Abstract
The nodavirus Flock house virus (FHV) has a bipartite, positive-sense RNA genome that is packaged into an icosahedral particle displaying T=3 symmetry. The high-resolution X-ray structure of FHV has shown that 10 bp of well-ordered, double-stranded RNA are located at each of the 30 twofold axes of the virion, but it is not known which portions of the genome form these duplex regions. The regular distribution of double-stranded RNA in the interior of the virus particle indicates that large regions of the encapsidated genome are engaged in secondary structure interactions. Moreover, the RNA is restricted to a topology that is unlikely to exist during translation or replication. We used electron cryomicroscopy and image reconstruction to determine the structure of four types of FHV particles that differed in RNA and protein content. RNA-capsid interactions were primarily mediated via the N and C termini, which are essential for RNA recognition and particle assembly. A substantial fraction of the packaged nucleic acid, either viral or heterologous, was organized as a dodecahedral cage of duplex RNA. The similarity in tertiary structure suggests that RNA folding is independent of sequence and length. Computational modeling indicated that RNA duplex formation involves both short-range and long-range interactions. We propose that the capsid protein is able to exploit the plasticity of the RNA secondary structures, capturing those that are compatible with the geometry of the dodecahedral cage.
High-resolution X-ray analysis of icosahedral viruses has provided detailed insights into the organization of the viral capsid and the structure of the individual coat protein subunits. The encapsidated nucleic acid, on the other hand, is rarely visualized, in part because it does not conform to the icosahedral symmetry of the virus particle. There are exceptions to this, however, as exemplified by the plant viruses bean pod mottle virus (BPMV) (3) and satellite tobacco mosaic virus (STMV) (10) and the invertebrate nodaviruses Pariacoto virus (PaV) (20) and Flock house virus (FHV) (7). In these viruses, regions of the encapsidated single-stranded RNA genome interact with coat protein subunits at symmetrically equivalent positions, and an average structure of these ordered regions has been visualized at high resolution.
BPMV, a T=3 icosahedral virus, has a bipartite positive-sense single-stranded RNA genome, with each RNA molecule packaged in a separate particle. In the crystal structure of particles containing RNA2 (∼3.6 kb), seven well-ordered ribonucleotides were visible near the icosahedral threefold axes of the virion (3). In addition, a total of 660 ribonucleotides corresponding to almost 20% of the packaged RNA could be modeled into the electron density. The overall structure of the ordered RNA is a single-stranded helix, which approximates that found for one strand of an A-type RNA duplex (3, 4). Because the RNA density in the X-ray map represents an average of the densities at symmetrically equivalent positions, the nucleotide sequence of the visualized RNA remains unknown. STMV, a T=1 icosahedral virus, packages a single strand of RNA containing 1,059 nucleotides. Up to 624 nucleotides, or 59% of the genome, were visualized in the 1.8-Å structure of the particle (9). In contrast to BPMV, these nucleotides form double-helical segments that structurally approximate A-form RNA. Using predictions for the position of the remaining nucleotides, a model of the packaged genome was proposed in which the RNA is folded into a series of stem-loops that are in intimate contact with the protein shell (9, 11).
The invertebrate viruses FHV and PaV are T=3 icosahedral viruses that contain a bipartite RNA genome. The single-stranded RNA1 and RNA2 molecules contain roughly 3,100 and 1,400 nucleotides, respectively, and are packaged into a single virion. The high-resolution X-ray structure of FHV revealed 10-bp fragments of RNA forming A-type duplexes at the icosahedral twofold contacts of the particle (7). This RNA accounts for approximately 600 ribonucleotides, or 13% of the encapsidated genome. The RNA duplexes observed at equivalent positions in PaV were significantly longer, comprising a stretch of 25 bp (20). Modeling of this duplex RNA into the density at the 30 twofold contacts gave the impression of a dodecahedral RNA cage that sits directly beneath the protein shell. This cage provides an average view of ∼35% of the encapsidated RNA.
The structural data confirmed earlier Raman spectral analyses of other viruses, which had indicated that as many as ∼60% of the bases in packaged, single-stranded RNA genomes are involved in hydrogen bonding (12). Given the fact that large portions of the encapsidated RNA are engaged in secondary structure interactions, the question arises whether these interactions are simply dictated by the primary sequence of the RNA or whether exogenous factors contribute to the final organization of the RNA in the virion. Thermodynamic considerations suggest that unassisted folding of the genome prior to assembly is highly unlikely, because it would give rise to numerous conformations, few of which would have the properties observed in the crystal structures of the particles. It is more likely that the folding process is guided by interaction with a cofactor, presumably the capsid protein.
In the work described here we have begun to address these issues by investigating the RNA structure in native and mutant FHV particles. Although assembly of FHV particles requires RNA, it is not dependent on the presence of viral RNAs 1 and 2. Particles containing heterologous RNAs can be generated using recombinant baculovirus expression vectors (16). We used electron cryomicroscopy (cryoEM) and image reconstruction to analyze four types of FHV particles that differed in their protein and RNA content. We found that encapsidated viral and heterologous RNAs have virtually identical dodecahedral structures adjacent to the capsid protein, indicating that the FHV coat protein and not the sequence of the nucleic acid controls the organization of this region of the packaged RNA. An N-terminal basic segment of the coat protein known to interact with the packaged genome is not required for this function.
MATERIALS AND METHODS
Preparation of virus particles.
Wild-type (wt) FHV was propagated in Drosophila melanogaster cells (Schneider's line 1) following standard procedures (18). Briefly, cells were suspended at 4 × 107 cells/ml in Schneider's insect medium supplemented with 15% fetal bovine serum. Gradient-purified FHV was added to a multiplicity of 5 to 10 PFU per cell and allowed to attach for 1 h at room temperature with occasional mild agitation. Infected cells (108; 2.5 ml) were then distributed into 100-mm tissue culture dishes containing 12.5 ml of growth medium. After incubation at 26°C for 48 h, cells were lysed with Nonidet P-40 and the cell debris was removed by low-speed centrifugation. Virus in the supernatant was pelleted through a 30% (wt/wt) sucrose cushion followed by sedimentation through a 10 to 40% (wt/wt) sucrose gradient. FHV particles were collected from the gradient tube by inserting a needle below the viral band and aspirating the material into a 1-ml syringe. Δ31 FHV was generated by cotransfecting Drosophila cells with wt RNA1 and in vitro-synthesized capped transcripts of RNA2 that lacked the coding sequence for amino acids 2 to 31. Details of this procedure have been described elsewhere (13). After incubation for 24 h at 26°C, the cells were lysed by freezing and thawing and cell debris was removed by low-speed centrifugation. Virus particles in the supernatant were used to infect fresh cells, and progeny Δ31 virions were isolated by sucrose gradient centrifugation after 48 h as described above. Virus-like particles (VLPs) of FHV containing either the wt coat protein or the Δ31 coat protein were generated in Spodoptera frugiperda cells (line IPLB-Sf21) using recombinant baculovirus vectors as described elsewhere (6, 16).
cryoEM and image analysis.
Preparation of frozen-hydrated specimens, cryoEM, and image analysis were performed as described previously (14, 21). Selected micrographs were digitized at a 21-μm interval using a Zeiss microdensitometer, corresponding to 4.7 Å on the specimen. A total of 450 to 650 images from several micrographs for each of the frozen samples were used for the reconstructions. The previously published map of FHV (5) at 22 Å resolution was used as the starting model for the Polar Fourier transform method (1). To optimize the search procedure, calculations were performed using only a portion of the Fourier transform of the masked image (between 1/70 Å−1 and 1/35 Å−1) to remove both low- and high-frequency noise. During iterative refinement, the high-frequency cutoff was progressively increased. The final reconstructions of the virions and their corresponding mutants were based on 122 to 137 particles. To assess the resolution of the maps, each data set was divided in half to compute two independent reconstructions. Cross-correlation analysis indicated that the effective resolution limit was 22 to 24Å, using a cutoff of 0.5. This was a conservative estimate, since the final map was computed from twice as many particles. All surface-shaded representations were visualized using AVS software (19). Contour levels were chosen to include the volume occupied by the capsid shell calculated from the number of the copies and the molecular weight of the coat protein. The volume occupied by the RNA was calculated on the basis of the sum of the molecular weights for the 4,500 nucleotides in RNA1 and RNA2. Contour levels were chosen to represent the volume of anhydrous RNA (∼1.4 × 106 Å3, using a partial specific volume of 0.55 cm3/g of dry RNA) rather than hydrated RNA (∼2.95 × 106 Å3, using a volume of 655 Å3/hydrated nucleotide).
RESULTS
RNA packaging phenotype of FHV particles.
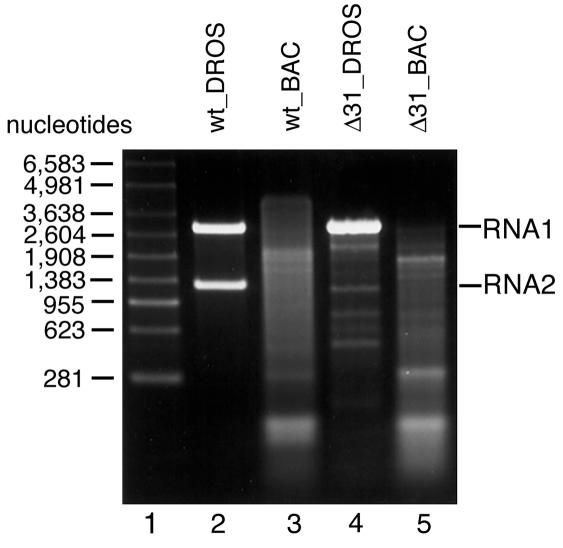
Four different types of FHV particles were examined in this study (Table 1). wt_DROS particles are native FHV particles purified from infected D. melanogaster cells. Native particles have T=3 icosahedral lattice symmetry and contain one copy of RNA1, the polymerase gene, and one copy of RNA2, the coat protein gene (Fig. 1, lane 2). wt_BAC particles were obtained by expression of wt FHV coat protein in S. frugiperda (Sf21) cells using a baculovirus vector. In this expression system, FHV coat protein spontaneously assembles into VLPs (16) whose protein capsid is crystallographically indistinguishable from that of native virions (unpublished results). However, the RNA content of wt_BAC particles consisted primarily of cellular RNAs rather than viral RNAs 1 and 2 (Fig. 1, lane 3). Only minor amounts of the coat protein message, a derivative of RNA2, were contained in this mixture (8, 16). Δ31_DROS particles were assembled from a mutant coat protein that lacks N-terminal residues 2 to 31. These residues, which are disordered in the FHV X-ray structure, comprise a basic region thought to interact with the encapsidated RNA (6). The Δ31_DROS particles were generated by transfection of Drosophila cells with wt RNA1 and mutated RNA2, which encoded the N-terminally deleted capsid protein. Although the Δ31 coat protein assembled into particles whose structure is similar to that of wt_DROS particles, their RNA packaging phenotype differed from that of native virions (13). As shown in Fig. 1 (lane 4), Δ31_DROS particles contained normal levels of RNA1 but subnormal amounts of RNA2. Moreover, the particles contained RNA species not normally observed in FHV. Some of these represent cellular RNAs, and others represent defective interfering RNAs of RNA1 and RNA2 (13). Despite the difference in RNA packaging, the Δ31_DROS and wt_DROS particles sediment at a similar rate on sucrose gradients and band at the same density on CsCl gradients (13), indicating that the amount of RNA packaged per virion is similar to that of native particles. This is also suggested by the fact that the ratio of absorbance at 260 and 280 nm is indistinguishable from that of wt particles (13). Δ31_BAC particles were obtained by expression of the Δ31 protein in Sf21 cells using a recombinant baculovirus vector. In this expression system, the Δ31 protein assembles into multiple types of VLPs, a significant portion of which have the same size and shape as native virions (6). These particles were isolated from the mixture by sucrose gradient sedimentation and used in the present study. As in the case of wt_BAC particles, Δ31_BAC particles contained primarily cellular RNAs (Fig. 1, lane 5).
TABLE 1.
Protein and RNA contents of FHV particles used in the present study
| Particle | Capsid protein | RNA contents |
|---|---|---|
| wt_DROS | wt | FHV RNA1 and RNA2 |
| wt_BAC | wt | Mostly cellular RNA and minor amounts of RNA2 |
| Δ31_DROS | Lacks residues 2 to 31 | FHV RNA1 and minor amounts of RNA2 and other RNAs |
| Δ31_BAC | Lacks residues 2 to 31 | Mostly cellular RNA and minor amounts of Δ31 RNA2 |
FIG. 1.
Electrophoretic analysis of RNA extracted from the four types of FHV particles used in this study. RNA was extracted with phenol-chloroform from gradient-purified particles, and 1.0 to 1.5 μg was separated by electrophoresis through a nondenaturing 2% agarose-Tris-acetate-EDTA gel. Nucleic acids were visualized with ethidium bromide. Lane 1, RNA molecular size markers; lane 2, RNA from wt FHV particles (wt_DROS); lane 3, RNA from wt VLPs (wt_BAC); lane 4, RNA from Δ31 FHV particles (Δ31_DROS); lane 5, RNA from Δ31 VLPs (Δ31_BAC).
cryoEM and image reconstruction.
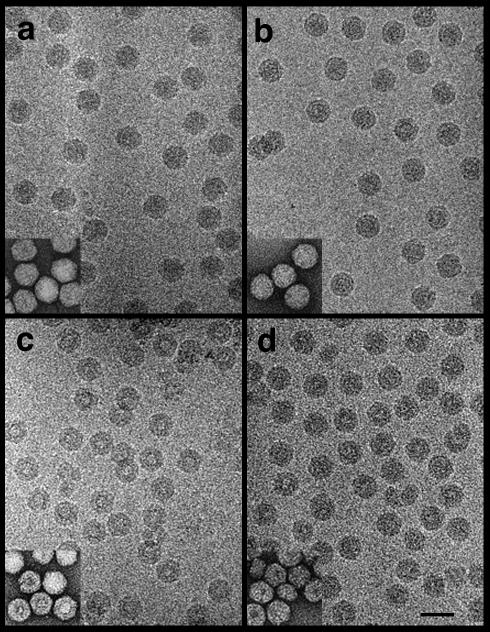
cryoEM of frozen-hydrated samples showed that the particles in each group had the expected hexagonal and circular profiles and a diameter of ∼340 Å (Fig. 2). This value was in good agreement with the maximum diameter of native FHV particles (350 Å) as determined by X-ray crystallography (7). In contrast to wt_DROS and wt_BAC particles, the corresponding Δ31 mutants showed some variation in the hexagonal profiles, suggesting that they might be less stable. In addition, the center of many Δ31 particles was considerably less dense than that of virions containing wt coat protein, consistent with the Δ31 particles being more stain permeable than particles containing the full-length coat protein (Fig. 2, insets).
FIG. 2.
Electron cryomicrographs of frozen-hydrated wt_DROS (a), wt_BAC (b), Δ31_DROS (c), and Δ31_BAC (d) particles. Note the constant diameter of the particles and distinctive circular and hexagonal profiles. Δ31_DROS and Δ31_BAC particles show an absence of density in the center of the particles. Inset: particles from the same preparation negatively stained with uranyl acetate. Bar = 500 Å.
Three-dimensional (3D) image reconstructions at 22 to 24 Å resolution showed a striking conservation in overall size, shape, and surface topography of the four different types of particles (Fig. 3). Closer inspection, however, revealed that the Δ31_BAC particles were somewhat smoother than the other types of particles, possibly due to increased conformational flexibility and associated lower resolution. This would agree with previous observations, which revealed that changes in the RNA content of FHV particles can affect their dynamic behavior (2). For example, wt_BAC particles, which contain mostly cellular RNA, are more dynamic in solution than wt_DROS particles containing FHV RNAs 1 and 2 (2), even though there are no differences in the high-resolution X-ray structures of the respective protein capsids in the crystalline state (unpublished results). Since Δ31_BAC particles are formed by mutant coat protein and random RNA, their structural flexibility might be further increased.
FIG. 3.
3D surface-shaded reconstructions of wt_DROS (green), Δ31_DROS (yellow), wt_BAC (blue), and Δ31_BAC (red) reveal solid protein capsids with nearly identical surface topographies. The particles appear hexagonal when viewed down the twofold vertex positions with a radius of ∼173 Å. Bar = 100 Å.
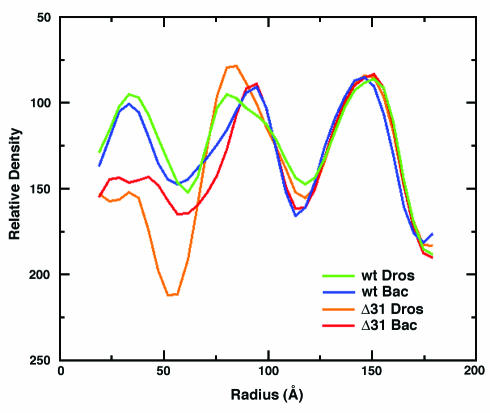
Spherically averaged, radial density plots computed from the 3D reconstructions revealed variations in the number of peaks for each particle. While the profiles of wt_DROS and wt_BAC particles contained three peaks of density, those of Δ31_DROS and Δ31_BAC contained only two peaks (Fig. 4). When the plots were superimposed, there was close correspondence in the peaks centered at a radius of 142 Å, suggesting little if any variance in the overall architecture of the protein shell as expected from the 3D maps (Fig. 3). All four types of particles also contained a conserved shell of density ascribed to RNA adjacent to that of the capsid. The contour level of the interior density was adjusted to correspond to the volume of 4,500 nucleotides (∼1.4 × 106 Å3), assuming that RNA1 and RNA2 were in the anhydrous state. The enclosed volume would be even larger if contoured for hydrated RNA. Thus, the observed density suggested full occupancy by the encapsidated RNA.
FIG. 4.
Spherically averaged radial density plots of the 3D reconstructions of wt_DROS, wt_BAC, and the corresponding Δ31 mutants. All four particles have almost the same outer diameter (∼340 Å). The peaks centered at ∼32 and ∼90 Å are ascribed to RNA, whereas the peak centered at ∼142 Å corresponds to the protein shell. The central RNA peak is much broader for wt_DROS and Δ31_DROS (green and yellow lines) than for wt_BAC and Δ31_BAC (blue and red lines). Particles containing wt coat protein (wt_DROS and wt_BAC; green and blue lines) show additional RNA centered at ∼32 Å. This was not observed for the corresponding Δ31 mutants.
There was significant variation in the thickness and position of the peaks representing RNA adjacent to that of the capsid. For example, in wt-BAC and Δ31_BAC, the peaks were comparatively narrow and the maximum density value was observed at a radius of ∼95 Å. However, for wt_DROS and Δ31_DROS the peaks were noticeably broader and the maximum density was shifted towards a radius of ∼85 Å. Most surprisingly, particles assembled from wt coat protein, i.e., wt_DROS and wt_BAC, showed an additional shell of density centered at ∼32Å. No density was detected for the corresponding Δ31 particles at this radius. We ascribe this interior density to additional ordered RNA, which is dependent on the presence of the flexible N terminus of the capsid protein.
Sectioned views of density.
Sectioned views of the density maps (Fig. 5) showed a distinct gap between most of the capsid density and the RNA density. The overall distribution of RNA density was similar, but scrutiny of the maps revealed differences. For instance, the RNA density in the DROS particles was more angular than for the BAC particles. The more spherical BAC RNA distribution gave rise to sharper peaks in the spherically averaged radial density profiles (Fig. 4). In contrast, the more angular RNA density in the DROS particles gave rise to broader peaks in the radial density profiles (Fig. 4) due to the larger distance along the twofold axes of symmetry compared with the fivefold axes of symmetry. There were also differences in the connectivity for the DROS versus the BAC particles (Fig. 5). The RNA density in the DROS particles was furrowed and disconnected, whereas the equivalent locations in the corresponding BAC particles displayed smoother and continuous tubes of density.
FIG. 5.
Surface-shaded, cross-sectional views of wt_DROS (green), Δ31_DROS (yellow), wt_BAC (blue), and Δ31_BAC (red). The internal density shown in gray scale is contoured to correspond to anhydrous RNA. Particles formed with wt capsid subunits contain two concentric shells of RNA, whereas particles formed from Δ31 subunits show a single shell of RNA. The absence of this internal RNA shell gives rise to particles that appear hollow in the electron micrographs (Fig. 2C and D). Particles formed from wt subunits also show close contacts with the RNA at the twofold symmetry axes (*). These contacts are absent in particles formed from the Δ31 mutant subunits. The RNA from particles assembled in Drosophila cells has a larger radius of ∼112 Å at the twofold symmetry axes and ∼84 Å at the fivefold symmetry axes, compared with ∼107 and ∼81 Å, respectively, for particles assembled in Sf21 cells using baculovirus expression vectors. Bar = 100Å.
Close contacts between the protein shell and the RNA were observed at the twofold symmetry axes in particles containing wt capsid protein but not in particles formed from the Δ31 subunits. In agreement with the radial density profiles (Fig. 4), a central shell of density was only visible in wt_DROS and wt_BAC particles. The lack of this internal density in the corresponding Δ31 particles accounts for the hollow appearance in the electron micrographs (Fig. 2C and D).
RNA organization.
Density that corresponded to the outer capsid shell was computationally removed from all four maps to reveal the internal density ascribed to encapsidated RNA. Remarkably, despite the considerable variation in the composition of the encapsidated nucleic acids, all four particles displayed tubes of density that formed a dodecahedral cage that we ascribed to well-ordered duplex RNA (Fig. 6). The rods of density were located at each of the 30 icosahedral twofold axes, a site where 10 bp of double-stranded RNA were visible in the high-resolution X-ray structure of native FHV (7). Compared to wt_DROS and Δ31_DROS particles, the rods forming the cage in wt_BAC and Δ31_BAC particles appeared thicker, especially where they merged at the threefold axes.
FIG. 6.
Surface-shaded representations of the internal density ascribed to RNA. The capsid density was removed to reveal internal density in all four maps ascribed to the encapsidated RNA. The solid contours correspond to the anhydrous RNA volume of 4,500 nucleotides (1.4 × 106 Å3), assuming a partial specific volume of 0.55 cm3/g for dry RNA. The rods of density are interpreted as ordered duplex RNA located at each of the 30 twofold axes. Additional rings of ordered density reside beneath the fivefold vertices. Bar = 50Å.
The cryoEM maps also showed RNA density beneath the fivefold vertices. With the exception of the Δ31_BAC particles, this density had the appearance of a solid or perforated pentagon. In wt_BAC and Δ31_DROS particles, threads of density extended to the RNA rods at the threefold axes. In contrast, the Δ31_BAC particles showed a distinct reorganization, with the density distributed into five spokes that radiated from the center toward the threefold axes. The heterogeneity of RNAs selected for packaging and the capacity of the N-terminally deleted coat protein to reorganize heterologous RNA into a distinct form may explain its propensity to assemble into multiple types of particles, most of which have ill-defined geometries and are smaller than native T=3 virions (6).
DISCUSSION
Nodavirus RNA is folded as a dodecahedral cage.
We have used four different types of FHV particles that differed in protein and/or RNA content to investigate the organization of the packaged RNA at low resolution by cryoEM and image reconstruction. All particles, regardless of their RNA content, exhibited tubes of density connecting neighboring threefold axes of symmetry leading to the appearance of a dodecahedral cage of RNA. This cage has also been observed in the cryoEM reconstruction of the nodavirus PaV (20). The notion that the tube-like density represents double-stranded RNA is based on the high-resolution X-ray structures of native FHV and PaV, which show well-defined duplex RNA at equivalent positions. In FHV, disconnected 10-bp fragments reside at the center of the twofold contacts (7), whereas 25 bp extending from threefold to threefold could be modeled for PaV (20). In the latter case, this accounts for ∼35% of the packaged RNA. Although the RNA density conforms well to the geometry expected for an A-type RNA helix, the possibility that the helices contain noncanonical base pairs or unpaired nucleotides cannot be excluded at this point. Moreover, even though the cryoEM reconstructions imply that the RNA at the threefold junctions is continuous, topological considerations and modeling studies (see below) indicate that this is not possible and that the RNA must deviate to the particle interior at some of these positions.
The reconstructions also revealed density ascribed to RNA at the center of the pentameric rings that form the RNA cage, but it was not obvious what conformation the nucleic acid strands assume at these locations. The density appeared subtly different in wt_BAC and Δ31_DROS particles when compared to wt_DROS and differed significantly in the Δ31_BAC particles. Overall, this suggests that changes in nucleic acid structure are more readily tolerated at this location than along the twofold contacts where the RNA tubes were a constant feature. Notably, in wt_BAC and Δ31_DROS particles the central density was connected to the cage by thin threads. This might provide a clue about the path that the RNA is taking from the cage to other locations.
Duplex RNA structure is independent of sequence and length.
One of the most surprising results of our study was the observation that the RNA packaging arrangement adjacent to the coat protein in wt_BAC particles, which contain a random collection of cellular RNAs of variable length, was almost identical to that observed for native particles containing one copy of RNA1 and RNA2. This strongly implies that the observed organization of the nucleic acid is independent of its primary sequence and length and that it does not result from spontaneous folding of the RNA prior to assembly. Instead, the RNA appears to assume this organization during condensation and encapsidation by the coat protein.
Although the structural plasticity of RNA makes it possible for nonviral RNAs to conform to the topological constraints imposed during nodavirus assembly, it is reasonable to assume that the viral RNAs have adapted particularly well to these constraints. As a result, they are likely to present an optimal framework for assembly and provide the most favorable contacts for the coat protein subunits. Two observations support this notion. First, wt_BAC particles have distinct biophysical properties when compared to wt_DROS particles. For example, they exhibit increased dynamic behavior (“particle breathing”), which is reflected by greater susceptibility to protease digestion (2). This most likely results from less-suitable interactions between RNA and coat protein subunits giving rise to an overall loss of structural stability. Second, FHV coat protein preferentially encapsidates viral RNAs over cellular RNAs in competition experiments (15). While this preference is probably in part controlled by the existence of specific encapsidation sites on the viral RNAs to which the coat protein binds with high affinity, the ability of the viral RNAs to adopt the required secondary and tertiary structures consistent with the T=3 capsid symmetry is likely to contribute.
The structural plasticity of RNA permits dodecahedral packaging.
Our experimental results pose a dilemma: how is it possible for FHV to package different RNAs into a well-defined dodecahedral cage whose tertiary structure is essentially independent of the size and sequence of the RNA? Different RNAs will almost certainly have substantially different preferred secondary structures, and it is very unlikely that any such secondary structure would contain double helices of appropriate lengths and connectivities to guarantee compatibility with the restrictions imposed by the dodecahedral cage. The solution of this dilemma probably lies in the observation that RNA secondary structure is dynamic. A given RNA has a range of secondary structures that are energetically accessible, particularly considering that the binding of capsid proteins can stabilize less-favorable secondary structures. Larson and McPherson (11) argued that the lowest-energy secondary structure for STMV RNA is unlikely to be folded in such a way as to match the geometry required of the RNA in the mature virus. They hypothesized that protein binding could provide the energy required to reorganize the RNA into a series of local stem-loops that would allow formation of the final icosahedral structure. We believe that FHV and PaV exploit the plasticity of RNA secondary structure in a similar fashion and that the binding of capsid proteins captures those elements of secondary and tertiary structure that form the dodecahedral cage, promoting the final protein-protein interactions that define the mature icosahedral capsid. In contrast with STMV, however, it is very likely that the nodavirus RNA secondary structures include substantial amounts of base pairing between regions that are quite far apart in the primary sequence, as discussed below.
Preliminary model for the nodaviral RNA structure.
The crystal structures of FHV and PaV reveal about 16 and 35%, respectively, of the RNA genomes, and each of these forms a dodecahedral cage. There are three major problems that must be solved in generating a model for the remainder of the RNA. First, the dodecahedral symmetry must be broken, because the RNA sequence does not contain that high level of symmetry and because a perfectly symmetric dodecahedral structure would require 12 interlinked closed-circular single-stranded RNAs (20). Symmetry breaking is also required to allow connection of the RNA in the outer cage with the RNA around the fivefold axes and lower radii. Disorder in the PaV X-ray density at the three-way junctions comprising the vertices of the dodecahedral cage suggests that some vertices may be the locations of “stalactites,” where the RNA penetrates the interior (20). A second problem is that the stalactites must be placed on the vertices in such a way as to permit the RNA to cover each edge of the dodecahedron twice, once in each direction, to give antiparallel helices. Finally, it can be shown that the RNA must be organized so that the strand that drops downward through a given stalactite must eventually return back up through that same stalactite to be consistent with the branched secondary structures of all known RNAs and to avoid knotting. One can break the strand once, since the nodaviral genomes have two single-stranded RNA molecules, but we have chosen not to specify the point of such a break in this preliminary model.
Our analysis suggests that there are many possible secondary structures and many possible arrangements of stalactites on the vertices that would be compatible with the observed density maps. It is worthwhile to describe one solution, because it illustrates that the problems described in the previous paragraph can be solved.
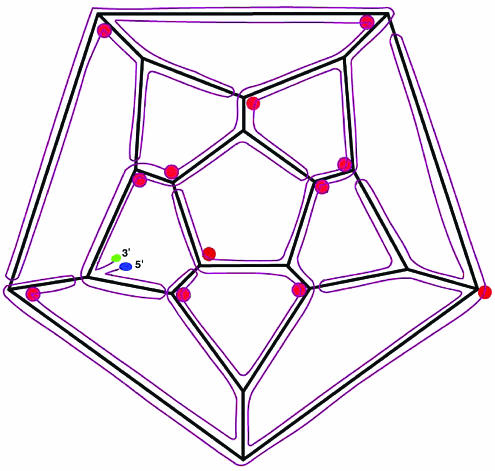
Different arrangements of the RNA can be accommodated at the vertices of the dodecahedral cage. Possible arrangements at vertices that do not have stalactites include the following: a three-way junction similar to those described in the original PaV model (20); a bent duplex that enters along one edge and exits along another, with the third edge being occupied by a stem-loop; and three stem-loops, each approaching along a different edge, with the loops all pointing toward the vertex. A similar variety of arrangements is possible at those vertices that do have stalactites. Icosahedral symmetry is imposed on the density maps and so there is no reason to favor one particular set of arrangements over another. The model we have developed is one of the simplest arrangements (Fig. 7). In this model, two of the vertices (upper left and lower right in Fig. 7) have four-way junctions and the other 18 have three-way junctions. Of the latter, 7 have all three arms radiating out from the junction along the edges of the cage, whereas the other 11 have two arms on the cage and the third arm forms a stalactite. This particular model requires that 11 edges are occupied by stem-loops, with the RNA approaching the vertex with the stalactite. The RNA then doubles back to form the duplex along the same edge, rather than passing through the vertex.
FIG. 7.
Path of model RNA around the edges of the dodecahedral cage and into the interior of the PaV capsid. The cage has been flattened into a 2D projection for graphic simplicity. A single-stranded RNA with 4,322 nucleotides (the sum of nucleotides in PaV RNA1 and RNA2) has been arranged so that every one of the 30 edges of the dodecahedral cage is occupied by an antiparallel duplex, as observed in the crystal structure. The strand runs from the 5′ end (solid blue circle), along all the edges of the cage, drops down into (and returns from) the interior of the capsid at 12 stalactites (solid red circles), and ends at the 3′ end (solid green circle). Two of the vertices (upper left and lower right) have four-way junctions, and the other 18 have three-way junctions. Of the latter, 7 have all three arms radiating out from the junction along the edges of the cage, whereas the other 11 have two arms on the cage and the third arm forms a stalactite. Eleven edges are occupied by stem-loops, with the RNA approaching the vertex with the stalactite, but then doubling back along the same edge, rather than passing through the vertex.
The electron density at the vertices of the dodecahedral cage in PaV is high enough that, regardless of the number of stalactites, it is impossible to generate models that have only local stem-loops of the kind that have been proposed for STMV (11). STMV is substantially smaller than the nodaviruses, so the RNA is largely found in a layer immediately inside the capsid, and the local folding model provides a simple and elegant solution to the arrangement of STMV RNA. The larger nodavirus genomes preclude such a simple arrangement and require base-pairing interactions between regions that are relatively far apart in the primary structure.
Dependence of RNA folding and packaging on the N and C termini of the capsid protein.
Several key observations led us to assume that the region of the coat protein most likely to be required for imposing the observed RNA structure is located at the N terminus, specifically within residues 1 to 30, which contain five arginine residues. This region is required for specific encapsidation of FHV RNA2 (13), suggesting the existence of a high-affinity recognition site to which the N terminus binds at the beginning of assembly. Secondly, although this region of the protein is not visible in the FHV X-ray structure, residues 7 to 30 are visible in some of the coat protein subunits of PaV and are known to be closely associated with the RNA duplex at the twofold contacts (20). Similar interactions are presumably established in FHV but lack the precise icosahedral symmetry. Lastly, deletion of residues 2 to 31 leads to aberrant assembly in the baculovirus expression system (6). (In our study, we specifically isolated Δ31_BAC T=3 particles for analysis.) Surprisingly, our results showed that deletion of the N terminus did not greatly affect the organization of the RNA adjacent to the capsid. This was particularly evident in the Δ31_DROS particles, in which the RNA structure was almost identical. Slight differences were observed in the Δ31_BAC particles. While the tubes of density at the twofold contacts were conserved, the connection to the central RNA was altered, giving the appearance of spokes radiating from the fivefold axis. This made the existence of the dodecahedral cage less obvious. Biochemically, the salient difference between the two types of particles was the presence of FHV RNA1 in each Δ31_DROS capsid, whereas Δ31_BAC particles contained only random cellular RNA. The fact that the RNA pattern in Δ31_DROS capsids was unaltered thus reinforces the notion that the viral RNAs are particularly well adapted to assuming the required secondary structure during assembly. It is tempting to speculate that the RNA cage may be primarily formed by FHV RNA1 in these particles. In contrast, when nonviral RNAs are packaged, the lack of the N terminus has more significant ramifications in that the RNA cannot be properly positioned under the pentameric contacts of the polymerizing coat protein subunits, and this is associated with an overall decrease in the precision of assembly. Our previous results have shown that the majority of Δ31_BAC particles are defective, exhibiting aberrant geometries and a smaller size than native T=3 particles (6). The geometry of the particles appears to be controlled to some degree by the length of the RNA that is packaged, since numerous small, oval-shaped particles are produced with tRNA (6).
As noted, the images of the Δ31_DROS and Δ31_BAC particles that lacked N-terminal amino acids 2 to 31 had a hollow appearance (Fig. 2C and D) and the 3D reconstructions lacked a central core of density, as was seen in the wt_DROS and wt_BAC particles (Fig. 5). Flexibility of the arginine-rich N terminus may allow it to extend into the interior of the particle to organize the central RNA core. The volume for the interior density in the wt_DROS and wt_BAC reconstructions accounted for 540 to 1,300 amino acids, depending on the selected contour and assuming 110 Da/amino acid and a partial specific volume of 0.74 cm3/g. However, the central density is ∼80 Å from the inner edge of the protein shell, and spanning this distance would account for much of the 30 N-terminal residues. If just the N terminus extends to the center of the particle and the central density represents RNA, the volume could account for ∼230 to 570 nucleotides, assuming that 4,500 nucleotides occupy 1.4 × 106 Å3.
Since the N terminus does not appear to be required for positioning double-stranded RNA at the twofold contacts during assembly, another region must have this function. A likely candidate is the C terminus which, like the N terminus, has been shown to play a role in specific encapsidation of the viral RNA. Although the portion of the C terminus that is required for specific packaging, residues 402 to 407, is not visible in the X-ray structure, two lysine residues at positions 371 and 375 are known to contact the sugar-phosphate backbone of the double-stranded RNA at the twofold contacts. Most significantly, deletion of the C-terminal 44 amino acids, including these lysine residues, completely inhibits assembly (17). Thus, we hypothesize that the C terminus of the coat protein plays an important role not only in the initial selection of the RNA for packaging but also in positioning it correctly between the interacting subunits for formation of the protein shell.
Acknowledgments
This work was supported by National Institutes of Health grants GM53491 (A.S.), GM066087 (M.Y.), GM34220 (J.E.J.), and RR12255 (S.C.H.). During the course of this work, M.Y. was an Established Investigator of the American Heart Association and Bristol-Myers Squibb and is now the recipient of a Clinical Scientist Award in Translational Research from the Burroughs Wellcome Fund.
Footnotes
Manuscript 15959-MB from the Scripps Research Institute.
REFERENCES
- 1.Baker, T. S., and R. H. Cheng. 1996. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol. 116:120-130. [DOI] [PubMed] [Google Scholar]
- 2.Bothner, B., A. Schneemann, D. Marshall, V. Reddy, J. E. Johnson, and G. Siuzdak. 1999. Crystallographically identical virus capsids display different properties in solution. Nat. Struct. Biol. 6:114-116. [DOI] [PubMed] [Google Scholar]
- 3.Chen, Z., C. Stauffacher, Y. Li, T. Schmidt, W. Bomu, G. Kamer, M. Shanks, G. Lomonossoff, and J. E. Johnson. 1989. Protein-RNA interactions in an icosahedral virus at 3.0 Å resolution. Science 245:154-159. [DOI] [PubMed] [Google Scholar]
- 4.Chen, Z., C. V. Stauffacher, and J. E. Johnson. 1990. Capsid structure and RNA packaging in comoviruses. Semin. Virol. 1:453-466. [Google Scholar]
- 5.Cheng, R. H., V. S. Reddy, N. H. Olson, A. J. Fisher, T. S. Baker, and J. E. Johnson. 1994. Functional implications of quasi-equivalence in a T=3 icosahedral animal virus established by cryo-electron microscopy and X-ray crystallography. Structure 2:271-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong, X. F., P. Natarajan, M. Tihova, J. E. Johnson, and A. Schneemann. 1998. Particle polymorphism caused by deletion of a peptide molecular switch in a quasi-equivalent virus. J. Virol. 72:6024-6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisher, A. J., and J. E. Johnson. 1993. Ordered duplex RNA controls capsid architecture in an icosahedral animal virus. Nature 361:176-179. [DOI] [PubMed] [Google Scholar]
- 8.Krishna, N. K., D. Marshall, and A. Schneemann. 2003. Analysis of RNA packaging in wild-type and mosaic protein capsids of flock house virus using recombinant baculovirus vectors. Virology 305:10-24. [DOI] [PubMed] [Google Scholar]
- 9.Larson, S. B., J. Day, A. Greenwood, and A. McPherson. 1998. Refined structure of satellite tobacco mosaic virus at 1.8 Å resolution. J. Mol. Biol. 277:37-59. [DOI] [PubMed] [Google Scholar]
- 10.Larson, S. B., S. Koszelak, J. Day, A. Greenwood, J. A. Dodds, and A. McPherson. 1993. Double helical RNA in satellite tobacco mosaic virus. Nature 361:179-182. [DOI] [PubMed] [Google Scholar]
- 11.Larson, S. B., and A. McPherson. 2001. Satellite tobacco mosaic virus RNA: structure and implications for assembly. Curr. Opin. Struct. Biol. 11:59-65. [DOI] [PubMed] [Google Scholar]
- 12.Li, T., Z. Chen, J. E. Johnson, and G. J. Thomas, Jr. 1990. Structural studies of bean pod mottle virus, capsid, and RNA in crystal and solution states by laser Raman spectroscopy. Biochemistry 29:5018-5026. [DOI] [PubMed] [Google Scholar]
- 13.Marshall, D., and A. Schneemann. 2001. Specific packaging of nodaviral RNA2 requires the N-terminus of the capsid protein. Virology 285:165-175. [DOI] [PubMed] [Google Scholar]
- 14.Opalka, N., M. Tihova, C. Brugidou, A. Kumar, R. N. Beachy, C. M. Fauquet, and M. Yeager. 2000. Structure of native and expanded sobemoviruses by electron cryo-microscopy and image reconstruction. J. Mol. Biol. 303:197-211. [DOI] [PubMed] [Google Scholar]
- 15.Schneemann, A. 1992. Studies on assembly and maturation of flock house virus, a small insect ribovirus. Ph.D. dissertation. University of Wisconsin, Madison.
- 16.Schneemann, A., R. Dasgupta, J. E. Johnson, and R. R. Rueckert. 1993. Use of recombinant baculoviruses in synthesis of morphologically distinct virus-like particles of flock house virus, a nodavirus. J. Virol. 67:2756-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneemann, A., and D. Marshall. 1998. Specific encapsidation of nodavirus RNAs is mediated through the C terminus of capsid precursor protein alpha. J. Virol. 72:8738-8746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schneemann, A., W. Zhong, T. M. Gallagher, and R. R. Rueckert. 1992. Maturation cleavage required for infectivity of a nodavirus. J. Virol. 66:6728-6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheehan, B., S. D. Fuller, M. E. Pique, and M. Yeager. 1996. AVS software for visualization in molecular microscopy. J. Struct. Biol. 116:99-106. [DOI] [PubMed] [Google Scholar]
- 20.Tang, L., K. N. Johnson, L. A. Ball, T. Lin, M. Yeager, and J. E. Johnson. 2001. The structure of Pariacoto virus reveals a dodecahedral cage of duplex RNA. Nat. Struct. Biol. 8:77-83. [DOI] [PubMed] [Google Scholar]
- 21.Tihova, M., K. A. Dryden, A. R. Bellamy, H. B. Greenberg, and M. Yeager. 2001. Localization of membrane permeabilization and receptor binding sites on the VP4 hemagglutinin of rotavirus: implications for cell entry. J. Mol. Biol. 314:985-992. [DOI] [PubMed] [Google Scholar]