SUMMARY
Enterohemorrhagic E. coli harbors a pathogenicity island encoding a type 3 secretion system used to translocate effector proteins into the cytosol of intestinal epithelial cells and subvert their function. The structural proteins of the translocon are encoded in a major espADB mRNA processed from a precursor. The translocon mRNA should be highly susceptible to RNase E cleavage because of its AU-rich leader region and monophosphorylated 5'-terminus, yet it manages to avoid rapid degradation. Here, we report that the espADB leader region contains a strong Shine-Dalgarno element (SD2) and a translatable mini-ORF of six codons. Disruption of SD2 so as to weaken ribosome binding significantly reduces the concentration and stability of esp mRNA, whereas codon substitutions that impair translation of the mini-ORF have no such effect. These findings suggest that occupancy of SD2 by ribosomes, but not mini-ORF translation, helps to protect espADB mRNA from degradation, likely by hindering RNase E access to the AU-rich leader region.
INTRODUCTION
Enterohemorrhagic Escherichia coli (EHEC) serotype O157:H7 is a food-borne human pathogen causing either bloody or non-bloody diarrhea and in a minor proportion of patients the more serious condition known as hemolytic uremic syndrome (HUS) (Griffin et al., 1988; Kaper et al., 2004; Schmidt, 2010). EHEC and related pathogens such as enteropathogenic E. coli (EPEC) carry in their genome a ca. 35 kb pathogenicity island called the locus of enterocyte effacement (LEE) (McDaniel et al., 1995). Through a type 3 secretion system (T3SS) encoded in the LEE, the bacterium injects effector proteins into the host cell. These proteins alter a myriad of cell functions, resulting in the tight attachment of EHEC cells to the gut epithelium and the loss or effacement of the microvilli (Tree et al., 2009). The external part of the T3SS (referred to here as the “translocon”) acts as a conduit for the delivery of the effector proteins and is made up primarily of the EspA, EspD and EspB proteins. EspA forms large filaments capped by a complex composed of EspD and EspB that connects the T3SS to the host cell (Knutton et al., 1998; Sekiya et al., 2001; Shaw et al., 2001; Luo and Donnenberg, 2011). While both EspB and EspD are proposed to form a pore in the eukaryotic cell (Kresse et al., 1999; Ide et al., 2001; Dasanayake et al., 2011), EspB is also an effector protein (Iizumi et al., 2007; Hamada et al., 2010). These translocator proteins are encoded in the LEE4 operon downstream of the gene encoding SepL, which is proposed to control the temporal switch between the secretion of translocators and effectors (O'Connell et al., 2004; Wang et al., 2008).
A promoter upstream of sepL drives the synthesis of a precursor transcript that is processed by RNase E at a cluster of sites in an AU-rich region near the end of sepL, generating a ca. 3 kb espADB mRNA (Kresse et al., 2000; Roe et al., 2003; Lodato and Kaper, 2009). This mRNA has a leader region of variable length (107–152 bases) and a monophosphorylated 5' end, while the 3' end is likely generated by intrinsic transcription termination downstream of the espB gene and inside the cesD2 coding sequence (see Fig 1) (Lodato and Kaper, 2009). Processing of the precursor mRNA seems to be a mechanism to differentially control the production of the SepL and EspADB proteins, as has been proposed in the regulation of other operons (Baga et al., 1988; Nilsson and Uhlin, 1991; Cam et al., 1996; Kokoska and Steege, 1998; Tamura et al., 2006; Bardey et al., 2005; Sawers, 2005). However, the physiological rationale of this mechanism for the functioning of the EHEC T3SS is not currently understood.
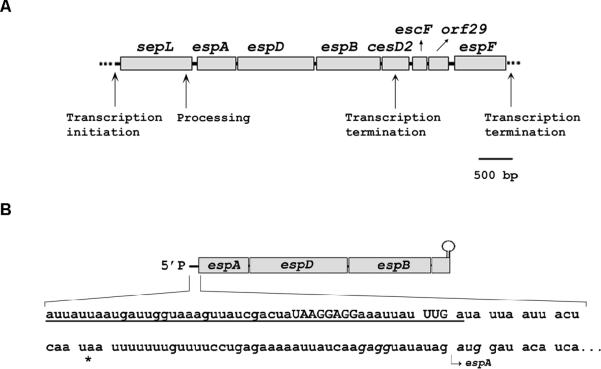
Figure 1. The LEE4 operon and the translocon-encoding espADB mRNA in EHEC.
(A) The LEE4 operon. The mechanisms that generate the LEE4 transcripts, including the major espADB mRNAs, are shown.
(B) espADB mRNA (main species, see Lodato and Kaper (2009)). The underlined sequence marks the portion of the leader region derived from the 3'-terminus of sepL. The leader region contains a mini-ORF with a Shine-Dalgarno element (SD2, capital letters), a UUG start codon (capital letters), and a UAA stop codon (asterisk).
Among the well-established properties of RNase E are its propensity to cleave RNA in AU-rich regions but with no defined sequence specificity (McDowall et al., 1994; Huang et al., 1998; Walsh et al., 2001) and its preference in vitro and in vivo for substrates with unpaired 5' ends that are monophosphorylated (Bouvet and Belasco, 1992; Mackie, 1998; Baker and Mackie, 2003; Jiang and Belasco, 2004; Celesnik et al., 2007). Consistent with prior biochemical studies, the crystal structure of the RNase E catalytic domain revealed a 5' sensor pocket where only a monophosphate, but not a triphosphate, can fit (Callaghan et al., 2005; Koslover et al., 2008; Kime et al., 2010). In addition, it was determined that the conversion of a triphosphorylated to a monophosphorylated 5' end by the RNA pyrophosphohydrolase RppH accelerates the degradation of hundreds of transcripts in E. coli (Deana et al., 2008). Collectively, these results established a mechanism for RNA degradation by RNase E that depends on the characteristics of the RNA 5' end. An additional mechanism, referred to as “direct entry” or “internal entry”, is independent of the substrate 5' end and is sometimes aided by the C-terminal half of the enzyme (Kime et al., 2010; Anupama et al., 2011; Bouvier and Carpousis, 2011). The two mechanisms are not mutually exclusive, and both can contribute to the degradation of some RNA targets (Garrey et al., 2009; Garrey and Mackie, 2011).
Besides the action of RppH, monophosphorylated 5' ends can be generated by endonucleolytic cleavage of an RNA precursor, as is the case for espADB mRNA. The abundance of this translocon-encoding mRNA suggests the existence of a mechanism by which it avoids rapid degradation. In this study we describe a novel mechanism in E. coli that can increase the stability of mRNA by protecting the AU-rich leader region.
RESULTS
The leader region of esp mRNAs encodes a mini-ORF
Sequence analysis of the espADB mRNA leader region suggested the presence of a six-codon mini-ORF with a strong Shine-Dalgarno element (SD2) and a noncanonical start codon (TTG) partially overlapping the sepL stop codon (TGA) (Fig. 1).
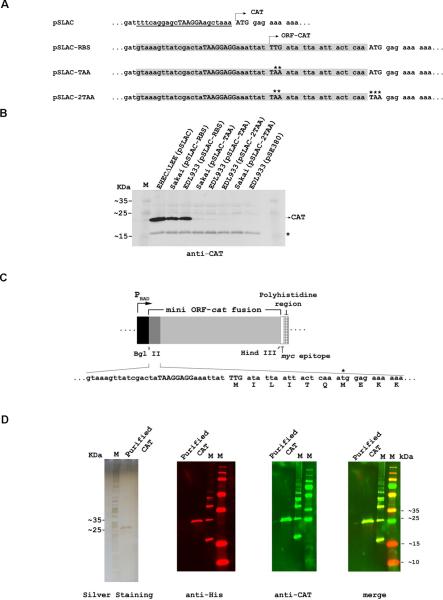
To test whether the mini-ORF was translated, we fused the SD2 sequence and the mini-ORF in frame to the coding region of chloramphenicol acetyltransferase (CAT). In this way, if SD2 and the initiation codon TTG were functional, an ORF-CAT fusion protein with six additional amino acids at the N-terminus would be synthesized. We modified a plasmid encoding CAT (pSLAC, a derivative of pSE380) to generate the plasmid pSLAC-RBS, in which the native cat ribosome binding site was replaced by SD2 and the mini-ORF (Fig. 2A). This plasmid was introduced into two EHEC strains (EDL933 and Sakai), and CAT synthesis was evaluated by Western blot analysis. Additionally, resistance to chloramphenicol (Cm) was confirmed by streaking the strains onto LB agar containing Cm. The fusion protein ORF-CAT was produced in both EHEC (pSLAC-RBS) strains (Fig. 2B), which were also resistant to Cm (Fig. S1A). However, when the start codon (TTG) was mutated to a TAA stop codon (plasmid pSLAC-TAA), the ORF-CAT protein was barely detectable (Fig. 2B), and the strains were sensitive to Cm (Fig. S1A). Only after several days at room temperature did the strains show growth on the Cm plates, indicating that very low CAT levels were being produced (Fig. S1B). The possibility that the residual CAT activity could be due to CAT synthesis starting at its native ATG codon downstream of the fusion site (Fig. 2A) was tested by additionally mutating this ATG codon to a TAA stop codon (plasmid pSLAC-2TAA). In this case, CAT was undetectable by immunoblotting (Fig. 2B), and the strains were sensitive to Cm even after several days of incubation (Fig. S1B).
Figure 2. Production of the ORF-CAT fusion protein.
(A) Plasmid sequences. SD2 and the first six codons of the mini-ORF (highlighted) were fused in-frame to the cat gene of pSLAC so as to replace the original SD element of cat. Asterisks mark the mutations subsequently introduced into the mini-ORF-cat gene fusion.
(B) Western blot analysis of ORF-CAT protein synthesis in EHEC transformed with the indicated plasmids. The EHEC strains ΔLEE(pSLAC) and EDL933(pSE380) were used as positive and negative controls, respectively. A non-specific E. coli protein (*) that adventitiously crossreacted with the antibodies was used as a loading control.
(C) Relevant sequence of the 5' region of the mini-ORF-CAT fusion in plasmid pBAD-CAT. The start codon and SD2 are shown in capital letters. The original cat start codon is labeled with an asterisk.
(D) The mini ORF-CAT fusion protein was purified from EHEC cell extracts by metal affinity chromatography on Ni2+-NTA, and its N-terminal sequence was determined (shown in 2C). The purified protein was subjected to SDS-PAGE (left panel) and detected by Western blotting with antisera, as indicated.
To determine the N-terminal sequence of the fusion protein, the SD2-mini-ORF-CAT fusion sequences were re-cloned to add a C-terminal His6 tag (Fig. 2C). The resultant ORF-CAT-His6 protein was purified by affinity chromatography on Ni2+-NTA (Fig. 2D, panel on the left). This purified protein was recognized by antisera against both the His6 tag and CAT (Fig. 2D), and its N-terminus bore six additional amino acids corresponding to the expected mini-ORF sequence (Fig. 2C).
The interaction between a Shine-Dalgarno sequence in an mRNA leader region and the anti Shine-Dalgarno at the 3' end of the 16S ribosomal RNA (rRNA) is an important determinant of translation initiation (Shine and Dalgarno, 1974; Steitz and Jakes, 1975; Kozak, 2005). The sequence of the SD2 element is coincident with the optimal E. coli Shine-Dalgarno sequence (TAAGGAGG). To test if the SD2 element affects mini ORF translation, its sequence was changed from TAAGGAGG to TAACGACG (plasmid pBAD-CAT-mutSD2) (Fig. S2A). Western blot analysis revealed that this mutation drastically reduced the concentration of the ORF-CAT fusion protein in EHEC (Fig. S2B).
To obtain further evidence that the mini-ORF is translated in its native sequence context, we fused the entire sepL gene and the mini-ORF in frame to lacZ (plasmid pLacZ-wt). Thus, β-galactosidase would be synthesized from this gene fusion only if the SD2 element and the mini-ORF were functional. In addition, the SD2 element was changed from TAAGGAGG to TAACGACG (plasmid pLacZ-mutSD2), or the mini-ORF start codon (TTG) was changed to TAA (plasmid pLacZ-TTG) (Fig. S3A). These plasmids were introduced into E. coli K-12 MC4100, and β-galactosidase activity was measured in cell extracts. β-galactosidase was high in E. coli (pLaz-wt), drastically reduced when the SD2 element was mutated, and almost undetectable when the mini-ORF start codon was changed to a stop codon (Figure S3B–C).
Together, these results demonstrate that a translatable mini-ORF starts at the end of sepL and extends into the intergenic region (IR). Processing of the sepL-espADB precursor produces espADB mRNA, whose leader region contains the SD2 element and the mini-ORF (Fig. 1).
Effect of SD2 and SDA disruption on esp mRNA and protein levels
The first three genes of the LEE4 operon - sepL, espA and espD – were cloned in plasmid pACYC184 under the control of the tet promoter (plasmid pPL1) (Lodato and Kaper, 2009). To analyze whether the mini-ORF could influence the expression of the downstream genes, the SD2 sequence was changed from TAAGGAGG to TAACGACG in plasmid pPL1 (plasmid pmut-SD2) (Fig. 3A). This plasmid was introduced into EHECΔLEE, and transcript levels were assayed by Northern blotting. Disruption of the SD2 element reduced the concentration of espAD mRNA (Fig. 3B). To determine whether the espA Shine-Dalgarno element (SDA) had a similar effect, its sequence was changed from GAGG to GACG (plasmid pmut-SDA). In this case also, the concentration of espAD mRNA was reduced (Fig. 3B, lane 3). Furthermore, the mRNA was barely detectable by Northern blotting when both SD2 and SDA were disrupted (Fig. 3B, lane 4) suggesting that ribosome binding to both regions protects the esp mRNA from degradation.
Figure 3. Effect of SD2 or SDA disruption on esp mRNA and protein levels.
(A) Mutations introduced into SD2 and/or SDA in plasmid pPL1.
(B – C) Northern blot analysis of (B) espAD and (C) espADB mRNA with oligonucleotide probes for espA (EspA4 or EspA3, respectively). In (C) RNA was extracted from EHEC strains at different OD600 values. Plasmid pmut-SD2-SDA has mutations in both SD2 and SDA. EHEC-mutSD2 has the SD2 mutations in the chromosome.
(D) Western blot analysis of EspA and EspB proteins. The immunoblot (left panel) was stained with Brilliant Blue R after EspA and EspB detection to show similar loading in each lane (right panel).
We introduced other mutations into the mini-ORF sequence of plasmid pPL1 to ascertain whether expression of the hexapeptide had an impact on mRNA levels (Fig. 4). The mini-ORF start codon TTG was changed to a stop codon (TAA) or to the more efficient start codon ATG (plasmids pmut-TTG and pmut-TTG-2, respectively). In addition, the third codon of the mini-ORF was changed to a stop codon TAA (plasmid pORF-TAA). Unlike the SD2 mutation, these ORF mutations did not significantly affect the concentration of esp mRNA (Fig. 4B and C). Additionally, processing of the sepL-espADB transcript was not altered when EHEC was transformed with a plasmid that overexpresses the mini-ORF in trans (Fig. S4). Therefore, transcript stability seems to depend on the presence of the mini-ORF ribosome binding site rather than on translation of the mini-ORF or the expression of a particular peptide.
Figure 4. Effect of mini-ORF mutations on esp mRNA concentration.
(A) Sequences of the mini-ORF and part of the intergenic region in the wild-type plasmid pPL1 and its mutated derivatives. The mutations, which are depicted in capital letters and underlined, were introduced into the mini-ORF region and include: the SD2 element (pmut-SD2, same as Figure 3), start codon (pmut-TTG and pmut-TTG-2), and the third codon (pORF-TAA). The mini-ORF SD2 and start codon are shown in uppercase letters.
(B) Analysis by Northern blotting of esp mRNA levels in EHECΔLEE transformed with plasmid pPL1 and its derivatives. Total RNA was extracted from cells grown in DMEM 0.2% glucose. The blot was hybridized with an espA-specific probe (EspA3), then stripped and re-hybridized with a probe for 16S rRNA.
(C) Quantification of esp transcript concentrations from Northern blots as shown in (B). For each lane, the esp band intensity was normalized to that of 16S rRNA to compensate for loading variations. Each ratio (± SD) is expressed relative to the normalized transcript level in EHECΔLEE(pPL1). The asterisk denotes that the difference in transcript levels is statistically significant (one sample, two sided t test, α= 0.05, P=0.0043).
In EHECΔLEE transformed with pPL1 or its derivatives, the processed RNA carries the mini-ORF and espAD but not espB. Therefore, the SD2 mutations were also introduced directly into the chromosome of EHEC strain EDL933 (EHEC-mutSD2) to analyze their effect on the whole esp transcript. Total RNA was extracted from cultures of the wild-type and mutant strains at several cell densities and analyzed by Northern blotting. The concentration of espADB mRNA was reduced in the EHEC-mutSD2 strain compared to the wild-type strain (Fig. 3C), confirming the importance of the SD2 element. Furthermore, the concentration of the EspA and EspB proteins was reduced in extracts from EHEC-mutSD2 compared to the wild-type strain, as determined by immunoblotting (Fig. 3D).
In the absence of a functional SD2 sequence RNase E can cleave downstream of SD2
A ribonuclease protection assay (RPA) was used to examine the sepL-espA intergenic region at higher resolution than is possible by Northern blotting (Fig. 5). In EHECΔLEE(pmut-SD2), we observed a minor RNA (labeled * in Fig. 5A, left panel) that corresponds to the main processed product in EHECΔLEE(pPL1) (see also Fig. 3 in Lodato and Kaper, 2009). However, in EHECΔLEE(pmut-SD2) shorter forms predominated (labeled ** and ***), some of which are also present in EHECΔLEE(pPL1) but at a low concentration. An identical pattern of RNA species was observed in the chromosomal mutant strain EHEC-mutSD2 (Fig. S5).
Figure 5. Effect of SD2 disruption on the 5' end of esp mRNA.
(A) Ribonuclease protection analysis (RPA) of processed esp mRNAs. Upper panel: Region complementary to the 382-nucleotide probe used for RPA. The arrow marks the approximate site of endonucleolytic processing by RNase E. Lower left panel: RPA of RNA from EHECΔLEE cells containing plasmid pPL1 (wild-type SD2 element) or pmut-SD2 (mutated SD2 element) (see Fig 3A). Total RNA from yeast was used as a negative control. Asterisks mark some of the processed transcripts. A triangle marks a processed species that is almost absent when SD2 is disrupted. A dot marks the unprocessed precursor. Lower right panel: E. coli K-12 carrying either a wild-type (wt) or temperature-sensitive (ts) RNase E allele was transformed with plasmid pmut-SD2. RNA samples were collected at a non-restrictive temperature (time 0) and at different time intervals after shifting the culture to the restrictive temperature and subjected to RPA.
(B) Location of the 5' ends of processed esp mRNAs in EHECΔLEE(pPL1-SD2). The products of 5' RLM-RACE were cloned and sequenced. *** indicates a cluster of sites where most of the 5' ends were located (18 out of 26 clones, highlighted in gray), corresponding to RPA products 130–140 bases long. ** indicates 5' ends (1 out of 26 clones for each underlined nucleotide) that correspond to RPA products 179–186 bases long. * indicates 5' ends (1 out of 26 clones for each underlined nucleotide) that correspond to RPA products 206–210 bases long. The 5' end represented by another clone corresponds to an upstream thymidine (triangle) and approximately matches the size of the RPA product labeled with a triangle in A. Two other clones have 5' ends located in the espA coding region downstream of the sequence shown in the figure. The SD2 and SDA sequences and start codons are shown in capital letters. The stop codons for sepL (TGA) and for the mini-ORF (TAA) are marked with a line above them. The arrows delimit the maximum region complementary to the RPA probe.
To analyze whether these shorter RNAs result from RNase E cleavage, we transformed pmut-SD2 into E. coli strain N3431, which produces a temperature-sensitive form of RNase E (rne-ts), and the isogenic wild-type strain N3433 (rne-wt) (Apirion, 1978; McDowall et al., 1993). Cultures of each strain were grown at a non-restrictive temperature (time 0) and then switched to the restrictive temperature, and RNA was extracted at time intervals. The shorter processed forms were observed in the rne-wt strain at all time points, while in the rne-ts mutant their abundance was greatly reduced at the restrictive temperature (Fig. 5A, right panel), indicating that the shorter forms are produced by RNase E cleavage. Next, the 5' ends of these shorter species were mapped by 5' RACE. We found that most of their 5' ends map to a cluster of thymidines downstream of the SD2 element in the intergenic region, and a minor fraction are located upstream of the SD2 element (Fig. 5B). The expected sizes of the RNAs starting at these positions are consistent with the RPA results. In contrast, our prior studies have shown that most of the 5' ends of espADB or espAD mRNA are located upstream of the SD2 sequence in wild-type EHEC or in EHECΔLEE(pPL1) (Lodato and Kaper, 2009).
Together, these results suggest that a ribosome bound to the SD2 element acts as a barrier to RNase E, which prefers to cut upstream of SD2 unless ribosome binding there is disrupted.
Disruption of the SD2 element reduces the half-life of the esp mRNA
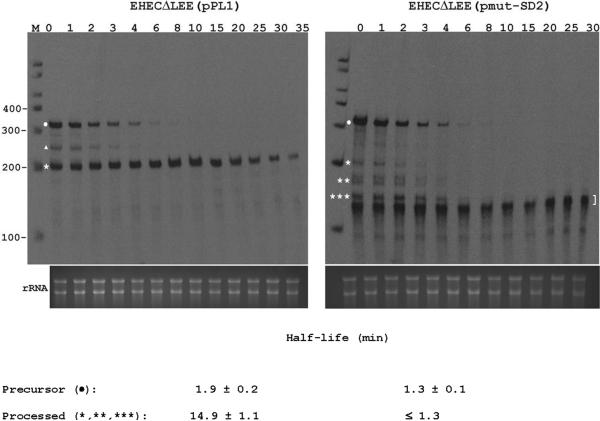
To test directly whether the disruption of the SD2 element reduces the stability of the esp transcript, we compared the half-life of the wild-type espAD mRNA (pPL1) and its SD2-mutated derivative (pmut-SD2) in EHECΔLEE. Exponential-phase cultures were treated with rifampicin to inhibit transcription, and RNA isolated at time intervals was analyzed by RPA.
Over the first 8 min after transcription inhibition, the wild-type precursor transcript (sepL-espAD) underwent processing with a half-life time of 1.9 min (Fig. 6, left panel). During this period, the degradation of the processed product (espAD) was partially compensated by its continued generation from the precursor. Thereafter, the wild-type-processed mRNA decayed slowly with a half-life of 14.9 min. In contrast, the mutant-processed species disappeared synchronously with its precursor, indicating that their half-life was no longer than the 1.3-min half-life of its precursor (Fig. 6, right panel). These results show that the mutant-processed species are much less stable than the wild-type counterpart. We conclude that disruption of the SD2 element markedly accelerates the degradation of the processed esp mRNA but has little effect on its rate of formation by precursor processing.
Figure 6. Effect of SD2 disruption on the decay of espAD mRNA.
EHECΔLEE was transformed with pPL1 (wild-type espAD) or its derivative pmut-SD2 (espAD with a mutated SD2 element (see Fig 3A)). Exponential-phase cultures were treated with rifampicin to inhibit transcription. RNA was isolated at time intervals (minutes after addition of rifampicin are shown at the top of each panel) and analyzed by RPA. Precursor mRNA is marked with a dot, while processed mRNA species are marked by a triangle and asterisks. A bracket denotes partially digested probe. An aliquot of each hybridization reaction was also examined by gel electrophoresis and ethidium staining (bottom of each panel). Mean half-life values ± SD are shown. The upper limit on the half-life of the mutant processed species (≤ 1.3 min for *, **, and *** derived from pmut-SD2) is based on the observation that these intermediates disappeared from rifampicin-treated cells at the same rate as their precursor, which decayed with a half-life of 1.3 ± 0.1 min.
RNase E inactivation prolongs the lifetime of espADB mRNA
If ribosome binding to the SD2 element in the leader region can protect processed esp mRNA from attack by RNase E, it follows that the espADB mRNAs should be more stable in an rne-ts strain than in the isogenic wild-type strain at the restrictive temperature. Measurement of the rate of esp mRNA degradation requires that it be synthesized independently of RNase E processing in order to decouple the two processes. Therefore, we cloned a fragment containing the leader region, espADB, cesD2, escF and ORF29 into a pBAD plasmid under the control of an arabinose-inducible promoter (plasmid pEsp). Upon induction by arabinose, the espADB mRNA encoded by pEsp is produced as a direct product of transcription initiation rather than by processing, with the only difference being its 5' phosphorylation state, i.e. the 5' end should be triphosphorylated instead of monophosphorylated because it is produced by transcription initiation and not cleavage. E. coli strains N3431 (rne-ts) and N3433 (rne-wt) containing plasmid pEsp were grown to early-log phase at a permissive temperature, the esp genes were induced by arabinose and then the cultures were shifted to a restrictive temperature. After 15 min, rifampicin was added to inhibit transcription, and RNA was isolated at time intervals. Northern blot analysis revealed a major espADB mRNA with the expected size of ca. 3 kb and a minor species of ca. 4 kb that resulted from transcription termination at a vector site downstream of ORF29 (Fig. 7 and data not shown). The half-life of espADB mRNA was 1.6 min in the rne-wt strain but 4.8 min in the rne-ts strain, indicating that it is degraded by RNase E (Fig. 7).
Figure 7. Effect of RNase E on the decay of espADB mRNA.
E. coli K-12 carrying either a wild-type (A) or temperature-sensitive RNase E (B) allele was transformed with plasmid pEsp, which encodes wild-type espADB as a primary product of transcription. The strains were grown to early-log phase at 32 °C, the esp genes were induced by 0.2% arabinose and then the cultures were shifted to 43 °C. After 15 min, rifampicin was added, and RNA samples were extracted at time intervals and analyzed by Northern blotting with an espA-specific probe (EspA4). 16S rRNA served as an internal standard. Mean half-life values ± SD are shown. The difference in half-life values between the wild-type and mutant strains is statistically significant (t test, α= 0.05, P value <0.0001)
Mutations in SD2 do not affect the intrinsic susceptibility of esp mRNA to degradation
In principle, mutations in SD2 could destabilize espADB mRNA either by acting directly to facilitate RNase E-mediated degradation or by preventing ribosomes from binding and shielding the RNA from cleavage. To distinguish between these possibilities, we tested whether mutations in SD2 alter the intrinsic susceptibility of esp mRNA to RNase E-mediated degradation in the absence of ribosomes and other cellular factors. For these experiments, two monophosphorylated RNAs were synthesized by in vitro transcription: IR-wtSD2, which resembled the 5'-terminal segment of espADB mRNA, and IR-mutSD2, which was identical except for a mutated SD2 element (Fig. 8A). In the presence of the purified N-terminal catalytic domain of RNase E (N-RNase E), there was no difference in the degradation of the two substrates (Fig. 8B, left panel). That the degradation of the in vitro transcribed substrates was due to N-RNase E activity and not to the presence of a contaminant was demonstrated by showing that a catalytically inactive form of N-RNase E (Callaghan et al., 2005) was unable to degrade these substrates (Fig. 8B, right panel). These findings indicate that the effect of the SD2 mutations on the concentration of the processed transcripts in E. coli is due not to a higher intrinsic susceptibility of espADB mRNA to degradation by RNase E but to an extrinsic influence such as ribosome binding.
Figure 8. In vitro degradation of esp RNA by N-RNase E.
(A) Sequences of the in vitro transcribed RNAs (IR-wtSD2 and IR-mutSD2) used as substrates in the assays shown in (B). The previously mapped 5' end of wild-type espADB mRNA in vivo is underlined. The 5' ends observed for the SD2 mutant in vivo are labeled with a dot. Both substrates have an additional guanosine at the 5' end as a result of transcription initiation by T7 RNA polymerase. G→C substitutions in the mutated SD2 element are indicated.
(B) The RNA substrates, which were monophosphorylated and internally radiolabeled, were treated with equal amounts of purified N-RNase E for the indicated times. Right panel: Treatment with catalytically inactive N-RNase E-D346A, which lacks an essential aspartate residue. Mean values ± SD are shown.
DISCUSSION
Here we report that the leader region of espADB mRNA in EHEC contains a short mini-ORF whose strong SD element (SD2) enhances translocon protein synthesis by protecting the message from degradation. This mRNA is produced by RNase E processing of a sepL-espADB precursor in an AU-rich region located near the 3' end of sepL, whose inefficient translation may facilitate processing by exposing RNase E cleavage sites (Lodato and Kaper, 2009 and our unpublished results).
Our data demonstrate that the SD2 element is able to drive mini-ORF translation irrespective of its sequence context (Fig. 2, Fig. S1, Fig. S2 and Fig. S3). G→C mutations in the SD2 element greatly impair mini-ORF translation (Fig. S2 and Fig. S3) and reduce the concentration and stability of processed esp mRNA (Fig. 3B–C and Fig 6, respectively).
The sepL-espADB precursor is processed by RNase E predominantly at sites located upstream of the mini-ORF to generate the main espADB mRNA species (labeled * in Fig. 5, Fig. 6 and Fig. S4) and a longer processing product (labeled ▲in Fig. 5, Fig. 6 and Fig. S4) (Fig. 9A). These processing products can then be degraded further by RNase E. This conclusion is supported by two observations. First, espADB mRNA is stabilized in a temperature-sensitive rne mutant at the restrictive temperature (Fig. 7). Second, RNase E cuts the SD2 mutant downstream of the mini-ORF, generating processed mRNA species that are shorter (labeled **, ***) (Fig. 5). Apparently, ribosomes that bind the SD2 element can protect espADB mRNA from degradation by competing with RNase E for access to AU-rich sequences located in the leader region and close to the mini-ORF (Fig. 9B, right). Whether RNase E mediated-degradation is assisted by the 5'-terminal monophosphate of espADB mRNA is not yet clear (Fig. 9B left and C).
Figure 9. Proposed mechanism for the protection of espADB mRNA from degradation by RNase E.
The precursor sepL-espADB mRNA is processed near the end of sepL to generate espADB mRNA with a monophosphorylated 5' end. A translatable six-codon mini-ORF (black diamond) is located in the leader region upstream of the Shine-Dalgarno element for espA (SDA). A strong SD element (SD2) (striped square) enables mini-ORF translation (A). RNase E can degrade the transcript through a 5' end-dependent mechanism (grey solid arrow) or by direct interaction with AU-rich sequences in the leader region (grey dashed arrow) (B left and C). However, ribosome binding to SD2 acts as a barrier to RNase E by reducing the access of RNase E either to the monophosphorylated 5' end or to downstream cleavage sites (B right). RNase E cleavage is represented by scissors.
In addition to protecting processed espADB mRNA from degradation, ribosome binding to SD2 appears to restrict processing of the sepL-espADB precursor primarily to sites upstream of the mini-ORF. The shorter mRNA species observed for the SD2 mutant could be generated as direct products of sepL-espADB processing (labeled •) (Fig 5) or by further cleavage of the longer processed species.
The difference between the half-lives of espAD mRNA and espADB mRNA (14.9 min vs 1.6, respectively) could be due to differences in the experimental conditions in which the stabilities were measured. The espADB mRNA half-life was measured in heat shocked cells while the espAD mRNA half-life was measured at 37°C. On the other hand, the espB gene may contain one or more important RNase E cleavage sites. Further studies will be needed to determine whether the espB gene or another region is important for destabilizing espADB mRNA.
Other mutations introduced into the mini-ORF sequence, including the substitution of TAA for the third codon of the mini-ORF or the replacement of its TTG start codon with TAA or ATG, did not significantly affect esp mRNA levels. Likewise, expression of the mini-ORF-encoded hexapeptide in trans did not affect the processing of the sepL-espADB precursor. These results imply that neither mini-ORF translation nor the hexapeptide itself regulates RNA processing or degradation. An analysis of mini-ORF sequences in other EHEC serotypes and other microorganisms carrying the LEE pathogenicity island shows that while the SD2 element is highly conserved, the length and sequence of the peptide encoded by the mini-ORF is variable (Fig. S6). These observations support our conclusion that the hexapeptide itself does not regulate RNA processing or degradation.
Recently, a mini-gene encoding only a dipeptide was reported to be present in the leader region of the EHEC LEE1 operon, which encodes Ler – a positive transcriptional regulator of LEE – as well as other components of the T3SS (Islam et al., 2012). The expression of an in-frame ler-lacZ fusion is abrogated if the mini-gene is disrupted. The LEE1 mini-gene is located close to the mRNA 5' end in a ~70% AU-rich leader region that could potentially be a target for RNase E cutting. As it is unlikely that a peptide of only two amino acids has any regulatory function, ribosome binding to that mini-gene could protect the LEE1 mRNA from degradation by RNase E by a mechanism similar to the one we have proposed for esp mRNA. Further studies of LEE1 mRNA levels in wild-type and mini-gene mutants will help to clarify the role of the mini-gene in LEE1 gene expression.
mRNA stabilization by SD elements and/or mini-ORFs located in leader regions could be widespread among both Gram-negative and Gram-positive bacteria despite significant differences in the mechanisms by which they degrade mRNA (Mathy et al., 2007; Bechhofer, 2009). In Bacillus thurigiensis, the leader region of a toxin-encoding mRNA harbors a strong SD element (STAB-SD) that increases mRNA stability. The same authors also found similar STAB-SD elements in the 5' region of other transcripts from other species, mainly in Gram-positive bacteria (Agaisse and Lereclus, 1996). Such elements that involve ribosome binding along with the well documented occurrence of 5'-terminal stem-loop structures can contribute to transcript stability in both Gram-negative and Gram-positive bacteria (Bouvet and Belasco, 1992; Emory et al., 1992; Bricker and Belasco, 1999; Hambraeus et al., 2002; Baker and Mackie, 2003; Sharp and Bechhofer, 2005; Richards et al., 2011).
While transcriptional regulation of virulence in EHEC has been widely studied (Mellies et al., 2007; Jimenez et al., 2010), only recently have the factors involved in posttranscriptional regulation started to be elucidated (Campellone et al., 2007; Bhatt et al., 2009; Hansen and Kaper, 2009; Shakhnovich et al., 2009; Kendall et al., 2011). Previously, quantification of espA mRNA and EspA filaments showed that their abundance increases from early to late log phase (Roe et al., 2003; Walters and Sperandio, 2006). The increase in esp mRNA concentration with the phase of the culture appears to be regulated at the transcriptional level through Ler (Nakanishi et al., 2006). Whether the stability of the espADB mRNA is also regulated by environmental conditions or the culture phase is currently unknown and deserves further consideration. On the other hand, EspA filament production was detected in a subpopulation of bacteria ranging from 1 to 95% depending on the strain and culture conditions (Roe et al., 2003). The mechanism reported here could contribute to such variability because any factors modulating the ability of RNase E to access cleavage sites in espADB mRNA could impact the concentration of the translocon proteins, and ultimately the construction of the T3SS.
EXPERIMENTAL PROCEDURES
Strains, plasmids and oligonucleotides
The strains and plasmids employed are described in Supporting information. The oligonucleotide sequences and their usage are described in Table S1.
RNA extraction
EHEC was grown aerobically at 37 °C with agitation in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen, catalog number 11885) supplemented with 0.2% glucose. The inoculum was grown overnight in Luria-Bertani (LB) medium at 37 °C. Media were supplemented with chloramphenicol (20 μg ml−1) or ampicillin (100 μg ml−1) when appropriate. Total RNA was isolated by one of two methods: RNeasy Midi extraction kit (Qiagen) or with a modified hot-phenol method (Ko and Lee, 2006). Briefly, cells from 1–6 ml culture were pelleted and resuspended in 500 μl of RNase free water (Invitrogen). RNA was extracted by adding 500 μl of acid phenol:chloroform (Invitrogen) containing 0.1 vol of 10× RNA extraction buffer (200 mM sodium acetate, pH 5.5, 5% SDS, and 10 mM EDTA). Samples were incubated at 65 °C for 5 min and centrifuged to collect the aqueous phase which was cleaned with one volume of chloroform. RNA was recovered by ethanol precipitation and extensively treated with TURBO DNase (Invitrogen) following the manufacturer's recommendation. RNA was extracted again with phenol:chloroform and ethanol precipitated.
Half-life determinations and Northern blots
E. coli strains N3431 and N3433 transformed with plasmid pEsp were grown in LB at 32 °C until early log phase (OD600 ~0.22–0.26) at which point esp genes were induced for 30 min by adding arabinose at 0.2% final concentration. The cultures were switched to 43 °C for 15 min, and then transcription was inhibited by addition of rifampicin (500 μg ml−1). Samples were taken at time intervals after one minute of antibiotic addition (time 0). RNA was extracted as described above and analyzed by Northern blot. In the experiments with the strains EHECΔLEE(pPL1) and EHECΔLEE(pmut-SD2), cultures were grown in LB at 37°C until log-phase (OD600 ~0.33–0.37) at which point transcription was inhibited by rifampicin addition (500 μg ml−1) and samples taken at time intervals for RNA isolation and RPA analysis. Northern Blots were performed as described previously (Lodato and Kaper, 2009). The ULTRAhyb-Oligo buffer (Invitrogen) was used for the overnight hybridizations at 42 °C and the membranes were washed as recommended by the manufacturer. The oligonucleotide probes were labeled with [γ32P]-ATP (PerkinElmer) and PNK (NEB). Blots were stripped and re-probed with a specific oligonucleotide for the 16S rRNA to correct for differences in lane loading. Radioactive bands were detected with a Storm 820 PhosphorImager (Molecular Dynamics) and their intensities quantified with ImageQuant software (Molecular Dynamics). The results shown in Fig. 4 are the average of 3–5 independent experiments. RNA half-lives were calculated by linear regression analysis of data of 3–4 independent experiments in Fig. 7. The statistical analysis was done with the NCSS (Kaysville, Utah) or GraphPad Prism programs.
RPA
RPA was performed with the RPA III Ribonuclease Protection Assay kit (Life Technologies) as described previously (Lodato and Kaper, 2009). Primer pairs F-T3LEE4/R-T7LEE4 were employed to PCR amplify the template used for in vitro transcription to synthesize the RNA probe. The RNA probe (382 bases) spans the 3' end of sepL, the intergenic region and the 5' end of espA and contains non-complementary sequences at the ends to discriminate a fully protected fragment from non-digested probe. For the analysis of the SD2 mutant, the probe was synthesized to include the G→C mutations. The probe was labeled using Biotin-16-UTP (Roche) to UTP ratio of 40:60. Hybridizations were done overnight at 42 °C. We previously established that the 5' end product of processing, i.e. the 3' end of sepL mRNA, is not detected in this assay probably because it is highly susceptible to degradation (see Fig. 3 and Fig.7 in Lodato and Kaper, 2009). Thus, only fragments corresponding to the 5' end of the espADB or espAD mRNA are detected.
The detection of the protected fragments was done as described previously (Lodato and Kaper, 2009) except that the blocking of membranes was done for 20 min and the washes were 5 min. The quantification of the mRNA bands in the films (Fig. 6) was done with the Image-Pro Plus software (Media Cybernetics). The hybridizations were done with 12μg of total RNA for the wild-type plus 360 pg of probe and 30μg of total RNA for the mutant plus 600 pg of probe. After overnight hybridizations, an aliquot of the hybridization reactions (corresponding to 2μg or 4μg of total RNA) was run in a formaldehyde-agarose gel and the 23S rRNA was quantified by densitometry analysis of gels. The 23S rRNA values were used to normalize for differences in total RNA among samples. In parallel to the analysis of half-live experiments, RPAs were also done with different concentrations of total RNA (1.5, 3, 6 and 12μg for the wild-type and 3.75, 7.5, 15, 30μg for the mutant). These samples were developed together with the half-life samples to ensure that the RNA quantifications were in a linear range. To obtain the reported half-life measurements, the normalized half-life values were multiplied by a correction factor that is the slope of the ln-ln graph of pixels vs RNA concentration. RNA half-lives were calculated by linear regression analysis of data of two independent experiments.
Western Blot
Western blots were done as previously described (Lodato and Kaper, 2009). For mini-ORF-CAT protein analysis, cell extracts were run in a Criterion Precast Gel 10–20% Tris-HCl (Bio-Rad). CAT was detected with anti-CAT polyclonal antibodies (Sigma) which were adsorbed three times with acetone powders to eliminate most of cross reacting proteins. The adsorbed antibodies were diluted 1:10,000 in LiCor blocking buffer with 0.1% Tween 20 before use. The His6 tag was detected with Tetra-His Antibody (Qiagen) diluted 1:2,000 in LiCor blocking buffer with 0.1% Tween 20. Detection of His6 and CAT were done in the 700 and 800 channel of the Licor scanner, respectively.
5' RACE
An RNA oligonucleotide (900 ng) was ligated with T4 RNA ligase (NEB) to 10 μg of total RNA isolated from EHECΔLEE(pmut-SD2). The RNA was reverse transcribed with SuperScript III First-Strand (Invitrogen) using random hexamers. The ligation products were amplified by PCR and cloned using a Topo TA Cloning Kit (Invitrogen).
Purification of hexahistidine-tagged mini-ORF-CAT
The mini-ORF-CAT-His6 protein was purified using Ni2+-NTA agarose resin (Qiagen) following the manufacturer's recommendations. Sakai(pBad-CAT) was grown in 400 ml of LB medium at 37 °C until log phase (OD600 = 0.57) at which point the synthesis of ORF-CAT-His6 was induced for 1 hr by adding 0.2% arabinose. The cells were collected, resuspended in 30 ml of binding buffer (100 mM NaH2PO4, 10 mM Tris-HCl, 8 M urea, 20 mM imidazol, pH 8.0) and broken using a French Press. After centrifugation of the lysate, the supernatant was mixed with 500 μl of resin previously pre-incubated with binding buffer. The slurry was incubated at room temperature for 1 hr, centrifuged, and washed 5 times with washing buffer (100 mM NaH2PO4, 10 mM Tris-HCl, 8 M urea, 300 mM NaCl, 0.5% Triton X-100, pH 6.3). The ORF-CAT-His6 protein was eluted with elution buffer (100 mM NaH2PO4, 10 mM Tris-HCl, 8 M urea, pH 4.5) and concentrated with Centricon Ultracel YM-3 (Millipore). The concentrate was run in a Criterion Precast Gel 10–20% Tris-HCl (Bio-Rad), and a band corresponding to the ORF-CAT-His6 expected size was cut from the gel. The protein was eluted overnight from the gel matrix with 400 μl of elution solution (50 mM Tris-HCl, 150 mM NaCl, 0.1 mM EDTA, pH 5.0) and then concentrated with Centricon Ultracel YM-3. The concentrated protein was run in a polyacrylamide gel, and transferred to a PVDF membrane (Millipore). The membrane was stained with Brilliant Blue R 250 to visualize the band. The band in the PVDF membrane was sent to Stanford PAN facility (Stanford School of Medicine) to determine its N-terminal sequence by Edman analysis.
Purification of hexahistidine-tagged N-RNase E by affinity chromatography
Wild-type and catalytically inactive forms of N-RNase E bearing a carboxy-terminal (His)6 tag were produced in E. coli strain BL21 (DE3) rne-131 Δrna containing plasmid pNRNE7001 or pNRNE7001-D346A. The synthesis of these proteins was induced by adding IPTG (1 mM) to a log-phase culture (A600 = 0.5) growing in LB medium at 37°C. After 3–4 hr, the cells were pelleted, resuspended in buffer A (10 mM Tris-Cl [pH 7.5], 1 mM EDTA, 0.1 mM dithiothreitol, 5% glycerol, 0.5% Genapol X-080, and Complete EDTA-free protease inhibitor (Roche)), and disrupted in EmulsiFlex-C3 high pressure homogenizer (Avestin). The cell lysate was cleared twice by centrifugation for 15 min at 10,000 rpm (4°C), and the N-RNase E in the resulting supernatant was purified by affinity chromatography on BD TALON metal affinity resin (Clontech) eluted with 150 mM imidazole in buffer A. The peak fractions were pooled, and N-RNase E was further purified to near homogeneity by anion-exchange chromatography on an UNO-Q column (Bio-Rad) eluted with a salt gradient (50 to 600 mM NaCl in buffer A). The peak fractions were pooled and concentrated into buffer A by ultrafiltration using an Amicon Ultra centrifugal filter (MWCO: 50kDa, Millipore). The purified proteins were stored at −80°C and diluted just before use in cleavage assays.
RNA synthesis
To examine the cleavage of espADB RNA by RNase E, a DNA fragment comprising a T7 RNA polymerase promoter fused to a DNA fragment encoding the 5'-terminal segment of espADB mRNA (IR-wtSD2 or IR-mutSD2, without and with mutations in the SD2 element, respectively) was amplified by PCR, gel purified, and used as a template for synthesizing internally radiolabeled 5'- monophosphorylated RNA by in vitro transcription with T7 RNA polymerase. The reaction mixture (20 μl) contained DNA template (0.2 μg), ATP (0.5 mM), UTP (0.5 mM), GTP (0.5 mM), CTP (0.05 mM), [α-32P]CTP (30 mCi), GMP (15 mM), RNasin (20 U; Promega), and T7 RNA polymerase (40 U, NEB) in 1× RNA polymerase reaction buffer (NEB). After incubation for 16 h at 37°C, transcripts were purified by phenol/chloroform extraction followed by ethanol precipitation.
In vitro RNA cleavage assays
RNA cleavage by purified N-RNase E was carried out at 30°C in a reaction mixture (30 μl) containing Tris Cl (25 mM, pH 7.5), MgCl2 (10 mM), KCl (60 mM), NH4Cl (100 mM), dithiothreitol (0.1 mM), glycerol (5% [vol/vol]), internally radiolabeled RNA (160 pM), and N-RNase E (200 nM). Reaction samples (5 μl) were removed at time intervals and quenched by the addition of 10 μl of loading buffer (22 mM Tris borate [pH 7.5], 40 mM EDTA, 90% formamide, 0.1% xylene cyanol). After denaturation at 95°C for 5 min, the samples were subjected to electrophoresis on an 8% polyacrylamide gel containing 8 M urea. Radioactive bands were visualized and quantified with a Molecular Dynamics Storm 820 PhosphorImager. Cleavage rates were calculated by linear regression analysis of data from 3 independent experiments.
Supplementary Material
ACKNOWLEDGEMENTS
Supported by NIH grants AI21657 and DK58957 to J.B.K and GM35769 to J.G.B. We thank Dr. W. Blackwelder and Dr. Y. Wu for their help in the statistical analysis showed in Fig. 4; Dr. F. Obata and A. Vozenilek for the use of the program Image-Pro Plus.
REFERENCES
- Agaisse H, Lereclus D. STAB-SD: a Shine-Dalgarno sequence in the 5' untranslated region is a determinant of mRNA stability. Mol Microbiol. 1996;20:633–643. doi: 10.1046/j.1365-2958.1996.5401046.x. [DOI] [PubMed] [Google Scholar]
- Anupama K, Leela JK, Gowrishankar J. Two pathways for RNase E action in Escherichia coli in vivo and bypass of its essentiality in mutants defective for Rho-dependent transcription termination. Mol Microbiol. 2011;82:1330–1348. doi: 10.1111/j.1365-2958.2011.07895.x. [DOI] [PubMed] [Google Scholar]
- Apirion D. Isolation, genetic mapping and some characterization of a mutation in Escherichia coli that affects the processing of ribonuleic acid. Genetics. 1978;90:659–671. doi: 10.1093/genetics/90.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baga M, Goransson M, Normark S, Uhlin BE. Processed mRNA with differential stability in the regulation of E. coli pilin gene expression. Cell. 1988;52:197–206. doi: 10.1016/0092-8674(88)90508-9. [DOI] [PubMed] [Google Scholar]
- Baker KE, Mackie GA. Ectopic RNase E sites promote bypass of 5'-end-dependent mRNA decay in Escherichia coli. Mol Microbiol. 2003;47:75–88. doi: 10.1046/j.1365-2958.2003.03292.x. [DOI] [PubMed] [Google Scholar]
- Bardey V, Vallet C, Robas N, Charpentier B, Thouvenot B, Mougin A, et al. Characterization of the molecular mechanisms involved in the differential production of erythrose-4-phosphate dehydrogenase, 3-phosphoglycerate kinase and class II fructose-1,6-bisphosphate aldolase in Escherichia coli. Mol Microbiol. 2005;57:1265–1287. doi: 10.1111/j.1365-2958.2005.04762.x. [DOI] [PubMed] [Google Scholar]
- Bechhofer DH. Messenger RNA decay and maturation in Bacillus subtilis. Prog Mol Biol Transl Sci. 2009;85:231–273. doi: 10.1016/S0079-6603(08)00806-4. [DOI] [PubMed] [Google Scholar]
- Bhatt S, Edwards AN, Nguyen HT, Merlin D, Romeo T, Kalman D. The RNA binding protein CsrA is a pleiotropic regulator of the locus of enterocyte effacement pathogenicity island of enteropathogenic Escherichia coli. Infect Immun. 2009;77:3552–3568. doi: 10.1128/IAI.00418-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet P, Belasco JG. Control of RNase E-mediated RNA degradation by 5'-terminal base pairing in E. coli. Nature. 1992;360:488–491. doi: 10.1038/360488a0. [DOI] [PubMed] [Google Scholar]
- Bouvier M, Carpousis AJ. A tale of two mRNA degradation pathways mediated by RNase E. Mol Microbiol. 2011;82:1305–1310. doi: 10.1111/j.1365-2958.2011.07894.x. [DOI] [PubMed] [Google Scholar]
- Bricker AL, Belasco JG. Importance of a 5' stem-loop for longevity of papA mRNA in Escherichia coli. J Bacteriol. 1999;181:3587–3590. doi: 10.1128/jb.181.11.3587-3590.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaghan AJ, Marcaida MJ, Stead JA, McDowall KJ, Scott WG, Luisi BF. Structure of Escherichia coli RNase E catalytic domain and implications for RNA turnover. Nature. 2005;437:1187–1191. doi: 10.1038/nature04084. [DOI] [PubMed] [Google Scholar]
- Cam K, Rome G, Krisch HM, Bouche JP. RNase E processing of essential cell division genes mRNA in Escherichia coli. Nucleic Acids Res. 1996;24:3065–3070. doi: 10.1093/nar/24.15.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Roe AJ, Lobner-Olesen A, Murphy KC, Magoun L, Brady MJ, et al. Increased adherence and actin pedestal formation by dam-deficient enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol. 2007;63:1468–1481. doi: 10.1111/j.1365-2958.2007.05602.x. [DOI] [PubMed] [Google Scholar]
- Celesnik H, Deana A, Belasco JG. Initiation of RNA decay in Escherichia coli by 5' pyrophosphate removal. Mol Cell. 2007;27:79–90. doi: 10.1016/j.molcel.2007.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasanayake D, Richaud M, Cyr N, Caballero-Franco C, Pittroff S, Finn RM, et al. The N-terminal amphipathic region of the Escherichia coli type III secretion system protein EspD is required for membrane insertion and function. Mol Microbiol. 2011;81:734–750. doi: 10.1111/j.1365-2958.2011.07727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deana A, Celesnik H, Belasco JG. The bacterial enzyme RppH triggers messenger RNA degradation by 5' pyrophosphate removal. Nature. 2008;451:355–358. doi: 10.1038/nature06475. [DOI] [PubMed] [Google Scholar]
- Emory SA, Bouvet P, Belasco JG. A 5'-terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes Dev. 1992;6:135–148. doi: 10.1101/gad.6.1.135. [DOI] [PubMed] [Google Scholar]
- Garrey SM, Blech M, Riffell JL, Hankins JS, Stickney LM, Diver M, et al. Substrate binding and active site residues in RNases E and G: role of the 5'-sensor. J Biol Chem. 2009;284:31843–31850. doi: 10.1074/jbc.M109.063263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrey SM, Mackie GA. Roles of the 5'-phosphate sensor domain in RNase E. Mol Microbiol. 2011;80:1613–1624. doi: 10.1111/j.1365-2958.2011.07670.x. [DOI] [PubMed] [Google Scholar]
- Griffin PM, Ostroff SM, Tauxe RV, Greene KD, Wells JG, Lewis JH, Blake PA. Illnesses associated with Escherichia coli O157:H7 infections. A broad clinical spectrum. Ann Intern Med. 1988;109:705–712. doi: 10.7326/0003-4819-109-9-705. [DOI] [PubMed] [Google Scholar]
- Hamada D, Hamaguchi M, Suzuki KN, Sakata I, Yanagihara I. Cytoskeleton-modulating effectors of enteropathogenic and enterohemorrhagic Escherichia coli: a case for EspB as an intrinsically less-ordered effector. FEBS J. 2010;277:2409–2415. doi: 10.1111/j.1742-4658.2010.07655.x. [DOI] [PubMed] [Google Scholar]
- Hambraeus G, Karhumaa K, Rutberg B. A 5' stem-loop and ribosome binding but not translation are important for the stability of Bacillus subtilis aprE leader mRNA. Microbiology. 2002;148:1795–1803. doi: 10.1099/00221287-148-6-1795. [DOI] [PubMed] [Google Scholar]
- Hansen AM, Kaper JB. Hfq affects the expression of the LEE pathogenicity island in enterohaemorrhagic Escherichia coli. Mol Microbiol. 2009;73:446–465. doi: 10.1111/j.1365-2958.2009.06781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Liao J, Cohen SN. Poly(A)- and poly(U)-specific RNA 3' tail shortening by E. coli ribonuclease E. Nature. 1998;391:99–102. doi: 10.1038/34219. [DOI] [PubMed] [Google Scholar]
- Ide T, Laarmann S, Greune L, Schillers H, Oberleithner H, Schmidt MA. Characterization of translocation pores inserted into plasma membranes by type III-secreted Esp proteins of enteropathogenic Escherichia coli. Cell Microbiol. 2001;3:669–679. doi: 10.1046/j.1462-5822.2001.00146.x. [DOI] [PubMed] [Google Scholar]
- Iizumi Y, Sagara H, Kabe Y, Azuma M, Kume K, Ogawa M, et al. The enteropathogenic E. coli effector EspB facilitates microvillus effacing and antiphagocytosis by inhibiting myosin function. Cell Host Microbe. 2007;2:383–392. doi: 10.1016/j.chom.2007.09.012. [DOI] [PubMed] [Google Scholar]
- Islam MS, Shaw RK, Frankel G, Pallen MJ, Busby SJ. Translation of a minigene in the 5' leader sequence of the enterohaemorrhagic Escherichia coli LEE1 transcription unit affects expression of the neighbouring downstream gene. Biochem J. 2012;441:247–253. doi: 10.1042/BJ20110912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Belasco JG. Catalytic activation of multimeric RNase E and RNase G by 5'-monophosphorylated RNA. Proc Natl Acad Sci U S A. 2004;101:9211–9216. doi: 10.1073/pnas.0401382101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez R, Cruz-Migoni SB, Huerta-Saquero A, Bustamante VH, Puente JL. Molecular characterization of GrlA, a specific positive regulator of ler expression in enteropathogenic Escherichia coli. J Bacteriol. 2010;192:4627–4642. doi: 10.1128/JB.00307-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- Kendall MM, Gruber CC, Rasko DA, Hughes DT, Sperandio V. Hfq virulence regulation in enterohemorrhagic Escherichia coli O157:H7 strain 86-24. J Bacteriol. 2011;193:6843–6851. doi: 10.1128/JB.06141-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kime L, Jourdan SS, Stead JA, Hidalgo-Sastre A, McDowall KJ. Rapid cleavage of RNA by RNase E in the absence of 5' monophosphate stimulation. Mol Microbiol. 2010;76:590–604. doi: 10.1111/j.1365-2958.2009.06935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutton S, Rosenshine I, Pallen MJ, Nisan I, Neves BC, Bain C, et al. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 1998;17:2166–2176. doi: 10.1093/emboj/17.8.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko JH, Lee Y. RNA-conjugated template-switching RT-PCR method for generating an Escherichia coli cDNA library for small RNAs. J Microbiol Methods. 2006;64:297–304. doi: 10.1016/j.mimet.2005.05.011. [DOI] [PubMed] [Google Scholar]
- Kokoska RJ, Steege DA. Appropriate expression of filamentous phage f1 DNA replication genes II and X requires RNase E-dependent processing and separate mRNAs. J Bacteriol. 1998;180:3245–3249. doi: 10.1128/jb.180.12.3245-3249.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koslover DJ, Callaghan AJ, Marcaida MJ, Garman EF, Martick M, Scott WG, Luisi BF. The crystal structure of the Escherichia coli RNase E apoprotein and a mechanism for RNA degradation. Structure. 2008;16:1238–1244. doi: 10.1016/j.str.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene. 2005;361:13–37. doi: 10.1016/j.gene.2005.06.037. [DOI] [PubMed] [Google Scholar]
- Kresse AU, Beltrametti F, Muller A, Ebel F, Guzman CA. Characterization of SepL of enterohemorrhagic Escherichia coli. J Bacteriol. 2000;182:6490–6498. doi: 10.1128/jb.182.22.6490-6498.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kresse AU, Rohde M, Guzman CA. The EspD protein of enterohemorrhagic Escherichia coli is required for the formation of bacterial surface appendages and is incorporated in the cytoplasmic membranes of target cells. Infect Immun. 1999;67:4834–4842. doi: 10.1128/iai.67.9.4834-4842.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato PB, Kaper JB. Post-transcriptional processing of the LEE4 operon in enterohaemorrhagic Escherichia coli. Mol Microbiol. 2009;71:273–290. doi: 10.1111/j.1365-2958.2008.06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Donnenberg MS. Interactions and predicted host membrane topology of the enteropathogenic Escherichia coli translocator protein EspB. J Bacteriol. 2011;193:2972–2980. doi: 10.1128/JB.00153-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie GA. Ribonuclease E is a 5'-end-dependent endonuclease. Nature. 1998;395:720–723. doi: 10.1038/27246. [DOI] [PubMed] [Google Scholar]
- Mathy N, Benard L, Pellegrini O, Daou R, Wen T, Condon C. 5'-to-3' exoribonuclease activity in bacteria: role of RNase J1 in rRNA maturation and 5' stability of mRNA. Cell. 2007;129:681–692. doi: 10.1016/j.cell.2007.02.051. [DOI] [PubMed] [Google Scholar]
- McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci U S A. 1995;92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowall KJ, Hernandez RG, Lin-Chao S, Cohen SN. The ams-1 and rne-3071 temperature-sensitive mutations in the ams gene are in close proximity to each other and cause substitutions within a domain that resembles a product of the Escherichia coli mre locus. J Bacteriol. 1993;175:4245–4249. doi: 10.1128/jb.175.13.4245-4249.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowall KJ, Lin-Chao S, Cohen SN. A+U content rather than a particular nucleotide order determines the specificity of RNase E cleavage. J Biol Chem. 1994;269:10790–10796. [PubMed] [Google Scholar]
- Mellies JL, Barron AM, Carmona AM. Enteropathogenic and enterohemorrhagic Escherichia coli virulence gene regulation. Infect Immun. 2007;75:4199–4210. doi: 10.1128/IAI.01927-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi N, Abe H, Ogura Y, Hayashi T, Tashiro K, Kuhara S, et al. ppGpp with DksA controls gene expression in the locus of enterocyte effacement (LEE) pathogenicity island of enterohaemorrhagic Escherichia coli through activation of two virulence regulatory genes. Mol Microbiol. 2006;61:194–205. doi: 10.1111/j.1365-2958.2006.05217.x. [DOI] [PubMed] [Google Scholar]
- Nilsson P, Uhlin BE. Differential decay of a polycistronic Escherichia coli transcript is initiated by RNase E-dependent endonucleolytic processing. Mol Microbiol. 1991;5:1791–1799. doi: 10.1111/j.1365-2958.1991.tb01928.x. [DOI] [PubMed] [Google Scholar]
- O'Connell CB, Creasey EA, Knutton S, Elliott S, Crowther LJ, Luo W, et al. SepL, a protein required for enteropathogenic Escherichia coli type III translocation, interacts with secretion component SepD. Mol Microbiol. 2004;52:1613–1625. doi: 10.1111/j.1365-2958.2004.04101.x. [DOI] [PubMed] [Google Scholar]
- Richards J, Liu Q, Pellegrini O, Celesnik H, Yao S, Bechhofer DH, et al. An RNA pyrophosphohydrolase triggers 5'-exonucleolytic degradation of mRNA in Bacillus subtilis. Mol Cell. 2011;43:940–949. doi: 10.1016/j.molcel.2011.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe AJ, Yull H, Naylor SW, Woodward MJ, Smith DG, Gally DL. Heterogeneous surface expression of EspA translocon filaments by Escherichia coli O157:H7 is controlled at the posttranscriptional level. Infect Immun. 2003;71:5900–5909. doi: 10.1128/IAI.71.10.5900-5909.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawers RG. Evidence for novel processing of the anaerobically inducible dicistronic focA-pfl mRNA transcript in Escherichia coli. Mol Microbiol. 2005;58:1441–1453. doi: 10.1111/j.1365-2958.2005.04915.x. [DOI] [PubMed] [Google Scholar]
- Schmidt MA. LEEways: tales of EPEC, ATEC and EHEC. Cell Microbiol. 2010;12:1544–1552. doi: 10.1111/j.1462-5822.2010.01518.x. [DOI] [PubMed] [Google Scholar]
- Sekiya K, Ohishi M, Ogino T, Tamano K, Sasakawa C, Abe A. Supermolecular structure of the enteropathogenic Escherichia coli type III secretion system and its direct interaction with the EspA-sheath-like structure. Proc Natl Acad Sci U S A. 2001;98:11638–11643. doi: 10.1073/pnas.191378598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakhnovich EA, Davis BM, Waldor MK. Hfq negatively regulates type III secretion in EHEC and several other pathogens. Mol Microbiol. 2009;74:347–363. doi: 10.1111/j.1365-2958.2009.06856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp JS, Bechhofer DH. Effect of 5'-proximal elements on decay of a model mRNA in Bacillus subtilis. Mol Microbiol. 2005;57:484–495. doi: 10.1111/j.1365-2958.2005.04683.x. [DOI] [PubMed] [Google Scholar]
- Shaw RK, Daniell S, Ebel F, Frankel G, Knutton S. EspA filament-mediated protein translocation into red blood cells. Cell Microbiol. 2001;3:213–222. doi: 10.1046/j.1462-5822.2001.00105.x. [DOI] [PubMed] [Google Scholar]
- Shine J, Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz JA, Jakes K. How ribosomes select initiator regions in mRNA: base pair formation between the 3' terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proc Natl Acad Sci U S A. 1975;72:4734–4738. doi: 10.1073/pnas.72.12.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura M, Lee K, Miller CA, Moore CJ, Shirako Y, Kobayashi M, Cohen SN. RNase E maintenance of proper FtsZ/FtsA ratio required for nonfilamentous growth of Escherichia coli cells but not for colony-forming ability. J Bacteriol. 2006;188:5145–5152. doi: 10.1128/JB.00367-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tree JJ, Wolfson EB, Wang D, Roe AJ, Gally DL. Controlling injection: regulation of type III secretion in enterohaemorrhagic Escherichia coli. Trends Microbiol. 2009;17:361–370. doi: 10.1016/j.tim.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Walsh AP, Tock MR, Mallen MH, Kaberdin VR, von GA, McDowall KJ. Cleavage of poly(A) tails on the 3'-end of RNA by ribonuclease E of Escherichia coli. Nucleic Acids Res. 2001;29:1864–1871. doi: 10.1093/nar/29.9.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters M, Sperandio V. Autoinducer 3 and epinephrine signaling in the kinetics of locus of enterocyte effacement gene expression in enterohemorrhagic Escherichia coli. Infect Immun. 2006;74:5445–5455. doi: 10.1128/IAI.00099-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Roe AJ, McAteer S, Shipston MJ, Gally DL. Hierarchal type III secretion of translocators and effectors from Escherichia coli O157:H7 requires the carboxy terminus of SepL that binds to Tir. Mol Microbiol. 2008;69:1499–1512. doi: 10.1111/j.1365-2958.2008.06377.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.