Abstract
Simian virus 40 large T antigen (TAg) is a viral oncoprotein that can promote cellular transformation. TAg's transforming activity results in part by binding and inactivating key tumor suppressors, including p53 and the retinoblastoma protein (pRb). We have identified a TAg-associated 185-kDa protein that has significant homology to the cullin family of E3 ubiquitin ligases. TAg binds to an SCF-like complex that contains p185/Cul7, Rbx1, and the F box protein Fbw6. This SCF-like complex binds to an N-terminal region of TAg. Several p185/Cul7-binding-deficient mutants of TAg were generated that retained binding to pRb and p53 and were capable of overcoming Rb-mediated repression of E2F transcription. Despite binding to pRb and p53, these p185/Cul7-binding-defective mutants of TAg were unable to transform primary mouse embryo fibroblasts. Cells expressing p185/Cul7-binding-defective mutants of TAg were unable to grow to high density or grow in an anchorage-independent manner as determined by growth in soft agar. Considering the significance of other TAg-interacting proteins in regulation of the cell cycle, p185/Cul7 may also regulate an important growth control pathway.
Simian virus 40 (SV40) large tumor antigen (TAg) is a powerful oncoprotein capable of transforming a variety of cell types and inducing tumor formation in animal models. The transforming activity of TAg is thought to be dependent, at least in part, upon binding and inactivating certain key regulators of the cell cycle, such as the tumor suppressors p53 and pRb as well as the pRb-related proteins p107 and p130 (1). The study of mechanisms by which TAg inactivates and exploits the cell cycle regulatory factors has led to a better understanding of the regulation of cell cycle.
TAg binding to pRb disrupts the ability of pRb to repress the E2F family of transcription factors. During the G1 phase of the cell cycle or under growth arrest conditions including serum starvation, pRb is underphosphorylated and bound to specific members of the E2F family. pRb recruits transcriptional corepressors to the complex, resulting in repression of promoters that contain E2F binding sites. In response to growth signals, pRb becomes phosphorylated in late G1 and dissociates from E2F during the S and G2 phases of the cell cycle. When dissociated from pRb, E2F promotes the expression of many genes required for entry into the S phase of the cell cycle. TAg binding to underphosphorylated pRb results in dissociation of pRb from the E2F transcription factors and loss of pRb repression. By inactivating pRb, TAg can promote entry into the cell cycle under conditions when cells would have normally remained in a growth-arrested state. This activity contributes to the growth of cells in an anchorage-independent manner and to growth under low-serum conditions. The LxCxE motif (residues 103 to 107) of TAg contributes to pRb binding. Mutation or deletion of the LxCxE motif disables TAg binding to pRb and renders TAg incapable of transformation. The LxCxE motif of TAg also binds to the pRb-related proteins p107 and p130 and contributes to the inactivation of their growth-suppressing function (66, 67).
A second transforming domain of TAg is contained within the N-terminal 82 residues and forms a DnaJ domain. The DnaJ domain of TAg binds specifically to Hsc70 and stimulates the ATPase hydrolysis activity of Hsc70. The DnaJ domain of TAg contributes to the inactivation of the pRb family (7, 47). The DnaJ domain cooperates with the LxCxE motif to release pRb family members from E2F and override the repression of E2F transcriptional activity. Mutation of the DnaJ domain reduces the ability of TAg to inactivate the growth-suppressing activities of pRb family members. The DnaJ domain of TAg also contributes to the ability of TAg to replicate SV40 ori-containing DNA, although the specific mechanism is not well understood.
A third transforming domain of TAg serves to bind to the p53 tumor suppressor. The p53 binding domain is bipartite, formed by residues 351 to 450 and 532 to 627 (28). TAg binds to the DNA binding domain of p53, resulting in inactivation of p53's ability to serve as a transcription factor (5, 16, 57). TAg binding to p53 results in decreased expression of p53-dependent genes, which contributes to growth arrest and apoptosis (24, 50). Binding to p53 also contributes to TAg's ability to immortalize primary mouse embryo fibroblasts (MEFs) and increases the life span of a variety of primary cell types (28, 49). The combination of pRb and p53 inactivation by TAg results in cell cycle entry and decreased sensitivity to growth arrest signals and apoptosis.
It has been suggested that in addition to inactivation of p53 and the pRb family, TAg has additional activities that contribute to transformation (10, 48). TAg binds to the coactivators p300 and CBP and perturbs their ability to modulate transcription. The p300/CBP binding domain of TAg overlaps with the p53 binding domain. Although, it has not been determined whether interaction with p300 and CBP contributes to TAg transformation (3, 15). TAg has also been reported to bind to the DNA damage response proteins Rad50, Mre11, and Nbs1 (14, 34). However, the contribution to cellular transformation by TAg has not been defined for these factors.
In mammalian cells, at least six cullin species have been reported to date, including Cul1, -2, -3, -4a, -4b, and -5. A seventh member of the mammalian cullin family, Cul7, was recently identified (2, 13). Cullins form the scaffold for multisubunit complexes that promote the ubiquitination of targeted substrates (17). Cul1 forms a specific complex known as SCF with Skp1 and one of several F box proteins (4, 36, 38, 39). The SCF complex with the F box protein β-TrCP (Fbw1) targets β-catenin and IκK for ubiquitination and subsequent degradation by the proteasome (18, 25, 65). Cul1 can also bind to the F box protein Skp2 that recruits the CDK inhibitor p27 and the pRb-related protein p130 for ubiquitination and degradation (8, 55, 58, 63). Cul1 also binds to Fbw7, which promotes the degradation of cyclin E (29, 40, 51). Cullins are covalently modified by Nedd8, a ubiquitin-like molecule that contributes to recruitment of the RING-containing protein Rbx1/Roc1 (32, 44). Rbx1 recruits a ubiquitin-conjugating enzyme (E2) to the SCF complex (27).
Kohrman and Imperiale first reported the association of TAg with an unknown 185-kDa cellular protein (30). Using various deletion constructs of TAg, they determined that the N-terminal 121 residues of TAg contributed to p185 binding. Field and colleagues later cloned a TAg-associated 193-kDa protein that could bind to the N-terminal 147 residues of TAg. They proposed that p193 contained a Bcl-2 homology domain (BH3) that behaved as a proapoptotic factor (62). We demonstrate that the p185 TAg-associated protein is identical to the 193-kDa previously reported TAg-associated protein and to the recently identified Cul7. In addition, we identify specific mutant constructs of TAg that lose binding to p185/Cul7 and are unable to transform primary MEFs. The mutant TAgs define a new transforming domain that is linked to p185/Cul7 binding.
MATERIALS AND METHODS
Plasmids.
The TAg cDNA-expressing vectors pSG5-T, K1, H42Q, and Δ434-444 have been described previously (9, 52, 66). Mutations in SV40 large TAg were generated by PCR-based site-directed mutagenesis, cloned into pSG5 (Stratagene), and verified by sequencing. Epitope-tagged versions of Fbw6 and Cul7 were generated by PCR and verified by sequencing.
The luciferase promoter reporter plasmids containing 3xE2F (31) or mutant E2F (3xmE2F) sites (41) have been described previously. Luciferase assays were performed using the dual-luciferase reporter assay kit according to the manufacturer's recommendations (Promega).
Cells.
All cells were cultured in complete medium containing Dulbecco's modified Eagle's medium with 10% Fetal Clone I serum (FCIS; HyClone). Primary MEFs were prepared from 13.5-day C57BL/6 mouse embryos (Taconic) or from Cul7/p185 knockout (−/−) mice (2). Subconfluent 10-cm plates of MEFs, passage 2 to 5, were transfected with plasmids pSG5-TAg and pEpuro in a 5:1 ratio (5 μg of total DNA) using Plus/Lipofectamine (GIBCO) (66). Three hours following transfection, the medium was replaced with complete medium and then 48 h later with fresh medium containing puromycin (2 μg/ml). The cells were selected in the presence of puromycin for 2 to 3 weeks. Colonies were pooled and expanded for further analysis. The retroviral construct pBABE-puro-T containing a cDNA for large TAg was also used to express TAg in MEFs.
A2P2 cells stably expressing hemagglutinin (HA)-tagged human p185 were generated by transfecting U2-OS cells (American Type Culture Collection) with pcDNA3-HA-p185 and pEpuro, followed by selection in puromycin. Several clones were obtained and selected for the highest levels of p185 expression.
Antibodies.
For immunoprecipitation and Western blot analysis, the following antibodies were used: for TAg, PAb419 and PAb430 (23); for p130, C-20 (Santa Cruz); for p53, 240 (Neomarkers); for pRb, XZ55 (Neomarkers); for vinculin, hVIN1 (Sigma); for ROC-1, Ab-1 (Neomarkers); and T7 tag (Novagen).
To generate a p185/Cul7 monoclonal antibody, BALB/cByJ-Rb(8.12)5Bnr mice (JAX Research Systems) were immunized with a glutathione S-transferase (GST) fusion protein containing the first 127 residues of human p185/Cul7. Splenocytes were fused with NS-1 myeloma cells according to standard protocols. The hybridoma SA12 (IgG1κ) was subcloned by limiting dilution.
Protein analysis.
GST fusion proteins of TAg were generated in pGEX2T and expressed in the BL21 strain of Escherichia coli. The fusion protein was isolated on GST-Sepharose, cleaved with thrombin, and purified by high-performance liquid chromatography in GCB buffer (50 mM Tris HCl [pH 8.0], 200 mM NaCl, 2 mM EDTA, 10 mM β-mercaptoethanol, 1.0% Triton X-100). Approximately 108 NIH 3T3 cells (American Type Culture Collection) were lysed in EBC (50 mM Tris-HCl [pH 8.0], 120 mM NaCl, and 0.5% NP-40) containing protease inhibitors (Calbiochem), mixed with 1.0 μg of purified T1-135, immunoprecipitated with PAb419, and separated in a sodium dodecyl sulfate-6% polyacrylamide gel electrophoresis (SDS-PAGE). A Coomassie-stained band of 185 kDa was analyzed at the Harvard Microchemistry Facility by tandem mass spectrometry on a Finnigan LCQ quadrupole ion trap mass spectrometer.
For immunoprecipitations, cells were lysed in NET-N buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, 0.5% NP-40). Lysates were incubated in primary antibody and 20 μl of 50% Sepharose-protein A (Pharmacia) at 4°C for 2 h. The immunoprecipitates were washed four times with NET-N buffer and then boiled in sample buffer and separated in an SDS-PAGE.
For Western blotting, nitrocellulose membranes were blocked in TBS-T buffer (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.05% Tween-20) containing 5% milk for 30 min. Following blocking, the membrane was incubated in the primary antibody in 1% bovine serum albumin in TBS-T for 2 h at room temperature or overnight at 4°C. After washing with TBS-T, the membranes were incubated with horseradish peroxidase-conjugated secondary antibody for 45 min and developed using Super Signal West Pico (Pierce).
Transformation assays.
Anchorage-independent growth of MEFs was determined in 35-mm wells. Each well was layered with 2 ml of complete medium containing 0.8% agarose. Cells were trypsinized and resuspended into complete medium containing 10% FCIS and 0.4% agarose at 38°C, layered on top of the 0.8% agarose, and allowed to solidify at room temperature. Cells were incubated at 37°C and fed every 4 to 5 days with complete medium containing 0.4% agarose for 2 to 3 weeks. Clusters of cells containing four or more cells were scored as colonies. For assays of growth to high density, 50,000 cells were seeded in 60-mm plates in triplicate and fed every other day for 15 to 20 days. Cells were trypsinized and counted in a Coulter Counter.
SV40 virus.
The pSV-B3 genomic DNA construct in pBR322 was excised with BamHI and subcloned into pBluescript SK(+/−) (Stratagene). The K1, F98A, and Δ98-99 mutations were introduced into the wild-type SV40 genome by replacing a PflM1-BsgI fragment. The ligation mixture was transfected into CV-1P cells with Plus/Lipofectamine. Infected cells were fed every 2 to 3 days until cytopathic effect was visible (10 to 12 days) (61). Supernatant from infected cells was collected, filtered through a 0.45-μm-pore-size filter, and used to reinfect CV-1P cells.
RESULTS
Identification of the TAg-associated 185-kDa protein.
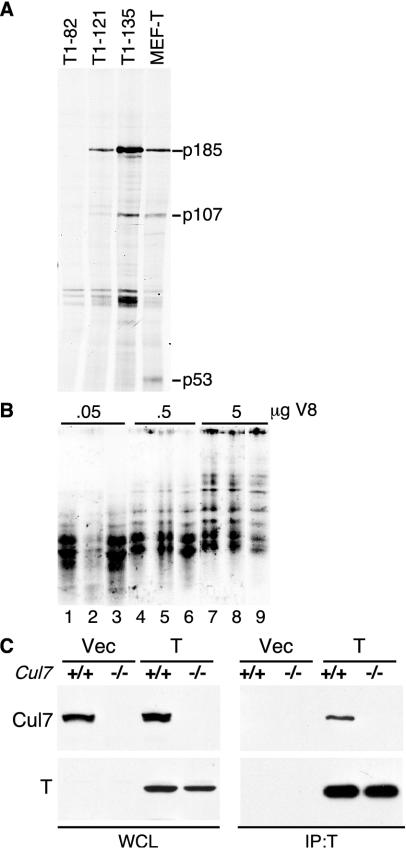
A 185-kDa protein was reported to be coprecipitated by SV40 TAg that required the N terminus of TAg for binding (30). It was noted that when TAg was added to cell lysates, it could coprecipitate p185, indicating that the complex formed in vitro (30). To identify p185, 35S-labeled NIH 3T3 cell lysates were mixed with purified N-terminal fragments of large TAg or with lysates from unlabeled MEFs stably expressing full-length TAg (66). As shown in Fig. 1A, full-length TAg as well as TAg fragments containing the N-terminal 135 (T1-135) or 121 (T1-121) residues coprecipitated a 185-kDa protein as well as the pRb-related protein p107. In contrast, the TAg fragment containing only the first 82 residues (T1-82) was unable to coprecipitate p185. These results support the earlier report indicating that the N-terminal 121 residues of TAg were sufficient to bind to p185 (30).
FIG. 1.
Identification of p185/Cul7 in a TAg immuno-complex. (A) Lysates were prepared from 35S-labeled NIH 3T3 cells, incubated with purified N-terminal fragments of TAg or with unlabeled lysates from MEFs stably expressing wild-type TAg, and immunoprecipitated with TAg monoclonal antibody PAb419. A 185-kDa protein coprecipitated with full-length TAg and the fragments 1-121 and 1-135. TAg-associated proteins, p107 and p53, are identified. (B) The 185-kDa protein was in vitro translated from mouse p185 cDNA (lanes 1, 4, and 7) or immunoprecipitated from 35S-labeled NIH 3T3 cell lysates with either anti-p185 monoclonal antibody SA12 alone (lanes 2, 5, and 8) or with anti-TAg PAb419 in the presence of T1-135 (lanes 3, 6, and 9). The three samples were separated by SDS-PAGE, and the 185-kDa band was excised and digested with three different concentrations of V8 protease. The resulting peptides were separated in an SDS-PAGE and autoradiographed. (C) pBABE-puro-TAg or pBABE-vector (Vec) was stably expressed in wild-type (+/+) or Cul7 knockout (−/−) MEFs. Lysates were immunoprecipitated using anti-TAg antibody PAb419 and Western blotted with anti-p185/Cul7 monoclonal antibody SA12 (top panel) or anti-TAg monoclonal antibody PAb419 (bottom panel).
A preparative-scale immunoprecipitation with lysates of NIH 3T3 cells and T1-135 was used to obtain sufficient amounts of p185 for sequencing by tandem mass spectrometry. Microsequencing identified three peptides that matched residues 12 to 27, 743 to 752, and 1501 to 1509 of a human gene, KIAA0076 (42). A homologous gene was cloned from a mouse cDNA library that shared 77% overall identity with the human gene. Sequence comparison revealed that the mouse p185 was identical to the 193-kDa TAg-associated protein described previously (62).
To confirm the identity of p185 as the TAg-associated protein, partial V8 protease digestion of p185 generated from in vitro translation of a cDNA clone was compared to that performed with p185 directly immunoprecipitated with the monoclonal antibody SA12 or p185 that was coprecipitated by TAg from MEF-T cells that had been labeled with [35S]methionine. Comparison of the partial proteolysis patterns revealed no significant differences in the digestion patterns (Fig. 1B), supporting the conclusion that the 185-kDa TAg-associated protein was the mouse homologue of KIAA0076 (30, 42, 62).
To determine if TAg bound specifically to p185/Cul7, MEFs were prepared from embryos with Cul7 homozygously deleted and that were infected with a retrovirus expressing large TAg (Fig. 1C) (2). Immunoprecipitation for TAg revealed that T could specifically coprecipitate p185/Cul7. These results confirm that TAg binds specifically to the p185/Cul7 protein.
Functional characterization of p185.
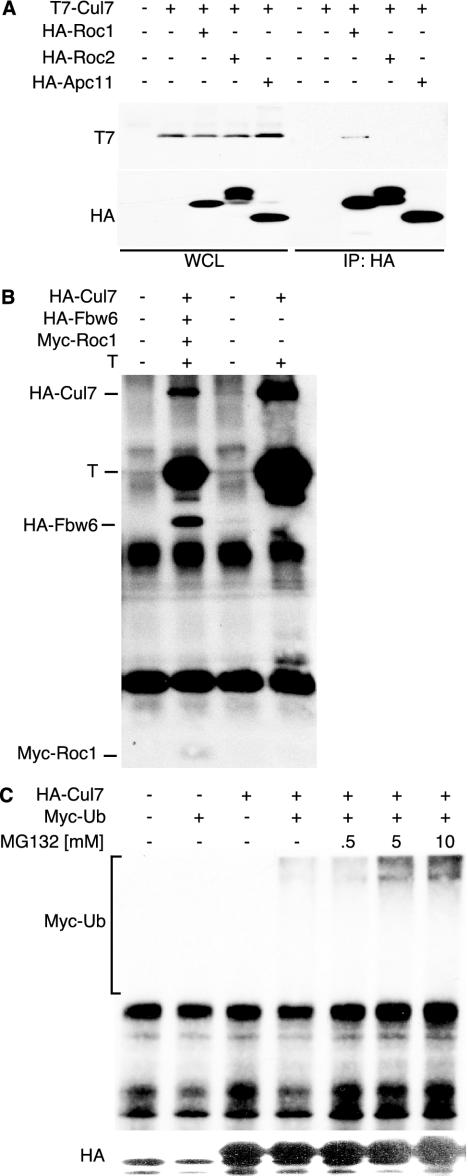
A standard BLAST search with p185 revealed significant homology between the C-terminal region of p185 and members of the cullin family of E3 ubiquitin ligases (2, 21). Given the strong homology to the E3 ubiquitin ligases, we sought to determine if p185 could bind to other known components of the cullin complexes. The region of p185 that contains the highest homology to the cullin family contains the binding site for the RING finger protein, Rbx1/Roc1 (2, 19, 20, 37, 43). The RING finger protein is an essential core component of the SCF complexes and may serve to recruit the E2 ubiquitin-conjugating enzyme for the ubiquitination function (19). To determine whether p185 could bind specifically to a RING-containing protein, T7-tagged p185 was cotransfected with HA-tagged versions of Roc1/Rbx1, Roc2, and the related Apc11. While each RING protein was expressed well, only Roc1/Rbx1 was able to coprecipitate p185 (Fig. 2A).
FIG. 2.
p185/Cul7 has properties of a cullin. (A) T7-tagged p185/Cul7 was transiently transfected with HA-tagged Roc1, Roc2, or APC11 into NIH 3T3 cells. Cell lysates were immunoprecipitated using anti-HA antibody 12CA5 and separated in an SDS-PAGE, followed by blotting with T7 and HA antibodies. (B) TAg was cotransfected with HA-p185/Cul7, HA-Fbw6, and myc-Roc1. The cell lysates were immunoprecipitated with anti-TAg (PAb419) antibody and blot-ted with anti-HA (12CA5), anti-TAg (PAb419), and anti-myc (9E10) antibodies. (C) HA-p185/Cul7 and myc-ubiquitin were transfected into NIH 3T3 cells. At 24 h after transfection, MG132 was added to the medium for 24 h. Lysates were immunoprecipitated with anti-HA antibody 12CA5, separated in an SDS-PAGE, and blotted with anti-HA (12CA5; top panel) or anti-myc (9E10; bottom panel) antibody.
Recently, p185 has been shown to form a specific complex with the F box protein Fbx29, also known as Fbw6 (2, 13, 29, 64). To determine whether TAg could bind to a Cul7 complex that contained Fbw6 and Roc1, NIH 3T3 cells were cotransfected with TAg and epitope-tagged p185, Fbw6, and Rbx1/Roc1. Immunoprecipitation for TAg revealed coprecipitation of p185/Cul7 along with Fbw6 and Rbx1/Roc1 (Fig. 2B).
To determine if Cul7 contained ubiquitinating activity, HA-tagged Cul7 was cotransfected with myc-tagged ubiquitin. Immunoprecipitation for HA-p185 followed by Western blotting for myc-ubiquitin revealed that p185 coprecipitated a ubiquitin ladder represented by multiple bands that extended to the top of the gel and that intensified in the presence of the proteasome inhibitor MG132 (Fig. 2C) (12). This result indicates that p185/Cul7 or an associated protein can become polyubiquitinated.
p185/Cul7 binding domain of TAg.
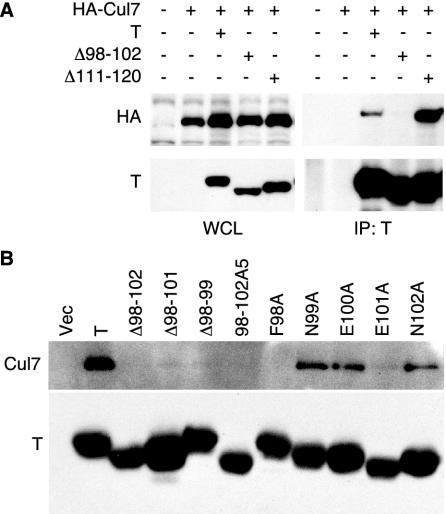
Immunoprecipitation of TAg suggested that the N-terminal 121 residues of TAg were sufficient to coprecipitate p185/Cul7 (Fig. 1A). To identify the region of TAg required for p185/Cul7 binding, small in-frame deletion constructs were generated in the N terminus of TAg and cotransfected with HA-tagged p185 in NIH 3T3 cells. Immunoprecipitation for TAg followed by Western blotting for HA revealed that wild-type TAg as well as a full-length construct with an in-frame deletion of residues 111 to 120 (Δ111-120) could coprecipitate p185/Cul7 (Fig. 3A). In addition, deletion of residues 107 to 111 (PVU-0) did not decrease the ability of TAg to bind to HA-p185 (data not shown) (66). In contrast, deletion of residues 98 to 102 (Δ98-102) reduced TAg's ability to coprecipitate p185/Cul7 (Fig. 3A).
FIG. 3.
TAg residues 98 to 102 are required for p185/Cul7 binding. (A) NIH 3T3 cells were transfected with HA-tagged Cul7 and wild-type TAg (T), deletion mutant Δ98-102, or Δ111-120. At 48 h following transfection, lysates were immunoprecipitated with TAg antibody PAb419, separated by SDS-PAGE, and blotted with PAb419 and HA antibody 12CA5. (B) Wild-type TAg and the indicated mutants of TAg within the region 98 to 102 were transfected in NIH 3T3 cells. Immunoprecipitation for TAg was followed by blotting with antibody against p185/Cul7 (SA12).
To further define the residues of TAg necessary for binding to p185/Cul7, additional deletions and alanine substitution mutations were generated in residues 98 to 102. These TAg mutant constructs were transiently transfected into NIH 3T3 cells. Cell lysates were immunoprecipitated with a TAg monoclonal antibody and Western blotted for coprecipitated endogenous p185/Cul7. As shown in Fig. 3B, deletion of residues 98 to 102, 98 to 101, and 98 to 99 as well as substitution of residues 98 to 102 with five alanines (98-102A5) reduced p185/Cul7 binding to TAg. Alanine substitution of residue 98 (F98A) also reduced p185/Cul7 binding, while alanine substitution of residues 99, 100, and 102 did not affect p185/Cul7 binding. Alanine substitution of E101 (E101A) greatly reduced p185/Cul7 binding. These results support the role of residues 98 to 102 and in particular F98 in p185/Cul7 binding.
Since residues 98 to 102 of TAg were in close proximity to the pRb binding region known as the LxCxE motif (residues 103 to 107), it was important to test if mutations in this region affected binding to pRb. Primary MEFs were stably transfected with wild-type TAg, a pRb-binding-defective mutant K1 (E107K), F98A, or Δ98-99. Pools of selected colonies were expanded and stained for TAg by indirect immunofluorescence, revealing that each of these TAg constructs was expressed in the nucleus of more than 90% of cells (data not shown).
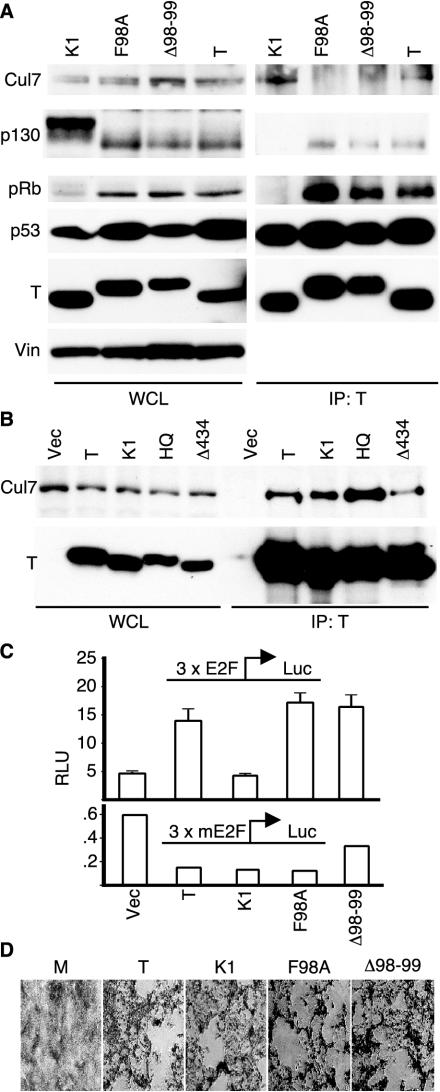
As shown in Fig. 4A (left panels), expression levels for each of the TAg constructs were similar in each of these cell lines. In addition, the expression levels of p53 were similar, reflecting the ability of TAg to increase the stability of p53. The lysates were also blotted for pRb-related proteins, including pRb and p130. Our laboratory has reported that wild-type TAg is capable of reducing the phosphorylation state of p130 and p107 (52, 53). Furthermore, an intact LxCxE motif and an N-terminal J domain were required for TAg to affect the phosphorylation status of p130 and p107. As shown in Fig. 4A, the phosphorylation pattern of p130 was reduced in the presence of wild-type TAg as well as the mutant constructs F98A and Δ98-99. In contrast, the LxCxE mutant K1 did not reduce the phosphorylation levels of p130. The ability of F98A and Δ98-99 to affect p130 phosphorylation indirectly supports the notion that these mutants retain an intact DnaJ domain and LxCxE motif.
FIG. 4.
p185/Cul7 binding mutants of TAg. (A) Whole-cell lysates (left panels) of MEFs stably expressing wild-type TAg, F98A, Δ98-99, or K1 were separated in an SDS-PAGE or immunoprecipitated for TAg with PAb419 antibody (right panels). Western blotting was performed for p185/Cul7 (SA12), p130 (C20), pRb (XZ55), p53 (240), and TAg (PAb419), with vinculin (Vin) as a loading control. (B) NIH 3T3 cells were transiently transfected with TAg, K1, H42Q, or Δ434-444. Lysates were immunoprecipitated with PAb419 and blotted for p185/Cul7 (SA12; top panel) or TAg (PAb419; bottom). (C) NIH 3T3 cells were cotransfected with the indicated TAg plasmids and a reporter expressing the luciferase gene driven by a wild-type (3xE2F, upper panel) or mutated (3xmE2F, lower panel) promoter. To correctfor transfection efficiency, a third plasmid constitutively expressing the Renilla luciferase was included in all transfections. Cells were harvested 48 h following transfection, and the relative luciferase units were measured as described in Materials and Methods. The results are presented as the means of triplicates ± standard deviations. (D) CV-1P cells were transfected with wild-type SV40 genome or a mutant genome containing the K1, F98A, or Δ98-99 mutation. The cells were kept under observation for approximately 12 days until cytopathic effect was observed. When plaques became evident, the cultures were fixed and stained with crystal violet.
To determine whether the mutation of residues 98 and 99 affected TAg's ability to bind p53 and the pRb-related proteins, lysates were immunoprecipitated for TAg. Similar to wild-type TAg, F98A and Δ98-99 were able to coprecipitate p53 as well as all members of the pRb family, including pRb and p130 (Fig. 4A, right panels). As expected, the LxCxE mutant K1 was unable to coprecipitate the pRb family members. In contrast, wild-type TAg and K1 were able to coprecipitate p185/Cul7, but neither F98A nor Δ98-99 could bind to p185/Cul7. These results suggest that the p185/Cul7 binding region of TAg can be distinguished from the pRb binding, LxCxE motif.
Since pRb binding by TAg could be distinguished from p185/Cul7 binding, we were curious whether other transforming domains of TAg contributed to p185/Cul7 binding. We tested the ability of the DnaJ mutant H42Q and the p53 binding mutant Δ434-444 to coprecipitate p185/Cul7 (9, 28, 52). As shown in Fig. 4B, wild-type TAg as well as the mutants K1, H42Q, and Δ434-444 could coprecipitate p185/Cul7 with comparable efficiency. From these results, it is apparent that the mutants F98A and Δ98-99 remain capable of binding to the well-known tumor suppressors pRb and p53 and that at least some mutants of TAg that fail to bind to pRb or p53 retain their ability to bind to p185/Cul7.
In addition to binding to the pocket proteins, TAg can inactivate the ability of the pRb family members to repress E2F-dependent transcription. To determine whether mutants of TAg that failed to bind Cul7 retained their ability to relieve pRb-mediated repression of E2F, an E2F promoter reporter assay was performed. NIH 3T3 cells were cotransfected with wild-type or mutant E2F promoter luciferase reporter plasmid and TAg. As shown in Fig. 4C, expression of wild-type TAg led to an increase in the E2F promoter activity compared to the vector control. In contrast, the pRb binding mutant K1 was unable to override the repression of the E2F promoter. Similar to wild-type TAg, expression of either F98A or Δ98-99 was able to override repression of the E2F promoter. A reporter construct that was identical except for mutation of the E2F sites failed to respond to any of the TAg constructs when tested under similar conditions, supporting the specificity of the E2F activity. These results, taken together with the pRb binding data, suggest that TAg binding to p185/Cul7 can be distinguished from binding and inactivation of the pRb family of proteins.
Since large TAg participates in many functions required for SV40 viral replication, we tested the ability of F98A and Δ98-99 to promote SV40 ori-dependent replication. Cotransfection of wild-type TAg or the p185/Cul7 binding mutants with an ori-containing plasmid revealed that these p185/Cul7 binding mutants were able to support replication of the ori-containing plasmid as efficiently as the wild-type TAg (data not shown) (26). In addition, complete SV40 viruses containing the mutations F98A or Δ98-99 were generated and found to replicate and produce a cytopathic effect in permissive CV-1P cells (Fig. 4D). The mutant viruses produced similar amounts large TAg protein as wild-type virus when examined by Western blotting (data not shown) and induced the cytopathic effect within a similar time frame as wild-type virus, suggesting that the mutants had no gross defects in the viral life cycle. Given these results, the two mutant TAg constructs, F98A and Δ98-99, appear to retain all of the activity necessary for viral replication.
p185/Cul7-binding mutants of TAg are transformation defective.
TAg is capable of transforming a variety of primary rodent cells (1, 66). Once it was established that the two TAg mutants F98A and Δ98-99 were intact in terms of pRb and p53 binding activities, we tested these mutants for their ability to transform cells. Expression of wild-type TAg, K1, F98A, or Δ98-99 led to immortalization of MEFs with no evidence for crisis or senescence (6, 59, 66). The ability of large TAg to immortalize MEFs has been linked most strongly to its ability to bind and inactivate p53 (28). Given that the mutants F98A and Δ98-99 can bind to p53 and can immortalize MEFs, it supports the model that these p185 binding mutants of TAg retain the ability to inactivate p53 function.
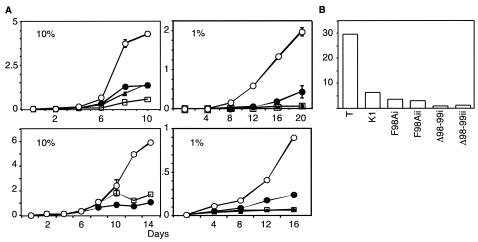
To test the ability to grow to high density, stable pools of TAg-expressing MEFs were plated at 104 cells per 60-mm dish and cultured in medium containing either 10% or 1% serum. Wild-type TAg-expressing MEFs were able to sustain growth to high density in 10% serum while the pRb binding mutant K1 had a significantly reduced growth rate (Fig. 5A, left panels). Notably, the F98A- or Δ98-99-expressing MEFs were also unable to grow as efficiently as MEFs expressing wild-type TAg. Furthermore, when cultured in 1% serum, MEFs expressing F98A, Δ98-99, and K1 were unable to grow as well as those expressing wild-type TAg (Fig. 5A, right panels).
FIG. 5.
TAg transformation of MEFs. (A) Equal numbers of primary MEFs stably expressing wild-type TAg (open circle), K1 (square), F98A (closed triangle), or Δ98-99 (closed circle) were seeded in 60-mm dishes. The cells were fed on alternate days with medium containing either 10% or 1% FCIS. The results in the top two and lower two panels are from two independently generated pools of stably transfected MEFs. The results are presented as means of total number of cells (105) on four 60-mm plates ± standard deviations. (B) An equal number of the above-mentioned MEFs was seeded in soft agar and allowed to grow for 3 weeks. Clusters of cells containing four or more cells were scored as colonies. Total colonies and single cells were counted. The results are shown as the percentage of colonies observed for each TAg clone. Results from two separate pools (i and ii) of F98A and Δ98-99 MEFs are presented.
TAg has the ability to induce anchorage-independent growth of MEFs. This ability of TAg has been reported to be dependent on both pRb and p53 binding (7, 9, 60). To test the ability of the TAg mutants to support growth in an anchorage-independent manner, MEFs expressing the various TAg constructs were seeded in semisolid medium containing 0.4% agarose and 10% serum. Clusters of cells containing four or more cells were counted as colonies after 3 weeks. Using these criteria, nearly 30% of the wild-type TAg-expressing cells were found to form colonies, whereas less than 10% of K1, F98A, or Δ98-99 formed colonies in soft agar (Fig. 5B). Therefore, loss of binding to pRb (K1) or p185/Cul7 (F98A and Δ98-99) reduces the ability of TAg to support anchorage-independent growth.
DISCUSSION
Study of SV40 T-Ag cellular transformation has led to the identification of several key growth control pathways. For example, p53 was first identified as a coprecipitating protein of TAg (33, 35). When the regions of TAg that contribute to binding to pRb and p53 were mutated, it was recognized that interaction with these tumor suppressors was required for transformation (11, 28). In this report, we identify a novel N-terminal domain of SV40 TAg that contributes to binding to the p185/Cul7 cullin. Certain mutations within this domain of TAg abolish p185/Cul7 binding yet retain binding to p53 and pRb family members. Evidence that the mutants F98A and Δ98-99 inactivate pRb and p53 was demonstrated by their ability to override pRb-mediated repression of an E2F promoter reporter and the ability to efficiently immortalize primary MEFs. However, these p185/Cul7-defective mutants of TAg failed to fully transform primary MEFs as evidenced by their inability to grow to high density and the reduced ability to grow in an anchorage-independent manner. These results suggest that residues 98 and 99 of TAg form part of a new transforming domain of TAg and that binding to p185/Cul7 may be an essential component of TAg-transforming activity.
In a recent paper from Hahn and colleagues, the transforming properties of SV40 large TAg in primary human cell strains were attributed to inactivation of pRb and p53 (22). Transformation of primary human cells was induced by expression of SV40 large and small TAgs, hTERT, and H-Ras. Large TAg could be substituted in this assay by overexpression of cyclin D and a dominant-active CDK4-R24C to inactivate pRb and a dominant mutant of p53 to inactivate p53. Similarly, large TAg could be substituted with expression of human papillomavirus (HPV) E7, known to inactivate pRb, and HPV E6 that inactivates p53. These results suggested that the sole transforming targets of large TAg were pRb and p53. However, it is possible that overexpression of cyclin D1 and R24C CDK4 may have transforming activities in addition to the inactivation of pRb. Similarly, a dominant-negative form of p53 may have additional transforming activities in addition to inactivation of p53. HPV E7 and E6 are known to interact with many other cellular proteins that have been implicated in transformation. It remains a possibility that p185 is a target of TAg in transformation of primary human cells.
TAg binding to p185/Cul7 can be separated from pRb and p53 inactivation and viral replication.
The LxCxE motif and DnaJ domain of TAg cooperate to bind pRb, override pRb-mediated repression of E2F reporters, and reduce the phosphorylation levels of p130 and p107 (54, 67). As shown in Fig. 4, F98A and Δ98-99 were able to efficiently coprecipitate pRb, p107, and p130, to alter the phosphorylation status of p107 and p130, and to activate expression from the E2F-dependent promoter reporter. These results strongly support the notion that these mutant TAg constructs retain the ability to inactivate pRb family members. Conversely, mutation of the LxCxE motif (K1; E107K), the DnaJ domain (H42Q), or the p53 binding domain (Δ434-444) did not reduce the ability of TAg to bind to p185/Cul7 (Fig. 4B).
TAg is known to affect the level and the status of p130 and p107 phospho-forms. In the presence of wild-type TAg, certain phospho-forms of p130 and p107 are reduced (53). This effect is dependent on the presence of an intact DnaJ domain and LxCxE motif. To investigate whether p185/Cul7 is involved in this effect, we compared the levels and phosphorylation status of p130 in MEFs stably expressing the wild-type or the p185/Cul7 binding-deficient TAg. The expression levels and phosphorylation status of p130 were comparable in the presence of either wild-type or p185/Cul7 binding-defective mutants of TAg. It appears, therefore, that binding to p185/Cul7 is not necessary for TAg to affect the phosphorylation levels of p130. Furthermore, we have no evidence that the potential ubiquitinating activity of associated p185/Cul7 affects the stability or phosphorylation state of the pRb-related proteins.
The p185/Cul7 binding mutants, F98A and Δ98-99, of TAg also bind to p53. TAg binding leads to an increase in the half-life of p53. This effect of TAg on the stability of p53 is due, at least in part, to reduced transcription of p53-dependent genes, most notably Mdm2. Mdm2 expression is induced in response to p53 activation and serves to promote the nuclear export, ubiquitination, and subsequent degradation of p53. In addition, TAg binding to p53 promotes the immortalization of wild-type MEFs and inhibits the onset of replicative senescence and crisis. The F98A and Δ98-99 mutants were as effective as wild-type TAg in immortalizing MEFs. Given that F98A and Δ98-99 can bind to p53, increase its level of expression, and promote the immortalization of MEFs, they appear to be similar to wild-type TAg in inactivating p53 function.
TAg plays an important role in the viral replication and assembly in the SV40 virus life cycle. The p185/Cul7 mutants of TAg showed no defect in mediating DNA replication of an SV40 ori-containing plasmid (data not shown). These mutations, when introduced into the SV40 genome, were also found not to disrupt viral replication and cytopathic effect.
E3 ligase function of p185/Cul7.
Structurally and functionally, p185/Cul7 appears to be closely related to a class of proteins known as cullins that form the scaffold of an E3 ligase complex involved in ubiquitination-degradation of regulatory cellular proteins. Similar to other well-studied cullins in yeast and higher eukaryotic organisms, p185/Cul7 can bind specifically to the RING finger protein, Rbx1/Roc1, and may serve to recruit E2 ubiquitin-conjugating activity. Consistent with this, p185/Cul7 or an associated protein appeared to be polyubiquitinated when immunoprecipitated in the presence of a proteasome inhibitor (Fig. 2C) (13). In addition, p185/Cul7 binds specifically to the F box protein Fbw6 (2, 13). p185/Cul7 is a unique member of the cullin family, because it also contains a region with strong homology to the Apc10/Doc1 component of the anaphase-promoting complex. Despite these homologies, it is not known if p185/Cul7 and Fbw6 normally target a specific substrate for ubiquitination and degradation.
Other small DNA virus proteins interact with components of the ubiquitination pathway and affect protein stability. The human papillomavirus E6 targets p53 for ubiquitination and degradation by recruiting the E6AP ubiquitin ligase (56). Adenovirus E1B 55K and E4 Orf6-dependent degradation of p53 involves Cul5 (46). Despite these similarities, we found no evidence that TAg binding to p185/Cul7 participated in the stabilization of p53 or any of the pRb-related proteins. It appears therefore that the TAg-Cul7 interaction, unlike the E6-E6AP or E4Orf6-Cul5 interaction, may not control the stability of associated p53.
It was reported that p193 (p185/Cul7) might have a proapoptotic activity that could be antagonized by TAg (45). It was also reported that deletion of a potential BH3-homology domain in p185/Cul7 led to loss of apoptotic effect. A caveat concerning the observations made by Tsai et al. may be that the BH3 deletion mutant of p193/p185/Cul7 disrupts the cullin domain and may have affected Roc1/Rbx1 binding (62). It is therefore possible that the loss of apoptotic function in the p185/Cul7 mutant observed resulted from a defect in the E3 ligase function, leading to the stabilization of a potential substrate and manifesting as an antiapoptotic effect. We have not observed any specific pro- or antiapoptotic effect upon expression of wild-type TAg or the p185-binding TAg mutants.
TAg binding to p185/Cul7 may lead to inhibition of its associated ubiquitinating activity, inhibition of the degradation of a specific substrate, or perhaps redirecting its activity to a different substrate. It is expected that p185/Cul7 in association with Fbw6 targets at least one specific substrate for ubiquitination and subsequent degradation. However, the identity of this substrate or the specific E3 ubiquitin ligase activity of p185/Cul7 is only speculative at this time. Considering that SV40 TAg binding to p185/Cul7 may be necessary for full cellular transformation, it would be an important goal to determine if the associated ubiquitination activity of p185/Cul7 contributes to TAg transformation.
Acknowledgments
We gratefully acknowledge the assistance of Jianmin Gan for generation of monoclonal antibody SA12 and William Lane of Harvard Microchemistry Facility for protein microsequencing. Gifts of plasmids for Myc-ubiquitin from Daniel Finley, KIAA0078 from Takahiro Nagase, Myc-Roc1 from Joan Conaway, pSV-B3 from Jim Pipas, and HA-Roc1, HA-Roc2, and HA-Apc11 from Yue Xiong are gratefully acknowledged.
This work was supported in part by National Institutes of Health grants RO1CA093804 and PO1CA50661.
REFERENCES
- 1.Ali, S. H., and J. A. DeCaprio. 2001. Cellular transformation by SV40 large T antigen: interaction with host proteins. Semin. Cancer Biol. 11:15-23. [DOI] [PubMed] [Google Scholar]
- 2.Arai, T., J. S. Kasper, J. R. Skaar, S. H. Ali, C. Takahashi, and J. A. DeCaprio. 2003. Targeted disruption of p185/Cul7 gene results in abnormal vascular morphogenesis. Proc. Natl. Acad. Sci. USA 100:9855-9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avantaggiati, M. L., M. Carbone, A. Graessmann, Y. Nakatani, B. Howard, and A. S. Levine. 1996. The SV40 large T antigen and adenovirus E1a oncoproteins interact with distinct isoforms of the transcriptional co-activator, p300. EMBO J. 15:2236-2248. [PMC free article] [PubMed] [Google Scholar]
- 4.Bai, C., P. Sen, K. Hofmann, L. Ma, M. Goebl, J. W. Harper, and S. J. Elledge. 1996. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86:263-274. [DOI] [PubMed] [Google Scholar]
- 5.Bargonetti, J., P. N. Friedman, S. E. Kern, B. Vogelstein, and C. Prives. 1991. Wild-type but not mutant p53 immunopurified proteins bind to sequences adjacent to the SV40 origin of replication. Cell 65:1083-1091. [DOI] [PubMed] [Google Scholar]
- 6.Bates, M. P., S. R. Jennings, Y. Tanaka, M. J. Tevethia, and S. S. Tevethia. 1988. Recognition of simian virus 40 T antigen synthesized during viral lytic cycle in monkey kidney cells expressing mouse H-2Kb- and H-2Db-transfected genes by SV40-specific cytotoxic T lymphocytes leads to the abrogation of virus lytic cycle. Virology 162:197-205. [DOI] [PubMed] [Google Scholar]
- 7.Campbell, K. S., K. P. Mullane, I. A. Aksoy, H. Stubdal, J. Zalvide, J. M. Pipas, P. A. Silver, T. M. Roberts, B. S. Schaffhausen, and J. A. DeCaprio. 1997. DnaJ/hsp40 chaperone domain of SV40 large T antigen promotes efficient viral DNA replication. Genes Dev. 11:1098-1110. [DOI] [PubMed] [Google Scholar]
- 8.Carrano, A. C., E. Eytan, A. Hershko, and M. Pagano. 1999. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat. Cell Biol. 1:193-199. [DOI] [PubMed] [Google Scholar]
- 9.Chao, H. H., A. M. Buchmann, and J. A. DeCaprio. 2000. Loss of p19(ARF) eliminates the requirement for the pRB-binding motif in simian virus 40 large T antigen-mediated transformation. Mol. Cell. Biol. 20:7624-7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, J., G. J. Tobin, J. M. Pipas, and T. Van Dyke. 1992. T-antigen mutant activities in vivo: roles of p53 and pRB binding in tumorigenesis of the choroid plexus. Oncogene 7:1167-1175. [PubMed] [Google Scholar]
- 11.DeCaprio, J. A., J. W. Ludlow, J. Figge, J.-Y. Shew, C.-M. Huang, W.-H. Lee, E. Marsilio, E. Paucha, and D. M. Livingston. 1988. SV40 large T antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell 54:275-283. [DOI] [PubMed] [Google Scholar]
- 12.de Graaf, P., N. A. Little, Y. F. Ramos, E. Meulmeester, S. J. Letteboer, and A. G. Jochemsen. 2003. Hdmx protein stability is regulated by the ubiquitin ligase activity of Mdm2. J. Biol. Chem. 278:38315-38324. [DOI] [PubMed] [Google Scholar]
- 13.Dias, D. C., G. Dolios, R. Wang, and Z. Q. Pan. 2002. CUL7: a DOC domain-containing cullin selectively binds Skp1. Fbx29 to form an SCF-like complex. Proc. Natl. Acad. Sci. USA 99:16601-16606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Digweed, M., I. Demuth, S. Rothe, R. Scholz, A. Jordan, C. Grotzinger, D. Schindler, M. Grompe, and K. Sperling. 2002. SV40 large T-antigen disturbs the formation of nuclear DNA-repair foci containing MRE11. Oncogene 21:4873-4878. [DOI] [PubMed] [Google Scholar]
- 15.Eckner, R., J. W. Ludlow, N. L. Lill, E. Oldread, Z. Arany, N. Modjtahedi, J. A. DeCaprio, D. M. Livingston, and J. A. Morgan. 1996. Association of p300 and CBP with simian virus 40 large T antigen. Mol. Cell. Biol. 16:3454-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farmer, G., J. Bargonetti, H. Zhu, P. Friedman, R. Prywes, and C. Prives. 1992. Wild-type p53 activates transcription in vitro. Nature 358:83-86. [DOI] [PubMed] [Google Scholar]
- 17.Feldman, R. M., C. C. Correll, K. B. Kaplan, and R. J. Deshaies. 1997. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 91:221-230. [DOI] [PubMed] [Google Scholar]
- 18.Fuchs, S. Y., A. Chen, Y. Xiong, Z. Q. Pan, and Z. Ronai. 1999. HOS, a human homolog of Slimb, forms an SCF complex with Skp1 and Cullin1 and targets the phosphorylation-dependent degradation of IκB and beta-catenin. Oncogene 18:2039-2046. [DOI] [PubMed] [Google Scholar]
- 19.Furukawa, M., Y. Zhang, J. McCarville, T. Ohta, and Y. Xiong. 2000. The CUL1 C-terminal sequence and ROC1 are required for efficient nuclear accumulation, NEDD8 modification, and ubiquitin ligase activity of CUL1. Mol. Cell. Biol. 20:8185-8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furukawa, Y., S. Iwase, J. Kikuchi, M. Nakamura, H. Yamada, and M. Matsuda. 1999. Transcriptional repression of the E2F-1 gene by interferon-alpha is mediated through induction of E2F-4/pRB and E2F-4/p130 complexes. Oncogene 18:2003-2014. [DOI] [PubMed] [Google Scholar]
- 21.Grossberger, R., C. Gieffers, W. Zachariae, A. V. Podtelejnikov, A. Schleiffer, K. Nasmyth, M. Mann, and J. M. Peters. 1999. Characterization of the DOC1/APC10 subunit of the yeast and the human anaphase-promoting complex. J. Biol. Chem. 274:14500-14507. [DOI] [PubMed] [Google Scholar]
- 22.Hahn, W. C., S. K. Dessain, M. W. Brooks, J. E. King, B. Elenbaas, D. M. Sabatini, J. A. DeCaprio, and R. A. Weinberg. 2002. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol. Cell. Biol. 22:2111-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harlow, E., L. V. Crawford, D. C. Pim, and N. M. Williamson. 1981. Monoclonal antibodies specific for simian virus 40 tumor antigens. J. Virol. 39:861-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang, D., A. Srinivasan, G. Lozano, and P. D. Robbins. 1993. SV40 T antigen abrogates p53-mediated transcriptional activity. Oncogene 8:2805-2812. [PubMed] [Google Scholar]
- 25.Jiang, J., and G. Struhl. 1998. Regulation of the Hedgehog and Wingless signalling pathways by the F-box/WD40-repeat protein Slimb. Nature 391:493-496. [DOI] [PubMed] [Google Scholar]
- 26.Kalderon, D., and A. E. Smith. 1984. In vitro mutagenesis of a putative DNA binding domain of SV40 large-T. Virology 139:109-137. [DOI] [PubMed] [Google Scholar]
- 27.Kawakami, T., T. Chiba, T. Suzuki, K. Iwai, K. Yamanaka, N. Minato, H. Suzuki, N. Shimbara, Y. Hidaka, F. Osaka, M. Omata, and K. Tanaka. 2001. NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 20:4003-4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kierstead, T. D., and M. J. Tevethia. 1993. Association of p53 binding and immortalization of primary C57BL/6 mouse embryo fibroblasts by using simian virus 40 T-antigen mutants bearing internal overlapping deletion mutations. J. Virol. 67:1817-1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koepp, D. M., L. K. Schaefer, X. Ye, K. Keyomarsi, C. Chu, J. W. Harper, and S. J. Elledge. 2001. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science 294:173-177. [DOI] [PubMed] [Google Scholar]
- 30.Kohrman, D. C., and M. J. Imperiale. 1992. Simian virus 40 large T antigen stably complexes with a 185-kilodalton host protein. J. Virol. 66:1752-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krek, W., D. M. Livingston, and S. Shirodkar. 1993. Binding to DNA and the retinoblastoma gene product promoted by complex formation of different E2F family members. Science 262:1557-1560. [DOI] [PubMed] [Google Scholar]
- 32.Lammer, D., N. Mathias, J. M. Laplaza, W. Jiang, Y. Liu, J. Callis, M. Goebl, and M. Estelle. 1998. Modification of yeast Cdc53p by the ubiquitin-related protein rub1p affects function of the SCFCdc4 complex. Genes Dev. 12:914-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lane, D. P., and L. V. Crawford. 1979. T antigen is bound to a host protein in SV40-transformed cells. Nature 278:261-263. [DOI] [PubMed] [Google Scholar]
- 34.Lanson, N. A., Jr., D. B. Egeland, B. A. Royals, and W. C. Claycomb. 2000. The MRE11-NBS1-RAD50 pathway is perturbed in SV40 large T antigen-immortalized AT-1, AT-2 and HL-1 cardiomyocytes. Nucleic Acids Res. 28:2882-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Linzer, D. I. H., and A. J. Levine. 1979. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell 17:43-52. [DOI] [PubMed] [Google Scholar]
- 36.Lisztwan, J., A. Marti, H. Sutterluty, M. Gstaiger, C. Wirbelauer, and W. Krek. 1998. Association of human CUL-1 and ubiquitin-conjugating enzyme CDC34 with the F-box protein p45(SKP2): evidence for evolutionary conservation in the subunit composition of the CDC34-SCF pathway. EMBO J. 17:368-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo, J., F. Su, D. Chen, A. Shiloh, and W. Gu. 2000. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature 408:377-381. [DOI] [PubMed] [Google Scholar]
- 38.Lyapina, S. A., C. C. Correll, E. T. Kipreos, and R. J. Deshaies. 1998. Human CUL1 forms an evolutionarily conserved ubiquitin ligase complex (SCF) with SKP1 and an F-box protein. Proc. Natl. Acad. Sci. USA 95:7451-7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michel, J. J., and Y. Xiong. 1998. Human CUL-1, but not other cullin family members, selectively interacts with SKP1 to form a complex with SKP2 and cyclin A. Cell Growth Differ. 9:435-449. [PubMed] [Google Scholar]
- 40.Moberg, K. H., D. W. Bell, D. C. Wahrer, D. A. Haber, and I. K. Hariharan. 2001. Archipelago regulates Cyclin E levels in Drosophila and is mutated in human cancer cell lines. Nature 413:311-316. [DOI] [PubMed] [Google Scholar]
- 41.Neuman, E., E. K. Flemington, W. R. Sellers, and W. G. J. Kaelin. 1994. Transcription of the E2F-1 gene is rendered cell cycle dependent by E2F DNA-binding sites within its promoter. Mol. Cell. Biol. 14:6607-6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nomura, N., T. Nagase, N. Miyajima, T. Sazuka, A. Tanaka, S. Sato, N. Seki, Y. Kawarabayasi, K. Ishikawa, and S. Tabata. 1994. Prediction of the coding sequences of unidentified human genes. II. The coding sequences of 40 new genes (KIAA0041-KIAA0080) deduced by analysis of cDNA clones from human cell line KG-1. DNA Res. 1:251-262. [DOI] [PubMed] [Google Scholar]
- 43.Ohta, T., J. J. Michel, A. J. Schottelius, and Y. Xiong. 1999. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol. Cell 3:535-541. [DOI] [PubMed] [Google Scholar]
- 44.Osaka, F., H. Kawasaki, N. Aida, M. Saeki, T. Chiba, S. Kawashima, K. Tanaka, and S. Kato. 1998. A new NEDD8-ligating system for cullin-4A. Genes Dev. 12:2263-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pasumarthi, K. B., and L. J. Field. 2002. Cardiomyocyte cell cycle regulation. Circ. Res. 90:1044-1054. [DOI] [PubMed] [Google Scholar]
- 46.Querido, E., P. Blanchette, Q. Yan, T. Kamura, M. Morrison, D. Boivin, W. G. Kaelin, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 15:3104-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rushton, J. J., D. Jiang, A. Srinivasan, J. M. Pipas, and P. D. Robbins. 1997. Simian virus 40 T antigen can regulate p53-mediated transcription independent of binding p53. J. Virol. 71:5620-5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sachsenmeier, K. F., and J. M. Pipas. 2001. Inhibition of Rb and p53 is insufficient for SV40 T-antigen transformation. Virology 283:40-48. [DOI] [PubMed] [Google Scholar]
- 49.Schmieg, F. I., and D. T. Simmons. 1988. Characterization of the in vitro interaction between SV40 T antigen and p53: mapping the p53 binding site. Virology 164:132-140. [DOI] [PubMed] [Google Scholar]
- 50.Segawa, K., A. Minowa, K. Sugasawa, T. Takano, and F. Hanaoka. 1993. Abrogation of p53-mediated transactivation by SV40 large T antigen. Oncogene 8:543-548. [PubMed] [Google Scholar]
- 51.Strohmaier, H., C. H. Spruck, P. Kaiser, K. A. Won, O. Sangfelt, and S. I. Reed. 2001. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature 413:316-322. [DOI] [PubMed] [Google Scholar]
- 52.Stubdal, H., J. Zalvide, K. S. Campbell, C. Schweitzer, T. M. Roberts, and J. A. DeCaprio. 1997. Inactivation of pRB-related proteins p130 and p107 mediated by the J domain of simian virus 40 large T antigen. Mol. Cell. Biol. 17:4979-4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stubdal, H., J. Zalvide, and J. A. DeCaprio. 1996. Simian virus 40 large T antigen alters the phosphorylation state of the RB-related proteins p130 and p107. J. Virol. 70:2781-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sullivan, C. S., P. Cantalupo, and J. M. Pipas. 2000. The molecular chaperone activity of simian virus 40 large T antigen is required to disrupt Rb-E2F family complexes by an ATP-dependent mechanism. Mol. Cell. Biol. 20:6233-6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sutterluty, H., E. Chatelain, A. Marti, C. Wirbelauer, M. Senften, U. Muller, and W. Krek. 1999. p45SKP2 promotes p27Kip1 degradation and induces S phase in quiescent cells. Nat. Cell Biol. 1:207-214. [DOI] [PubMed] [Google Scholar]
- 56.Talis, A. L., J. M. Huibregtse, and P. M. Howley. 1998. The role of E6AP in the regulation of p53 protein levels in human papillomavirus (HPV)-positive and HPV-negative cells. J. Biol. Chem. 273:6439-6445. [DOI] [PubMed] [Google Scholar]
- 57.Tan, T. H., J. Wallis, and A. J. Levine. 1986. Identification of the p53 protein domain involved in formation of the simian virus 40 large T-antigen-p53 protein complex. J. Virol. 59:574-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tedesco, D., J. Lukas, and S. I. Reed. 2002. The pRb-related protein p130 is regulated by phosphorylation-dependent proteolysis via the protein-ubiquitin ligase SCF(Skp2). Genes Dev. 16:2946-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tevethia, M. J., H. A. Lacko, and A. Conn. 1998. Two regions of simian virus 40 large T-antigen independently extend the life span of primary C57BL/6 mouse embryo fibroblasts and cooperate in immortalization. Virology 243:303-312. [DOI] [PubMed] [Google Scholar]
- 60.Thompson, D. L., D. Kalderon, A. E. Smith, and M. J. Tevethia. 1990. Dissociation of Rb-binding and anchorage-independent growth from immortalization and tumorigenicity using SV40 mutants producing N-terminally truncated large T antigens. Virology 178:15-34. [DOI] [PubMed] [Google Scholar]
- 61.Tremblay, J. D., K. F. Sachsenmeier, and J. M. Pipas. 2001. Propagation of wild-type and mutant SV40. Methods Mol. Biol. 165:1-7. [DOI] [PubMed] [Google Scholar]
- 62.Tsai, S. C., K. B. Pasumarthi, L. Pajak, M. Franklin, B. Patton, H. Wang, W. J. Henzel, J. T. Stults, and L. J. Field. 2000. Simian virus 40 large T antigen binds a novel Bcl-2 homology domain 3-containing proapoptosis protein in the cytoplasm. J. Biol. Chem. 275:3239-3246. [DOI] [PubMed] [Google Scholar]
- 63.Tsvetkov, L. M., K. H. Yeh, S. J. Lee, H. Sun, and H. Zhang. 1999. p27(Kip1) ubiquitination and degradation is regulated by the SCF(Skp2) complex through phosphorylated Thr187 in p27. Curr. Biol. 9:661-664. [DOI] [PubMed] [Google Scholar]
- 64.Winston, J. T., D. M. Koepp, C. Zhu, S. J. Elledge, and J. W. Harper. 1999. A family of mammalian F-box proteins. Curr. Biol. 9:1180-1182. [DOI] [PubMed] [Google Scholar]
- 65.Yaron, A., A. Hatzubai, M. Davis, I. Lavon, S. Amit, A. M. Manning, J. S. Andersen, M. Mann, F. Mercurio, and Y. Ben-Neriah. 1998. Identification of the receptor component of the IκBα-ubiquitin ligase. Nature 396:590-594. [DOI] [PubMed] [Google Scholar]
- 66.Zalvide, J., and J. A. DeCaprio. 1995. Role of pRb-related proteins in simian virus 40 large-T-antigen-mediated transformation. Mol. Cell. Biol. 15:5800-5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zalvide, J., H. Stubdal, and J. A. DeCaprio. 1998. The J domain of simian virus 40 large T antigen is required to functionally inactivate RB family proteins. Mol. Cell. Biol. 18:1408-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]