Abstract
Avian infectious bronchitis virus (IBV) defective RNAs (D-RNAs) have been used for the expression of heterologous genes in a helper-virus-dependent expression system. The heterologous genes were expressed under the control of an IBV transcription-associated sequence (TAS) derived from gene 5 of IBV Beaudette. However, coronavirus D-RNA expression vectors display an inherent instability following serial passage with helper virus, resulting in the eventual loss of the heterologous genes. The use of the picornavirus encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES) sequence to initiate gene translation was investigated as an alternative method to the coronavirus-mediated TAS-controlled heterologous gene expression system. IBV D-RNAs containing the chloramphenicol acetyltransferase (CAT) reporter gene, under EMCV IRES control, were assessed for IRES-mediated CAT protein translation. CAT protein was detected from T7-derived IBV D-RNA transcripts in a cell-free protein synthesis system and in situ in avian chick kidney (CK) cells following T7-derived D-RNA synthesis from a recombinant fowlpox virus expressing the bacteriophage T7 DNA-dependent RNA polymerase. However, CAT protein was not detected in CK cells from IRES-containing IBV D-RNAs, in which the IRES-CAT construct was inserted at two different positions within the D-RNA, in the presence of helper IBV. Northern blot analysis demonstrated that the IRES-containing D-RNAs were not rescued on serial passage with helper virus, indicating that the EMCV IRES sequence had a detrimental effect on IBV D-RNA rescue.
Avian infectious bronchitis virus (IBV), a group 3 member of the genus Coronavirus (order Nidovirales, family Coronaviridae), is a highly infectious pathogen of domestic fowl that replicates primarily in the respiratory tract but also in epithelial cells of the gut, kidney, and oviduct (10, 13, 14). Genetically very similar coronaviruses cause disease in turkeys and pheasants (11, 12). Coronaviruses are enveloped viruses that replicate in the cell cytoplasm and contain an unsegmented, 5′-capped and 3′-polyadenylated, single-stranded, positive-sense RNA genome of 28 to 32 kb (16, 25, 38). During the replication cycle, coronaviruses produce a 3′-coterminal nested set of polycistronic subgenomic mRNAs (sgmRNAs) synthesized by a discontinuous transcription process during the synthesis of the negative strand (36, 37). The sgmRNAs contain identical 5′ ends due to the addition of a leader sequence derived from the 5′ end of the genomic RNA (gRNA). Preceding the body sequence of each sgmRNA sequence on the gRNA is a consensus sequence, the transcription-associated sequence (TAS) (20), involved in the acquisition of the leader sequence. All coronavirus envelopes contain at least three membrane proteins, the spike glycoprotein, a small membrane protein, and an integral membrane protein. In addition, the coronavirus virion also contains a nucleocapsid protein that interacts with the gRNA.
Coronavirus defective RNAs (D-RNAs), which lack large parts of the genome, are produced following virus passage at a high multiplicity of infection. While all D-RNAs contain cis-acting sequences necessary for replication, only a subset of D-RNAs contain sequences necessary for packaging into virions in the presence of a helper virus. Coronavirus D-RNAs have been used for site-directed mutagenesis of virus genomes (29) and for the expression of heterologous genes in helper-dependent expression systems (1, 2, 15, 17, 18, 21, 24, 26, 27, 40, 41, 44, 45). Heterologous gene expression by coronavirus D-RNAs has required an appropriate TAS for the synthesis of a D-RNA-derived sgmRNA, which is subsequently translated in a cap-dependent manner. We have routinely used the IBV Beaudette gene 5 TAS for the expression of heterologous genes from IBV-derived D-RNAs. The Beaudette gene 5 TAS was chosen because it has the shortest sequence between the 3′ end of the TAS and the AUG of open reading frame (ORF) 5a and because the Beaudette sgmRNA 5 is one of the most abundantly expressed sgmRNAs (40). The IBV Beaudette gene 5 TAS consists of two canonical consensus sequences (CUUAACAA) in a tandem repeat and has been used to express the chloramphenicol acetyltransferase (CAT) and luciferase (Luc) reporter genes (17, 40) and chicken gamma interferon (IFN-γ) (18).
Internal ribosome entry site (IRES) sequences allow translation to occur internally within an RNA via a cap-independent mechanism by recruiting the eukaryotic initiation factors, initiator tRNA, ribosomal subunits, and other cellular factors (5, 19, 28). The first IRES was identified in the poliovirus genome (31), resulting in the development of bicistronic expression vectors usually utilizing an IRES sequence from either poliovirus or encephalomyocarditis virus (EMCV) (22). The inclusion of an IRES to initiate heterologous gene translation within a coronavirus D-RNA should obviate the need to generate specific sgmRNAs for the expression of heterologous genes by allowing translation directly from the D-RNA. In this study, we describe experiments to determine the feasibility of utilizing EMCV IRES-mediated internal initiation for CAT protein synthesis from IBV D-RNAs during IBV helper virus-dependent rescue.
MATERIALS AND METHODS
Cells and viruses.
IBV Beaudette was grown in 11-day-old embryonated specific-pathogen-free domesticated fowl eggs, harvested from allantoic fluid 20 h postinfection, and used as helper virus for the rescue of IBV D-RNAs (41). IBV was passaged in primary chick kidney (CK) cells (32). Fowlpox virus FPV/T7, a recombinant expressing the bacteriophage T7 DNA-dependent RNA polymerase under the control of the vaccinia virus P7.5 early-late promoter (8), was grown in chicken embryonic fibroblasts (17).
Oligonucleotides.
All oligonucleotides used in this work were obtained from MWG-Biotech and are listed on Table 1.
TABLE 1.
Oligonucleotides used for construction of D-RNAs
| Oligonucleotide | Sequence (5′-3′) | Polarity | Positionb |
|---|---|---|---|
| BGL2TAS5 | GAAGATCTGTGTTTTACTTAACAAAAACTTAACAAAa | + | 25452-25479 |
| CAT-PAC | GGCTTAATTAAATGGAGAAAAAAATCACTGG | + | NAc |
| CAT-XHO | GGGCTCGAGAAAATTACGCAAAGCCCTGC | − | NA |
| EMCV-1 | GGGTCGCGAGCGCTTCCGCCCCTCCCCCTCCCCCCCCCTAACGT | + | NA |
| EMCV-2 | GGGTCGCGACTCGAGCATCGATGATATCTTAATTAAGATCTAGCGCTGTTTTTCAAAGGAAAACC | − | NA |
| Gene5+ve | GTGTTTTACTTAACAAAAACTTAACAAATACGGACG | + | 25452-25487 |
| Gene5−ve | CGTCCGTATTTGTTAAGTTTTTGTTAAGTAAAACAC | − | 25452-25487 |
Recombinant DNA techniques.
Recombinant DNA techniques used standard procedures (3, 35) or were used according to the manufacturers' instructions. All IBV-related nucleotide and amino acid residue numbers refer to the positions in the IBV Beau-R genome (9), GenBank accession no. AJ311317.
Construction of D-RNAs.
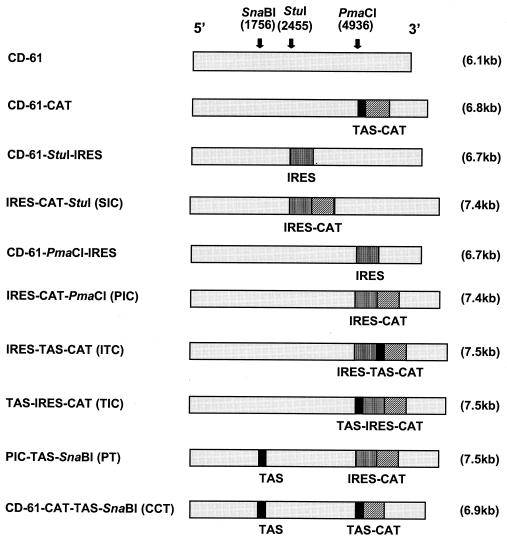
All of the D-RNAs (Fig. 1) used were constructed by using the IBV D-RNA CD-61 cDNA sequence in pIBV-Vec (17). The CD-61 cDNA in pIBV-Vec is under the control of a T7 promoter (T7ψ) and is terminated downstream of the poly(A) tail by a hepatitis delta antigenome ribozyme (HδR) and a T7 terminator (T7φ) sequence (17). The CAT gene, under the control of the IBV Beaudette gene 5 TAS (TAS-CAT) (40), was ligated into the PmaCI site of CD-61 in pIBV-Vec, producing pCD-61-CAT. The EMCV IRES sequence (a gift from Graham Belsham, IAH Pirbright Laboratory) was amplified by PCR with oligonucleotides EMCV-1 and EMCV-2 and ligated into pTarget (Promega), producing pIRES. Oligonucleotide EMCV-1 added NruI and Eco47III restriction sites on the 5′ end of the EMCV IRES sequence, and oligonucleotide EMCV-2 added Eco47III, BglII, PacI, EcoRV, XhoI, and NruI restriction sites on the 3′ end of the EMCV IRES sequence. The NruI-digested IRES sequence from pIRES was inserted into the StuI and PmaCI sites of CD-61 in pIBV-Vec, producing pCD-61-StuI-IRES and pCD-61-PmaCI-IRES, respectively. The CAT gene was amplified by PCR with oligonucleotides CAT-PAC and CAT-XHO and inserted into pTarget, producing pCAT. Oligonucleotides CAT-PAC and CAT-XHO added PacI and XhoI restriction sites to the 5′ and 3′ ends of the CAT sequence, respectively. PacI-XhoI-digested CAT sequence from pCAT was directionally inserted into the PacI and XhoI sites, downstream of the EMCV IRES sequence in pCD-61-StuI-IRES and pCD-61-PmaCI-IRES, producing pStuI-IRES-CAT (SIC) and pPmaCI-IRES-CAT (pPIC), respectively. The TAS-CAT gene cassette was amplified by PCR with oligonucleotides BGL2TAS5 and CAT-XHO, in which oligonucleotide BGL2TAS5 added a BglII restriction site to the 5′ end of the TAS. The BglII-TAS-CAT-XhoI-digested PCR product was inserted into pPIC, producing pIRES-TAS-CAT (pITC).
FIG. 1.
Schematic diagrams of D-RNAs. All of the D-RNAs were constructed with the IBV D-RNA CD-61 cDNA in pIBV-Vec (17) by insertion of the appropriate sequence in the restriction sites SnaBI, StuI, and PmaCI. The positions of the three restriction sites are shown in the CD-61 sequence (light grey). The sizes of the various D-RNAs are displayed in parentheses. The boxes represent the IBV Beaudette gene 5 TAS (TGTTTTACTTAACAAAAACTTAACAAA) (black), the EMCV IRES (vertical lines), and the CAT reporter gene (diagonal lines).
Plasmid pPIC was digested with KpnI (within CD-61) and SpeI (within EMCV IRES) to remove a 1,744-bp fragment comprising 1,286 bp of CD-61 and 458 bp of the EMCV IRES, which was inserted into pBluescript, producing pBluescript-PIC. Oligonucleotides gene5+ve and gene5−ve were annealed to create a 36-bp adapter, consisting of the IBV Beaudette gene 5 TAS and sequence between the TAS and the ORF 5a start codon, which was inserted into the Eco47III site proximal to the IRES sequence in pBluescript-PIC, producing pBluescript-TAS-PIC. A 1,780-bp SpeI-KpnI fragment was removed from pBluescript-TAS-PIC and used to replace the 1,744-bp SpeI-KpnI fragment in pPIC, producing pTAS-IRES-CAT (pTIC). The 36-bp adapter was inserted into the SnaBI site of pCD-61-CAT and pPIC, producing pCD-61-CAT-TAS-SnaBI (pCCT) and pPIC-TAS-SnaBI (pPT), respectively. Eco47III-digested EMCV IRES from pCD-61-PmaCI-IRES was religated back into the D-RNA sequence in the antisense orientation, producing pCD-61-IRESneg. Sequence analysis was used to confirm that the various sequences had been inserted correctly.
In vitro transcription/translation.
An in vitro transcription/translation reaction was performed with a rabbit reticulocyte lysate-coupled transcription and translation cell-free protein synthesis system (Promega). Plasmid DNA (1 μg) was added to 25 μl of rabbit reticulocyte lysate, 2 μl of reaction buffer, 1 μl of T7 RNA polymerase (15 U/μl), 2 μl of 1 mM amino acid mixture, and 1 μl of RNasin RNase inhibitor (40 U/μl) in a final reaction volume of 50 μl. Each transcription/translation reaction mixture was incubated at 30°C for 1 h.
In situ transcription/translation.
CK cells (70 to 80% confluent) in 25-cm2 flasks were infected with FPV/T7 at a multiplicity of infection of 10 and incubated at 37°C for 2 h. The initial inoculum was replaced with 3 ml of CK cell media, and the cells were incubated at 37°C for an additional 2 h. The cells were transfected with 2 μg of plasmid DNA, with Lipofectin transfection reagent (Invitrogen), and incubated in Opti-MEM serum-free medium (Invitrogen) at 37°C for 20 h.
Helper IBV rescue of D-RNAs.
In vitro T7-derived transcripts were synthesized from 1 μg of plasmid DNA and electroporated into IBV Beaudette-infected CK cells (P0) (41). Progeny virus (V1) and any D-RNA from 1 ml of the P0 CK cell medium were passaged on CK cells (P1); this was repeated for two more passages (P2 to P3).
Identification of IBV-derived RNAs.
Total cytoplasmic RNA was extracted from IBV-infected CK cells with RNeasy (Qiagen), electrophoresed in denaturing 1% agarose-2.2 M formaldehyde gels, and transferred to Hybond XL nylon membranes (Amersham). IBV-derived RNAs were detected by hybridization to a 309-bp IBV probe, covalently labeled with psoralen-biotin (BrightStar; Ambion), corresponding to the last 309 nucleotides at the 3′ end of the IBV genome (17). RNAs containing the EMCV IRES were detected by using a 573-bp Eco47III DNA fragment, covalently labeled with psoralen-biotin, corresponding to the EMCV IRES from pPIC. Hybridized probes were detected by using streptavidin-alkaline phosphatase as described in references 17 and 18.
Analysis of CAT protein.
The CAT protein was detected by enzyme-linked immunosorbent assay (ELISA) (Boehringer Mannheim). Following rescue of D-RNAs in CK cells, approximately 2 × 106 cells were scraped from the plastic into the cell culture medium, pelleted by centrifugation at 2,500 rpm (Beckman GS-15R), and lysed in 1 ml of lysis buffer (Boehringer Mannheim) (40). For detection of CAT protein in the cell-free reactions, 150 μl of lysis buffer was added to each 50 μl of the in vitro reaction mixture sample. Following the addition of lysis buffer, all of the samples were incubated at room temperature for 30 min and the CAT protein was detected by ELISA (40). Serial dilutions of the samples were made, and the amounts of CAT protein were determined by comparison to standard amounts of CAT protein.
RESULTS
Insertion of EMCV IRES into IBV D-RNAs.
IBV D-RNA CD-61 has been utilized as an IBV-based expression vector following the insertion of heterologous genes, CAT, Luc, and chicken IFN-γ, under the control of the IBV Beaudette gene 5 TAS (17, 18, 40). Due to the instability of coronavirus D-RNAs as expression vectors, we decided to investigate an alternative expression system in which a picornavirus IRES sequence was chosen to initiate translation of a heterologous gene without the requirement of a D-RNA-derived sgmRNA. The EMCV IRES sequence was selected for its efficiency to initiate translation in vitro and ability to initiate translation in various cell types (6, 7).
A series of D-RNAs, based on IBV CD-61, were produced to investigate (i) the stability of EMCV IRES-containing D-RNAs and (ii) IRES-mediated heterologous gene translation from IBV D-RNAs. All of the IBV D-RNA sequences (Fig. 1) within pIBV-Vec (17) were constructed by using three restriction sites in CD-61, SnaBI and StuI within domain II and PmaCI within domain III (33, 40). The SnaBI and StuI sites are within the D-RNA-specific ORF (33). Previous results had demonstrated that the expression of reporter genes from the SnaBI site did not affect the stability of the D-RNA any more than expression of reporter genes from the PmaCI site within domain III, which is not within the D-RNA-specific ORF (18, 40). We have expressed a variety of heterologous genes, CAT (40), Luc (17) and chicken IFN-γ (18), from the PmaCI site under the control of TAS-directed coronavirus transcription. The introduction of these sequences did affect the stability of the D-RNAs. However, we were able to detect expression of the genes up to passage 10 (17, 18, 40). The IRES-CAT sequences were inserted into the two different positions (StuI and PmaCI) within D-RNA CD-61 to determine whether there was any position-specific effect of the IRES sequence on either expression of CAT or stability of the resultant D-RNA. D-RNA CD-61-CAT, based on pIBV-Vec, was essentially identical to the D-RNA described in reference 40, which was shown to be capable of initiating CAT gene expression during D-RNA rescue, except that the IBV D-RNA cDNA was terminated by the HδR-T7φ sequence. D-RNAs StuI-IRES-CAT (SIC) and PmaCI-IRES-CAT (PIC) contained the CAT gene, under control of the EMCV IRES, in the StuI and PmaCI sites of CD-61, respectively. D-RNAs IRES-TAS-CAT (ITC) contained the IBV TAS between the IRES and CAT gene sequences, and TAS-IRES-CAT (TIC) contained the IBV TAS immediately proximal to the EMCV IRES sequence. D-RNAs ITC and TIC were constructed to ascertain whether an IRES sequence, in conjunction with an IBV TAS, could initiate CAT protein expression and whether expression of CAT, under both IRES and TAS control, would be more efficient then either alone. D-RNAs CD-61-CAT-TAS-SnaBI (CCT) and PIC-TAS-SnaBI (PT) contained a TAS sequence in the SnaBI site upstream of the TAS-CAT and IRES-CAT sequences, respectively, and were constructed to ascertain the effect of a distant distal TAS on CAT gene expression.
In vitro assessment of IRES-mediated CAT translation.
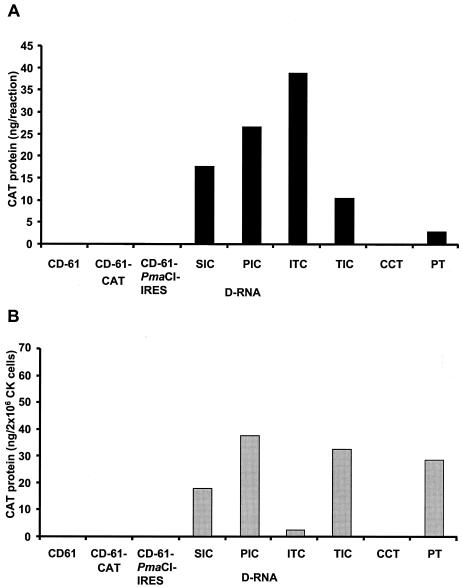
In vitro cell-free rabbit reticulocyte lysate-coupled transcription and translation assays were performed to determine whether IRES-mediated CAT translation could occur from IBV D-RNAs. Plasmids representing the IRES-containing D-RNAs SIC, PIC, ITC, TIC, and PT all resulted in detectable levels of CAT protein (Fig. 2A). In contrast, plasmids representing D-RNAs CD-61-CAT and CCT, containing the TAS-CAT cassette, and plasmids representing D-RNAs CD-61 and CD-61-PmaCI-IRES, which did not contain the CAT gene, did not, as expected, produce CAT protein (Fig. 2A). This verified that the CAT protein expressed from D-RNAs SIC, PIC, ITC, TIC, and PT was not due to ribosomal scanning from the 5′ end of the T7-derived D-RNA transcripts. CAT protein expressed from the in vitro T7-transcribed D-RNAs resulted from EMCV IRES-mediated translation.
FIG. 2.
Detection of CAT protein expressed from D-RNAs. (A) The IBV D-RNAs were transcribed and translated in vitro with the rabbit reticulocyte-coupled TNT cell-free protein synthesis system. (B) Plasmid DNAs, representing the various IBV D-RNAs, were transfected into FPV/T7-infected CK cells for the in situ synthesis of the T7-derived D-RNA transcripts and concomitant translation of CAT protein. CAT protein was detected by ELISA and quantified with standard amounts of CAT protein. The amounts of CAT protein detected in vitro were from 1 μg of plasmid DNA. The amounts of CAT protein detected in situ were determined following the potential transfection of 2 × 106 CK cells.
In situ assessment of IRES-mediated CAT translation.
To determine whether the EMCV IRES could function in primary CK cells, plasmids representing the various D-RNAs were transfected into FPV/T7-infected CK cells for the in situ expression of T7-derived D-RNA transcripts. CAT protein was detected from plasmids representing the IRES-containing D-RNAs, SIC, PIC, ITC, TIC, and PT (Fig. 2B). In contrast, no detectable levels of CAT protein were identified, as expected, from plasmids representing D-RNAs CD-61, CD-61-CAT, CD-61-PmaCI-IRES, and CCT, or from mock-infected CK cells transfected with pPIC or pCD-61-CAT (Fig. 2B). This demonstrated that detection of CAT protein in situ did not result from ribosomes binding to the 5′ end of the T7-derived D-RNA transcripts and scanning through to the CAT gene. These results confirmed the in vitro cell-free expression results and demonstrated that the EMCV IRES sequence was capable of initiating CAT protein translation from within IBV D-RNAs in avian CK cells. The presence of the IBV TAS in D-RNAs TIC, ITC, and PT did not prevent IRES-mediated CAT translation in situ (Fig. 2B). However, the amounts of CAT protein detected in CK cells from D-RNA ITC were 6.1-, 16.3-, 12.8-, and 12.4-fold lower than the amounts of CAT protein detected from D-RNAs SIC, PIC, TIC, and PT, respectively. This observation indicated that the IBV TAS, between the IRES and the CAT gene, had some detrimental effect on IRES-mediated translation in CK cells when compared to CAT expression following in vitro translation (Fig. 2A).
Assessment of IRES-mediated CAT translation following IBV helper virus-dependent rescue of the D-RNAs.
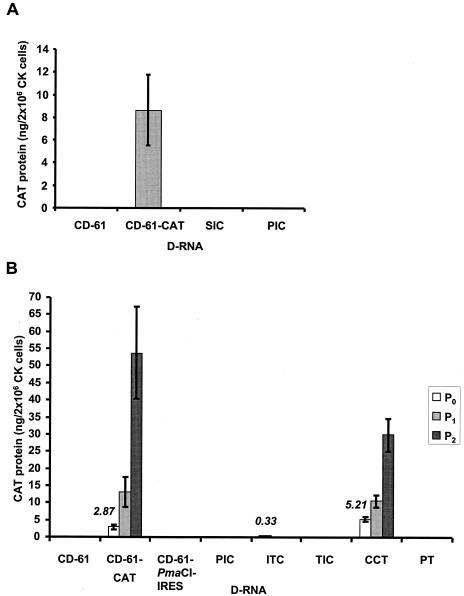
Both in vitro and in situ translation of the IRES-CAT-containing D-RNAs had demonstrated expression of CAT protein, indicating that the EMCV IRES was functional within a T7-derived transcript of the D-RNAs. Therefore, in vitro T7-derived transcripts of D-RNAs CD-61, CD-61-CAT, SIC, and PIC were electroporated into IBV Beaudette-infected CK cells (P0), and cell lysates were assayed for the presence of CAT protein. Only D-RNA CD-61-CAT resulted in the expression of CAT protein; no CAT protein was expressed from the IRES-CAT-containing D-RNAs SIC and PIC (Fig. 3A). This observation was in contrast to the in vitro and in situ expression results in which no CAT protein was detected from the CAT gene under the control of the IBV TAS but CAT, under the control of the EMCV IRES, was detected (Fig. 2). This indicated that EMCV IRES-mediated translation may not occur in the presence of IBV. The observation that neither D-RNA SIC or PIC resulted in IRES-mediated CAT translation indicated that there was no position-specific effect of the IRES within the D-RNA sequence.
FIG. 3.
Detection of CAT protein in IBV-infected CK cells following rescue of D-RNAs. (A) IBV-infected CK cells (P0) were electroporated with in vitro T7-derived D-RNAs transcripts, lysed, and analyzed for the presence of CAT protein. The bars represent the means of the results from five separate transfection reactions (± standard deviations). (B) IBV-infected CK cells (P0) were electroporated with in vitro T7-derived D-RNAs transcripts, and progeny virus and any packaged D-RNAs were serially passaged twice on CK cells (P1 to P2). The cells were lysed and analyzed for the presence of CAT protein. The histogram bars represent P0 (open bar), P1 (light grey), and P2 (dark grey) CK cells, with each value representative of the mean of the results from three electroporation reactions (± standard deviation). The amounts of CAT protein detected in P0 CK cells containing D-RNAs CD-61-CAT, ITC, and CCT are indicated in italics. CAT protein was detected and quantified by ELISA following the potential infection and electroporation of 2 × 106 CK cells.
To further investigate IRES-mediated CAT translation in the presence of IBV helper virus, in vitro T7-transcribed D-RNAs were electroporated into Beaudette-infected CK cells and any progeny virus and D-RNA were serially passaged twice in CK cells (P1 and P2). CK cell lysates (P0 to P2) were assayed for the presence of CAT protein. With the exception of D-RNA ITC, none of the IRES-containing D-RNAs produced detectable amounts of CAT protein in the presence of helper IBV (Fig. 3B). The two TAS-CAT-containing D-RNAs, CD-61-CAT and CCT, resulted in the expression of increasing amounts of CAT protein following rescue (replication and packaging) on serial passage of the D-RNAs as previously observed (40). D-RNA ITC, containing the IBV TAS sequence between the IRES and CAT gene sequences, resulted in expression of CAT protein only in P0 CK cells (Fig. 3B). The amounts of CAT protein detected from D-RNA ITC in P0 CK cells were 8.7- and 15.8-fold lower than the amounts of CAT protein detected in P0 CK cells from D-RNAs CD-61-CAT and CCT, respectively. This indicated that although some replication of D-RNA ITC may have occurred in the P0 CK cells, either insufficient amounts of D-RNA were produced for a successful rescue event or the IRES sequence affected rescue of the D-RNA. No IRES-mediated CAT translation was detected in IBV-infected CK cells electroporated with D-RNA PIC at any passage, indicating that potential amplification, following replication, did not result in the expression of CAT protein from this D-RNA, confirming the P0 results (Fig. 3A). Overall, the results indicated that IRES-mediated CAT translation did not occur, from two different positions within the D-RNA sequence, in the presence of IBV helper virus.
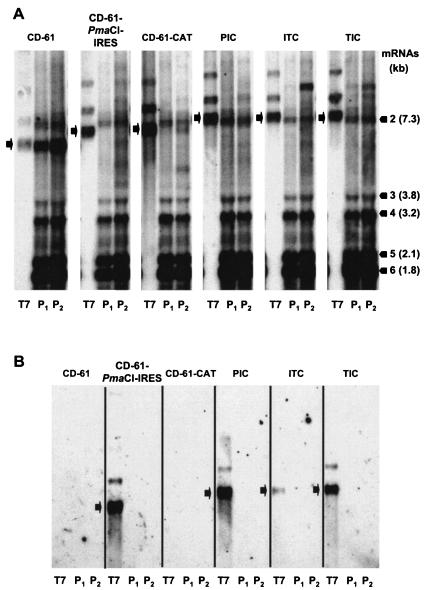
The failure of IRES-mediated CAT translation may have resulted from inhibition of replication of the IRES-containing D-RNAs. The observation that D-RNA ITC did result in the expression of some CAT protein in P0 CK cells indicated either that some replication of the D-RNA occurred or that TAS-mediated sgmRNA synthesis, from the TAS immediately proximal to the CAT gene, may have occurred. To determine whether the IRES-containing D-RNAs replicated in the presence of helper IBV, RNA was isolated from P1 and P2 CK cells potentially containing D-RNAs CD-61, CD-61-CAT, CD-61-PmaCI-IRES, PIC, ITC, and TIC and analyzed by Northern blot analysis. The presence of D-RNAs following rescue of the D-RNAs would be indicative of D-RNA replication.
D-RNA CD-61 was detected in RNA from P1 and P2 CK cells and D-RNA CD-61-CAT in RNA from P2 CK cells (Fig. 4A). This demonstrated that both D-RNAs were rescued by helper IBV, although CD-61-CAT was rescued less efficiently, due to the presence of the CAT gene, as previously observed (40). None of the IRES-containing D-RNAs, CD-61-PmaCI-IRES, PIC, ITC, and TIC, were detected in P1 and P2 CK cells. All of the RNA samples analyzed contained the IBV sgmRNAs, confirming that helper IBV was present. However, analysis of the in vitro T7-transcribed D-RNAs PIC, ITC, and TIC, used for the electroporation of IBV-infected CK cells, showed that they were of a similar size to IBV sgmRNA 2. Any potential rescue of the D-RNAs could have been masked by the IBV sgmRNA. Therefore, the RNAs were analyzed with the EMCV IRES-specific probe. The in vitro T7-derived D-RNA transcripts containing the EMCV IRES sequence were detected, but D-RNAs CD-61-PmaCI-IRES, PIC, ITC, and TIC were not detected in the P1 and P2 CK cells (Fig. 4B). These results showed that none of the IRES-containing D-RNAs were rescued by helper IBV, indicating that the IRES sequence was detrimental to rescue of the D-RNAs, most likely by inhibition of D-RNA replication.
FIG. 4.
Northern blot analysis of RNA extracted from IBV-infected CK cells following rescue of D-RNAs. IBV-infected CK cells (P0) were electroporated with in vitro T7-derived D-RNAs transcripts, and progeny virus and any packaged D-RNAs were serially passaged twice on CK cells (P1 to P2). RNA from the CK cells and samples of the in vitro T7-transcribed D-RNAs were separated by electrophoresis in 1% denaturing agarose gels and transferred to nylon membranes. RNA was hybridized with either an IBV-specific probe capable of detecting IBV gRNA, sgmRNAs, and D-RNAs (A) or an EMCV IRES-specific probe (B). The black bars indicate the positions and sizes of the IBV Beaudette sgmRNAs. The black arrows indicate the positions of either the in vitro T7-transcribed D-RNAs or the IBV-rescued D-RNAs in P1 and P2 CK cells. The RNAs detected between sgmRNAs 4 and 5 are observed routinely for all strains of IBV, as originally identified by Stern and Kennedy (39), and are of unknown origin. The T7-derived D-RNA transcripts used for electroporation (lanes T7) of IBV-infected CK cells contained larger RNA species of lower intensity, attributed to T7 polymerase read-through of the T7φ sequence (4, 30).
The observation that CD-61-PmaCI-IRES was not rescued by helper IBV indicated that presence of the IRES sequence alone, rather than as a translational unit, prevented helper-virus-dependent rescue of the IRES-containing D-RNAs. Detection of CAT protein in P0 CK cells containing D-RNA ITC most probably resulted from TAS-controlled CAT expression via the synthesis of a CAT sgmRNA from the D-RNA. Our results indicated that none of the other IRES-containing D-RNAs were replicated; therefore, it is unlikely that ITC was replicated. There was no evidence that D-RNA TIC expressed CAT protein, even though this was similar to ITC, except that the TAS was proximal to the IRES sequence rather than to the CAT gene sequence. This indicated that the IRES sequence may affect TAS-controlled sgmRNA synthesis as well as D-RNA replication.
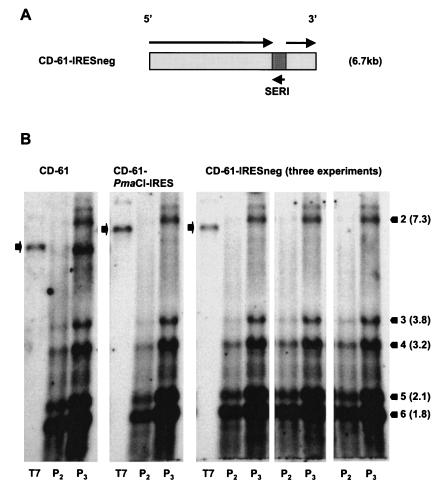
Rescue of D-RNA CD-61-IRESneg containing the EMCV IRES sequence in antisense orientation.
The above results showed that the IRES sequence was deleterious for D-RNA helper-virus-dependent rescue. This effect may have resulted from the IRES sequence destabilizing the D-RNA; the presence of heterologous sequences in coronavirus D-RNAs result in their eventual loss (2, 17, 18, 26, 27, 40, 44, 45). Alternatively, the IRES secondary structure may have affected replication of the D-RNA by inhibiting or reducing the efficiency of the IBV replicase complex, or recruitment of ribosomes by the IRES may have affected the IBV replicase, resulting in insufficient amounts of D-RNA to effect a successful rescue event.
A D-RNA, CD-61-IRESneg, containing the EMCV IRES in an antisense orientation was constructed (Fig. 5A). The EMCV IRES is unable to initiate translation in an antisense orientation (23, 27, 34). CD-61-IRESneg was used to determine whether the potential effect on the IBV replicase was due to an intrinsic property of the IRES sequence or whether the recruitment of ribosomes to the D-RNA was preventing efficient replication of the IRES-containing D-RNAs.
FIG. 5.
Analysis of the rescue of D-RNA CD-61-IRESneg. (A) Schematic diagram of D-RNA CD-61-IRESneg containing the EMCV IRES sequence in an antisense orientation (SERI). Black arrows represent the orientations of the respective sequences. (B) Northern blot analysis of RNA following rescue of the D-RNAs. The rescue of CD-61-IRESneg was performed in triplicate. IBV-infected CK cells (P0) were electroporated with in vitro T7-derived D-RNA transcripts, and progeny virus and any packaged D-RNAs were serially passaged three times (P1 to P3). RNA from P2 to P3 CK cells and in vitro T7-transcribed D-RNAs (lanes T7) were separated by electrophoresis in a 1% denaturing agarose gel, transferred to nylon membranes, and hybridized with an IBV-specific probe. The black bars indicate the positions and sizes of the IBV Beaudette sgmRNAs. The black arrows indicate the positions of the in vitro T7-transcribed D-RNAs and the potential positions of the rescued D-RNAs in P2 and P3 CK cells.
In vitro T7-derived transcripts of D-RNAs CD-61, CD-61-PmaCI-IRES, and CD-61-IRESneg were electroporated into IBV-infected CK cells. Progeny virus and any replicated D-RNAs were passaged three times (P1 to P3) in CK cells, and RNA was extracted from P2 and P3 cells and analyzed by Northern blot analysis. Analysis of the RNAs (Fig. 5B) showed that D-RNA CD-61 was rescued, with the highest amounts coming from P3 CK cells. As anticipated, D-RNA CD-61-PmaCI-IRES was not rescued. D-RNA CD-61-IRESneg was not detected in RNAs from P2 and P3 CK cells, performed in three separate experiments (Fig. 5B), demonstrating that IBV was unable to rescue a D-RNA with the EMCV IRES sequence in the antisense orientation. These results demonstrated that the recruitment of ribosomes by the IRES sequence was not responsible for affecting rescue of the IRES-containing D-RNAs. Therefore, the inability of IBV to rescue the IRES-containing D-RNAs must have resulted from some intrinsic property of the IRES sequence, most likely by having a detrimental effect on replication of the D-RNAs.
DISCUSSION
Expression of a heterologous gene from an IBV D-RNA, under the control of an IBV TAS, during helper-virus-dependent rescue requires D-RNA replication and synthesis of a D-RNA-derived sgmRNA and resulted in maximum protein expression by P4 (15, 17, 18, 40). Several rounds of passage were required to achieve maximal expression of the gene, but this resulted in eventual loss of the D-RNA. The expression of two heterologous genes, CAT (40) and chicken IFN-γ (18), was not affected by the position of the insert within the sequence of D-RNA CD-61, indicating that there was no position-specific effect of the heterologous genes at the two positions used. One of the positions, SnaBI, is within domain II of CD-61 and interrupts the D-RNA-specific ORF, resulting from in-frame deletions during the generation of CD-61. The second site, PmaCI, is within domain III and does not interrupt the CD-61-specific ORF. The PmaCI site has been our preferred site for the insertion of heterologous genes. We decided to explore an alternative expression system by using the EMCV IRES to initiate internal translation from within a D-RNA. Such an approach would circumvent the necessity of having to generate TAS-mediated D-RNA-derived sgmRNAs for the expression of the required protein and would recruit the D-RNA as a template for translation. This would potentially increase initial expression levels without dependence on the efficiency of D-RNA-derived sgmRNA synthesis. The EMCV IRES has been used for cap-independent translation of many proteins and offers the opportunity to express proteins without the requirement of specialized transcription initiation sequences and signals.
The EMCV IRES was demonstrated to be capable of initiating CAT protein translation from T7-transcribed IBV D-RNAs both in a mammalian-derived cell-free protein synthesis system and in situ in FPV/T7-infected CK cells. This indicated that the IRES was capable of initiating translation in the context of an IBV D-RNA and was functional in avian cells. However, no IRES-mediated CAT protein synthesis was observed in the presence of helper IBV in CK cells from two independent positions within D-RNA CD-61. The loss of IRES-mediated translation during IBV helper virus rescue could result from several factors: (i) an intrinsic property of the IRES sequence preventing replication of the D-RNAs, possibly due to the secondary structure of the IRES inhibiting or reducing the replication efficiency of the IBV replicase complex; (ii) the position of the IRES sequence within the D-RNA may be critical for successful rescue of the D-RNA; (iii) the D-RNA was capable of being replicated, but the IRES sequence was inactivated, possibly resulting from perturbation of the IRES secondary structure by the IBV replicase complex; (iv) recruitment of ribosomes by the IRES sequence for initiation of protein synthesis, preventing replication of the D-RNA by the IBV replicase complex; and (v) requirement of an IBV TAS to delineate D-RNAs between replication and transcription. To determine which of these scenarios could have resulted in the lack of CAT expression following helper-virus-dependent rescue of the IRES-containing D-RNA, several different D-RNAs were produced and analyzed for CAT expression.
The expression of heterologous genes from coronavirus D-RNAs requires replication of the D-RNA and a TAS for synthesis of an sgmRNA. The mechanism of D-RNA replication is unclear. Previous results on the rescue of IBV D-RNAs utilizing different strains of IBV demonstrated that the D-RNAs are not replicated by reiterative replication but are replicated in a manner analogous to sgmRNA synthesis (41). The D-RNAs acquired their leader sequence from the helper virus genome, a process termed leader switching. Consequently, if coronavirus D-RNAs are replicated in a manner analogous to sgmRNA synthesis, it is unclear how sgmRNA synthesis occurs from the D-RNAs; negative-sense copies of sgmRNAs are not thought to be templates for smaller sgmRNAs. Naturally derived coronavirus D-RNAs do not contain TASs, therefore the insertion of TAS-controlled genes within a coronavirus D-RNA may be one reason why D-RNAs expressing a heterologous gene are intrinsically unstable. The levels of D-RNA-derived sgmRNAs have been found to be lower in comparison to the sgmRNAs synthesized from gRNA. Therefore, the presence of a TAS within the D-RNA may predestine some D-RNA molecules for sgmRNA synthesis, a possible reason for the low amounts of D-RNA-derived sgmRNAs, with the majority of D-RNA molecules being packaged into virions and unavailable for sgmRNA synthesis. Therefore, the loss of the TAS as a consequence of replacement with an IRES sequence may result in all of the D-RNAs being packaged and, in effect, translationally inactive. The inclusion of a TAS together with the IRES sequence might result in a proportion of such D-RNAs being translationally active. To test this hypothesis, a series of D-RNAs containing both the EMCV IRES sequence and the Beaudette gene 5 TAS were produced.
Initially, two D-RNAs, ITC with the TAS between the IRES and CAT gene and TIC with the TAS proximal to the IRES, were constructed. Secondly, a D-RNA, PT, with the TAS inserted 3.2 kb upstream of the IRES-CAT sequence, was constructed. Only D-RNA ITC was capable of expressing detectable levels of CAT protein in IBV-infected P0 CK cells but was not rescued by helper IBV. The amounts of CAT protein expressed were approximately 8.7- and 15.8-fold lower then the amounts from D-RNAs CD-61-CAT and CTT, respectively, both of which contained the CAT gene under the control of a TAS. D-RNA CTT contained a second TAS 3.2 kb upstream of the TAS-CAT sequence, indicating that the presence of a second TAS did not have a detrimental effect on TAS-controlled CAT expression. The inability to detect little or no CAT protein expression from D-RNAs ITC, TIC, and PT in the presence of helper IBV indicated that inclusion of a TAS with the IRES did not provide a mechanism for the expression of CAT protein.
The inability of helper IBV to rescue any of the IRES-containing D-RNAs indicated that the presence of the IRES sequence was detrimental to replication. This conclusion was supported by the inability to rescue CD-61-IRESneg, which contained the IRES sequence in the antisense orientation, eliminating recruitment of ribosomes by the IRES sequence, as being responsible for preventing replication of the D-RNAs. Therefore, the most probable reason for the inability to rescue the IRES-containing D-RNAs was inhibition of helper IBV-dependent replication of the D-RNAs as a result of some intrinsic property of the IRES sequence. As indicated above, this could be due to the IRES secondary structure inhibiting the IBV replicase complex for generation of either negative- or positive-sense transcripts or a combination of both. The alternative explanation, that IBV replicase complex could perturb the IRES structure preventing IRES-mediated translation, is unlikely, as this would have resulted in replication of the D-RNA without translation of CAT protein, whereas no replication of the IRES-containing D-RNAs was observed. We observed no position-specific effects of the IRES sequence; the IRES-CAT construct was found to be inactive in IBV-infected cells irrespective of its position in the CD-61 sequence.
From the results presented in this study, the most likely explanation for the lack of IRES-mediated CAT translation is the inability of the IBV replicase complex to replicate the IRES-containing D-RNAs. Previous coronavirus D-RNA expression vectors have displayed various levels of instability depending on the nature of the inserted sequence. Expression of the Luc reporter gene was more unstable and had a lower expression efficiency then β-glucuronidase or CAT reporter gene expression in transmissible gastroenteritis virus (TGEV) and IBV D-RNAs, respectively (21, 40). Similarly, expression of hemagglutinin-esterase and murine IFN-γ, when expressed from MHV D-RNAs, was not detected beyond P3 and P4, respectively (26, 44). These observations would imply that some aspect of any heterologous sequence, either the primary sequence, secondary structure, or some tertiary interaction, can adversely affect the stability of the D-RNA or simply that the D-RNAs need to revert to their most stable form. The loss of an IBV D-RNA-expressing chicken IFN-γ following serial passage was due to the production of new and presumably more stable D-RNAs (18). A similar observation was made for TGEV D-RNAs (1, 2). In all previous examples of the rescue of coronavirus D-RNAs containing heterologous gene sequences, some degree of replication was observed, indicating that the insertion sites of the heterologous sequences within the D-RNAs did not prevent replication of the D-RNA. There was no indication of any rescue of the IRES-containing D-RNAs or expression of CAT, except for D-RNA ITC, indicating that the IRES sequence did not simply have a destabilizing effect on replication but caused complete abolition of D-RNA replication.
The lack of IRES-controlled gene translation from IBV D-RNAs is contradictory to the results of Lin and Lai (27), who detected EMCV IRES-initiated CAT protein expression from the MHV D-RNA DissF during helper-virus-dependent rescue at both P0 and P1, although the D-RNA rescue efficiency was low. Following rescue of the D-RNA, CAT activity decreased 20-fold, accredited to a reduction in D-RNA packaging efficiency. However, another explanation, in light of the experiments conducted in this study, is that the IRES sequence was also detrimental to helper-virus-dependent replication of the MHV D-RNA. Lin and Lai (27) used an in situ approach for the rescue of the IRES-containing D-RNA utilizing vaccinia virus (vTF7-3) expressing T7 RNA polymerase. It was possible that vTF7-3 resulted in sufficient levels of the MHV D-RNA, rather than MHV-mediated replication; for low-efficiency rescue in P1 cells, there was no report as to whether the D-RNA was detected and expressed CAT in P2 cells.
Although our results indicate that the EMCV IRES had a detrimental effect on helper virus rescue of IBV D-RNAs, an equine arteritis virus (EAV) replicon (a self-replicating RNA), containing an EMCV IRES, has been shown to replicate and initiate translation of downstream genes (42, 43). EAV is an arterivirus belonging to a group of viruses related to coronaviruses and within the order Nidovirales which have a similar mechanism to coronaviruses for generating sgmRNAs. Interestingly, no replication of the EAV replicon was detected when the IRES and associated gene were inserted in the antisense orientation (43). A flavivirus (Kunjin) replicon, containing CAT under the control of the EMCV IRES, expressed CAT but replicated with reduced efficiency, 10- to 20-fold less, compared to a Kunjin replicon lacking the IRES CAT sequence (23). Insertion of the IRES CAT sequence in the antisense orientation did not affect replication, compared to the control, but did not express CAT protein. Although the Flaviviridae family and Nidovirales order have different replication strategies, it appears that the EMCV IRES in the antisense orientation can have different effects on replication of different positive-sense RNA viruses. The observations that the EMCV IRES functions when part of an arterivirus replicon but prevents replication of coronavirus D-RNAs indicates that the IRES sequence may have to be part of the gRNA or replicon RNA for successful replication and IRES-mediated translation. It will be interesting to establish whether an EMCV IRES will be tolerated in a coronavirus gRNA or replicon.
Acknowledgments
This work was supported by the Department of Environment, Food, and Rural Affairs (DEFRA) project code OD0712 and the Biotechnology and Biological Sciences Research Council (BBSRC). B.D. was the holder of an Institute for Animal Health Research Studentship.
REFERENCES
- 1.Alonso, S., A. Izeta, I. Sola, and L. Enjuanes. 2002. Transcription regulatory sequences and mRNA expression levels in the coronavirus transmissible gastroenteritis virus. J. Virol. 76:1293-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso, S., I. Sola, J. P. Teifke, I. Reimann, A. Izeta, M. Balasch, J. Plana-Duran, R. J. Moormann, and L. Enjuanes. 2002. In vitro and in vivo expression of foreign genes by transmissible gastroenteritis coronavirus-derived minigenomes. J. Gen. Virol. 83:567-579. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley and Sons, Inc., New York, N.Y.
- 4.Ball, A. L. 1992. Cellular expression of a functional nodavirus RNA replicon from vaccinia virus vectors. J. Virol. 66:2335-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belsham, G. J., and N. Sonenberg. 1996. RNA-protein interactions in regulation of picornavirus RNA translation. Microbiol. Rev. 60:499-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borman, A. M., J.-L. Bailly, M. Girard, and K. M. Kean. 1995. Picornavirus internal ribosome entry segments: comparison of translation efficiency and the requirements for optimal internal initiation of translation in vitro. Nucleic Acids Res. 23:3656-3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borman, A. M., P. Le Mercier, M. Girard, and K. M. Kean. 1997. Comparison of picornaviral IRES-driven internal initiation of translation in cultured cells of different origins. Nucleic Acids Res. 25:925-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Britton, P., P. Green, S. Kottier, K. L. Mawditt, Z. Pénzes, D. Cavanagh, and M. A. Skinner. 1996. Expression of bacteriophage T7 RNA polymerase in avian and mammalian cells by a recombinant fowlpox virus. J. Gen. Virol. 77:963-967. [DOI] [PubMed] [Google Scholar]
- 9.Casais, R., V. Thiel, S. G. Siddell, D. Cavanagh, and P. Britton. 2001. Reverse genetics system for the avian coronavirus infectious bronchitis virus. J. Virol. 75:12359-12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cavanagh, D. 2001. A nomenclature for avian coronavirus isolates and the question of species status. Avian Pathol. 30:109-115. [DOI] [PubMed] [Google Scholar]
- 11.Cavanagh, D., K. Mawditt, M. Sharma, S. E. Drury, H. L. Ainsworth, P. Britton, and R. E. Gough. 2001. Detection of a coronavirus from turkey poults in Europe genetically related to infectious bronchitis virus of chickens. Avian Pathol. 30:365-378. [DOI] [PubMed] [Google Scholar]
- 12.Cavanagh, D., K. Mawditt, D. d. B. Welchman, P. Britton, and R. E. Gough. 2002. Coronaviruses from pheasants (Phasianus colchicus) are genetically closely related to coronaviruses of domestic fowl (infectious bronchitis virus) and turkeys. Avian Pathol. 31:81-93. [DOI] [PubMed] [Google Scholar]
- 13.Cavanagh, D., and S. Naqi. 2003. Infectious bronchitis, p. 101-119. In Y. M. Saif, H. J. Barnes, J. R. Glisson, A. M. Fadly, L. R. McDougald, and D. E. Swayne (ed.), Diseases of poultry, 11th ed. Iowa State University Press, Ames.
- 14.Cook, J. K., J. Chesher, W. Baxendale, N. Greenwood, M. B. Huggins, and S. J. Orbell. 2001. Protection of chickens against renal damage caused by a nephropathogenic infectious bronchitis virus. Avian Pathol. 30:423-426. [DOI] [PubMed] [Google Scholar]
- 15.Dalton, K., R. Casais, K. Shaw, K. Stirrups, S. Evans, P. Britton, T. D. Brown, and D. Cavanagh. 2001. cis-Acting sequences required for coronavirus infectious bronchitis virus defective-RNA replication and packaging. J. Virol. 75:125-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Vries, A. A. F., M. C. Horzinek, P. J. M. Rottier, and R. J. de Groot. 1997. The genome organisation of the Nidovirales: similarities and differences between Arteri-, Toro- and Coronaviruses. Semin. Virol. 8:33-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans, S., D. Cavanagh, and P. Britton. 2000. Utilizing fowlpox virus recombinants to generate defective RNAs of the coronavirus infectious bronchitis virus. J. Gen. Virol. 81:2855-2865. [DOI] [PubMed] [Google Scholar]
- 18.Hackney, K., D. Cavanagh, P. Kaiser, and P. Britton. 2003. In vitro and in ovo expression of chicken gamma interferon by a defective RNA of avian coronavirus infectious bronchitis virus. J. Virol. 77:5694-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hellen, C. U., and P. Sarnow. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 15:1593-1612. [DOI] [PubMed] [Google Scholar]
- 20.Hiscox, J. A., K. L. Mawditt, D. Cavanagh, and P. Britton. 1995. Investigation of the control of coronavirus subgenomic mRNA transcription by using T7-generated negative-sense RNA transcripts. J. Virol. 69:6219-6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Izeta, A., C. Smerdou, S. Alonso, Z. Penzes, A. Mendez, J. Plana-Durán, and L. Enjuanes. 1999. Replication and packaging of transmissible gastroenteritis coronavirus-derived synthetic minigenomes. J. Virol. 73:1535-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jang, S. K., H.-G. Kräusslich, M. J. H. Nicklin, G. M. Duke, A. C. Palmenberg, and E. Wimmer. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khromykh, A. A., and E. G. Westaway. 1997. Subgenomic replicons of the flavivirus Kunjin: construction and applications. J. Virol. 71:1497-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krishnan, R., R. Y. Chang, and D. A. Brian. 1996. Tandem placement of a coronavirus promoter results in enhanced mRNA synthesis from the downstream-most initiation site. Virology 218:400-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai, M. M., and D. Cavanagh. 1997. The molecular biology of coronaviruses. Adv. Virus Res. 48:1-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liao, C.-L., X. Zhang, and M. M. C. Lai. 1995. Coronavirus defective-interfering RNA as an expression vector: the generation of a pseudorecombinant mouse hepatitis virus expressing hemagglutinin-esterase. Virology 208:319-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin, Y. J., and M. M. C. Lai. 1993. Deletion mapping of a mouse hepatitis virus defective interfering RNA reveals the requirement of an internal and discontiguous sequence for replication. J. Virol. 67:6110-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinez-Salas, E., R. Ramos, E. Lafuente, and S. Lopez de Quinto. 2001. Functional interactions in internal translation initiation directed by viral and cellular IRES elements. J. Gen. Virol. 82:973-984. [DOI] [PubMed] [Google Scholar]
- 29.Masters, P. S. 1999. Reverse genetics of the largest RNA viruses. Adv. Virus Res. 53:245-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pattnaik, A. K., L. A. Ball, A. W. LeGrone, and G. W. Wertz. 1992. Infectious defective interfering particles of VSV from transcripts of a cDNA clone. Cell 69:1011-1020. [DOI] [PubMed] [Google Scholar]
- 31.Pelletier, J., and N. Sonenberg. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334:320-325. [DOI] [PubMed] [Google Scholar]
- 32.Pénzes, Z., K. Tibbles, K. Shaw, P. Britton, T. D. K. Brown, and D. Cavanagh. 1994. Characterization of a replicating and packaged defective RNA of avian coronavirus infectious bronchitis virus. Virology 203:286-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pénzes, Z., C. Wroe, T. D. Brown, P. Britton, and D. Cavanagh. 1996. Replication and packaging of coronavirus infectious bronchitis virus defective RNAs lacking a long open reading frame. J. Virol. 70:8660-8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roberts, L. O., and G. J. Belsham. 1997. Complementation of defective picornavirus internal ribosome entry site (IRES) elements by the coexpression of fragments of the IRES. Virology 227:53-62. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 36.Sawicki, S. G., and D. L. Sawicki. 1990. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J. Virol. 64:1050-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sawicki, S. G., and D. L. Sawicki. 1998. A new model for coronavirus transcription. Adv. Exp. Med. Biol. 440:215-219. [DOI] [PubMed] [Google Scholar]
- 38.Siddell, S. G. 1995. The Coronaviridae, p. 1-10. In S. G. Siddell (ed.), The Coronaviridae. Plenum Press, New York, N.Y.
- 39.Stern, D. F., and S. I. T. Kennedy. 1980. Coronavirus multiplication strategy. I. Identification and characterization of virus-specified RNA. J. Virol. 34:665-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stirrups, K., K. Shaw, S. Evans, K. Dalton, R. Casais, D. Cavanagh, and P. Britton. 2000. Expression of reporter genes from the defective RNA CD-61 of the coronavirus infectious bronchitis virus. J. Gen. Virol. 81:1687-1698. [DOI] [PubMed] [Google Scholar]
- 41.Stirrups, K., K. Shaw, S. Evans, K. Dalton, D. Cavanagh, and P. Britton. 2000. Leader switching occurs during the rescue of defective RNAs by heterologous strains of the coronavirus infectious bronchitis virus. J. Gen. Virol. 81:791-801. [DOI] [PubMed] [Google Scholar]
- 42.Tijms, M. A., L. C. van Dinten, A. E. Gorbalenya, and E. J. Snijder. 2001. A zinc finger-containing papain-like protease couples subgenomic mRNA synthesis to genome translation in a positive-stranded RNA virus. Proc. Natl. Acad. Sci. USA 98:1889-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Dinten, L. C., H. van Tol, A. E. Gorbalenya, and E. J. Snijder. 2000. The predicted metal-binding region of the arterivirus helicase protein is involved in subgenomic mRNA synthesis, genome replication, and virion biogenesis. J. Virol. 74:5213-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, X., D. R. Hinton, D. J. Cua, S. A. Stohlman, and M. M. Lai. 1997. Expression of interferon-gamma by a coronavirus defective-interfering RNA vector and its effect on viral replication, spread, and pathogenicity. Virology 233:327-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, X., D. R. Hinton, S. Park, B. Parra, C.-L. Liao, M. M. Lai, and S. A. Stohlman. 1998. Expression of hemagglutinin/esterase by a mouse hepatitis virus coronavirus defective-interfering RNA alters viral pathogenesis. Virology 242:170-183. [DOI] [PMC free article] [PubMed] [Google Scholar]