macrophages induced mild pulmonary inflammation at the steady-state and surprisingly precipitated mortality upon infection with S. pneumoniae. This accelerated mortality was associated with impaired bacterial clearance and inability to sustain macrophages locally. Overall, we uncovered critical roles for macrophage PPARγ in promoting resolution of inflammation and maintaining functionality in lung macrophages where it plays a pivotal role in supporting pulmonary host defense. Additionally, this work identifies specific macrophage populations as potential targets for the anti-inflammatory actions of PPARγ agonists.
Introduction
PPARγ is a ligand-controlled transcription factor of the nuclear receptor family capable of regulating gene expression by transactivation or transrepression (1). First discovered as the master regulator of the genetic program supporting adipocyte differentiation, PPARγ is involved in the regulation of a number of physiological processes such as the response to insulin, cell proliferation, cellular lipid metabolism, and inflammation (2). Thus, PPARγ activation is an attractive therapeutic target in a variety of diseases such as type 2 diabetes, cancer, atherosclerosis, and immune disorders. Activation of PPARγ can be achieved by natural fatty acid derivatives as well as synthetic ligands from the thiazolidinedione family, the latter being used clinically to improve insulin sensitivity in type 2 diabetic patients (3).
The anti-inflammatory role of PPARγ came to the forefront in the late 1990s, when 15-deoxy-delta-12,14-prostaglandin J2 (15d-PGJ2) and thiazolidinediones were shown to dampen macrophage activation in vitro by activating PPARγ(4, 5). Since then, the anti-inflammatory role of PPARγ agonists has been extensively documented in vitro and in vivo (1, 6). Indeed, PPARγ agonists suppress DSS-induced colitis (7), obesity-induced insulin resistance (8), and the progression of atherosclerosis (9). By contrast, deletion of PPARγ in macrophages exacerbates the development of atherosclerosis (10, 11), colitis (12) and obesity-induced insulin resistance (13). Based on these studies, a model emerges wherein macrophages are universally central targets of PPARγ modulation. However, it is not known whether all monocyte/macrophage populations express PPARγ or rely on its activation to maintain homeostasis or to carry out their functions in different organs during inflammation. Ultimately, the design and development of therapeutic strategies based on the use of PPARγ agonists to combat inflammatory diseases would benefit from the identification of the specific macrophage populations potentially responsive to these agonists.
In this context, we decided to profile the expression of PPARγ in a range of macrophage population extracted from different organs, delineate its preferential site of expression and examine the impact of its deficiency during the steady state and after inflammatory challenge in relevant tissues. We show in vivo that PPARγ is induced in monocytes recruited to sites of inflammation as they differentiate into macrophages, and its function is required to fully turn off inflammatory cell recruitment during resolution. In resting tissue macrophages, PPARγ expression was found to be restricted to specific populations, which are lung and splenic red pulp macrophages. In the lung, but not the spleen, deficiency of PPARγ in macrophages was associated with low-level, spontaneous inflammation in the steady state and profound alterations in macrophage gene expression. Challenge with S. pneumoniae revealed that deletion of PPARγ in lung macrophages impaired host defense, delaying bacterial clearance and thereby accelerating infection-induced mortality. Overall, these findings uncovered a key role of macrophage PPARγ in supporting resolution of inflammation, while pointing specifically to the lung as a central organ where the function of PPARγ goes beyond an anti-inflammatory role and extends critically into maintenance of host defense.
Materials and Methods
Animals and treatments
LysM-cre mice (C57BL/6J) and PPARγ floxed mice (C57BL/6J) were obtained from Jackson Laboratories and crossed in house to generate mice with PPARγ deficiency in myeloid cells (hereafter named LysM-Cre × PPARγflox/flox). LysM-cre × Rosa26-stopfloxEGFP reporter mice were bred in house. Csf2rb−/− Csf2rb2−/− and C57Bl/6J control mice were obtained from Jackson Laboratories.For acute inflammation and resolution experiments, peritonitis was induced by intraperitoneal injection of 1 ml sterile thioglycollate (Sigma, 3% wt/vol). Induction of inflammation in the spleen was achieved by intravenous lipopolysaccharide injection (Escherichia coli 026:B6, 20 μg/mouse). For infection experiments, mice were inoculated intranasally with 2.106 colony-forming units (cfu) or 5.105 cfu of Streptococcus pneumoniae serotype 3 (American Type Culture Collection, ATCC #6303) and survival was assessed every other day over a period of 12 days. Mice were housed in a specific pathogen-free environment and used in accordance with protocols approved by the Institutional Animal Care and Utilization Committee at Mount Sinai School of Medicine.
Microarray analysis
Monocytes were identified as CD115+ low side-scatter cells and sorted into two subsets based on Ly6-C expression as previously described (14, 15). All other microarrays on mononuclear phagocytes were carried out as part of the Immunological Genome Project (www.immgen.org) (16). The isolation procedures and corresponding flow plots for all cells can be found on the ImmGen website. Steady state macrophages from the peritoneum were sorted into two populations (17), including CD115+ F4/80hi MHC II− Ly6-C− B220− and CD115+ F4/80lo MHC II+ Ly6-C− B220− populations; inflamed peritoneal macrophages were CD115+ F4/80int Ly6-C− B220−, whereas neutrophils were sorted as Ly6-G+Ly6-Cint CD115− B220− cells. In the lung, macrophages were sorted as CD11c+ MHC IIlo SiglecF+ CD11b− cells (18), and lung DCs as CD11c+ MHC II+ cells that were either CD11b+ (CD11b+ DCs) or CD103+ (CD103+ DCs) (18), Jakubzick, 2008 #179}. Brain microglia were sorted as CD45lo CD11b+ F4/80+ cells (19). Gut macrophages were CD45+ CD11clo MHC II+ CD103− CD11b+ cells (20). In the spleen, red pulp macrophages were F4/80hi MHCint cells and DC subsets were CD11c+ MHC II+ cells that differentially expressed CD4 (CD11b+ CD4+ CD8−) or CD8 (CD11b− CD4− CD8+) (21). RNA was prepared from sorted populations from C57BL/6J mice after sorting directly into TRIzol reagent, amplified and hybridized on the Affymetrix Mouse Gene 1.0 ST. For data analysis using ImmGen datasets, raw data for all populations were normalized using the RMA algorithm. Extensive quality control documents are available on the Immgen website. All datasets have been deposited at National Center for Biotechnology Information/Gene Expression Omnibus under accession number GSE15907 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE15907). Microarrays on blood monocytes treated with a PPARγ agonist were performed as previously described (15) using Affymetrix GeneChip® 430 2.0 arrays. Corresponding datasets have been deposited at National Center for Biotechnology Information/Gene Expression Omnibus under accession number GSE32034 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE32034).
Blood and tissue sample preparation for flow cytometry
Mouse blood was collected by non-terminal submandibular or terminal cardiac puncture and red blood cells were lysed in hypotonic buffer (PharmLyse, BD Bioscience). Total leukocytes were quantitated by fresh blood dilution in Turk’s solution (Ricca Chemical Company). Lungs were harvested, minced, incubated in Hanks’ balanced saline solution containing 3% FBS and collagenase D for 1 h, passed through a 18-gauge needle to obtain homogeneous cell suspensions and filtered using a 100-μm cell strainer. Bronchoalveolar lavage was obtained by flushing the airways four times with Hanks’ balanced saline solution. Spleens were minced, placed into the cup portion of a cell strainer and then gently mashed and pushed through the cell strainer. Red blood cells were then lysed in hypotonic buffer. Peritoneal exudates were collected using cold Hanks’ balanced saline solution (HBSS). Cell suspensions were then stained with appropriate antibodies for 30 min on ice and data were acquired on a BD FACS Canto II Flow Cytometer (BD Biosciences) and analyzed with FlowJo software (Treestar).
Fluorescent conjugates of anti-mouse CD115 (AFS98), F4/80 (BM8), CD45 (30-F11), CD11c (N418), IA-IE (M5/114.15.2), CD4 (GK1.5), CD8 (53-6.7), CD45.2 (104) and CD45.1 (A20) were purchased from eBiosciences. Anti-mouse Gr-1 (Ly-6C/G, RB6-8C5) and CD36 (HM36) were purchased from Biolegend. Anti-mouse F4/80 (CI:A3-1) was purchased from Serotec. Anti-mouse CD36 (CRF D-2712), Ly6G (1A8) and siglec F (E50-2440) were purchased from BD Bioscience. Anti mouse FABP4 (BAF1443) was from R & D Systems.
Immunoblot analysis
FACS sorted cells were homogenized in lysis buffer containing protease inhibitors. Protein extracts were run on Criterion gels (Bio-Rad) and blotted onto nitrocellulose membranes. After blocking, immunoblots were incubated with primary antibodies against PPARγ and βActin (Cell signaling). Blots were then incubated with fluorescent secondary antibodies and proteins were detected using the fluorescence-based Odyssey Infrared Imaging System (LI-COR Biosciences).
Macrophage transfer
Peritoneal macrophages were retrieved by lavage from CD45.2 Lys-Cre × PPARγflox/flox and wild-type controls 5 days after thioglycollate instillation. Then, 5.106 macrophages were injected into the peritoneum of naïve CD45.1 wild-type mice and the number of recruited CD45.1+ neutrophils was assessed 24 hours later.
Monocyte labeling in vivo
Ly-6Clo monocytes were labeled in vivo by intravenous injection of 1-μm Fluoresbrite green fluorescent (YG) plain microspheres (Polysciences Inc.) diluted 1:4 in sterile PBS (22, 23). Ly-6Chi monocytes were labeled with beads using the same protocol, except that beads were administered 3 days after intravenous injection of clodronate-loaded liposomes (250 μl per mouse) (22). Labeling efficiency was verified by flow cytometry one and/or two days after labeling by analysis of blood collected i.v. through the submandibular vein. Clodronate was a gift from Roche and was incorporated into liposomes as previously described (24).
Analysis of gene expression by quantitative real time PCR (qPCR)
RNA samples were prepared using TRIzol reagent (Invitrogen) from thioglycollate-elicited macrophages isolated from mice at sacrifice. Each RNA preparation was hybridized with oligo dT (Invitrogen) and reverse-transcribed using Superscript III reverse transcriptase (Invitrogen). Quantitative real time PCR was performed using a LightCycler PCR System (Roche) as previously described (25). Expression data was analysed by crossing points calculated from the LightCycler data analysis software and corrected for PCR efficiencies of both the target and the reference gene.
Analysis of bacterial burden
Bacterial burden was quantified by plating 10-μl of lung homogenates serially diluted in trypticase soy broth (BD) on blood agar plates (Trypticase Soy Broth + 1.875% agar + 5% sheep blood). After incubating plates at 37°C for 18-24 hrs, colonies were counted.
Statistical analysis
Data are expressed as mean ± SEM. Statistical differences were assessed using a 2-tailed t test or ANOVA (with Tukey’s post-test analysis) with GraphPad Prism software. A P value of less than 0.05 was considered statistically significant.
Results
Differential expression of PPARγ and regulation of canonical PPARγ target genes among different tissue macrophage populations
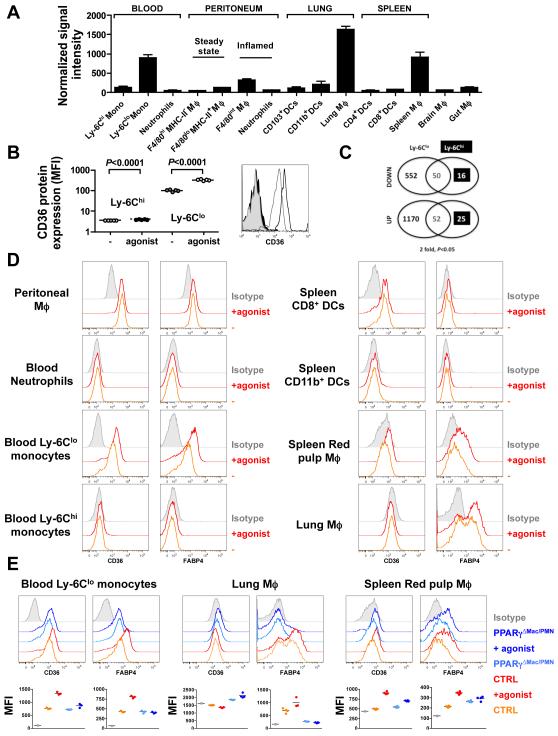
To better understand the role of PPARγ in mononuclear phagocytes, we first assessed PPARγ mRNA expression in blood monocytes, resident macrophages from different tissues including the lung, splenic red pulp, brain (microglia), gut and peritoneum, as well as in inflammatory peritoneal macrophages. For monocytes, we independently assessed the two major circulating subsets that in mice differentially express Ly6-C and which have counterparts in other species, including humans (14, 26). We compared these populations to spleen and lung conventional dendritic cell subsets as well as neutrophils. The populations were sorted (see http://www.immgen.org for detailed sorting strategies) and further analyzed by gene array. PPARγ mRNA was differentially expressed over several orders of magnitude in different mononuclear phagocytes (Fig. 1A). Macrophages from the steady-state peritoneum, brain, and gut expressed only low levels of PPARγ, equivalent to the signal intensity in Ly-6Chi blood monocytes and neutrophils. By contrast, high levels of PPARγ mRNA were observed in Ly-6Clo monocytes, splenic red pulp macrophages and pulmonary macrophages (Fig. 1A). Consistent with this, treatment of wild-type mice with the PPARγ agonist rosiglitazone induced further expression of the PPARγ-inducible CD36 protein at the cell surface of Ly-6Clo but not Ly-6Chi monocytes in wild-type animals, suggesting that only the Ly-6Clo but not Ly6-Chi monocytes were responsive to PPARγ activation (Fig. 1B). Indeed, PPARγ activation profoundly impacted the transcriptome of Ly-6Clo monocytes (602 genes down-regulated and 1222 genes up-regulated, 2-fold cut off) (Fig.1C, supplemental Table 1), and especially affected gene signatures such as “dendritic cell maturation”, “p53 signaling”, “NFAT and immune response” (unpublished data). By contrast, Ly-6Chi monocytes were largely unresponsive to the agonist (66 genes down-regulated and 77 genes up-regulated) (Fig. 1C, supplemental Table 2), suggesting that the levels of PPARγ in Ly-6Chi monocytes, and by extension in neutrophils, dendritic cells, steady-state peritoneum, brain, and gut macrophages are too low to confer significant responsiveness to PPARγ ligands under homeostatic conditions. This was further confirmed as neutrophils, dendritic cells, and peritoneal macrophage did not upregulate the expression of the two prototypic PPARγ target genes CD36 and FABP4 following PPARγ agonist treatment (pioglitazone), whereas other populations that express PPARγ upregulated CD36 and/or FAPB4 (Fig. 1D). Populations that upregulated CD36 and FABP4 in response to PPARγ agonists typically did so in a PPARγ-dependent manner (Fig. 1E), but diversity in expression of these canonical PPARγ targets was substantial. Lung macrophages did not express surface levels of CD36, even after PPARγ agonist treatment (Fig. 1E), but blood Ly6-Clo monocytes increased CD36 expression in response to pioglitazone in a PPARγ-dependent manner (Fig. 1E). FABP4 was differentially expressed among lung macrophages, raising the possibility of heterogeneity in this population, and its expression was completely dependent upon PPARγ, whether at baseline or after pioglitazone treatment. By contrast, basal FABP4 was not dependent upon PPARγ in spleen macrophages, though it was responsive to induction by pioglitazone in a PPARγ-dependent manner (Fig. 1E). Overall, these data point to a great diversity in PPARγ expression among resting differentiated macrophages, indicating that PPARγ upregulation is not necessarily an inevitable consequence of macrophage development (27), and reveal that the expression of putative PPARγ target genes are regulated somewhat differently in different tissue macrophage populations.
Figure 1. PPARγgene expression profiling and regulation of canonical PPARγtarget genes in mononuclear phagocytes.
(A) PPARγ mRNA expression was analyzed by gene array and depicted to show signal intensity in sorted myeloid cell populations. Data are derived from 3 separate analyses that are each derived from n=5 mice. (B) Cell surface expression of CD36 analyzed by flow cytometry on monocyte subsets from mice fed a regular chow diet (−) or a diet supplemented with the PPARγ agonist rosiglitazone (agonist) for a week (n=5 mice per group). (C) The number of genes regulated in monocyte subsets following PPARγ activation by rosiglitazone assessed through whole-genome array analysis. (D) Protein levels of CD36 and FABP4 in myeloid populations at the steady state and following PPARγ agonist treatment (agonist, pioglitazone) were monitored by flow cytometry. (E) Expression of CD36 and FABP4 in myeloid populations of LysM-Cre × PPARγflox/flox mice (PPARγΔMac/PMN) and controls (CTRL) at the steady state and following PPARγ agonist treatment (agonist, pioglitazone). Mean fluorescence intensity (MFI) is plotted (n=3-4 mice per group).
Acquisition of PPARγ expression by Ly-6Chi monocyte-derived inflammatory macrophages is necessary for full resolution of acute inflammation
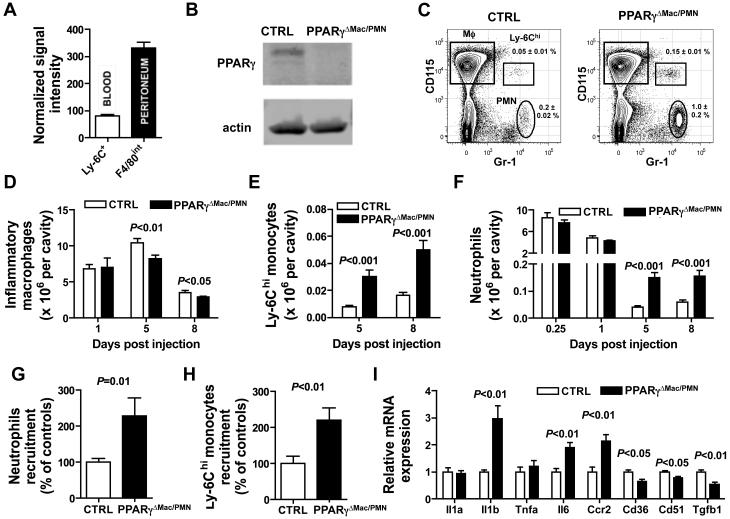
PPARγ activity has been associated with anti-inflammatory responses. In the inflammatory milieu of the thioglycollate-treated peritoneum, elicited macrophages from the peritoneal cavity expressed 3-fold higher PPARγ mRNA than blood Ly-6Chi monocytes (Fig. 2A) from which they derive (14, 28). PPARγ was functional in these cells as PPARγactivation using a synthetic ligand increased cell surface expression of the PPARγ-inducible protein CD36 (data not shown) and PPARγ was efficiently deleted in these cells in LysM-Cre × PPARγflox/flox mice (Fig. 2B). In wild-type mice, leukocytes accumulate for several days after thioglycollate injection, with a marked resolution phase between days 5 and 8 when inflammatory macrophage numbers decline to baseline levels (29). To examine whether PPARγ deficiency in Ly-6Chi monocyte-derived inflammatory peritoneal macrophages would alter the initiation and/or the resolution of thioglycollate-induced inflammation, we used LysM-Cre × PPARγflox/flox mice that here lack PPARγ expression specifically in macrophages, as neutrophils do not express PPARγ. Firstly, using LysM-Cre × Rosa26-stopfloxEGFP reporter mice to identify cells with use of the LysM promoter using GFP expression, we confirmed that more than 90% of inflammatory macrophages in the inflamed peritoneum would be targeted in LysM-Cre × PPARγflox/flox mice, in addition to macrophages in resting peritoneum and neutrophils (data not shown). In the steady state, the total numbers of the two peritoneal resident macrophage populations (17) (CD115+ F480hi MHC-II− or CD115+ F480lo MHC-II+) were unchanged in LysM-Cre × PPARγflox/flox mice as compared with controls, and the numbers of infiltrated neutrophils and Ly-6Chi monocytes were similarly very low in the presence or absence of PPARγ (data not shown).
Figure 2. PPARγ expression in peritoneal inflammatory macrophages favors the resolution of acute inflammation.
(A) Relative PPARγ mRNA expression in inflammatory macrophages from the peritoneum and their peripheral blood Ly-6Chi monocyte precursors. (B) Western blot analysis of PPARγ protein in cell-sorted thioglycollate-elicited peritoneal macrophages (C) FACS plot illustrating the gating strategy used for inflammatory peritoneal macrophages (Mφ; CD115+ Gr-1/Ly-6C−), Ly-6Chi monocytes (CD115+ Gr-1/Ly-6C+) and neutrophils (PMN; Gr-1/Ly-6G+ CD115lo/−) in the peritoneal cavity 5 days after initiation of peritonitis. (D) Inflammatory peritoneal macrophage number in LysM-C r e × P P A Rγflox/flox mice (PPARγΔMac/PMN) and controls (CTRL) during the course of thioglycollate-induced peritonitis (n=5-9 mice per group). (E) Ly-6Chi monocyte numbers in the peritoneal cavity at 5 and 8 days post induction of peritonitis (n=8-14 per group). (F) Neutrophil counts in the peritoneum at 0.25, 1, 5 and 8 days after peritonitis induction (n=4-12 mice per group). (G) Inflammatory peritoneal macrophages from LysM-Cre × PPARγflox/flox mice (PPARγΔMac/PMN) and controls (CTRL) (both CD45.2) were transferred into naïve CD45.1 recipients and recipient neutrophils recruitment was evaluated 24 hours later (n=6-8 mice per group). (H) Circulating Ly-6Chi monocytes were labeled i.v with latex fluorescent beads 3 days after induction of inflammation and the number of recruited beads-positive Ly-6Chi monocyte in the peritoneal was assessed 48 hours later in LysM-Cre × PPARγflox/flox mice (PPARγΔMac/PMN) and controls (CTRL) (n=6-7 mice per group). (I) Quantification of mRNA expression by peritoneal inflammatory macrophages recovered 5 days after induction of inflammation assessed by quantitative real-time PCR for select genes (n=5 mice per group).
During the course of peritonitis, early accumulation of CD115hi inflammatory macrophages (Fig 2C) in the peritoneum was unaltered by PPARγ deficiency 1 day after instillation of thioglycollate, but was slightly decreased after 5 and 8 days in LysM-Cre × PPARγflox/flox mice (Fig. 2D). However, we noted a 3-5-fold increase in the number of infiltrated Ly-6Chi monocytes at both day 5 and 8 compared to control mice (Fig. 2C and 2E), while circulating monocyte subset numbers remained similar over time in both LysM-Cre × PPARγflox/flox mice and controls (data not shown). As Ly-6C is retained only transiently after monocyte recruitment into tissues (26, 30, 31), these data revealed that monocyte recruitment to the peritoneal cavity did not fully shut down in LysM-Cre × PPARγflox/flox mice. Furthermore, while early accumulation of neutrophils (6 hours and 24 hours) was comparable between LysM-Cre × PPARγflox/flox mice and control animals, peritoneal neutrophil numbers were likewise elevated 3-4-fold in LysM-Cre × PPARγflox/flox mice during the usual resolution phase occurring between days 5 and 8 (Fig. 2F). Concomitantly, blood neutrophil counts were elevated at these later time points in LysM-Cre × PPARγflox/flox mice compared to controls (data not shown), marking systemic inflammation. Interestingly, the increase in peritoneal neutrophils and Ly-6Chi monocytes, as well as the decrease in inflammatory macrophages, were still evident at day 14 post-thioglycollate treatment, the latest time point examined (data not shown). Transfer of 5 × 106 thioglycollate-elicited macrophages, retrieved from donors during the early resolution period at day 5, to the peritoneum of resting mice led to recruitment of neutrophils (Fig. 2G) and monocytes (Fig. 2H), and these numbers were doubled when transferred macrophages lacked PPARγ. PPARγ-deficient thioglycollate-elicited macrophages, retrieved at day 5, had increased mRNA expression of Il1b, Il6 and Ccr2, and decreased levels of Cd36, Cd51 and Tgfb1 compared to controls (Fig. 2I). Collectively, these data show that PPARγ deficiency in myeloid cells has little impact on the early phases of an inflammatory response. However, macrophage PPARγ expression in thioglycollate-elicited inflammatory macrophages is necessary to bring about resolution. Indeed, its deficiency leads to a state of chronic low-grade inflammation, at least in part because macrophages retain a more pro-inflammatory phenotype.
PPARγ activation promotes macrophage-dependent cessation of neutrophil recruitment and favors resolution of acute inflammation
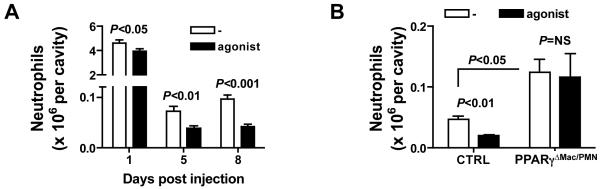
Given the data above indicating that the cessation of leukocyte recruitment that characterizes resolution of inflammation is impaired in LysM-Cre × PPARγflox/flox mice, we sought to determine whether treatment with PPARγ agonists would conversely favor the shut down of leukocyte recruitment in wild-type animals. Indeed, PPARγ agonist treatment reduced neutrophil counts in the peritoneum following thioglycollate administration at each time point studied, especially in the later phases of inflammation (Fig. 3A). This effect required PPARγ expression in macrophages because treatment with the PPARγ agonist failed to reduce neutrophil counts in the cavity of LysM-Cre × PPARγflox/flox mice (Fig. 3B). These data support the concept that PPARγ activation suppresses the recruitment of leukocytes in later phases of tissue injury in a macrophage PPARγ-dependent manner, promoting resolution of inflammation.
Figure 3. PPARγ activation favors the resolution of acute inflammation.
(A) Neutrophil counts in the peritoneum at 1, 5 and 8 days after peritonitis induction in wild-type mice fed a regular diet (−) or a diet containing the PPARγ agonist pioglitazone (agonist) (n=8-10 mice per group). (B) Peritoneal neutrophil counts 5 days after peritonitis induction in LysM-Cre × PPARγflox/flox mice (PPARγΔMac/PMN) fed a regular diet (−) or a diet containing the PPARγ agonist pioglitazone (agonist) (n=4-5 mice per group).
PPARγ deletion in macrophages leads to low-grade constitutive inflammation in the lung but not in the spleen
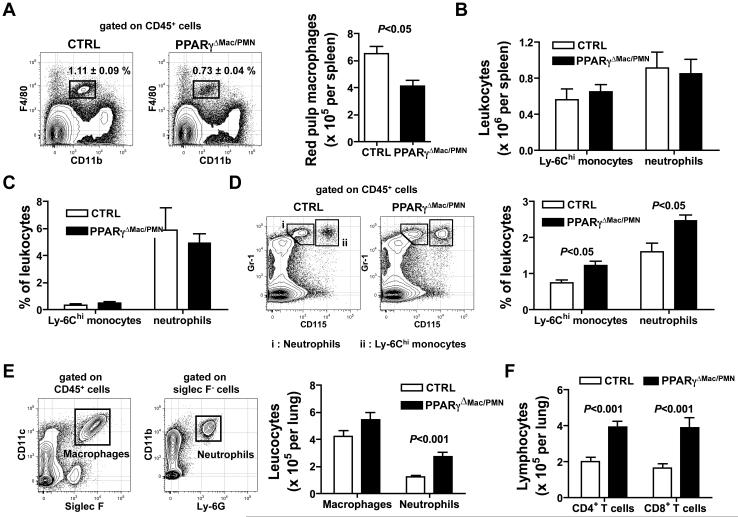
Considering our findings that PPARγ expression in inflammatory macrophages as well as its activation by pharmacological agonists favors resolution of both acute and chronic inflammation, we wondered whether deletion of PPARγ in resting macrophage populations that normally express high levels of PPARγ (lung and splenic red pulp macrophages) would promote inflammation. LysM-Cre × Rosa26-stopfloxEGFP reporter mice confirmed that resident lung and splenic red pulp macrophages would be targeted in LysM-Cre × PPARγflox/flox mice (data not shown). Total splenocyte numbers were similar in LysM-Cre × PPARγflox/flox mice and controls (data not shown), but red pulp macrophages (F4/80hi CD11blo) were approximately 1/3 less numerous in LysM-Cre × PPARγflox/flox spleens (Fig. 4A), possibly arguing for a role of PPARγ in the maintenance of this population. There were no signs of inflammation in the resting spleen of LysM-Cre × PPARγflox/flox mice as splenic Ly-6Chi monocyte and neutrophil numbers were comparable to controls (Fig. 4B). Consistent with peritoneal inflammation, macrophage PPARγ deficiency did not have an impact on the induction of inflammation in the spleen at an early time point after i.v. administration of LPS (day 1, Fig. 4C), while it led to increased neutrophils and Ly-6Chi monocytes recruitment to the spleen at a later time point (day 5, Fig. 4D), again arguing for a key role of PPARγ in resolution of inflammation.
Figure 4. Impact of PPARγ deletion in splenic red pulp and lung macrophage.
(A) Red pulp macrophage percentages and counts in the spleen of LysM-Cre × PPARγflox/flox mice (PPARγΔMac/PMN) and controls (CTRL) in the steady state (n=4 mice per group). (B) Neutrophil and Ly-6Chi monocyte counts in the spleen of LysM-Cre × PPARγflox/flox mice (PPARγΔMac/PMN) and controls (CTRL) in the steady state (n=4 mice per group). (C) Neutrophil and Ly-6Chi monocyte counts in the spleen of LysM-Cre × PPARγflox/flox mice (PPARγΔMac/PMN) and controls (CTRL) 24 hours after LPS was injected i.v. (n=3 mice per group) (D) FACS plot illustrating the gating strategy used for Ly-6Chi monocytes (CD115hi Gr-1/Ly-6C+) and neutrophils (Gr-1/Ly-6G+ CD115lo), and neutrophil and Ly-6Chi monocyte counts in the spleen of LysM-Cre × PPARγflox/flox mice (PPARγΔMac/PMN) and controls (CTRL) 5 days after i.v. administration of LPS (n=3 mice per group). (E) FACS plot illustrating the gating strategy used for lung macrophages (CD11c+ Siglec-F+) and neutrophils (CD11b+ Ly-6G+), and respective cell counts in the lung of LysM-Cre × PPARγflox/flox mice (PPARγΔMac/PMN) and controls (CTRL) in the steady state (n=6-8 mice per group). (F) CD4+ and CD8+ T lymphocyte counts in the lung LysM-Cre × PPARγflox/flox mice (PPARγΔMac/PMN) and controls (CTRL) in the steady state (n=6-8 mice per group).
When we examined the lung, we observed that PPARγ deletion in macrophages led to a low-grade inflammatory response without supplying an overt exogenous stimulus. Indeed, we observed increased leukocyte infiltration with elevated numbers of neutrophils (Fig. 4E), CD4+ and CD8+ T lymphocytes (Fig. 4F), while macrophage numbers were comparable to controls (Fig. 4E).
Overall, whereas PPARγ is expressed by both splenic red pulp and pulmonary macrophages, its deficiency only obviously had an impact on lung tissue homeostasis in the steady state, arguing for an interaction between tissue environment and the outcome of altered macrophage PPARγ signaling.
Altered gene expression and lipid homeostasis in lung macrophages deficient in PPARγ
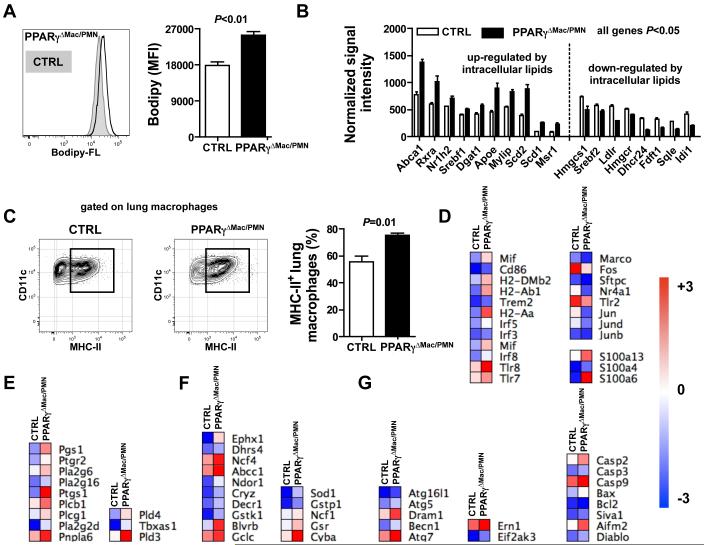
The low-grade inflammation observed only in the lung but not in the spleen suggested that the impact of PPARγ might be environment-dependent. The alveolar space is permanently filled with a surfactant made of lipids (90%) and proteins (10%) (32) and we noted increased cellular lipid content in lung macrophages lacking PPARγ as indicated by increased sterol staining using Bodipy FL (Fig. 5A), in line with previous work reporting the development of pulmonary alveolar proteinosis in these mice (33, 34). Then, in order to better understand the role of PPARγ in lung macrophages, micro-array analysis was performed on sorted lung macrophages from LysM-Cre × PPARγflox/flox mice and controls. This whole genome array analysis uncovered 721 genes that were down-regulated and 2088 genes whose expression was increased in lung macrophages lacking PPARγ, highlighting a profound alteration of their transcriptome (supplemental Table 3 and 4). In line with their increased intracellular sterol content, we found that PPARγ-deficient lung macrophages induced a number of mRNA transcripts associated with cellular lipid metabolism and in particular those associated with an increase in activity of the LXR transcription factor. Expression levels of Nr1h2 (also known as LXRβ), a sensor of intracellular sterol levels, and its partner Rxra were increased in lung macrophages obtained from LysM-Cre × PPARγflox/flox mice as compared to controls (Fig. 5B). Consequently, the expression levels of several target genes of the LXR/RXR heterodimer (Abca1, Srebf1, Apoe, Mylip, Abcg1, Scd2 and Scd1) were equally increased (Fig. 5B). Finally, the mRNA level of the scavenger receptor Msr1 and of the triacylglycerol synthesis enzyme Dgat1 were also enhanced (Fig. 5B). This expression profile was mirrored by decreased expression of genes involved in the cholesterol biosynthetic pathway (Hmgcs1, Srebf2, Hmgcr, Fdft1, Dhcr24, Sqle and Idi1) and in the uptake of extracellular cholesterol (Ldlr) (Fig. 5B). As the vast majority of these genes are not known to be under the direct control of PPARγ, it suggests that many of the genes regulated here are regulated indirectly. Since we observed an increase in the percentage of MHC-II+ lung macrophages in LysM-Cre × PPARγflox/flox mice (Fig. 5C), we sought to determine whether this was correlated with an increased expression of genes associated with macrophage activation. We found increased mRNA levels of genes encoding costimulatory molecules (Cd86, H2-DMb2, H2-Ab1 and H2-Aa), members of the IRF family of transcription factors (Irf3, Irf5 and Irf8), innate immune receptors (Tlr7, Tlr8 and Trem2) and the pro-inflammatory mediator Mif (Fig. 5D). Moreover, mRNA expression levels of members of the S100 protein family (S100a13, S100a4 and S100a6), known to mediate inflammatory signals, were up-regulated in lung macrophages from LysM-Cre × PPARγflox/flox mice (Fig. 5D). However, other genes involved in inflammation such as transcription factors (Fos, Nr4a1, Jun, Jund, Junb), the TLR receptor Tlr2, the scavenger receptor Marco and the surfactant opsonin Sftpc were down-regulated (Fig. 5D). Consistent with the increased intracellular lipid content observed in lung macrophages from LysM-Cre × PPARγflox/flox mice, we noted that the mRNA expression of several phospholipases (Pla2g6, Plcb1, Pnpla6, Pld3, Pld4) was increased in these cells as well as the expression of genes involved in prostanglandin and thromboxane synthesis (Pgs1, Ptgr2, Ptgs1 and Tbxas1) (Fig. 5E). We also noted that numerous genes regulated by the transcription factor Nrf2, a master regulator of the antioxidant response, were up-regulated in PPARγ-deficient pulmonary macrophages compared to controls, indicating increased oxidative stress in LysM-Cre × PPARγflox/flox mice (Fig. 5F). Finally, mRNA levels of mediators of autophagy (Atg5, Dram1, Becn1, Atg7) and apoptosis (Casp2, Casp9, Bax, Aifm2) were increased in lung macrophages lacking PPARγ compared to controls (Fig. 5G). Taken together, these findings reveal that PPARγ-deficient pulmonary macrophages present a markedly altered transcriptome, most likely secondary to the lipid loading, affecting several key pathways related to classical macrophage functions.
Figure 5. PPARγ is critical to preserve lung macrophage cellular homeostasis.
(A) Cellular lipid levels were assessed in resting lung macrophages from LysM-Cre × PPARγflox/flox mice (PPARγΔMac/PMN) and controls (CTRL) using Bofipy-FL staining (n=3 mice per group). (B) mRNA expression of genes modulated by intracellular lipid levels was determined by microarray. (C) Flow cytometry plot and quantification of cell surface MHC-II protein levels in lung macrophages from LysM-Cre × PPARγflox/flox mice (PPARγΔMac/PMN) and controls (CTRL) (n=3-4 mice per group). Heat maps representing mRNA levels of genes involved in macrophage activation (D), lipid signaling (E), oxidative stress signaling (F) and cell death/autophagy (G).
Impaired bacterial clearance in the lungs and accelerated mortality in mice lacking PPARγ in macrophages following S. pneumoniae infection
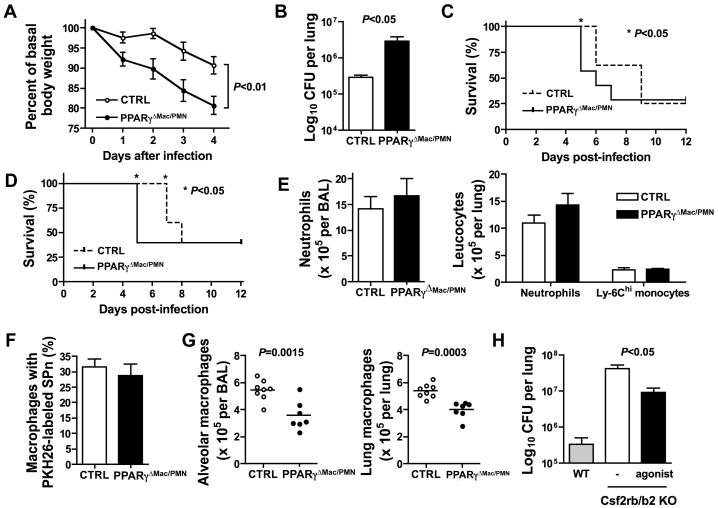
Given that the gene expression profile of lung macrophages deficient in PPARγ is profoundly altered, we next investigated whether infectious challenge of LysM-Cre × PPARγflox/flox mice would lead to a perturbed innate immune response to pathogens. Here, we found that LysM-Cre × PPARγflox/flox mice were more susceptible to infection with Streptococcus pneumoniae. Weight loss associated with infection was more pronounced in LysM-Cre × PPARγflox/flox compared to controls over a period of 4 days before death occurred (Fig. 6A). This increased susceptibility to S. pneumoniae infection was due to impaired bacterial clearance as bacterial burden was increased by approximately one log in the lung of LysM-Cre × PPARγflox/flox mice compared to controls 48 hours after infection (Fig. 6B). This correlated with faster dissemination of the bacteria into the bloodstream (data not shown) as well as accelerated death in these mice (Fig. 6C). LysM-Cre × PPARγflox/flox mice challenged with a lower dose of the pathogen similarly succumbed faster than controls. Indeed, while 100% of control mice were still alive 6 days after infection, only 40% of LysM-Cre × PPARγflox/flox mice survived to this time point (Fig. 6D). Surprisingly, we observed similar neutrophils and Ly-6Chi monocytes recruitment to the bronchoalveolar space and the lung 24 hours after infection in LysM-Cre × PPARγflox/flox mice and controls (Fig. 6E). Increased bacterial burden in LysM-Cre × PPARγflox/flox mice was not due to impaired phagocytosis as labeled-S. pneumoniae were taken up by PPARγ-deficient alveolar macrophages as efficiently as controls in vivo (Fig. 6F). However, resident alveolar and interstitial pulmonary macrophage counts were significantly decreased by approximately 50% and 35% respectively 24 hours after instillation of Streptococcus pneumoniae (Fig. 6G). Finally, the disease pulmonary alveolar proteinosis (PAP) is due to alterations in GM-CSF signaling, and it was recently shown that PPARγ expression in GM-CSF-deficient lung macrophages was low (35). Furthermore, viral vectors to restore PPARγ in GM-CSF KO mice led to reduced lipid accumulation and increased cholesterol efflux in lung macrophages (36). Since PAP is associated with increased susceptibility to infection, we sought to determine if PPARγactivation could improve bacterial clearance in Csf2rb−/− Csf2rb2−/− mice (37). Indeed, Csf2rb−/− Csf2rb2−/− mice, which also display alveolar proteinosis, have significantly higher bacterial burden (approximately 2 logs) than WT control mice and PPARγ activation by pioglitazone partially decreased this enhanced burden (Fig 6H). Therefore, these data now connect PPARγ to host defense and control of bacterial burden in the lung through maintenance of local macrophage functions.
Figure 6. PPARγ expression in lung macrophage is necessary to combat infection.
(A) Body weight loss was determined following infection in LysM-C r e × P P A Rγflox/flox mice (PPARγΔMac/PMN) and controls (CTRL) (n=9 mice per group). (B) Lung bacterial load was measured 48 hours after infection in LysM-Cre × PPARγflox/flox mice (PPARγΔMac/PMN) and controls (CTRL) (n=9 mice per group). Survival to infection was assessed over a period of 12 days following high dose (2.106 CFU) (C) and low dose (5.105 CFU) (D) S. pneumoniae inoculation in the lung of LysM-Cre × PPARγflox/flox mice (PPARγΔMac/PMN) and controls (CTRL) (n=5-8 mice per group). (E) Neutrophil and Ly-6Chi monocyte counts in the BAL and the lung of LysM-Cre × PPARγflox/flox mice (PPARγΔMac/PMN) and controls (CTRL) were determined 24 hours after infection (n=7-8 mice per group). (F) PKH26-labeled S. pneumoniae phagocytosis by resident alveolar macrophages was assessed by flow cytometry 30 minutes after inoculation (n=6 mice per group). (G) Alveolar and pulmonary resident macrophages counts in LysM-Cre × PPARγflox/flox mice (PPARγΔMac/PMN) and controls (CTRL) 24 hours after infection (n=7-8 mice per group). (H) Lung bacterial burden was determined 48 hours after S. pneumoniae inoculation in the lungs of wild-type mice, Csf2rb−/− Csf2rb2−/− mice and Csf2rb−/− Csf2rb2−/− mice with prior treatment with the PPARγagonist pioglitazone for 2 weeks (n=3-4 mice per group).
Discussion
The anti-inflammatory role of PPARγ in macrophages is well established. However, little is known regarding its impact on specific resting macrophage populations as well as on the dynamic of inflammation in vivo. It was recently recognized that establishing the expression profile of PPARγ in tissue macrophages in vivo would be helpful in clarifying its role in the regulation of inflammatory processes (38). Here, we unexpectedly revealed that many resident macrophages do not express substantial levels of PPARγ, including those in the brain, peritoneum and gut. The level of PPARγ in these cells was as low as in Ly-6Chi monocytes, which show no PPARγ activity after synthetic PPARγagonist administration in vivo. By contrast to these tissues and cells, Ly-6Clo blood monocytes, resting red pulp splenic and pulmonary macrophages expressed high levels of mRNA for PPARγ. In addition, PPARγ was induced in inflammatory macrophages differentiating from circulating Ly-6Chi monocytes entering an inflammatory site, albeit to a lower level than observed in the resting macrophages that were positive. In different tissues, the expression of canonical PPARγ target genes like CD36 and FABP4 was distinct even among those macrophages that were PPARγ+, highlighting the importance of context in regulation of PPARγ-related pathways and underscoring the diversity observed among macrophages from different organs.
As the ability of PPARγ to transrepress inflammatory genes has been thoroughly documented (1), we expected that macrophage loss of PPARγ during thioglycollate-mediated peritonitis would lead to a more proinflammatory phenotype. However, the absence of PPARγ in LysM-Cre × PPARγflox/flox mice did not impact the accumulation of leukocytes during the initial phase of the inflammatory response. This could be explained by the fact that immature and differentiating Ly-6Chi monocytes, which express negligible or low levels of PPARγ, were dominant at this time point. By contrast, persistent neutrophil and Ly-6Chi monocyte influx occurs in LysM-Cre × PPARγflox/flox mice during the later period when more differentiated inflammatory macrophages, that now express PPARγ, begin to dominate and when resolution is observed in control mice. These data suggest, therefore, that PPARγ plays especially important roles in the late stages and resolution of inflammation. These roles very likely include repression of proinflammatory genes, and indeed we observed that proinflammatory genes were elevated in PPARγ-deficient thioglycollate-elicited macrophages, but may also include impaired induction of genes associated with repair and healing. Previous studies have linked PPARγ with the development of alternatively activated macrophages (39) and with tissue repair in injured muscle (30), and IL-4 is known to promote the production of PPARγ ligands (40). An elegant in-depth study recently revealed that while PPARγ is not required for development of alternatively activated macrophages in C57BL/6J mice, there is synergy with IL-4 such that the transcription factor Stat6 that is critical for IL-4 signaling binds to the enhancer elements in PPARγ target genes and markedly augments the PPARγresponse (41). Our findings that PPARγ appears to play a bigger role in determining the rate/magnitude of contraction of the inflammatory response rather than the magnitude of earlier phases fits well with concepts of PPARγ playing a key role in tissue repair, healing, and overall resolution.
Future studies on the possible interface between PPARγ and lipids previously associated with resolution (42) seem in order. At present, resolvins are known not to serve as PPARγ ligands (42), but an intersection between PPARγ and the pathways that regulate such pro-resolution mediators may exist. Ligands for PPARγ during resolution may be limiting, because we observed that provision of synthetic ligands to mice hastened the shut down of neutrophil recruitment in a macrophage PPARγ-dependent manner during the terminal phases of thioglycollate-induced inflammation. This finding is in line with recent published data in a model of granulomatous disease (43) and supports the logic of therapeutically enhancing PPARγ activity to promote resolution of ongoing inflammation.
Highest expression of PPARγ mRNA among macrophages in the mouse, resting or inflamed, was observed in the lung. Analysis of FABP4 expression in lung macrophages suggests that there may be heterogeneity among lung macrophages with regard to expression or activity of PPARγ. We show that the absence of PPARγ in LysM-Cre × PPARγflox/flox mice induced mild lung inflammation in the absence of experimental challenge. This underlying inflammation may stem from a key role for PPARγ expression by macrophages to maintain cellular as well as tissue lipid homeostasis in the presence of pulmonary surfactant lipids. Indeed, previous work indicates that lipid surfactant accumulates in the alveoli of LysM-Cre × PPARγflox/flox mice (33, 34). Consistent with this observation, we found that expression of genes that regulate intracellular lipid homeostasis are markedly altered in pulmonary macrophages lacking PPARγ. Genes involved in sterol uptake and synthesis were downregulated while genes linked to cholesterol sensing and efflux were up-regulated, and in particular, mRNA transcripts controlled by LXR were induced. Likely, the enhanced sterol loading drives induction of the LXR pathway as a mechanism to deal with the high lipid loading. Additionally, we found that numerous pathways associated with a range of macrophage functions were altered in the absence of PPARγ in lung macrophages, and genes associated with cell death were upregulated, leading to the conclusion that disruption of PPARγ signaling profoundly altered their transcriptome. However, the changes in gene expression are complex and likely do not reflect changes only associated with direct PPARγ targets, as many of the effects observed as likely indirect changes that reflect a sequence of changes that occur in response to the loss of PPARγ in macrophages that usually express it in the lung.
With the expectation that the absence of PPARγ in lung macrophages would exacerbate inflammation in the context of infection and subsequently favor bacterial clearance, we infected control and LysM-Cre × PPARγflox/flox mice with S. pneumoniae. Bolstering our expectations that the inflammatory infiltrate may be increased in response to this infection were data in the literature indicating that mice lacking the cholesterol efflux gene Abcg1, and thus a gene expected to intersect functionally with PPARγ, manifest enhanced inflammation and increased bacterial clearance in response to infection in the lung (44). Following infection, weight loss and mortality were surprisingly accelerated in LysM-Cre × PPARγflox/flox mice. As in the acute model of sterile inflammation induced by thioglycollate, the number of infiltrating neutrophils and monocytes was not changed in the first days following infection. Further similar to the thioglycollate model, but far more pronounced, the number of mature macrophages was significantly reduced in LysM-Cre × PPARγflox/flox mice following infection with S. pneumoniae, although macrophage counts were similar to control mice in the steady state. These reduced macrophage numbers may account for the associated observation that clearance of S. pneumoniae was impaired under these conditions. Although we were unable to find an increased number of non-viable macrophages (using annexin V as a readout), the upregulation of cell death genes even in the steady state is consistent with this idea, and other explanations such as impaired phagocytosis of bacteria were eliminated. While future work will be required to be sure that macrophage death accounts for why LysM-Cre × PPARγflox/flox mice succumb to S. pneumoniae infection more than control mice, we believe that the observation that macrophage PPARγ deficiency impacts the outcome of infection is quite significant on its own. Patients with pulmonary alveolar proteinosis (PAP), a disease linked to impaired GM-CSF signaling, have an increased risk of super infection (32) and it is known that suppressed GM-CSF signaling leads to lower PPARγ levels in the lung (35). Moreover, Csf2rb−/− Csf2rb2−/− mice, a mouse model of PAP, are more susceptible to S. pneumoniae infection (37). While it was already recognized that increasing PPARγ in models of PAP might reverse aspects of the disease such as lipid accumulation in macrophages, we now linked the loss of PPARγ per se and increased susceptibility to infection in PAP. Importantly, down-regulation of PPARγ and/or impairment in PPARγ signaling is also observed in cystic fibrosis (45-47), and PPARγ agonist treatment has been recently shown to ameliorate the severity of the cystic fibrosis phenotype in mice (47). Since cystic fibrosis is also tightly associated with an increased susceptibility to lung infection (48), PPARγ may participate centrally in impacting susceptibility to infection there as well. Future studies to investigate this possibility will be very important.
In summary, through taking the approach that started with characterization of the diversity of macrophages with respect to expression of PPARγ, the present work illustrates that PPARγ acts at the cellular level to favor contraction of inflammation and in the steady state is expressed in specific macrophage populations, especially in lung macrophages where it is critically involved in the maintenance of host defense.
Supplementary Material
Acknowledgments
We thank Christophe Benoist, Jeffrey Ericson and Scott Davies for technical assistance with micro-array analysis through the Immunological Genome Project (R24 AI072073). We also thank the Flow Cytometry Core at the Mount Sinai School of Medicine for assistance of use of the facility for flow cytometric cell sorting. We are grateful to Andy Platt and Julie Helft for helpful discussion and reading of the manuscript.
Footnotes
Disclosures
None of the authors has a conflict of interest related to this work.
Support for this work included NIH grants AI061741 and AI049653 and American Heart Association (AHA) Established Investigator Award 0740052 to GJR, NIH HL086899 to M.M., DFG project grant SP71314-1 to R.S. and DFG projects Ha 1083/16-1 and 15-1 to AJH. ELG is supported by a fellowship from AHA (10POST4160140) and AC is funded by a fellowship from the NIH NHLBI 5F30HL099028-02.
References
- 1.Glass CK, Ogawa S. Combinatorial roles of nuclear receptors in inflammation and immunity. Nat Rev Immunol. 2006;6:44–55. doi: 10.1038/nri1748. [DOI] [PubMed] [Google Scholar]
- 2.Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARgamma. Annu Rev Biochem. 2008;77:289–312. doi: 10.1146/annurev.biochem.77.061307.091829. [DOI] [PubMed] [Google Scholar]
- 3.Yki-Jarvinen H. Thiazolidinediones. N Engl J Med. 2004;351:1106–1118. doi: 10.1056/NEJMra041001. [DOI] [PubMed] [Google Scholar]
- 4.Ricote M, Li AC, Willson TM, Kelly CJ, Glass CK. The peroxisome proliferator-activated receptor-gamma is a negative regulator of macrophage activation. Nature. 1998;391:79–82. doi: 10.1038/34178. [DOI] [PubMed] [Google Scholar]
- 5.Jiang C, Ting AT, Seed B. PPAR-gamma agonists inhibit production of monocyte inflammatory cytokines. Nature. 1998;391:82–86. doi: 10.1038/34184. [DOI] [PubMed] [Google Scholar]
- 6.Daynes RA, Jones DC. Emerging roles of PPARs in inflammation and immunity. Nat Rev Immunol. 2002;2:748–759. doi: 10.1038/nri912. [DOI] [PubMed] [Google Scholar]
- 7.Su CG, Wen X, Bailey ST, Jiang W, Rangwala SM, Keilbaugh SA, Flanigan A, Murthy S, Lazar MA, Wu GD. A novel therapy for colitis utilizing PPAR-gamma ligands to inhibit the epithelial inflammatory response. J Clin Invest. 1999;104:383–389. doi: 10.1172/JCI7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mukherjee R, Davies PJ, Crombie DL, Bischoff ED, Cesario RM, Jow L, Hamann LG, Boehm MF, Mondon CE, Nadzan AM, et al. Sensitization of diabetic and obese mice to insulin by retinoid X receptor agonists. Nature. 1997;386:407–410. doi: 10.1038/386407a0. [DOI] [PubMed] [Google Scholar]
- 9.Li AC, Brown KK, Silvestre MJ, Willson TM, Palinski W, Glass CK. Peroxisome proliferator-activated receptor gamma ligands inhibit development of atherosclerosis in LDL receptor-deficient mice. J Clin Invest. 2000;106:523–531. doi: 10.1172/JCI10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Babaev VR, Yancey PG, Ryzhov SV, Kon V, Breyer MD, Magnuson MA, Fazio S, Linton MF. Conditional knockout of macrophage PPARgamma increases atherosclerosis in C57BL/6 and low-density lipoprotein receptor-deficient mice. Arterioscler Thromb Vasc Biol. 2005;25:1647–1653. doi: 10.1161/01.ATV.0000173413.31789.1a. [DOI] [PubMed] [Google Scholar]
- 11.Chawla A, Boisvert WA, Lee CH, Laffitte BA, Barak Y, Joseph SB, Liao D, Nagy L, Edwards PA, Curtiss LK, et al. A PPAR gamma-LXR-ABCA1 pathway in macrophages is involved in cholesterol efflux and atherogenesis. Mol Cell. 2001;7:161–171. doi: 10.1016/s1097-2765(01)00164-2. [DOI] [PubMed] [Google Scholar]
- 12.Shah YM, Morimura K, Gonzalez FJ. Expression of peroxisome proliferator-activated receptor-gamma in macrophage suppresses experimentally induced colitis. Am J Physiol Gastrointest Liver Physiol. 2007;292:G657–666. doi: 10.1152/ajpgi.00381.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hevener AL, Olefsky JM, Reichart D, Nguyen MT, Bandyopadyhay G, Leung HY, Watt MJ, Benner C, Febbraio MA, Nguyen AK, et al. Macrophage PPAR gamma is required for normal skeletal muscle and hepatic insulin sensitivity and full antidiabetic effects of thiazolidinediones. J Clin Invest. 2007;117:1658–1669. doi: 10.1172/JCI31561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geissmann F, Jung S, Littman DR. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity. 2003;19:71–82. doi: 10.1016/s1074-7613(03)00174-2. [DOI] [PubMed] [Google Scholar]
- 15.Ingersoll MA, Spanbroek R, Lottaz C, Gautier EL, Frankenberger M, Hoffmann R, Lang R, Haniffa M, Collin M, Tacke F, et al. Comparison of gene expression profiles between human and mouse monocyte subsets. Blood. 2010;115:e10–19. doi: 10.1182/blood-2009-07-235028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heng TS, Painter MW. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 17.Ghosn EE, Cassado AA, Govoni GR, Fukuhara T, Yang Y, Monack DM, Bortoluci KR, Almeida SR, Herzenberg LA. Two physically, functionally, and developmentally distinct peritoneal macrophage subsets. Proc Natl Acad Sci U S A. 2010;107:2568–2573. doi: 10.1073/pnas.0915000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sung SS, Fu SM, Rose CE, Jr., Gaskin F, Ju ST, Beaty SR. A major lung CD103 (alphaE)-beta7 integrin-positive epithelial dendritic cell population expressing Langerin and tight junction proteins. J Immunol. 2006;176:2161–2172. doi: 10.4049/jimmunol.176.4.2161. [DOI] [PubMed] [Google Scholar]
- 19.Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bogunovic M, Ginhoux F, Helft J, Shang L, Hashimoto D, Greter M, Liu K, Jakubzick C, Ingersoll MA, Leboeuf M, et al. Origin of the lamina propria dendritic cell network. Immunity. 2009;31:513–525. doi: 10.1016/j.immuni.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waskow C, Liu K, Darrasse-Jeze G, Guermonprez P, Ginhoux F, Merad M, Shengelia T, Yao K, Nussenzweig M. The receptor tyrosine kinase Flt3 is required for dendritic cell development in peripheral lymphoid tissues. Nat Immunol. 2008;9:676–683. doi: 10.1038/ni.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tacke F, Ginhoux F, Jakubzick C, van Rooijen N, Merad M, Randolph GJ. Immature monocytes acquire antigens from other cells in the bone marrow and present them to T cells after maturing in the periphery. J Exp Med. 2006;203:583–597. doi: 10.1084/jem.20052119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Potteaux S, Gautier EL, Hutchison SB, van Rooijen N, Rader DJ, Thomas MJ, Sorci-Thomas MG, Randolph GJ. Suppressed monocyte recruitment drives macrophage removal from atherosclerotic plaques of Apoe-/-mice during disease regression. J Clin Invest. 2011;121:2025–2036. doi: 10.1172/JCI43802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Rooijen N, Sanders A. Liposome mediated depletion of macrophages: mechanism of action, preparation of liposomes and applications. J Immunol Methods. 1994;174:83–93. doi: 10.1016/0022-1759(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 25.Gautier EL, Huby T, Ouzilleau B, Doucet C, Saint-Charles F, Gremy G, Chapman MJ, Lesnik P. Enhanced immune system activation and arterial inflammation accelerates atherosclerosis in lupus-prone mice. Arterioscler Thromb Vasc Biol. 2007;27:1625–1631. doi: 10.1161/ATVBAHA.107.142430. [DOI] [PubMed] [Google Scholar]
- 26.Qu C, Edwards EW, Tacke F, Angeli V, Llodra J, Sanchez-Schmitz G, Garin A, Haque NS, Peters W, van Rooijen N, et al. Role of CCR8 and other chemokine pathways in the migration of monocyte-derived dendritic cells to lymph nodes. J Exp Med. 2004;200:1231–1241. doi: 10.1084/jem.20032152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore KJ, Rosen ED, Fitzgerald ML, Randow F, Andersson LP, Altshuler D, Milstone DS, Mortensen RM, Spiegelman BM, Freeman MW. The role of PPAR-gamma in macrophage differentiation and cholesterol uptake. Nat Med. 2001;7:41–47. doi: 10.1038/83328. [DOI] [PubMed] [Google Scholar]
- 28.Tacke F, Alvarez D, Kaplan TJ, Jakubzick C, Spanbroek R, Llodra J, Garin A, Liu J, Mack M, van Rooijen N, et al. Monocyte subsets differentially employ CCR2, CCR5, and CX3CR1 to accumulate within atherosclerotic plaques. J Clin Invest. 2007;117:185–194. doi: 10.1172/JCI28549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bellingan GJ, Caldwell H, Howie SE, Dransfield I, Haslett C. In vivo fate of the inflammatory macrophage during the resolution of inflammation: inflammatory macrophages do not die locally, but emigrate to the draining lymph nodes. J Immunol. 1996;157:2577–2585. [PubMed] [Google Scholar]
- 30.Arnold L, Henry A, Poron F, Baba-Amer Y, van Rooijen N, Plonquet A, Gherardi RK, Chazaud B. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J Exp Med. 2007;204:1057–1069. doi: 10.1084/jem.20070075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheong C, Matos I, Choi JH, Dandamudi DB, Shrestha E, Longhi MP, Jeffrey KL, Anthony RM, Kluger C, Nchinda G, et al. Microbial stimulation fully differentiates monocytes to DC-SIGN/CD209(+) dendritic cells for immune T cell areas. Cell. 2010;143:416–429. doi: 10.1016/j.cell.2010.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trapnell BC, Whitsett JA, Nakata K. Pulmonary alveolar proteinosis. N Engl J Med. 2003;349:2527–2539. doi: 10.1056/NEJMra023226. [DOI] [PubMed] [Google Scholar]
- 33.Bonfield TL, Thomassen MJ, Farver CF, Abraham S, Koloze MT, Zhang X, Mosser DM, Culver DA. Peroxisome proliferator-activated receptor-gamma regulates the expression of alveolar macrophage macrophage colony-stimulating factor. J Immunol. 2008;181:235–242. doi: 10.4049/jimmunol.181.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baker AD, Malur A, Barna BP, Ghosh S, Kavuru MS, Malur AG, Thomassen MJ. Targeted PPAR{gamma} deficiency in alveolar macrophages disrupts surfactant catabolism. J Lipid Res. 2010;51:1325–1331. doi: 10.1194/jlr.M001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomassen MJ, Barna BP, Malur AG, Bonfield TL, Farver CF, Malur A, Dalrymple H, Kavuru MS, Febbraio M. ABCG1 is deficient in alveolar macrophages of GM-CSF knockout mice and patients with pulmonary alveolar proteinosis. J Lipid Res. 2007;48:2762–2768. doi: 10.1194/jlr.P700022-JLR200. [DOI] [PubMed] [Google Scholar]
- 36.Malur A, Baker AD, McCoy AJ, Wells G, Barna BP, Kavuru MS, Malur AG, Thomassen MJ. Restoration of PPARgamma reverses lipid accumulation in alveolar macrophages of GM-CSF knockout mice. Am J Physiol Lung Cell Mol Physiol. 2011;300:L73–80. doi: 10.1152/ajplung.00128.2010. [DOI] [PubMed] [Google Scholar]
- 37.LeVine AM, Reed JA, Kurak KE, Cianciolo E, Whitsett JA. GM-CSF-deficient mice are susceptible to pulmonary group B streptococcal infection. J Clin Invest. 1999;103:563–569. doi: 10.1172/JCI5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szanto A, Nagy L. The many faces of PPARgamma: anti-inflammatory by any means? Immunobiology. 2008;213:789–803. doi: 10.1016/j.imbio.2008.07.015. [DOI] [PubMed] [Google Scholar]
- 39.Odegaard JI, Ricardo-Gonzalez RR, Goforth MH, Morel CR, Subramanian V, Mukundan L, Red Eagle A, Vats D, Brombacher F, Ferrante AW, et al. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature. 2007;447:1116–1120. doi: 10.1038/nature05894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang JT, Welch JS, Ricote M, Binder CJ, Willson TM, Kelly C, Witztum JL, Funk CD, Conrad D, Glass CK. Interleukin-4-dependent production of PPAR-gamma ligands in macrophages by 12/15-lipoxygenase. Nature. 1999;400:378–382. doi: 10.1038/22572. [DOI] [PubMed] [Google Scholar]
- 41.Szanto A, Balint BL, Nagy ZS, Barta E, Dezso B, Pap A, Szeles L, Poliska S, Oros M, Evans RM, et al. STAT6 transcription factor is a facilitator of the nuclear receptor PPARgamma-regulated gene expression in macrophages and dendritic cells. Immunity. 2010;33:699–712. doi: 10.1016/j.immuni.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Serhan CN. Novel lipid mediators and resolution mechanisms in acute inflammation: to resolve or not? Am J Pathol. 2010;177:1576–1591. doi: 10.2353/ajpath.2010.100322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fernandez-Boyanapalli R, Frasch SC, Riches DW, Vandivier RW, Henson PM, Bratton DL. PPARgamma activation normalizes resolution of acute sterile inflammation in murine chronic granulomatous disease. Blood. 2010;116:4512–4522. doi: 10.1182/blood-2010-02-272005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Draper DW, Madenspacher JH, Dixon D, King DH, Remaley AT, Fessler MB. ATP-binding cassette transporter G1 deficiency dysregulates host defense in the lung. Am J Respir Crit Care Med. 2010;182:404–412. doi: 10.1164/rccm.200910-1580OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ollero M, Junaidi O, Zaman MM, Tzameli I, Ferrando AA, Andersson C, Blanco PG, Bialecki E, Freedman SD. Decreased expression of peroxisome proliferator activated receptor gamma in cftr-/-mice. J Cell Physiol. 2004;200:235–244. doi: 10.1002/jcp.20020. [DOI] [PubMed] [Google Scholar]
- 46.Maiuri L, Luciani A, Giardino I, Raia V, Villella VR, D’Apolito M, Pettoello-Mantovani M, Guido S, Ciacci C, Cimmino M, et al. Tissue transglutaminase activation modulates inflammation in cystic fibrosis via PPARgamma down-regulation. J Immunol. 2008;180:7697–7705. doi: 10.4049/jimmunol.180.11.7697. [DOI] [PubMed] [Google Scholar]
- 47.Harmon GS, Dumlao DS, Ng DT, Barrett KE, Dennis EA, Dong H, Glass CK. Pharmacological correction of a defect in PPAR-gamma signaling ameliorates disease severity in Cftr-deficient mice. Nat Med. 2010;16:313–318. doi: 10.1038/nm.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grassme H, Becker KA, Zhang Y, Gulbins E. CFTR-dependent susceptibility of the cystic fibrosis-host to Pseudomonas aeruginosa. Int J Med Microbiol. 2010;300:578–583. doi: 10.1016/j.ijmm.2010.08.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.