Abstract
The circumstances under which unintegrated lentivirus DNA can persist and be a functional template for transcription and protein expression are not clear. We constructed and validated the first class I (nonpleiotropic) integrase (IN) mutants for a non-human lentivirus (feline immunodeficiency virus [FIV]) and analyzed both these and known class I human immunodeficiency virus type 1 IN mutants. The FIV IN mutants (D66V and D66V/D118A) had class I properties: Gag/Pol precursor expression, proteolytic processing, particle formation, and reverse transcriptase (RT) production were normal, while the transduction of dividing fibroblasts was prevented and integration was blocked. When injected into rat retinas, the wild-type (WT) vector produced extensive, persistent transgene expression, compared with only rare positive neuronal cells for the IN mutant vector. In contrast, both WT and mutant vectors produced entirely equivalent, effective transduction levels of primary rat neurons (retinal ganglion cells). By testing the hypothesis that the unexpected retinal neuron transduction was related to cell cycle status, we found that when fibroblasts were growth arrested, transduction and internally promoted transgene expression were not inhibited at all by the class I FIV or HIV-1 IN mutations. Cells were then transduced under aphidicolin arrest and were released from the block 48 h later. Vector expression was stable and durable during repeated passaging in WT vector-transduced cells, while the release of cells transduced with equivalent RT units of class I IN mutant FIV or HIV vector resulted in a steady decline of expression, from 97 to 0% of cells by day 10. Southern blot and PCR analyses showed a lack of integration, irrespective of cell cycle, for the class I mutants and an increase in one- and two-long terminal repeat circular and linear unintegrated DNAs in growth-arrested cells. We conclude that if cell division is prevented, unintegrated FIV and HIV-1 vector DNAs can produce high-level internally promoted transgene expression equivalent to WT vectors. The expression correlates with the unintegrated DNA levels. These observations may facilitate the study of the roles of IN and other preintegration complex components in preintegration phases of infection by (i) providing an alternative way to monitor unintegrated nuclear cDNA forms, (ii) restricting ascertainment to the transcriptionally functional subset of unintegrated DNA, (iii) enabling analysis in individual, nondividing cells, and (iv) uncoupling other potential functions of IN from integration.
Shortly after infection of a cell by a retrovirus, reverse transcription of the RNA genome yields a linear cDNA copy, which along with the viral integrase (IN) and other proteins comprises the preintegration complex (PIC), the functional precursor to integration (5). Certain features of the IN structure are conserved among retroelements, and conserved amino acid residues that are critical for catalysis have been identified. Retroviral INs have three domains: an N-terminal domain, a central catalytic core domain, and a C-terminal domain (26). The N-terminal domain contains a zinc finger-like sequence that influences IN oligomerization (9). The C-terminal domain, which is the most divergent, binds DNA in a sequence-independent manner (20, 58). IN has also been reported to play other roles in the lentiviral life cycle, in particular in nuclear import of the PIC (23).
The genetic analysis of IN functions is not straightforward because the enzyme is generated by viral protease-mediated cleavage from the Gag/Pol precursor. Many IN mutations produce pleiotropic effects on Gag/Pol-derived functions, including particle formation and reverse transcription (17). Accordingly, two types of IN mutants are generally recognized. Nonspecific phenotypes (which have been termed class II) result from deletions, truncations, and numerous single amino acid changes (17, 19, 56). In contrast, nonpleiotropic (class I) mutations affect only the DNA cleaving and joining reaction, while leaving intact other measurable aspects of the virus life cycle, such as Gag/Pol precursor processing, particle formation, virion morphogenesis, reverse transcription, and PIC nuclear import (17, 18, 28, 29). In human immunodeficiency virus type 1 (HIV-1), mutations of any of three residues that participate in the catalytic center (D64, D116, and E152) produce class I properties (15, 18, 29). These residues form a catalytic triad (DX39-58DX35E) that is broadly conserved in retroelement INs (5, 17). Experimentally, class I IN mutants enable control designs that compare the fates of integration-defective, structurally normal particles differing in only one amino acid in a single enzyme that comprises a very small molar fraction of virion protein molecules (17).
Few data exist for mutants of other lentivirus INs. Tomonaga et al. generated frameshift and truncation mutants of feline immunodeficiency virus (FIV) IN which had class II phenotypes (55). In vitro analyses of bacterially expressed FIV IN mutants established that the central core domain determines in vitro target site selection (49) and that a D118N mutation results in the loss of 3′-end processing and joining activities in vitro (50). Virus studies to establish class I properties have not been carried out for any non-HIV lentiviral IN.
While chromosomal integration is classically considered an obligate part of the retrovirus life cycle (47), evidence has accumulated for integration-independent expression of various HIV-1 proteins (1, 11, 19, 52, 56, 59), albeit at a variable and generally small fraction of wild-type (WT) levels. In at least one T-cell line (MT-4 cells), productive replication and serial passaging of class I mutant virus have even been demonstrated (36). In HeLa-CD4 β-galactosidase indicator (MAGI) cells (27), class I HIV-1 IN mutant viruses display measurable, reduced titers, ranging from 11.5 to 18% of that of the WT (1, 19, 56). However, MAGI cells function by reporting Tat transactivation and are capable of detecting low levels of early gene expression; HIV-1 structural proteins were not detected and productive infections did not occur in these studies. The infection of RD cells (35) and HeLa cells (43) with class I HIV-1 mutants having luciferase substituted for nef produced 0.2 and 3.6%, respectively, of the level of WT luciferase; in the latter case, deletion of Vpr reduced this to 0.6%, suggesting a role for this accessory protein in modulating unintegrated DNA long terminal repeat (LTR) transactivation. More recently, some expression of tat transcripts, but not Tat protein, Rev, or the Rev-dependent structural proteins, was found to occur from unintegrated class I IN mutant HIV-1 DNA in resting T cells (59). Nakajima et al. examined the properties of class I IN mutant HIV-1 in different T-cell lines and primary cells (36). When infected with a reporter virus expressing chloramphenicol acetyltransferase (CAT) from the nef open reading frame, three T-cell lines displayed 333- to 1,000-fold less reverse transcriptase (RT)-adjusted CAT activity for class I mutants than for WT IN virus, and one (Jurkat) showed no detectable CAT expression. However, two cell lines (MT-4 and C8166) showed spreading infections with a class I mutant virus, and MT-4 cells permitted serial passaging of virus. The mutant virus was over 4 log less infectious than the WT in these cells, leading to delayed replication kinetics yet robust peak RT activities comparable to those of the WT (36). In this and other studies (21, 56), these mutants did not productively replicate or express detectable Gag/Pol in primary T cells and macrophages.
The persistence of unintegrated lentiviral DNA in cells and its use for monitoring the progression of early events in the life cycle and in vivo replication are important issues that are controversial in some respects. Reverse-transcribed linear viral cDNAs have three principal fates besides bona fide integration, none of which are precursors to integration: the conversion to 2-LTR circles by the host cell nonhomologous DNA end-joining system (reference 31 and references therein); the formation of 1-LTR circles, probably by homologous recombination (22); and auto-integration to form defective, rearranged DNAs (51). 2-LTR circles arise only after PIC nuclear import and are readily detected unambiguously by PCR, which has led to the wide use of PCR for monitoring nuclear translocation of the PIC (7, 24, 30). By blocking the access of linear cDNAs to the integration end point, class I HIV-1 mutants also result in higher levels of 2-LTR circles than the WT virus (3, 13, 17, 19, 28, 36, 45). When IN is WT, abundant unintegrated DNA accumulation has generally been regarded as a feature of a high-multiplicity superinfection (3) that does not have clearly established pathogenic importance in vivo (5). Nevertheless, recent studies suggested that expression from this DNA has biological significance (59), and prevalent unintegrated HIV-1 DNA can be detected in vivo (12, 37, 54). Unintegrated DNA is in fact the most prevalent form of DNA (2 log more than integrated DNA) in resting and activated CD4+ T cells in vivo (12). Unintegrated DNA may be proapoptotic (14, 31).
2-LTR circular DNA has been considered a marker of recent infection (38, 39, 60), with the further application of identifying antiretroviral drug-refractory HIV-1 replication in vivo (48). This method is based on the premise that unintegrated lentiviral DNA is intrinsically unstable, presumably due to its susceptibility to cellular nucleases. However, recent analyses have concluded otherwise, namely that circular forms are stable intracellularly, undergo only dilutional attrition in direct proportion to cell division, and may therefore persist in vivo without ongoing HIV-1 replication (6, 8, 10, 40). Butler et al. examined cell cycle status as a variable in DNA persistence and found that 2-LTR circles are the principal persisting form of unintegrated HIV-1 vector DNAs, reach maximal abundance by 12 to 24 h, and are notably stable if cells are growth arrested with aphidicolin (10), which was important for the present study's focus on cell cycle status as a key variable. Transgene or virus expression was not examined. In general, the circumstances under which these DNAs are competent for transcription and expression and the dependence of this on the viral LTR versus internal promoters remain unclear. For a review of recent DNA quantification data, see reference 8.
Here we report the construction and evaluation of FIV IN D66V and a double mutant, D66V/D118A. Along with analogous HIV-1 IN mutant vectors, we examined these in the context of virus-promoted and internally (cytomegalovirus [CMV]) promoted vectors and in dividing and nondividing cells.
MATERIALS AND METHODS
Generation of D66V and D66V/D118A FIV IN mutants.
A 1.8-kb FIV pol fragment generated with PCR primers 5′-ATATACTAGTTCTAGAGAAGCCTGGGAATC-3′ and 5′-ATATGAATTCTCCGGAGGTAGCCTAG-3′ was subcloned by using the incorporated EcoRI and SpeI sites. The GAT codon (encoding aspartic acid) at IN residue 66, which is homologous to residue 64 in HIV-1 IN, was mutagenized to GTA (valine) by site-directed mutagenesis (QuikChange; Stratagene, La Jolla, Calif.) with oligonucleotides 5′-GCCTGGTATCTGGCAAATGGTATGCACACACTTTGATGGC-3′ and 5′-GCCATCAAAGTGTGTGCATACCATTTGCCAGATACCAGGC-3′. An internal fragment (BspE1-Bsu36I) was then removed and inserted between BspE1 and Bsu36I in packaging plasmids pCF1Δenv and pFP93 (32-34, 42, 46), generating pCF1Δenv.D66V and pFP93.D66V. A second round of mutagenesis in the D66V-mutagenized PCR fragment was performed to change the GAT codon encoding aspartic acid at position 118, which is homologous to residue 116 in HIV-1 IN, to GCA (alanine) by using mutagenesis primers 5′-AATGTTACTGAATTACAAACAGCAAATGGACCAAATTTTAAAAATC-3′ and 5′-GATTTTTAAAATTTGGTCCATTTGCTGTTTGTAATTCAGTAACATT-3′, again followed by insertion into pCF1Δenv and pFP93. The mutations and fidelity to the WT sequence in the remainder of the inserts were confirmed by sequencing.
HIV IN mutant packaging plasmid.
A double mutant (N/N) of HIV NL4-3 containing D64N and D116N mutations (36) was kindly provided by A. Engelman (Dana-Farber Cancer Institute, Boston, Mass.). A 2,737-bp ApaI-AflII fragment spanning the mutations was inserted in place of the WT fragment in the HIV-1 pNL4-3-based packaging construct pCMVΔR8.91 (61), and the presence of the mutation in this construct, pCMVΔR8.91D64N + D116N, was verified by DNA sequencing.
Cells.
Cell lines were cultured in Dulbecco's modified Eagle medium with 10% fetal calf serum. 293T cells were used as virus and subgenomic vector producer cells. HeLa, Crandell feline kidney (CrFK), human fibrosarcoma HT1080, and 293T cells were used for transduction experiments. Primary rat retinal ganglion cells were prepared by immunopanning (2). Retinas from 8-day-old Sprague-Dawley rats (Harlan, Indianapolis, Ind.) were dissociated in a 15-U/ml papain solution (Worthington Biochemical Corporation, Lakewood, N.J.). The tissue was titrated in a solution of 2-mg/ml ovomucoid (Roche Molecular Biochemicals, Indianapolis, Ind.), 0.004% DNase (Sigma, St. Louis, Mo.), and 1-mg/ml bovine serum albumin (Sigma) to obtain single-cell suspensions. Cells were incubated with an anti-rat macrophage antibody (1:100; Accurate Chemical & Scientific, Westbury, N.Y.). Cells were then filtered through a 15-μm-pore-size Nitrex mesh (Tetko Inc., Kansas City, Mo.), incubated on two sequential 150-mm-diameter anti-rabbit immunoglobulin G (IgG) (Jackson Immunoresearch Laboratories, Inc., West Grove, Pa.) panning plates (Falcon), and then incubated on one 100-mm-diameter anti-Thy1.1 plate (Antibody Core Facility, Mayo Clinic, Rochester, Minn.). Adherent retinal ganglion cells were collected and centrifuged at 200 × g for 15 min. Retinal ganglion cells were plated at 8,000 to 10,000 cells per well in a poly-d-lysine (10 μg/ml; Sigma)-coated 96-well plate and were cultured in serum-free Dulbecco's modified Eagle complete medium containing the following: Neurobasal with bovine serum albumin (Sigma), selenium (Sigma), putrescine (Sigma), thyroxine (Sigma), 3,3′,5-triido-l-thyronine (Sigma), transferrin (Sigma), progesterone (Sigma), pyruvate (Invitrogen Corporation, Grand Island, N.Y.), glutamine with basic fibroblast growth factor, BDNF, and CNTF (Peprotech, Rocky Hill, N.J.), and forskolin (5 μM; Sigma). RT activity-normalized WT and single-IN-mutant lacZ FIV vectors as well as a mock vector (described below) were added to cells 1 h after plating.
Preparation and titration of vector stocks.
All FIV vectors and viruses were produced in 293T cells by using a 5′ U3-substituted (CMV-promoted) system (41, 42). The transfer vector pCT26, containing an internally promoted (human CMV immediate-early gene promoter-enhancer) lacZ marker gene, was cotransfected with either the mutant or WT pCF1Δenv or pFP93 packaging construct and pMD.G by the calcium phosphate transient transfection method, as previously described (34). A mock vector was prepared by transient transfection into 293T cells of the same amounts of pCT26 DNA and pMD.G DNA used for the generation of the transducing vector, while omitting the packaging plasmid, followed by subsequent concentration as indicated below for packaged vectors. The plasmid transfection efficiency evaluated by X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining was >80% for the transfected 293T producer cells for both mock and packaged CT26 vectors. For an enhanced green fluorescent protein (GFP) vector, the transfer vector pGiNWF (32) was cotransfected with either the mutant or WT FP93 packaging construct and pMD.G. Two-chamber cell factories (CF2) were used to generate supernatants for concentrated stocks. Supernatants collected 48 h after replacement of the transfection mix with fresh medium were filtered through a 0.2-μm-pore-size filter, concentrated by one or two rounds of ultracentrifugation in a fixed-angle A621 rotor at 21,000 rpm for 2 h or in a swinging bucket SW28 rotor, divided into aliquots, and frozen at −80°C. For HIV-1 vectors, the transfer construct pHR′CMVlacZ (61) containing lacZ as the marker gene was cotransfected with either pCMVΔR8.91D64N + D116N or pCMVΔR8.91 and pMD.G, also in two-chamber cell factories (CF2), and the vectors were concentrated by the same method as that for FIV vectors. The IN-intact CT26, GiNWF, and PHR′CMVlacZ stocks were titrated on CrFK cells as described previously (32-34). Briefly, the cells were transduced with various dilutions of concentrated vector stock. The medium was changed after 6 h and cells were analyzed after 24 to 48 h as described in the text. Cells were fixed in 0.1% glutaraldehyde for 5 min at room temperature and were stained with 500 μl of X-Gal solution per well for 16 h. The X-Gal solution was washed off and replaced with 1% glutaraldehyde. Foci were counted to determine the percent transduction and the titers.
Preparation and titration of virus stocks.
Single (D66V) and double (D66V/D116A) mutant INs were inserted as BspE1-Bsu36I fragments into pCT5 (42) and pCT5efs (25). pCT5 enables the expression of full-length, infectious FIV 34TF10 (53) in human cells by using a fusion of the CMV immediate-early promoter to the viral R repeat (41, 42). pCT5efs (envelope frameshift) contains an env-frameshifting 29-bp insertion at nucleotide (nt) 7146 of pCT5 (25). 293T cells were transfected with pCT5 or the IN mutant CT5.D66V.D118A to generate full-length virus particles with native envelopes, as previously described (41, 42). Cotransfection of pCT5 with pMD.G was done to yield vesicular stomatitis virus glycoprotein (VSV-G)-pseudotyped full-length virions also possessing the native envelope. 293T cells were transfected with pCT5efs and pCT5efs.D66V along with pVSVG to generate VSV-G-pseudotyped FIV particles carrying full-length genomes but lacking the native envelope protein. The transfection mixtures were replaced with fresh medium after 14 h. Supernatants were collected 32 h after the transfection mix was washed off. The supernatants were syringe filtered and frozen in 500-μl aliquots. The WT intact IN viruses were titrated on CrFK cells by using a focal infectivity assay to detect the expression of Gag/Pol proteins as described previously (41). The RT activity was measured for all six viruses.
RT assay.
Virus stocks were characterized by an RT assay, which was used to normalize intact IN and mutant IN preparations. Ten-microliter samples of undiluted and diluted supernatants were assayed by mixing them with 50 μl of RT cocktail consisting of a poly(A) template (5 μg/ml) and a pd(t)12-18 oligonucleotide (1.57 μg/ml) in 50 mM Tris (pH 7.8), 7.5 mM KCl, 2 mM dithiothreitol, 5 mM MgCl2, 0.05% NP-40, and 1 μCi of [32P]TTP. Samples were incubated in a Falcon flexible plastic 96-well plate for 1.5 h at 37°C. Five microliters of the reaction was blotted onto a Whatman DE-81 ion-exchange filter-paper 96-well plate. After being dried in a 37°C oven for 15 min, the plate was washed with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and ethanol on a Filtermate Harvester. The plate was dried in a 37°C oven for 30 min. The bottom of the plate was sealed, and 35 μl of Microscint-O scintillation fluor was pipetted into each well. A TopCount NXT microplate luminescence-scintillation counter (Packard) was used to measure the radioactivity of each sample.
Immunoblotting.
293T cells were transiently transfected with CT5, CT5.D66V, and CT5.D66V.D118A. Cells were lysed in a solution containing 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate (SDS), 50 mM Tris (pH 8.0), and protease inhibitors (Complete Mini; Roche). After normalization for protein concentration, lysates were combined with β-mercaptoethanol, boiled, and separated in an SDS-10% polyacrylamide gel. The proteins were transferred to a polyvinylidene difluoride membrane (Immobilon-P; Millipore) by using a Bio-Rad Trans-Blot SD semidry electrophoretic transfer cell. The transferred blot was blocked in 0.2% I-block (Tropix)-0.1% Tween solution before being probed with Petaluma (FIV-infected cat) serum (1:1,000). Peroxidase-conjugated goat anti-feline IgG (1:1,000; ICN) was used as the secondary antibody. Blots were developed with a chemiluminescence kit (Amersham Pharmacia) before exposure to radiographic film.
Vector transduction of dividing and nondividing cells.
HT1080, HeLa, 293T, and CrFK cells were seeded in 24-well plates at 45,000 cells/well 24 h before treatment with 15 μg of aphidicolin per ml in medium. Cells were transduced 24 h after the aphidicolin block, which was replenished in the medium at the time of transduction or medium changes. Growth arrest was verified by flow cytometry with propidium iodide. Dividing cells were seeded the day before transduction, at the time of the aphidicolin treatment of cells scheduled for growth arrest. Dividing and nondividing cells were transduced with RT-normalized stocks of intact IN or class I mutant IN vectors. The transduction supernatant was removed after 6 to 8 h. At 2 days posttransduction, the cells transduced with the lacZ vector were fixed in 0.1% glutaraldehyde in phosphate-buffered saline (PBS) for 5 min at room temperature and were stained with 500 μl of X-Gal solution per well for 16 h. The X-Gal solution was replaced with 0.1% glutaraldehyde in PBS for short-term storage at 4°C, and positive and negative cells were counted to determine percentages of transduction. Cells transduced with the enhanced GFP vector were fixed in 1% formaldehyde and analyzed by fluorescence-activated cell sorting. For some experiments, cells were maintained in parallel to track transgene expression over time.
FIV infection of CrFK cells.
RT-normalized mutant and WT FIV stocks were used to infect dividing and aphidicolin-treated CrFK cells. Infections were done with fourfold serial dilutions of virus. After overnight incubation, the cells were washed three times with PBS and new medium was added. Cells were fixed in methanol at 42 h postinfection. Infected foci were detected by an immunoperoxidase assay using FIV-positive cat plasma (44). Titers were calculated for dividing and growth-arrested cells as the mean numbers of foci × the dilution factors.
Subretinal injections.
Right eyes of 7-day-old Sprague-Dawley rats (n = 10; Harlan Laboratories) were injected subretinally with WT vector, and left eyes were injected with a mutant IN vector (D66V). Anaesthetized animals were placed under an operating microscope and eyelids were opened gently with an instrument. A latex membrane with a 1.5-mm central slit was used to prolapse and secure the eye. An initial sclerotomy was made at the pars plana, 1 mm posterior to the limbus, with a 30-gauge needle. A custom needle (32-gauge, 12° bevel, 7-mm length) (Hamilton Co., Reno, Nev.) mounted on a 5-μl Hamilton microsyringe was directed tangentially through the sclerotomy between the retina and the sclera and was advanced to the subretinal space with the bevel of the needle facing the retina. Two microliters of 4.6 × 108 transinducing units (TU) of WT or IN mutant/ml was delivered by subretinal injection. Animals were sacrificed after 2 months. Enucleated eyes were fixed in 10% formalin at 4°C for 90 min, the corneas and lenses were removed, and eye cups were incubated in X-Gal reagent overnight at 37°C.
Southern blotting for integrated and unintegrated vector DNAs.
Genomic DNA was digested to completion with HindIII or SacI, as described in Results and the figure legends, and was separated in agarose gels before being blotted onto nitrocellulose. Total DNAs were collected by using a Qiagen Tissue DNeasy kit at different times posttransduction. Fifty micrograms of 48-h genomic DNA was digested to completion with SacI and divided into two aliquots per sample to be used with two different probes. Probe 1 is specific for the 3′ end of the reverse-transcribed vector cDNA. Probe 2 is an internal MluI fragment within the lacZ transgene of pCT26. Probes were randomly labeled with α-dCTP by using a Random Primers DNA labeling kit (Gibco BRL).
Quantification of 2-LTR circles.
Total DNAs from dividing and aphidicolin-arrested cells transduced with RT-normalized stocks of double mutant IN and WT lacZ vectors were collected by using a Qiagen Tissue DNeasy kit at 24 and 48 h posttransduction. Five hundred nanograms of total DNA from each sample was analyzed. 2-LTR circles were quantified by real-time PCR in a Roche LightCycler with primers specific to the 2-LTR circle junction by using the Roche FastStart DNA kit. A 2-LTR circle template was constructed by deleting virus sequences between the two LTRs of CT5. Tenfold dilutions of this template served as standards.
RESULTS
Initial characterization of FIV IN mutants in vitro and in vivo.
The D66 and D118 residues targeted in FIV IN by site-directed mutagenesis correspond to the D64 and D116 residues in the catalytic triad of HIV-1 (Fig. 1). IN mutants were tested in the context of single-round minimal FIV reporter vectors, which in their intact IN versions transduce nondividing cells and produce sustained gene expression in tissue culture, animal, and explanted human organ models (32, 34, 42, 46). Full-length FIV and VSV-G-pseudotyped FIV with or without a frame-shifting oligonucleotide insertion in FIV env were also tested. For the FIV vectors, we constructed single (D66V) and double (D66V/D118A) FIV packaging plasmids. For comparison to HIV-1, we constructed a double IN mutant (D64N/D116N) HIV packaging construct. For all comparisons, WT and mutant IN virions were generated in parallel, and RT activity-normalized stocks were compared in order to expose cells to an equivalent number of particles.
FIG. 1.
Alignment of HIV-1 and FIV IN proteins. The overall identity (asterisks) is 37%. Dots indicate conservative differences. The aspartic acid and glutamic acid residues in the DX39-58DX35E triad are shown in bold. The aspartic acid residues mutated in the present study are underlined, and amino- and carboxy-terminal regions flanking the catalytic core domain are shaded.
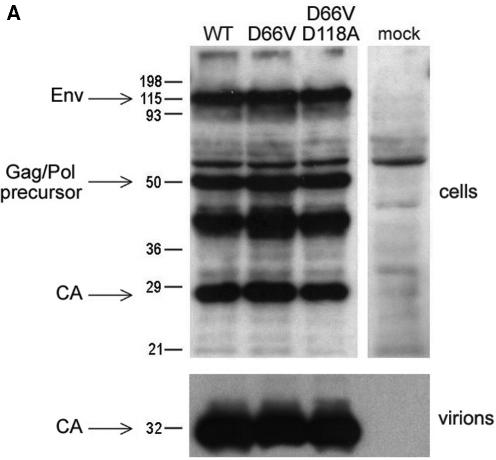
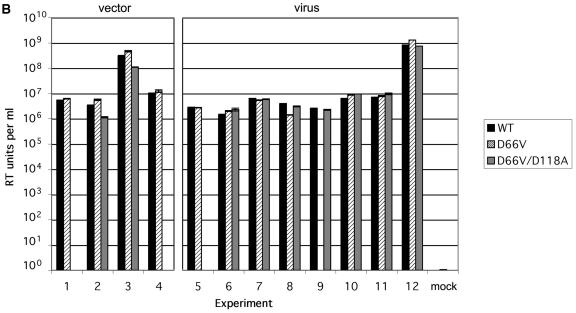
The single (D66V) and double (D66V/D118A) mutations did not alter FIV Gag/Pol precursor expression or proteolytic processing or envelope glycoprotein expression (Fig. 2A, top panel). Supernatant particle production was also equivalent for WT and mutant IN viruses or vectors (Fig. 2B). As expected, RT production varied 2 to 3 orders of magnitude between experiments due to variables such as transfection efficiency, but WT and mutant RT values correlated well within any one experiment (Fig. 2B). As seen previously with WT FIV (25), both D66V and D66V/D188A mutant IN viral particles contained predominantly the mature 27-kDa FIV capsid protein (CA); mature CA levels in supernatants and the degree of processing were equivalent to those of the WT (Fig. 2A, bottom panel).
FIG. 2.
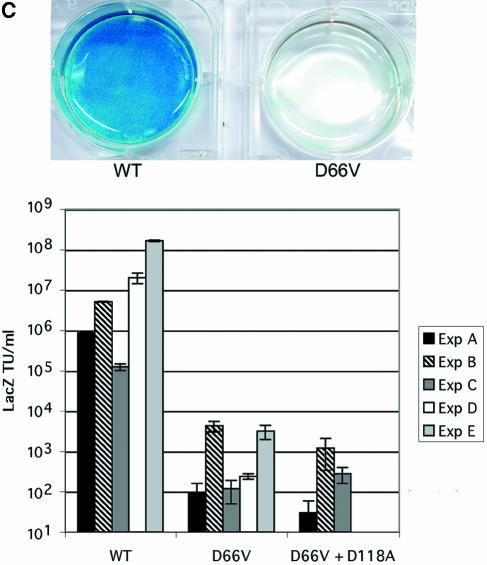
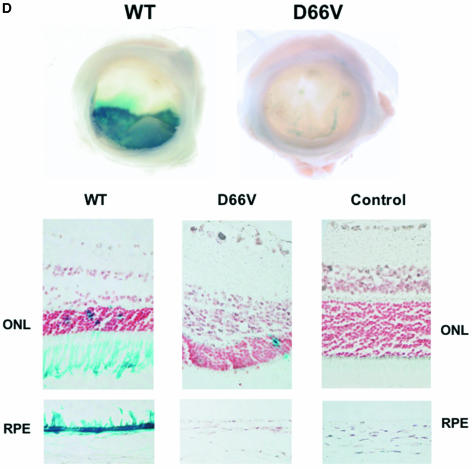
Characterization of FIV IN mutants: class I properties. (A) Comparison of viral protein expression and processing when IN is WT or mutant. 293T cells were transfected with WT pCT5 or pCT5 incorporating the indicated IN mutants. After 48 h, the cells (top) and sucrose cushion-pelleted virions (bottom) were lysed, and equal amounts of protein were separated by SDS-polyacrylamide gel electrophoresis. Immunoblotting was done with FIV-infected cat plasma (1:1,000) and a peroxidase-conjugated goat anti-feline IgG secondary antibody. (B) Virion production. 293T cells were transfected with WT or IN mutant viruses (derived from pCT5) or packaging plasmids (derived from pFP93). The results of consecutive experiments in which one or both mutants were transfected in parallel with the WT parent plasmid are shown for vector preparations on the left and for virus preparations on the right. Standard deviations of the triplicate RT activity measurements are denoted by bars and are too small to be appreciated graphically for some supernatants. (C) Transduction of dividing cells by RT activity-normalized WT and IN mutant vectors. The photographs over the plot show X-Gal-stained dishes of cells transduced with the WT (left, MOI = 5) and D66V (right, RT activity normalized to that of the WT) vectors used for panel B. Bars indicate standard deviations of triplicate measurements. (D) Retinal transduction. Rats were injected subretinally with 2 μl of 4.6 × 108 TU of WT FIV vector/ml (right eyes) and with the RT activity equivalent of the D66V IN mutant FIV vector (left eyes). Macroscopic views of whole eye cups (top) and histological sections (bottom) of a representative pair from 10 animals sacrificed at the earliest time point of 2 months postinjection are shown. Injection was done at the 6 o'clock position. Retinal sections are shown as composites of the neuroretina, with the ONL and juxtaposed retinal pigment epithelium (RPE), which artificially detached postmortem, labeled. The WT vector caused extensive transduction of the retinal pigment epithelium and some transduction of the ONL. There were only traces of beta-galactosidase expression in D66V vector-injected eyes, and this was restricted to cells of the ONL.
However, the transduction of dividing cells was prevented by the single and double IN mutations. CrFK cells transduced in log phase with RT-normalized mutant IN FIV lacZ vector and scored for transduction 48 h later showed a 3- to 5-log reduction in the number of β-galactosidase-positive cells compared to the WT IN vector (Fig. 2C). The drastic reductions in marker gene transduction in these dividing cell experiments were consistent with the lack of integration demonstrated by Southern blotting (see below) and were equivalent in magnitude in multiple cell lines, including CrFK, HT1080, 293T, and HeLa cells. Similarly, in neonatal rats injected subretinally with WT IN vector and the RT activity equivalent of the D66V IN vector, the WT vector caused extensive transduction of the retinal pigment epithelium and some transduction of the outer nuclear layer (ONL) at 2 months postinjection in all 10 animals (Fig. 2D). In contrast, there were only traces of β-galactosidase expression in D66V vector-injected eyes. Note that this was restricted to neuronal cells of the ONL.
Vector expression in nondividing cells.
In further experiments, it became evident that the internally promoted transgene expression of the class I IN mutant vectors could be strikingly effective in certain circumstances. D66V IN FIV vectors were repeatedly observed to transduce purified primary postmitotic rat retinal ganglion cells efficiently, at levels equivalent to or only slightly less than WT IN vectors, while control mock vectors prepared without the packaging plasmid but containing equivalent levels of β-galactosidase and VSV-G did not produce any transduction (Fig. 3).
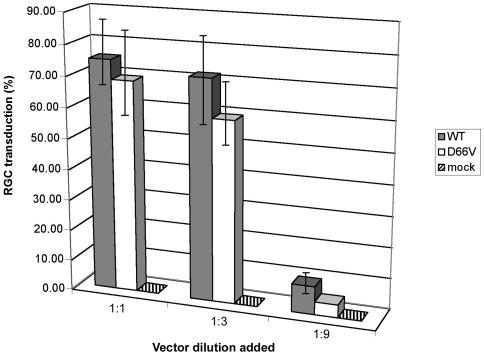
FIG. 3.
Retinal ganglion cell transduction. Rat retinal ganglion cells (RGCs) isolated by immunopanning were plated and transduced with three dilutions of RT-normalized lacZ WT and D66V IN mutant FIV vector as well as a mock vector. The percentage of β-galactosidase-expressing cells was counted at 48 h posttransduction. Bars indicate standard deviations of the means of triplicate well counts. WT versus D66V differences reached statistical significance only at the lowest dilution (P = 0.6, 0.06, and 0.02 for the 1:1, 1:3, and 1:9 dilutions, respectively). Repetition of the experiment three times with three different retinal ganglion cell preparations produced the same result.
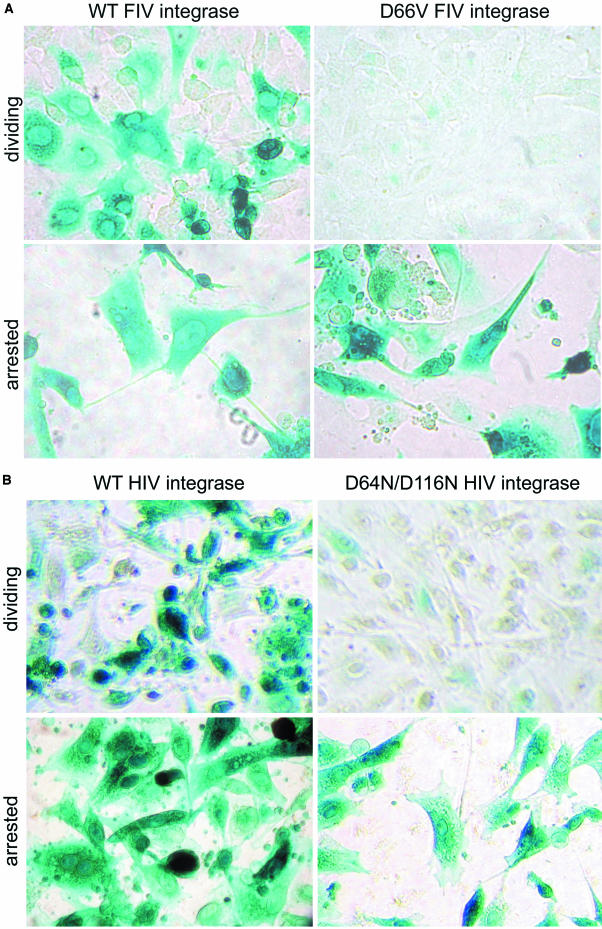
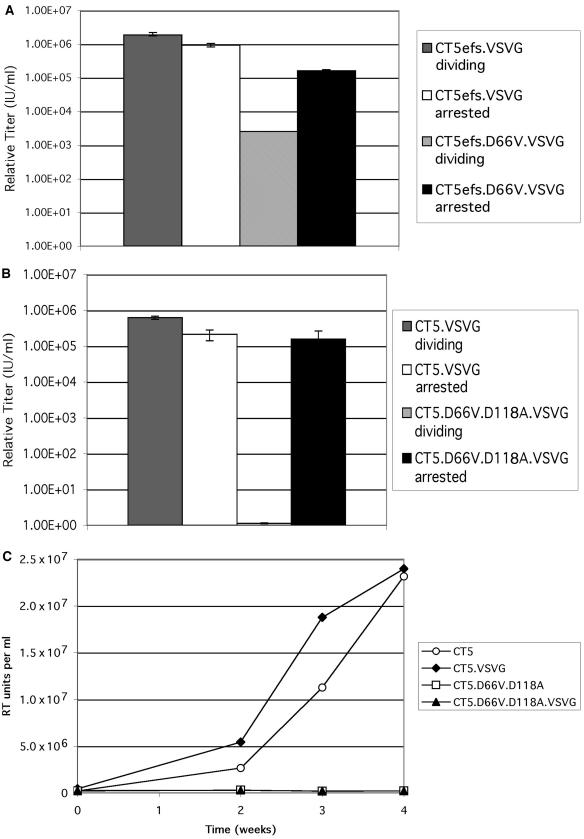
To test the hypothesis that the unexpected results in retinal ganglion cells were related to cell cycle status, we initially examined dividing and growth-arrested (with 15 μg of aphidicolin/ml) CrFK fibroblasts. In surprising contrast to the results shown in Fig. 2, we observed transgene expression by the D66V and D66V/D118A FIV IN mutants that was indistinguishable from that of the intact IN vector (Fig. 4A). At a multiplicity of infection (MOI) of 5, the WT vector and the RT-normalized D66V/D118A vector transduced 99 and 97% of cells, respectively. The level of β-galactosidase activity per cell, estimated by a visual inspection of the converted substrate, was also at least equivalent to, and in some experiments was more than, that for the WT vector in the nondividing cells (Fig. 4A). The mock vector produced no transduction. Similar cell-cycle-dependent transgene expression was seen with the IN mutant HIV-1 lacZ vector (Fig. 4B) and with an FIV gfp vector (data not shown). The same equivalence for the WT IN vector and the class I IN mutant vector in growth-arrested cells was seen with lower MOI infections, which as for the experiments shown in Fig. 4, caused percentages of transduction that were consistent with Poisson distribution predictions. Some variation was discernible, however. The percent transduction in growth-arrested HT1080 cells transduced with a double IN mutant FIV vector was equal to that for the WT FIV vector, while that of the HIV-1 double IN mutant vector was 38% that of the WT HIV-1 vector. Consistent with the previous results of others for class I IN mutant, internally promoted HIV-1 vectors in dividing cells, quite a low level of expression of the IN mutant FIV and HIV-1 vectors could be detected in dividing cells at the 48-h time point, but β-galactosidase activity was considerably lower in these cells than in nondividing cells (Fig. 4) and this disappeared completely after the cells were passaged again at 96 h (data not shown).
FIG.4.
Cell cycle status and vector expression. CrFK cells were treated with 15 μg of aphidicolin/ml or left untreated and were then transduced 24 h later (with replenishment of aphidicolin). Forty-eight hours after transduction (72 h into growth arrest), cells were fixed and stained with X-Gal. (A) FIV vector. Images with RT activity-normalized IN intact (left) or IN mutant (right) FIV vectors are shown (MOI = 5). The top panels show dividing cells, and the bottom panels show arrested cells. (B) HIV-1 vector. Images with RT-normalized IN intact (left) or IN mutant (right) HIV-1 vectors are shown (MOI = 5). The top panels show dividing cells, and the bottom panels show arrested cells.
Effects of release from growth arrest on transgene expression.
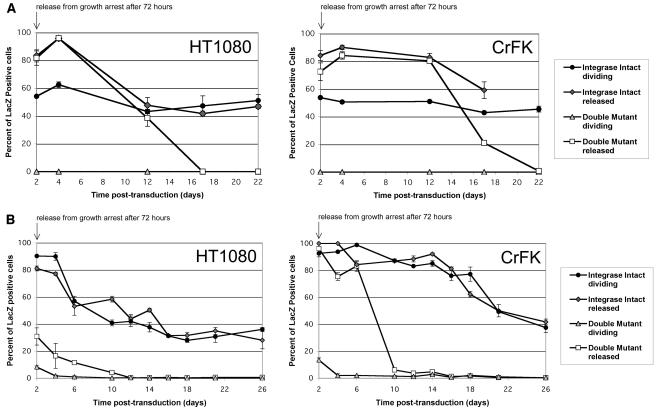
Having identified a marked effect by growth arrest, we then examined the effect of allowing cells to reenter the cell cycle (Fig. 5). Again, transgene expression mediated by the IN mutant vectors was equivalent to that of WT vectors. After 24 and 48 h, the relative titers were even 20 to 40% higher for IN mutants. However, when the aphidicolin-treated transduced cells were moved to aphidicolin-free medium 48 h after transduction, cell division resumed after approximately 36 h and the high percent transduction in cells transduced with IN mutant FIV and HIV-1 vectors declined steadily and was lost by day 10. In contrast, the transgene expression of intact IN FIV and HIV-1 vectors was stable and durable to repeated passaging (Fig. 5). In addition to confirming the WT-equivalent expression in growth-arrested cells, these experiments confirmed that the class I IN FIV mutants do not produce a stable, heritable level of transduction that survives mitosis. Reentry into the cell cycle resulted in the loss of expression by mutant but not WT IN vectors. We therefore proceeded to correlate the expression patterns with the integration states, forms, and quantities of viral cDNAs in cells.
FIG. 5.
Reentry into the cell cycle results in loss of expression for class I IN mutants. (A) FIV vector. The percent LacZ expression over time is shown. Growth-arrested CrFK and HT1080 cells were treated with 15 μg of aphidicolin/ml 24 h before transduction. Arrested and dividing cells were transduced with RT-normalized stocks of IN mutant (D66V/D118A) and IN intact FIV lacZ vector at an MOI of 5. (B) HIV-1 vector. For comparison, arrested and dividing cells were also transduced with RT-normalized stocks of IN mutant (D64N/D116N) and IN intact HIV-1 lacZ vector at an MOI of 5. Percentages of cells expressing the lacZ transgene were measured for 3 weeks by fixing and staining them with X-Gal. Bars represent standard deviations of triplicate experiments.
Analyses of viral DNA in cells transduced with IN mutant FIV vectors.
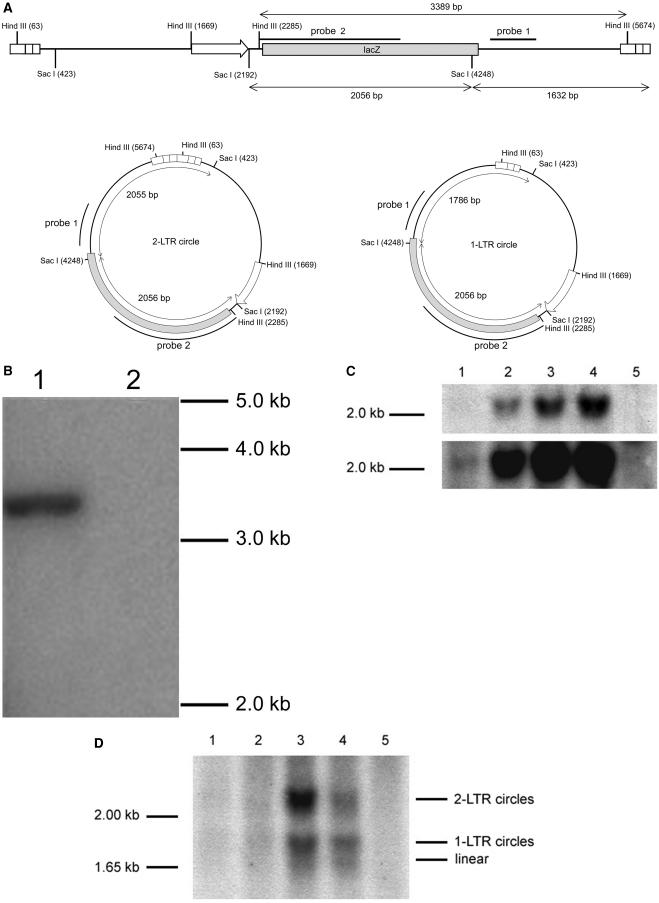
The ability of vectors to integrate in cells was first examined by Southern blotting of total cellular DNAs from cells transduced at an MOI of 5 with RT-normalized IN mutant (D66V) and WT FIV vectors (Fig. 6). The relevant restriction enzyme sites and probes used for these and subsequent experiments are shown in Fig. 6A. Total cellular DNAs were prepared at 37 days posttransduction from cells that were released from growth arrest at day 2 after transduction, digested with HindIII, and probed with a labeled MluI fragment (probe 1). Cells transduced with the WT IN vector showed stable persistence of the transgene (Fig. 6B, lane 1). In contrast, after release from growth arrest, cells transduced with the IN mutant vector did not (Fig. 6B, lane 2), which correlated with the loss of transgene expression seen in the experiments for Fig. 5 and confirmed the class I properties.
FIG.6.
Stable transgene integration: comparison of WT and IN mutant FIV vector. (A) Probes and restriction enzymes used for the data shown in panels B to D. (B) Southern blot. CrFK cells were transduced with RT-normalized stocks of WT and single IN mutant (D66V) FIV lacZ vectors. Genomic DNA was collected 37 days after transduction and was digested with HindIII. The blot was hybridized with probe 1, which detects a 3,389-bp internal HindIII vector fragment. Lane 1, WT IN vector; lane 2, D66V vector. (C) Characterization of unintegrated DNA by total vector DNA quantitation at 48 h. Dividing and growth-arrested CrFK cells were transduced with RT activity-normalized stocks of WT and double IN mutant (D66V/D118A) FIV lacZ vectors. The total cellular DNA was purified 48 h after transduction, digested to completion with SacI, and Southern blotted with a lacZ probe (probe 2) that detects a 2,056-bp internal SacI fragment present in all forms (integrated, linear, and circular) of vector cDNA. The bottom panel shows a more exposed version of the blot in the top panel. Lane 1, dividing cells transduced with D66V/D118A FIV vector; lane 2, dividing cells transduced with WT FIV vector; lane 3, growth-arrested cells transduced with D66V/D118A FIV vector; lane 4, growth-arrested cells transduced with WT FIV vector; lane 5, untransduced cells. (D) Characterization of unintegrated DNA by detection of different forms of unintegrated vector DNA at 48 h. CrFK cells were transduced with RT-normalized stocks of WT and double IN mutant (D66V/D118A) FIV lacZ vectors. The total DNA was collected 48 h after transduction and digested with SacI. The probe (probe 1) detects a 2,055-bp fragment from 2-LTR circles, a 1,786-bp fragment from 1-LTR circles, a 1,637-bp fragment from linear cDNA, and junction fragments larger than 1,637 bp from integrated cDNA. Lane 1, dividing cells, D66V/D118A vector; lane 2, dividing cells, WT vector; lane 3, growth-arrested cells, D66V/D118A vector; lane 4, growth-arrested cells, WT vector; lane 5, untransduced control cells.
Total viral cDNA levels in cells were then examined by Southern blotting at an earlier time point, 48 h after WT and IN mutant (D66V/D118A) FIV vector transduction (Fig. 6C). The probe used was an internal MluI fragment (Fig. 6A, probe 2) that detects all forms of vector cDNA, including integrated, linear, and 1- and 2-LTR circular forms, after digestion with SacI. Dividing cells transduced with the D66V/D188A FIV vector (Fig. 6C, lane 1) showed very little vector cDNA, whereas cells transduced with the WT vector (lane 2) gave a strong signal. In contrast, cells that had their growth arrested with aphidicolin and that were transduced with either the IN mutant (lane 3) or WT (lane 4) vector showed high total vector cDNA levels which were equal to or higher than the total DNA levels in dividing cells. These results indicated that class I IN mutant viral cDNA persists at markedly different levels depending on cell cycle status.
Transgene expression in cells transduced with the IN mutant FIV vector could arise from a variety of unintegrated viral cDNA forms, including 1-LTR circles, 2-LTR circles, and the linear form. Therefore, Southern blotting was used to analyze the relative amounts of the two circular forms and the linear form in the total cellular DNA in dividing and nondividing cells 48 h after transduction with RT activity-normalized double IN mutant and WT FIV vectors (Fig. 6D). Correlating with the high percentages of transduction and beta-galactosidase expression levels, unintegrated viral cDNA linear and circular forms persisted in nondividing cells but not in dividing cells. Linear, 1-LTR circle, and 2-LTR circle DNA forms accumulated to higher levels in growth-arrested cells transduced with the IN mutant FIV vector than in those with the WT vector, with 2-LTR circles being predominant. Both of these results are consistent with the requirement for growth arrest for IN mutant vector protein expression. A comparison of lanes 1 and 2 in Fig. 6D (dividing cells) also reveals a faint smear in lane 2 that is consistent with polyclonal integration junction bands of the expected 1.64-kb or larger size. To corroborate these results, we used real-time PCR to quantify the differences in 2-LTR circle levels present in arrested and dividing cells transduced with RT-normalized D66V/D118A IN mutant and WT IN FIV vectors (Table 1). 2-LTR circles were present at significantly higher levels in nondividing cells transduced with the IN mutant vector. Growth arrest led to a 2.9-fold increase in WT vector 2-LTR circle accumulation at 48 h. When the possibility of integration was also blocked (double IN mutant vector), growth arrest led to a 16.4-fold increase in 2-LTR circle accumulation at 48 h. The level of 2-LTR circles in nondividing cells transduced with the double IN mutant vector was >2.5 times higher than that for WT IN vector 2-LTR circles at this time point.
TABLE 1.
2-LTR circle quantification in dividing and nondividing cellsa
| Cell type | 2-LTR circles at 24 h
|
2-LTR circles at 48 h
|
||
|---|---|---|---|---|
| Wild type | Double mutant | Wild type | Double mutant | |
| Dividing | 7,236 ± 47 | 6,149 ± 39 | 5,245 ± 786 | 2,437 ± 244 |
| Nondividing | 6,948 ± 1,191 | 10,896 ± 87 | 15,112 ± 555 | 39,913 ± 1,370 |
Dividing and growth-arrested CrFK cells were transduced with RT-normalized stocks of wild-type and double integrase mutant (D66V/D118A) FIV lacZ vectors. Total DNA was collected 24 and 48 h after transduction. Values are copies of 2-LTR circle DNA per 500 ng of genomic DNA and are given as means ± standard deviations from three 10-cm dishes for each vector and time point. At 48 h, all values differed significantly from each of the other three (P < 0.02). For the D66V/D118A dividing/nondividing cell comparison at 48 h (16.4-fold increase), P = 0.005. At 24 h, the IN mutant nondividing cell value was significantly different than each dividing cell value (P < 0.01).
IN mutant full-length virus properties.
We next tested cell cycle-dependent expression from the viral LTR in the full-length virus instead of from internal vector promoters. Because the human CXCR4-interacting native FIV envelope is so rapidly and strongly fusogenic in 293T producer cells (41), FIV produced in this way typically has titers of 103 per ml or less. Therefore, VSV-G-pseudotyping in trans was also done during virus production to increase the titer in the first round of infection and thereby widen the dynamic range of the results. Titers of RT activity-normalized stocks of mutant IN and WT IN viruses were determined on dividing and nondividing CrFK cells by immunoperoxidase staining after serial dilution (Fig. 7A and B). This assay detects foci of FIV structural protein expression (44). As expected, the RT activity-adjusted IN mutant viruses produced 3- to 5-log fewer detectable foci than WT viruses in dividing CrFK cells. In contrast, growth-arrested cells infected with IN mutant viruses produced titers of 10 to 30% those of the WT. However, these did not reach or exceed WT levels as was seen with internally promoted transgene-expressing vectors. The IN mutant viruses with an intact FIV envelope did not productively replicate in dividing CrFK cells, while the WT virus did (Fig. 7C), indicating that the basic reproductive ratio (57) is <1 in the absence of integration in this cell line. This result is consistent with data for most cell lines with class I IN mutant HIV-1 (19, 36, 56).
FIG. 7.
Class I IN mutant and WT virus infection of CrFK cells. (A and B) Cells were infected with dilutions of class I mutant and WT viruses, which were RT normalized and tested in parallel on aphidicolin-treated (15 μg/ml) and untreated CrFK cells. Viruses were pseudotyped and experiments included IN mutant and WT versions of virus produced with pCT5efs (FIV env frame shifted) (A) and with pCT5 (FIV env intact) (B). (C) Virus replication time course. Infections were done with equal amounts of RT activity using both pseudotyped and unpseudotyped FIVs. RT activity was measured in supernatants at the indicated times.
DISCUSSION
We have constructed catalytic core FIV IN mutants and have shown that these mutants are functionally analogous in infected cells to previously described active site HIV-1 mutants that target the homologous residues. Integration was blocked, while viral protein processing and particle production were normal, and these class I properties were further validated by subsequent data showing WT-equivalent competence for the generation of reverse-transcribed DNA and the transgene expression in nondividing cells that correlates with stable unintegrated DNA.
Besides establishing class I mutant properties for a non-HIV-1 lentiviral IN for the first time and corroborating and extending studies of in vitro reactions with purified bacterially expressed FIV IN (50), the following four aspects of the present study are the most significant in light of previously published data about unintegrated retroviral DNA: (i) the key distinction observed between nondividing cells and dividing cells; (ii) the study of internally promoted expression from single-round lentiviral vectors; (iii) the focus on both expression and DNA persistence in the same study; and (iv) the comparative analysis of two divergent lentiviruses from different mammalian host groups. The data show that integration and productive replication are blocked by these class I mutants but that internally promoted transgene expression can be equivalent to that of WT lentivirus vector DNA if cells are growth arrested. Unintegrated HIV-1 and FIV cDNA forms do not reside in an intrinsically disfavored or inaccessible state with regard to transcription and protein expression.
The analysis of cell cycle dependence reported here was stimulated when we noted that an IN D66V FIV vector intended as an additional negative control did not perform similarly to a control mock vector in transductions of postmitotic rat retinal ganglion cells and unexpectedly yielded beta-galactosidase expression equivalent to that of the intact IN vector. This result focused our attention on determining the extent to which unintegrated virus DNA is accessible as a template for gene expression, how this phenomenon is revealed by an internal promoter, and its cell cycle dependence. Further experiments showed that despite a block to integration, the class I IN mutant FIV-based vector and similar HIV-1 vectors exhibited robust, WT-equivalent transgene expression, not only in these terminally differentiated neuronal cells, but also in growth-arrested cell lines. Expression was progressively lost upon reentry into the cell cycle, with a time course expected from dilution of the unintegrated DNA proportional to cell division. This high-level transgene expression is mediated by unintegrated linear or circular forms of vector cDNA, which we show accumulate only in growth-arrested cells and do so to higher levels than with WT virus. 2-LTR circles were more prevalent in growth-arrested cells than 1-LTR circles or the linear form, and both circular forms were more prevalent in growth-arrested cells than in dividing cells. The effect of dilutional attrition with cell division is consistent with the DNA quantification data of others (10, 40). The present data extend recent published findings about unintegrated lentiviral DNA by clarifying the circumstances under which the unintegrated DNA is available for transcription and expression and determining the dynamic range that is possible.
The persistence of unintegrated lentiviral DNA in cells and its role in identifying ongoing viral replication are important issues. The present data are consistent with a model in which dilutional attrition is the predominant means by which circular forms diminish in dividing cells (8, 10, 40). It cannot be determined from either our experiments or those of others which form of the unintegrated DNA mediates the expression observed in our growth-arrested cell experiments. Expression may be from circular or linear forms or both. One study suggested that linear forms may degrade faster than 2-LTR circles in cells (40). 2-LTR circles were more prevalent in our transduction experiments (Fig. 6). It is noteworthy that the most abundant form of HIV-1 DNA in resting and activated CD4+ T cells from patients with asymptomatic HIV-1 infections appears to be unintegrated DNA, which is roughly 100-fold more abundant than integrated DNA (12). Most of this DNA appears to be linear and lacks ends that are competent for integration; consistent with this, only a small fraction of it can produce replication-competent virus, as detected with cocultivation methods that involve cell cycling (12). Both 2-LTR and 1-LTR circles are also present in these cells, but in lower amounts than the linear form (12). Li et al. suggested that the double-stranded ends of unintegrated linear viral cDNA mimic chromosomal breaks and may induce apoptosis (31). In order to avoid death, the cell may rapidly convert linear viral cDNA into circles by nonhomologous DNA end-joining machinery or homologous recombination. This may explain why, when compared to the WT, class I IN mutant viruses generate higher levels of LTR circles.
The use of an internal promoter in single-round vectors appears to be a factor in the maximal expression of class I mutants in nondividing cells, and this may therefore be useful (see below). LTR-directed FIV expression was 3- to 10-fold lower than that of the WT (Fig. 7). Class I mutants of FIV did not productively replicate in a susceptible cell line in the present study, and the analogous HIV-1 mutants have been shown to do so only in one or two cell lines in tissue culture (36). It is important to emphasize that no class I IN mutant of any retrovirus has been shown to productively replicate in primary cells.
In contrast to the results with retinal ganglion cells and growth-arrested cell lines, the IN D66V FIV vector did not produce transgene expression persisting to 2 months in rat retinas, with the exception of some neurons in the ONL. One explanation is that the retinal pigment epithelium targeted by these other lentiviral vectors (16) may undergo cell cycling in response to disruption with subretinal injections and transduction. Alternatively, unintegrated DNA may not persist in this tissue for 2 months, the shortest postinjection duration studied, or as seen with HIV-1 LTR-directed expression of class I IN mutant HIV-1 (36), there are likely to be marked cell type-specific differences in the ability of the internally promoted vectors to produce high-level expression.
Class I IN mutant lentiviral vectors might conceivably be used for therapeutic gene delivery without the risk of insertional mutagenesis in postmitotic cell targets permissive to such expression. However, such an approach would discard the signature advantage of retroviral vectors, integration, and other vectors such as adenoviral vectors may be more appropriate for such goals. With the exception of the small numbers of outer nuclear retina layer cells noted above, we have not yet observed class I IN mutant vector transduction in vivo. For preclinical gene therapy experiments in animals, class I mutants will therefore continue to be useful for producing control vectors that document a requirement for integration, as shown here for retinal pigment epithelium and previously for the brain with HIV-1 vectors (4).
More directly, however, these results revealing robust expression from unintegrated lentiviral vector DNAs under particular conditions of cell type and growth arrest could facilitate basic lentivirological research. IN has been implicated in nuclear import of the PIC in growth-arrested cells, and the detection of 2-LTR circle DNA in bulk cell populations is frequently used to monitor PIC nuclear import. The methods described in the present report, which can be used both with quantitative enzyme (β-galactosidase or luciferase) assays and with single cell marking genes, could facilitate the study of the role of IN and other PIC components in preintegration phases of lentiviral infection by enabling the monitoring of transcriptionally competent lentiviral nuclear DNAs in the absence of integration. The important features for this purpose are the capabilities to simultaneously (i) specifically monitor unintegrated nuclear cDNA forms in individual cells, (ii) confine the analysis to nondividing cells, (iii) restrict ascertainment to that subset of unintegrated reverse-transcribed DNA that is transcriptionally functional, and (iv) uncouple other potential functions of IN from integration. While these results apply to both HIV-1 and FIV, the comparative simplicity of the FIV genome (vpr/vpx, nef, and vpu are not represented) may also provide some advantages for the investigation of singular lentiviral properties such as PIC nuclear import.
Acknowledgments
We thank Alan Engelman and Mark Muesing for providing HIV IN mutants, Margaret Barr for FIV-infected cat plasma, Wulin Teo for technical assistance with vector preparation, and other laboratory colleagues for scientific suggestions and help with experiments.
This work was supported by NIH grants AI47536 (E.M.P) and EY12798 (J.M.H.) and by a Pfizer Scholars Grant for New Faculty (E.M.P.).
REFERENCES
- 1.Ansari-Lari, M. A., L. A. Donehower, and R. A. Gibbs. 1995. Analysis of human immunodeficiency virus type 1 integrase mutants. Virology 211:332-335. [DOI] [PubMed] [Google Scholar]
- 2.Barres, B. A., B. E. Silverstein, D. P. Corey, and L. L. Chun. 1988. Immunological, morphological, and electrophysiological variation among retinal ganglion cells purified by panning. Neuron 1:791-803. [DOI] [PubMed] [Google Scholar]
- 3.Bergeron, L., and J. Sodroski. 1992. Dissociation of unintegrated viral DNA accumulation from single-cell lysis induced by human immunodeficiency virus type 1. J. Virol. 66:5777-5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bloemer, U., L. Naldini, T. Kafri, D. Trono, I. M. Verma, and F. H. Gage. 1997. Highly efficient and sustained gene transfer in adult neurons with a lentivirus vector. J. Virol. 71:6641-6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, P. O. 1999. Integration, p. 161-203. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 6.Brussel, A., D. Mathez, S. Broche-Pierre, R. Lancar, T. Calvez, P. Sonigo, and J. Leibowitch. 2003. Longitudinal monitoring of 2-long terminal repeat circles in peripheral blood mononuclear cells from patients with chronic HIV-1 infection. AIDS 17:645-652. [DOI] [PubMed] [Google Scholar]
- 7.Bukrinsky, M. I., N. Sharova, M. P. Dempsey, T. L. Stanwick, A. G. Bukrinskaya, S. Haggerty, and M. Stevenson. 1992. Active nuclear import of human immunodeficiency virus type 1 preintegration complexes. Proc. Natl. Acad. Sci. USA 89:6580-6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bushman, F. 2003. Measuring covert HIV replication during HAART: the abundance of 2-LTR circles is not a reliable marker. AIDS 17:749-750. [DOI] [PubMed] [Google Scholar]
- 9.Bushman, F. D., A. Engelman, I. Palmer, P. Wingfield, and R. Craigie. 1993. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc. Natl. Acad. Sci. USA 90:3428-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butler, S. L., E. P. Johnson, and F. D. Bushman. 2002. Human immunodeficiency virus cDNA metabolism: notable stability of two-long terminal repeat circles. J. Virol. 76:3739-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cara, A., F. Guarnaccia, M. S. Reitz, R. C. Gallo, and F. Lori. 1995. Self-limiting, cell type-dependent replication of an integrase-defective human immunodeficiency virus type 1 in human primary macrophages but not T lymphocytes. Virology 208:242-248. [DOI] [PubMed] [Google Scholar]
- 12.Chun, T. W., L. Carruth, D. Finzi, X. Shen, J. A. DiGiuseppe, H. Taylor, M. Hermankova, K. Chadwick, J. Margolick, T. C. Quinn, Y. H. Kuo, R. Brookmeyer, M. A. Zeiger, P. Barditch-Crovo, and R. F. Siliciano. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387:183-188. [DOI] [PubMed] [Google Scholar]
- 13.Colicelli, J., and S. P. Goff. 1985. Mutants and pseudorevertants of Moloney murine leukemia virus with alterations at the integration site. Cell 42:573-580. [DOI] [PubMed] [Google Scholar]
- 14.Daniel, R., R. A. Katz, and A. M. Skalka. 1999. A role for DNA-PK in retroviral DNA integration. Science 284:644-647. [DOI] [PubMed] [Google Scholar]
- 15.Drelich, M., R. Wilhelm, and J. Mous. 1992. Identification of amino acid residues critical for endonuclease and integration activities of HIV-1 IN protein in vitro. Virology 188:459-468. [DOI] [PubMed] [Google Scholar]
- 16.Duisit, G., H. Conrath, S. Saleun, S. Folliot, N. Provost, F. L. Cosset, V. Sandrin, P. Moullier, and F. Rolling. 2002. Five recombinant simian immunodeficiency virus pseudotypes lead to exclusive transduction of retinal pigmented epithelium in rat. Mol. Ther. 6:446-454. [DOI] [PubMed] [Google Scholar]
- 17.Engelman, A. 1999. In vivo analysis of retroviral integrase structure and function. Adv. Virus Res. 52:411-426. [DOI] [PubMed] [Google Scholar]
- 18.Engelman, A., and R. Craigie. 1992. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J. Virol. 66:6361-6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engelman, A., G. Englund, J. M. Orenstein, M. A. Martin, and R. Craigie. 1995. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J. Virol. 69:2729-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engelman, A., A. B. Hickman, and R. Craigie. 1994. The core and carboxyl-terminal domains of the integrase protein of human immunodeficiency virus type 1 each contribute to nonspecific DNA binding. J. Virol. 68:5911-5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Englund, G., T. S. Theodore, E. O. Freed, A. Engleman, and M. A. Martin. 1995. Integration is required for productive infection of monocyte-derived macrophages by human immunodeficiency virus type 1. J. Virol. 69:3216-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farnet, C. M., and W. A. Haseltine. 1991. Circularization of human immunodeficiency virus type 1 DNA in vitro. J. Virol. 65:6942-6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallay, P., T. Hope, D. Chin, and D. Trono. 1997. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. USA 94:9825-9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gulizia, J., M. P. Dempsey, N. Sharova, M. I. Bukrinsky, L. Spitz, D. Goldfarb, and M. Stevenson. 1994. Reduced nuclear import of human immunodeficiency virus type 1 preintegration complexes in the presence of a prototypic nuclear targeting signal. J. Virol. 68:2021-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kemler, I., R. Barraza, and E. M. Poeschla. 2002. Mapping of the encapsidation determinants of feline immunodeficiency virus. J. Virol. 76:11889-11903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khan, E., J. P. Mack, R. A. Katz, J. Kulkosky, and A. M. Skalka. 1991. Retroviral integrase domains: DNA binding and the recognition of LTR sequences. Nucleic Acids Res. 19:851-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J. Virol. 66:2232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leavitt, A. D., G. Robles, N. Alesandro, and H. E. Varmus. 1996. Human immunodeficiency virus type 1 integrase mutants retain in vitro integrase activity yet fail to integrate viral DNA efficiently during infection. J. Virol. 70:721-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leavitt, A. D., L. Shiue, and H. E. Varmus. 1993. Site-directed mutagenesis of HIV-1 integrase demonstrates differential effects on integrase functions in vitro. J. Biol. Chem. 268:2113-2119. [PubMed] [Google Scholar]
- 30.Lewis, P., M. Hensel, and M. Emerman. 1992. Human immunodeficiency virus infection of cells arrested in the cell cycle. EMBO J. 11:3053-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, L., J. M. Olvera, K. E. Yoder, R. S. Mitchell, S. L. Butler, M. Lieber, S. L. Martin, and F. D. Bushman. 2001. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 20:3272-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loewen, N., C. Bahler, W. Teo, T. Whitwam, M. Peretz, R. Xu, M. Fautsch, D. H. Johnson, and E. M. Poeschla. 2002. Preservation of aqueous outflow facility after second-generation FIV vector-mediated expression of marker genes in anterior segments of human eyes. Investig. Ophthalmol. Vis. Sci. 43:3686-3690. [PubMed] [Google Scholar]
- 33.Loewen, N., R. Barraza, T. Whitwam, D. Saenz, I. Kemler, and E. Poeschla. 2003. FIV vectors, p. 251-271. In M. Federico (ed.), Lentivirus gene engineering protocols. Humana Press, New York, N.Y. [DOI] [PubMed]
- 34.Loewen, N., M. Fautsch, M. Peretz, C. Bahler, J. D. Cameron, D. H. Johnson, and E. M. Poeschla. 2001. Genetic modification of human trabecular meshwork with lentiviral vectors. Hum. Gene Ther. 12:2109-2119. [DOI] [PubMed] [Google Scholar]
- 35.Masuda, T., V. Planelles, P. Krogstad, and I. S. Chen. 1995. Genetic analysis of human immunodeficiency virus type 1 integrase and the U3 att site: unusual phenotype of mutants in the zinc finger-like domain. J. Virol. 69:6687-6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakajima, N., R. Lu, and A. Engelman. 2001. Human immunodeficiency virus type 1 replication in the absence of integrase-mediated DNA recombination: definition of permissive and nonpermissive T-cell lines. J. Virol. 75:7944-7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pang, S., Y. Koyanagi, S. Miles, C. Wiley, H. V. Vinters, and I. S. Chen. 1990. High levels of unintegrated HIV-1 DNA in brain tissue of AIDS dementia patients. Nature 343:85-89. [DOI] [PubMed] [Google Scholar]
- 38.Panther, L. A., R. W. Coombs, S. A. Aung, C. dela Rosa, D. Gretch, and L. Corey. 1999. Unintegrated HIV-1 circular 2-LTR proviral DNA as a marker of recently infected cells: relative effect of recombinant CD4, zidovudine, and saquinavir in vitro. J. Med. Virol. 58:165-173. [DOI] [PubMed] [Google Scholar]
- 39.Pauza, C. D., P. Trivedi, T. S. McKechnie, D. D. Richman, and F. M. Graziano. 1994. 2-LTR circular viral DNA as a marker for human immunodeficiency virus type 1 infection in vivo. Virology 205:470-478. [DOI] [PubMed] [Google Scholar]
- 40.Pierson, T. C., T. L. Kieffer, C. T. Ruff, C. Buck, S. J. Gange, and R. F. Siliciano. 2002. Intrinsic stability of episomal circles formed during human immunodeficiency virus type 1 replication. J. Virol. 76:4138-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poeschla, E., and D. Looney. 1998. CXCR4 is required by a non-primate lentivirus: heterologous expression of feline immunodeficiency virus in human, rodent and feline cells. J. Virol. 72:6858-6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poeschla, E., F. Wong-Staal, and D. Looney. 1998. Efficient transduction of nondividing cells by feline immunodeficiency virus lentiviral vectors. Nat. Med. 4:354-357. [DOI] [PubMed] [Google Scholar]
- 43.Poon, B., and I. S. Chen. 2003. Human immunodeficiency virus type 1 (HIV-1) Vpr enhances expression from unintegrated HIV-1 DNA. J. Virol. 77:3962-3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Remington, K. M., B. Chesebro, K. Wehrly, N. C. Pedersen, and T. W. North. 1991. Mutants of feline immunodeficiency virus resistant to 3′-azido-3′-deoxythymidine. J. Virol. 65:308-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roth, M. J., P. Schwartzberg, N. Tanese, and S. P. Goff. 1990. Analysis of mutations in the integration function of Moloney murine leukemia virus: effects on DNA binding and cutting. J. Virol. 64:4709-4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saenz, D. T., and E. M. Poeschla. FIV: from lentivirus to lentivector. Gene Ther., in press. [DOI] [PubMed]
- 47.Sakai, H., M. Kawamura, J. Sakuragi, S. Sakuragi, R. Shibata, A. Ishimoto, N. Ono, S. Ueda, and A. Adachi. 1993. Integration is essential for efficient gene expression of human immunodeficiency virus type 1. J. Virol. 67:1169-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharkey, M. E., I. Teo, T. Greenough, N. Sharova, K. Luzuriaga, J. L. Sullivan, R. P. Bucy, L. G. Kostrikis, A. Haase, C. Veryard, R. E. Davaro, S. H. Cheeseman, J. S. Daly, C. Bova, R. T. Ellison III, B. Mady, K. K. Lai, G. Moyle, M. Nelson, B. Gazzard, S. Shaunak, and M. Stevenson. 2000. Persistence of episomal HIV-1 infection intermediates in patients on highly active anti-retroviral therapy. Nat. Med. 6:76-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shibagaki, Y., and S. A. Chow. 1997. Central core domain of retroviral integrase is responsible for target site selection. J. Biol. Chem. 272:8361-8369. [DOI] [PubMed] [Google Scholar]
- 50.Shibagaki, Y., M. L. Holmes, R. S. Appa, and S. A. Chow. 1997. Characterization of feline immunodeficiency virus integrase and analysis of functional domains. Virology 230:1-10. [DOI] [PubMed] [Google Scholar]
- 51.Shoemaker, C., J. Hoffman, S. P. Goff, and D. Baltimore. 1981. Intramolecular integration within Moloney murine leukemia virus DNA. J. Virol. 40:164-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stevenson, M., S. Haggerty, C. A. Lamonica, C. M. Meier, S. K. Welch, and A. J. Wasiak. 1990. Integration is not necessary for expression of human immunodeficiency virus type 1 protein products. J. Virol. 64:2421-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Talbott, R. L., E. E. Sparger, K. M. Lovelace, W. M. Fitch, N. C. Pedersen, P. A. Luciw, and J. H. Elder. 1989. Nucleotide sequence and genomic organization of feline immunodeficiency virus. Proc. Natl. Acad. Sci. USA 86:5743-5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Teo, I., C. Veryard, H. Barnes, S. F. An, M. Jones, P. L. Lantos, P. Luthert, and S. Shaunak. 1997. Circular forms of unintegrated human immunodeficiency virus type 1 DNA and high levels of viral protein expression: association with dementia and multinucleated giant cells in the brains of patients with AIDS. J. Virol. 71:2928-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tomonaga, K., S. I. Itagaki, H. Kashiwase, Y. Kawaguchi, Y. Inoshima, Y. Ikeda, and T. Mikami. 1998. Characterization of an integrase mutant of feline immunodeficiency virus. Arch. Virol. 143:1-14. [DOI] [PubMed] [Google Scholar]
- 56.Wiskerchen, M., and M. A. Muesing. 1995. Human immunodeficiency virus type 1 integrase: effects of mutations on viral ability to integrate, direct viral gene expression from unintegrated viral DNA templates, and sustain viral propagation in primary cells. J. Virol. 69:376-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wodarz, D., and M. A. Nowak. 2002. Mathematical models of HIV pathogenesis and treatment. Bioessays 24:1178-1187. [DOI] [PubMed] [Google Scholar]
- 58.Woerner, A. M., M. Klutch, J. G. Levin, and C. J. Marcus-Sekura. 1992. Localization of DNA binding activity of HIV-1 integrase to the C-terminal half of the protein. AIDS Res. Hum. Retrovir. 8:297-304. [DOI] [PubMed] [Google Scholar]
- 59.Wu, Y., and J. W. Marsh. 2001. Selective transcription and modulation of resting T cell activity by preintegrated HIV DNA. Science 293:1503-1506. [DOI] [PubMed] [Google Scholar]
- 60.Zazzi, M., L. Romano, M. Catucci, G. Venturi, A. De Milito, P. Almi, A. Gonnelli, M. Rubino, U. Occhini, and P. E. Valensin. 1997. Evaluation of the presence of 2-LTR HIV-1 unintegrated DNA as a simple molecular predictor of disease progression. J. Med. Virol. 52:20-25. [DOI] [PubMed] [Google Scholar]
- 61.Zufferey, R., D. Nagy, R. J. Mandel, L. Naldini, and D. Trono. 1997. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Bio/Technol. 15:871-875. [DOI] [PubMed] [Google Scholar]