Abstract
The avian and Nelson Bay reoviruses are two of only a limited number of nonenveloped viruses capable of inducing cell-cell membrane fusion. These viruses encode the smallest known membrane fusion proteins (p10). We now show that a region of moderate hydrophobicity we call the hydrophobic patch (HP), present in the small N-terminal ectodomain of p10, shares the following characteristics with the fusion peptides of enveloped virus fusion proteins: (i) an abundance of glycine and alanine residues, (ii) a potential amphipathic secondary structure, (iii) membrane-seeking characteristics that correspond to the degree of hydrophobicity, and (iv) the ability to induce lipid mixing in a liposome fusion assay. The p10 HP is therefore predicted to provide a function in the mechanism of membrane fusion similar to those of the fusion peptides of enveloped virus fusion peptides, namely, association with and destabilization of opposing lipid bilayers. Mutational and biophysical analysis suggested that the internal fusion peptide of p10 lacks alpha-helical content and exists as a disulfide-stabilized loop structure. Similar kinked structures have been reported in the fusion peptides of several enveloped virus fusion proteins. The preservation of a predicted loop structure in the fusion peptide of this unusual nonenveloped virus membrane fusion protein supports an imperative role for a kinked fusion peptide motif in biological membrane fusion.
The physical properties of phospholipid bilayers impose an energy barrier to spontaneous membrane fusion, thereby maintaining the compartmentalized nature of cells (48, 61). Nonetheless, regulated membrane merger is an essential cellular process. Cellular fusion proteins facilitate membrane merger for diverse processes, including intracellular vesicle transport and the formation of zygotes, multinucleated myotubes, and osteoclasts (44, 58, 59). Furthermore, the entry of all enveloped viruses involves membrane fusion induced by viral fusion proteins (29, 52, 58). Understanding the mechanisms of membrane fusion mediated by proteins from diverse sources, therefore, has broad implications.
The best-characterized fusion proteins are those involved in the entry of enveloped viruses. The fusion machinery of enveloped viruses frequently consists of a single large multimeric protein (18, 50). A hallmark feature of enveloped viral fusion proteins is the presence of two hydrophobic sequences, the transmembrane (TM) domain and the fusion peptide, that simultaneously anchor the protein in both the donor and target membranes (58). Numerous studies have demonstrated the significance of enveloped virus fusion peptides in destabilizing target membranes and favoring membrane merger (17, 25). Fusion peptides are short membrane-seeking motifs that frequently assume helical structures in association with membranes. These fusion peptides lie sequestered within the tertiary structure of the prefusion conformation of the fusion protein. Fusion protein activation follows receptor binding- or low pH-induced remodeling of the multimeric fusion protein, frequently involving the formation and rearrangement of coiled-coil structures that expose the buried fusion peptide for membrane interactions. In many viral fusion proteins, subsequent refolding events generate a stable six-helix bundle that positions the TM and fusion peptide motifs at the same end of the bundle (49). Various models propose that these complex protein-refolding events may serve to regulate exposure of the fusion peptide, draw the donor and target membranes into close apposition, and/or provide the energy required to promote lipid mixing and membrane fusion (4, 6, 10, 33, 38).
The avian reoviruses (ARV) and Nelson Bay reovirus (NBV) are two rare examples of nonenveloped viruses that induce syncytium formation (15). Both viruses express small, 10-kDa proteins (p10) within infected cells that promote successive cell-cell fusion events (45). These fusion-associated small transmembrane (FAST) proteins possess a number of unique properties that distinguish them from the well-characterized enveloped virus fusion proteins. Unlike enveloped viruses, the nonenveloped ARV and NBV do not require membrane fusion for entry into cells and therefore maintain p10 as a nonstructural protein (45). The sole function of p10 in the virus replication cycle appears to be the induction of syncytium formation following p10 expression in virus-infected cells, which in turn contributes to a lytic response and release of progeny virus particles (16). Furthermore, p10 fusion activity does not appear to be triggered by low pH or specific receptor interactions; p10 promotes the fusion of many cell types from different species, suggesting that it does not bind to specific receptors, and it functions at neutral pH (16, 39). In addition to these biological differences, the structural properties of the p10 FAST proteins contrast markedly with those of enveloped virus fusion proteins. Like the enveloped virus fusion proteins, p10 assumes an Nexo/Ccyt topology at the surfaces of reovirus-infected or p10-transfected cells (45). However, a central TM domain results in a very small (∼40-residue) ectodomain incapable of six-helix bundle formation and/or the extensive conformational changes that appear to be essential for the fusion activity of enveloped virus fusion proteins. Therefore, p10 may represent a rudimentary fusion protein with limited need for stability in harsh external environments, target membrane specificity, regulation of fusogenic activity, or maintenance of donor membrane integrity, as required with enveloped virus fusion proteins. The apparent simplicity of p10 relative to other biological fusion machinery suggests that p10 may serve as a promising candidate for understanding the minimal requirements of biological membrane fusion.
Determining the mechanism by which p10 induces membrane fusion requires a clearer understanding of the roles of specific motifs in this process. We now report on the essential role of a short, moderately hydrophobic region present in the small ectodomain of p10 which may function in a manner similar to that of enveloped virus fusion peptides, serving as a membrane interaction motif during the fusion process. Biophysical and mutagenic analyses suggest that the predicted p10 fusion peptide exists as a loop structure devoid of alpha-helical content. Our identification of and structural model for the first predicted nonenveloped virus fusion peptide highlights the conservation of a kinked fusion peptide structure, the importance of which has only recently been recognized.
MATERIALS AND METHODS
Plasmids and cells.
The continuous quail cell line QM5 was cultured as previously described (16). The 12CA5 anti-hemagglutinin (HA) mouse hybridoma cells were grown in RPMI 1640 containing 10% fetal bovine serum and penicillin-streptomycin as previously described (45) and were used for production of the HA monoclonal antibody. The eukaryotic expression vector pcDNA3 (Invitrogen) was used for expression of p10 and its derivatives.
Materials.
All lipids were purchased from Avanti Polar Lipids (Alabaster, Ala.). The p10 hydrophobic patch (HP) peptide (p10hp) was synthesized by Biosource (Hopkinton, Mass.) in the form acetyl-CNGATAVFGNVHC-amide. The extended p10 HP peptide (p10ehp) was synthesized by Waterloo Peptide Synthesis (Waterloo, Ontario, Canada) in the form acetyl-PGSCNGATAVFGNVHCQAA-amide with cystine residues. Peptides were purified to >95% purity by high-performance liquid chromatography. A simian immunodeficiency virus (SIV) fusion peptide (SIVfp), GVPVLGPLGPLA-amide, was included as a positive control in the lipid-mixing assays. The 12CA5 anti-HA (immunoglobulin G2b [IgG2b], kappa) antibodies were produced in-house from a mouse hybridoma, and the cell supernatant (∼0.12 μg of anti-HA immunoglobulin/μl) was used for surface staining either directly or following concentration (∼1.6 μg/μl) by precipitation with 35% ammonium sulfate. Alkaline phosphatase-, Texas red-, and fluorescein isothiocyanate-conjugated goat anti-mouse IgG antibodies and protein G-agarose were obtained from Life Technologies Inc. CellTracker Green CMFDA (5-chloromethylfluorescein diacetate) and CellTracker Blue CMAC (7-amino-4-chloromethylcoumarin) were purchased from Molecular Probes (Eugene, Oreg.). Lysosome and proteasome inhibitors were purchased from Sigma-Aldrich. The pSAAM protein sequence analysis and modeling program was used for secondary-structure predictions (12).
Cloning and transfection.
The protocols for construction of the HA-tagged p10 (HA-p10) expression construct, and the generation of all HA-p10 substitutions using three-primer PCR, have been described (45). The ectodomain constructs of HA-p10 (p10e and associated substituted constructs) were amplified by a single touchdown PCR using a forward primer corresponding to the N terminus of HA-p10 that contained the sequence encoding the signal peptide of influenza virus HA with a signal peptidase cleavage site (MLTIIALSYIFCLALGQ). The sequences of all constructs were confirmed. Plasmids were transfected into cells using Lipofectamine (Life Technologies Inc.), as previously described (45).
Immunocytochemical and immunofluorescent staining.
HA-p10 was detected within transfected cell monolayers by immunocytochemical staining using concentrated anti-HA antibodies diluted 1:800 as described previously (45). Stained cells were visualized and photographed on a Nikon Diaphot inverted microscope at ×100 magnification. Surface immunofluorescence was performed on live cells using a 2:3 dilution of anti-HA antibody supernatants as previously described (45). Controls for the specificity of the surface-staining protocol included a functional p10 construct with HA epitope tags in the p10 C-terminal endodomain. This construct exhibited strong intracellular staining but no surface staining, as previously reported (45).
Pulse-chase labeling. At 30 h posttransfection, cells were incubated at 37°C for 20 min with methionine-free Dulbecco's modified Eagle's medium (DMEM), followed by a 5-min incubation with 50 μCi of [35S]methionine/ml in methionine-free DMEM. To ensure the removal of unincorporated [35S]methionine, the cells were washed two times with DMEM containing a 10-fold concentration of cold methionine and six times with Hanks balanced salt solution (HBSS). The cells were incubated for 0, 30, 60, or 120 min with medium 199 at 37°C prior to lysis or collection of medium and immunoprecipitation.
Immunoprecipitation.
Radiolabeled cells were lysed in 1× final lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 1 μg each of aprotinin, leupeptin, and pepstatin/ml). For every 2 × 106 cells, 4 μl of concentrated anti-HA antibodies was preincubated with 15 μl of protein G-agarose for 1 h with shaking at room temperature (RT). Antibody-protein G-agarose complexes were washed once with lysis buffer and incubated with samples for 1 h at RT followed by two stringent washes with each of the following: lysis buffer, high-salt buffer (50 mM Tris-HCl, pH 7.5, 500 mM NaCl, 0.1% Nonidet P-40, 0.05% sodium deoxycholate), and low-salt buffer (50 mM Tris-HCl, pH 7.5, 0.1% Nonidet P-40, 0.05% sodium deoxycholate). Immune complexes were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and fluorography using 15% acrylamide gels.
Heterotypic cell fusion assay.
Sixteen hours following transfection with the HA-p10 constructs, QM5 cells were incubated with HBSS containing 10 μM (final concentration) CMAC for 45 min at 37°C, followed by washes with HBSS to remove excess probe. Nontransfected Hep2 cells were similarly labeled with 5 μM (final concentration) CMFDA. The labeled QM5 and Hep2 cells were suspended and reseeded at a ratio of 3:2 (QM5-HepG2). Forty-eight hours following the original transfection, the cells were immunofluorescently stained using concentrated HA monoclonal antibody (1:200) and goat anti-mouse IgG conjugated with Texas red (1:25) as previously described (45).
Triton X-114 partitioning assay.
Twenty-four hours posttransfection, cells were incubated for 30 min with methionine-free DMEM, followed by a 30-min incubation with 50 μCi of [35S]methionine/ml in methionine-free DMEM. The cells were incubated for 10 min on ice, washed with cold phosphate-buffered saline (PBS), and incubated on ice with a cold solution of 1% Triton X-114, 10 mM Tris-HCl (pH 7.4), and 150 mM NaCl for 15 min with rocking. The solubilized cells were centrifuged at 2,500 × g for 5 min at 4°C to remove debris and nuclei, followed by centrifugation at 15,000 × g for 15 min at 4°C to remove other insolubles. The supernatant was then incubated at 31°C for 10 min, and the aqueous and detergent phases were separated by centrifugation at 300 × g for 3 min at RT. Both fractions were brought up to a 1-ml volume with 5× lysis solution and water to produce a final 1× lysis solution and were immunoprecipitated as described above.
Membrane fractionation of transfected cells and sucrose gradient centrifugation.
Membranes of [35S]methionine-labeled transfected cells were purified exactly as previously described (45). To remove lumenal- and peripheral-membrane-associated proteins, the membranous pellet was resyringed into PBS and treated with 100 mM Na2CO3 (pH 11.3) or 500 mM NaCl for 30 min on ice prior to the recovery of membranes by centrifugation at 100,000 × g for 25 min. For sucrose gradient fractionation, the final membrane fractions were syringed 10 times through a 30-gauge syringe in a small volume of PBS and mixed with 80% sucrose-PBS to produce a 66% sucrose solution that was overlaid with equal volumes of 50 and 5% sucrose-PBS solutions. Centrifugation at 200,000 × g for 2 h separated the lipid fraction that floated above the 50% sucrose from protein aggregates that formed tight pellets. Each fraction was then immunoprecipitated.
CD.
The circular dichroism (CD) spectra were recorded using a model 61 DS CD instrument (AVIV Associates, Lakewood, N.J.). The sample was contained in a 1-mm-path-length cell that was maintained at 25°C in a cell holder with a thermostat. The CD data are expressed as the mean residue ellipticity. The secondary structure was estimated with the program Selcon3 (51). All CD runs were made with peptide dissolved in 10 mM sodium phosphate buffer containing 0.15 M NaF and 1 mM EDTA at pH 7.4. When CD runs were made under reducing conditions, the buffer also contained 4 mM dithiothreitol (DTT).
Lipid-mixing assay for membrane fusion.
A resonance energy transfer assay (53) was used to monitor membrane fusion. Large unilamellar vesicles (LUVs) containing dioleoylphosphatidylcholine (DOPC)-dioleoylphosphatidylethanolamine (DOPE)-cholesterol at a molar ratio of 1:1:1 were prepared. Two populations of LUVs were prepared, one unlabeled and one labeled with 2 mol% each of N-Rh-PE [N-(lissamine rhodamine B sulfonyl)phosphatidylethanolamine] and N-NBD-PE [N-(7-nitro-2,1,3-benzoxadiazol-4-yl)phosphatidylethanolamine]. A 9:1 molar ratio of unlabeled to labeled liposomes was used in the assay. Fluorescence was recorded at excitation and emission wavelengths of 465 and 530 nm, respectively, using a 490-nm cutoff filter placed between the cuvette and the emission monochromator, with 4-nm bandwidths, using an SLM Aminco Bowman AB-2 spectrofluorimeter. Siliconized glass cuvettes (1 cm2) were used with continuous stirring in a cuvette holder with a thermostat. Measurements were carried out using a buffer containing 5 mM HEPES, 5 mM morpholineethanesulfonic acid, 5 mM citric acid, 0.15 M NaCl, and 1 mM EDTA, pH 7.4. When working under reducing conditions, the buffer contained, in addition, 4 mM DTT. LUVs at a final lipid concentration of 50 μM were added to 2 ml of buffer in the cuvette at 37°C, and then the peptide was injected from a solution of dimethyl sulfoxide (DMSO). Fluorescence was recorded for several minutes, and then 20 μl of 10% Triton X-100 was added (final concentration, 0.1%). The initial residual fluorescence intensity prior to acidification, F0, was taken as zero. The maximum fluorescence intensity, Fmax, was obtained by dilution of the labeled lipids with 20 μl of 10% Triton X-100. The percent lipid mixing at time t is given by the following formula: [(Ft − F0)/(Fmax − F0)] × 100. All runs were done in duplicate and were found to be in close agreement. Controls were done using comparable volumes of DMSO in the absence of peptide. The intensity of maximal fluorescence with Triton was found to be close to that obtained when the mole fraction of labeled lipids was reduced 10-fold with unlabeled lipid and was taken as the fluorescence corresponding to 100% fusion.
RESULTS
Essential motifs in the ectodomain of p10.
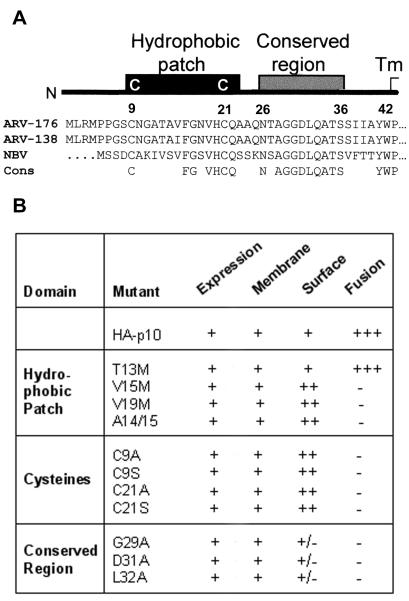
The Nexo/Ccyt topology of p10 suggests that the 39-residue N-terminal extracellular domain (ectodomain) is a possible candidate for interactions with proteins or lipids of target membranes. The ectodomain consists of two motifs which are preserved between the ARV and NBV p10 proteins; a region of moderate hydophobicity flanked by two conserved cysteines that we call the HP and an adjacent region of 11 highly conserved residues (the conserved region [CR]) (Fig. 1A). Previously, it was demonstrated that deletion of the HP or nonconservative substitutions at the flanking cysteine residues are detrimental to the function of p10 (45). However, the basis for the loss of fusion activity was not determined. To discern the roles of these motifs in p10-mediated membrane fusion, we conducted a more thorough substitution analysis of both the CR and HP. For ease of interpretation, a summary of results for all constructs is presented in Fig. 1B, and specific constructs and results are discussed below.
FIG. 1.
Sequence conservation and mutational analysis of the p10 ectodomain. (A) Schematic representation of the p10 ectodomain, including sequence and structural conservation between ARV strains 138 and 176 and NBV. The completely conserved residues (Cons) are indicated. (B) Summary of expression, membrane association, fusogenic activity (scored on a three-plus scale), and surface localization of ARV p10 containing two N-terminal HA epitopes (HA-p10) and various site-specific substitutions. The constructs are named using the single-letter amino acid code to indicate the identity of the authentic amino acid, its position, and the identity of the substituted residue.
Addition of two HA epitopes to the N termini of all p10 constructs permitted comparative assessments in immunoprecipitation and surface immunostaining protocols. Experiments demonstrated that the N-terminal HA epitopes delayed the kinetics of syncytium formation but did not prevent p10-mediated fusion (see below). Following transfection of altered HA-p10 constructs into quail fibroblasts, cell-cell fusion was monitored for 4 days, the endpoint for maximal syncytium formation, before it was concluded that a particular construct was incapable of inducing fusion (Fig. 1B). Previous studies demonstrated the extent of membrane fusion induced by authentic p10 and substituted p10 expression constructs using Giemsa-staining and immunostaining (45). Since the substituted p10 constructs examined in the present study were almost exclusively fusion minus, cell-cell fusion was reported using a qualitative immunostaining assay that allowed simultaneous detection of syncytia and single antigen-positive cells in the transfected monolayers, confirming that the absence of syncytia was not due to altered transfection efficiencies (Fig. 2A). The single-cell antigen-positive foci generated by the fusion-minus mutants contrasted markedly with the appearance of syncytia induced by authentic HA-tagged p10, which appeared as large areas of dark antigen-positive cytoplasmic staining surrounding clusters of unstained nuclei (Fig. 2A).
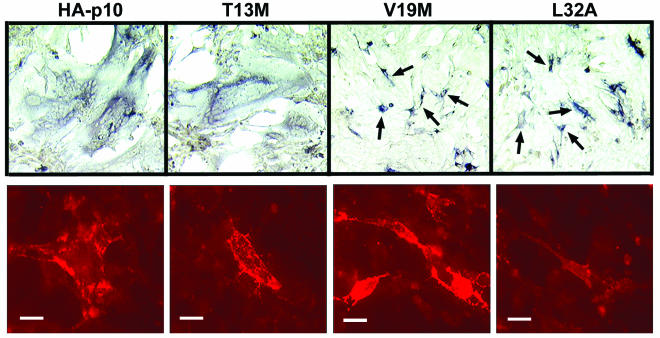
FIG. 2.
HP and conserved domain of p10 are necessary for fusion and surface localization, respectively. Immunohistochemical staining (top row) was used to identify antigen-positive syncytial foci (appearing as large areas of darkly staining cytoplasm surrounding clusters of unstained nuclei) or single antigen-positive cells (arrows) in cell monolayers transfected with HA-p10 or substituted HA-p10 constructs. The degree of cell-cell fusion was qualitatively assessed by microscopic examination of stained monolayers and scored on a three-plus scale based on the relative sizes and abundance of syncytial foci. Results are shown for HA-p10 and a subset of the substituted HA-p10 constructs (T13M, V19M, and L32A). Results from similar analyses for all of the constructs are summarized in Fig. 1B. Images were captured at ×100 magnification. Surface immunofluorescence (bottom row) with live cells was used to detect the relative levels of surface-localized HA-p10 or substituted HA-p10 constructs on transfected cells. Fluorescent images were photographed at ×630 magnification, and images were captured under identical parameters for comparison by intensity of fluorescence. Bars, 20 μm.
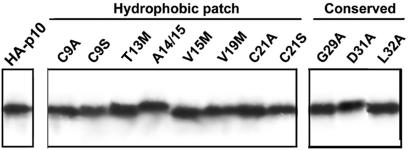
Previous studies demonstrated that p10 is an integral membrane protein that resides entirely within the membrane fractions of cells (45). Fractionation of transfected cells by differential centrifugation followed by immunoprecipitation of radiolabeled membrane-associated p10 was used to confirm that the substitutions did not affect membrane insertion or protein expression of the substituted p10 constructs (Fig. 3). Controls included examining the soluble supernatant for the absence of p10, and the absence of the soluble reovirus σC protein in the membrane fraction (data not shown).
FIG. 3.
Substitutions in the ectodomain do not alter p10 membrane insertion. The presence of authentic HA-p10 and the various substituted constructs in the membranes of cells was determined by immunoprecipitation of the solubilized membrane fraction from radiolabeled transfected cells following extraction of the membranes with alkaline buffer to remove peripheral membrane proteins. Equivalent cell volumes of the immunoprecipitates were resolved by SDS-PAGE (15% acrylamide) and detected by fluorography.
The low level of p10 surface expression precluded using fluorescence-activated cell sorter analysis to quantify surface expression of the substituted p10 constructs. Therefore, immunofluorescent staining of live cells was used to demonstrate the surface expression and orientation of p10 constructs (Fig. 2B). Surface immunofluorescence for all constructs was captured using identical parameters to permit qualitative comparison of surface expression between mutants and the authentic p10. Control experiments using p10 constructs carrying HA epitope tags in the C-terminal endodomain previously indicated the specificity of the surface-staining protocol for the N-terminally tagged p10 constructs (45).
Three substitutions were introduced within the CR (G29A, D31A, and L32A). All three CR mutations abolished p10-mediated fusion (Fig. 1 and 2A). However, in all three CR constructs, although expression, membrane integration, and protein stability were identical to those of the authentic p10 construct, surface localization was qualitatively diminished (Fig. 2B and 3). It is unlikely that these substitutions affected the orientation of p10, since the distribution of positively charged residues adjacent to the transmembrane domain, which remained identical for all CR constructs, is the primary determinant of protein topology (36). Reduced surface expression suggested that the CR might contribute to p10 traffic to, or retrieval from, the surfaces of cells. The extent of surface localization presumably impacts on the fusogenic potential of p10; therefore, these results did not address whether the CR might also be directly involved in the fusion reaction.
A direct role in the fusion activity of p10 was demonstrated for the HP. Replacing the valine at position 15 or 19 with the slightly less hydrophobic methionine residue (methionine substitutions provided a combination of the smallest incremental decrease in hydrophobicity with the least pronounced change in side chain volume) ablated p10-mediated fusion without affecting p10 expression or membrane association (Fig. 3). Abrogation of fusion activity by the V15M and V19M substitutions could not be explained by decreased surface expression, since surface localization was qualitatively increased for these constructs (Fig. 2B). In contrast, the polar threonine residue within the HP withstood mutation to methionine without affecting p10-induced cell-cell fusion (Fig. 2A). The insertion of an alanine between residues 14 and 15 within the p10 HP (A14/15), which would have minimal effect on overall hydrophobicity but would distort the residue arrangement within all secondary structures, eliminated the fusogenic activity of p10 (Fig. 1). In addition, the conserved ectodomain cysteines were essential for p10-mediated fusion. Replacement of either cysteine with alanine or serine eliminated membrane fusion without affecting p10 surface expression or topology (Fig. 1). The structural similarity between cysteine and serine residues is consistent with an essential role for cysteine residues, suggesting that the loss of fusion by C9S and C21S may reflect the requirement for disulfide bond formation.
Therefore, the ectodomain appears to have two essential and distinct motifs; the CR within p10 influences surface expression (and possibly fusion), while the HP is directly implicated in the fusion process independent of surface localization.
The ectodomain of p10 is not involved in cross-membrane pairing.
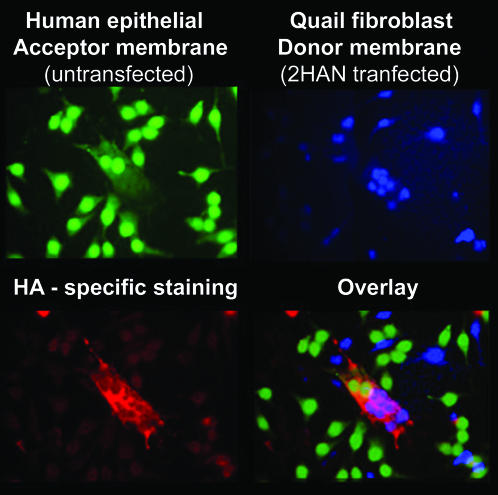
The significant differences between p10 and the fusion proteins of enveloped viruses precluded the assumption that p10 functions similarly to other viral fusion machinery. For example, it was conceivable that p10 might function more like the SNARE proteins involved in intracellular vesicle fusion (37, 44) by pairing with itself in opposing membranes. To address this possibility, we investigated the requirement for p10 in the target membrane. Cell tracker dyes were used to distinguish between two populations of cells, only one of which was transfected with a p10 expression plasmid. Untransfected Hep2 target cells (Fig. 4) were cocultured with p10-transfected QM5 quail fibroblasts (Fig. 4). The p10-induced syncytia present within the mixed population were identified by immunofluorescent staining with HA-specific monoclonal antibodies (Fig. 4). The overlay of the fluorescent images clearly indicated that syncytia contained nuclei from both p10-expressing and non-p10-expressing cells (Fig. 4, merge). Therefore, the ectodomain of p10 does not function to bridge p10 molecules between opposing membranes.
FIG. 4.
p10 is necessary only within the donor membrane. Untransfected Hep2 target cells labeled with CellTracker green (top left) were cocultured with HA-p10-transfected QM5 donor cells labeled with CellTracker blue (top right). The monolayers were fixed and immunostained using anti-HA monoclonal antibodies and Texas red-conjugated goat anti-mouse secondary antibodies to detect p10-induced syncytia (bottom left). The bottom right panel shows an overlay of the previous panels to reveal a p10-induced syncytium that contained nuclei of both QM5 and Hep2 origin.
The p10 HP associates with membranes.
Hydrophobic plots of the p10 HP, using the Wimley-White interfacial hydrophobicity scale (60), suggested that this region might partition at a bilayer interface, a property common to the fusion peptides of enveloped viruses (40, 54). However, the overall hydrophobicity of the p10 HP, as calculated using the normalized consensus hydrophobicity scale (20), is significantly less than those of other fusion peptides (0.29 for ARV p10 versus an average of 0.61 for enveloped virus fusion peptides) (45, 59).
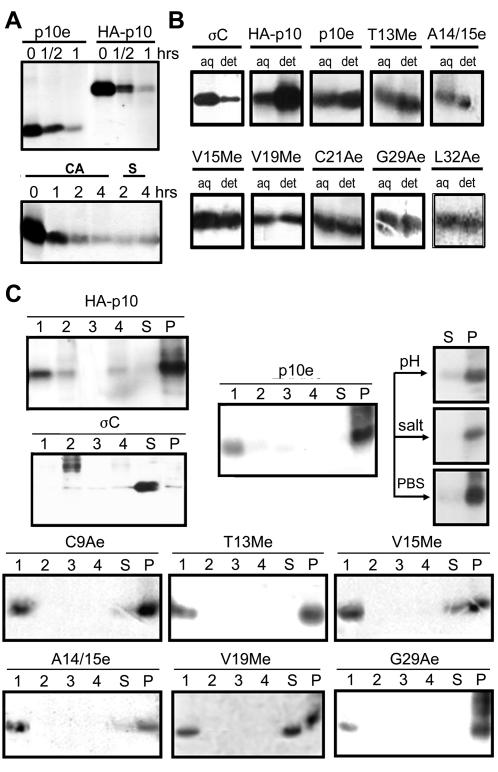
To address the hydrophobic and membrane interaction properties of the p10 HP, secretion constructs of the p10 ectodomain (p10e) were created. A 17-residue signal peptide of influenza A virus HA containing the signal peptidase cleavage site was included to target the p10 ectodomain to the endoplasmic reticulum (ER) lumen, where it normally resides. This p10e construct displayed the standard degradation profile of full-length p10 (Fig. 5A, top),which is extensively degraded to a residual, stable population of p10 (M. Shmulevitz and R. Duncan, unpublished data). The secretion and size of the secreted ER-targeted ectodomain (Fig. 5A, bottom) confirmed cotranslational removal of the HA signal peptide, as previously reported (7). The delay and limited quantity of secreted p10e were predicted to reflect the rapid degradation and membrane-seeking qualities described below.
FIG. 5.
HP has membrane association properties. (A) (Top) Pulse-chase analysis (see Materials and Methods) revealed that the p10 ectodomain construct (p10e) and HA-p10 exhibit the same rapid-degradation profile characteristic of p10. (Bottom) The secreted form (S) of the p10e construct displayed the same gel mobility as cell-associated (CA) p10e, confirming the cleavage and removal of the HA signal peptide used to direct the p10e construct into the ER lumen. (B) Triton X-114 partitioning analysis of radiolabeled cells transfected with soluble ARV sigma C (σC), full-length p10 (HA-p10), or ectodomain constructs with specific substitutions. The substituted constructs are named as in Fig. 1, with an “e” to indicate ectodomain only. The cells were solubilized in 1% Triton X-114 on ice, followed by separation of the detergent (det) and aqueous (aq) phases at elevated temperatures. The presence of p10 in each phase was detected by immunoprecipitation, SDS-PAGE, and fluorography. (C) The transfected cells were separated into the membranous pellet (P) and soluble supernatant (S) fractions as in Fig. 3 to determine the location of the indicated radiolabeled p10 construct. The membrane fraction from the cells was further extracted using high pH (Na2CO3; pH 11.3), high salt (500 mM NaCl), or PBS, and the membrane and soluble fractions were reisolated by centrifugation, as shown for the p10e construct. The final membrane pellet fraction was then centrifuged on sucrose gradients, and the gradients were fractionated to assess the distribution of the indicated p10 proteins in the pelleted protein aggregate fraction (lanes 4), in the soluble protein fractions containing 66 or 50% sucrose (lanes 2 and 3), and in the membrane-associated fraction at the 5-50% sucrose interface (lanes 1).
Triton X-114 phase-partitioning analysis (9) was used to assess the relative preference of the p10 ecotodomain for hydrophobic environments. QM5 cells expressing the soluble ARV σC protein, the HA-p10 protein, the p10 ectodomain (p10e), or p10 ectodomains containing single-amino-acid substitutions (T13Me, A14/15e, V15Me, V19Me, C21Ae, G29Ae, and L32Ae) were solubilized in the presence of cold 1% Triton X-114, followed by separation of the detergent and aqueous phases at elevated temperatures. As expected, the cytoplasmic reovirus sigma C protein associated almost exclusively with the aqueous fraction (Fig. 5B). In contrast, the transmembrane domain-bearing p10 preferentially associated with the detergent fraction. The p10e construct, lacking the TM domain and endodomains, partitioned approximately equally to both the aqueous and detergent phases, as expected for a peptide predicted to orient at a bilayer interface. Modifications to the primary sequence of the p10 CR, as well as small changes to the hydrophobicity of the p10 HP, did not translate into noticeable effects on the preferential distribution between the aqueous and detergent fractions, as T13Me, A14/15e, V15Me, V19Me, C21Ae, G29Ae, and L32Ae all behaved similarly to p10e. Therefore, although the ectodomain of p10 clearly demonstrates hydrophobic properties, slight changes to the hydrophobicity did not significantly alter the partitioning properties in spite of the profound effects of these substitutions on the fusogenic activity of p10.
The membrane interaction potential of the ectodomain of p10 was confirmed by fractionation of cells using differential centrifugation (Fig. 5C). Analysis of protein distribution revealed that the soluble sigma C protein was found almost exclusively in the soluble fraction, while full-length HA-p10 was entirely present within the membranous pellet fraction of cells. The p10 ectodomain construct also associated primarily with the membrane fraction of cells, and this association was resistant to extraction with either high salt or high pH, suggesting that the membrane association was avid and dependent on forces other than ionic interactions. The ectodomain displayed the same membrane association properties when translated in vitro in the presence of canine microsomal membranes (data not shown). The membrane fractions from disrupted cells were further subjected to sucrose gradient centrifugation to establish whether the p10 present in the membrane pellet reflected membrane association or formation of insoluble protein aggregates. The ectodomain of p10 floated at the interface between the 50 and 5% sucrose layers (Fig. 5C, fraction 1), with little protein evident in the insoluble protein pellet (Fig. 5C, fraction 4). These results implied that the p10 ectodomain possesses inherent membrane interaction potential, a property shared with the fusion peptides of enveloped viruses.
The V15M and V19M substitutions that eliminated p10 fusogenic activity, when present in the context of just the p10 ectodomain (V15Me and V19Me), decreased but did not eliminate ectodomain membrane association (Fig. 5C). The insertion of alanine within the HP (A14/15e) also displaced a portion of the ectodomain peptide from the membrane fraction, consistent with the proposal that the positioning of the residues within the HP is important for p10 membrane interactions and fusion activity. Conversely, the T13Me construct or p10 constructs containing alterations in the CR (i.e., G29Ae) displayed no such change in their membrane associations. Replacement of the conserved cysteine with alanine (C9Ae) had only a minimal effect on the association of the ectodomain with cellular membranes, suggesting that disulfide bonds are not essential for association of the ectodomain with membranes. Although membrane association was not eliminated by substitutions that abrogated fusion activity, modifications to the p10 HP may have more drastic effects on the characteristics of membrane interactions, such as depth and angle of insertion, which may reflect the loss of fusion activity with these mutants.
Liposome fusion activity of the p10 HP.
Further evidence that the p10 HP may function as a fusion peptide was obtained by using a liposome fusion assay and synthetic peptides based on the HP. The two peptides corresponded either to the core region of the p10 HP bounded by the two conserved cysteine residues (p10hp) or extended by three residues beyond each cysteine (p10ehp). The longer peptide was synthesized by using cystines to promote disulfide bond formation and contained additional flanking residues to promote loop formation rather than a cyclic peptide. The previously characterized fusion peptide of SIV was used for comparison of liposome fusion activity.
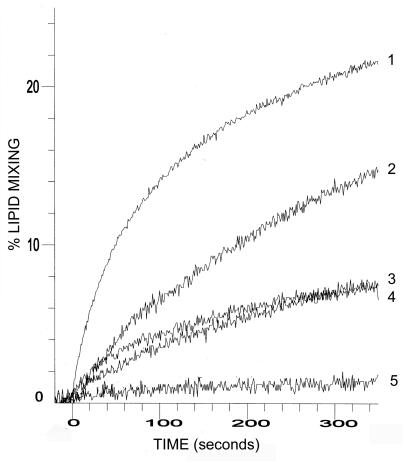
The p10hp, p10ehp, or SIV peptide was added to LUVs composed of DOPC-DOPE-cholesterol (molar ratio, 1:1:1), and the extent of lipid mixing was quantified by resonance energy transfer (53). No lipid mixing was found between vesicles in the absence of peptide (Fig. 6, DMSO control). Conversely, the p10hp and p10ehp peptides promoted lipid mixing, inducing ∼7 and 15% lipid mixing, respectively, at a 10 μM concentration of peptide. By comparison, the moderately fusogenic, and considerably more hydrophobic, SIV fusion peptide at a concentration of 5 μM resulted in ∼22% lipid mixing. Furthermore, the liposome fusion activity of p10 fusion peptides was dose dependent (Fig. 6, p10ehp, 10 μM versus 5 μM). The induction of lipid mixing by the p10 peptides appeared to be specific and not merely a reflection of their hydrophobic nature, since the additional residues that flank the cysteines in the p10ehp peptide decreased the overall hydrophobicity of the peptide (from 0.32 to 0.26 average hydrophobicity) while inducing approximately twice the level of lipid mixing as the smaller, more hydrophobic p10hp peptide. In addition, treatment of the p10ehp peptide with reducing agents decreased lipid mixing to near background levels (see Fig. 8A), implying that the lipid mixing is specific. The membrane interaction potential of the p10 ectodomain and the ability of the p10 HP peptides to induce fusion of liposomes support the designation of this region as a potential fusion peptide motif.
FIG. 6.
The HP of p10 induces liposome fusion. Resonance energy transfer was used to follow the time course of lipid mixing induced by 5 μM concentrations of the SIV fusion peptide (1); 10 (2) or 5 (3) μM concentrations of the p10ehp extended HP peptide; or a 10 μM concentration of the p10hp HP peptide (4) using 50 μM LUVs composed of DOPC-DOPE-cholesterol (1:1:1). A DMSO control was included in the assays (5).
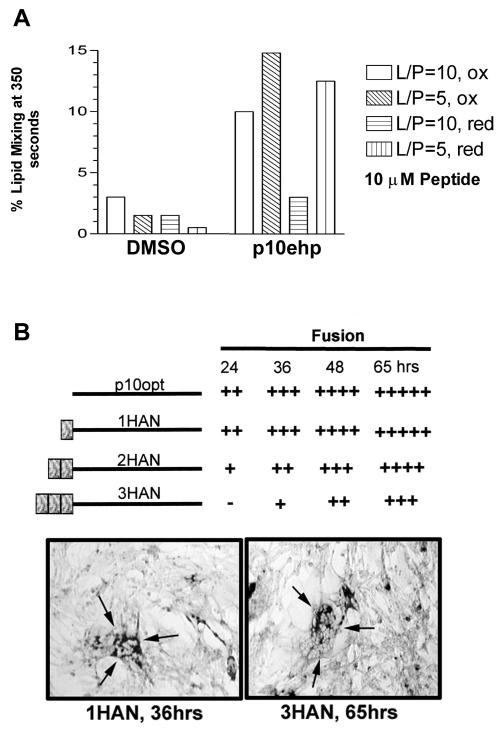
FIG. 8.
Reducing agents alter lipid-mixing activity of the internal p10 fusion peptide. (A) The percent lipid mixing induced by DMSO or 10 μM concentrations of the p10hp or p10ehp peptides were quantified at 350 s using LUVs composed of DOPC-DOPE-cholesterol (1:1:1). Liposome fusion at a lipid-to-peptide ratio of 5 (L/P = 5) or 10 (L/P = 10) were assessed under oxidizing (ox) or reducing (4 mM DTT; red) conditions as indicated. (B) Authentic p10 containing an optimized translation initiation site (p10opt) or p10opt modified with one (1HAN), two (2HAN), or three (3HAN) HA epitopes at the N terminus was assessed for fusogenic activity in transfected quail fibroblasts by immunostaining, as described in the legend to Fig. 2. The extent of fusion was assessed qualitatively by the relative numbers and sizes of syncytia at 24, 36, 48, and 65 h posttransfection. The sizes of syncytia at 36 h for cells transfected with 1HAN were comparable to the extent of syncytium induced by 3HAN by 65 h, showing that additions to the N terminus delayed, but did not prevent, p10-mediated fusion, consistent with p10 HP functioning as an internal fusion peptide.
Structural characteristics of the p10 fusion peptide.
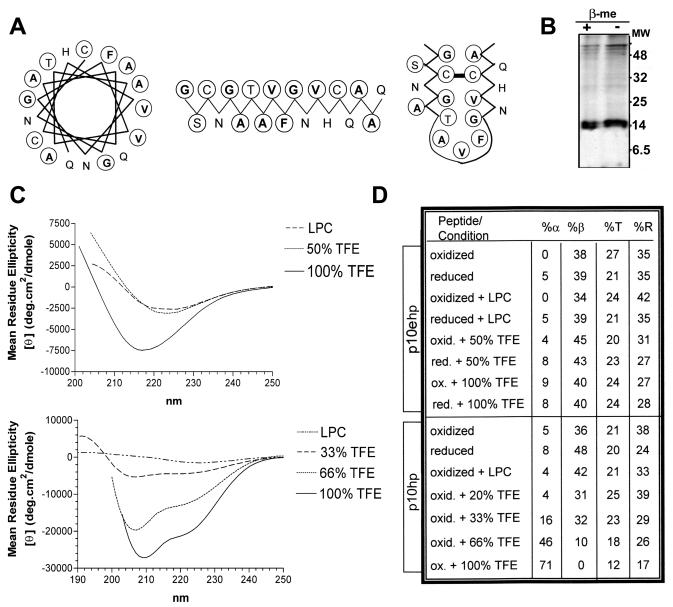
Having established that the p10 HP may function as a fusion peptide motif, we conducted further biochemical and biophysical studies to characterize this motif. Fusion peptides are structurally polymorphic, although an alpha-helical structure may be preferred in the membrane-associated, fusion-active conformation (32, 34, 55). Although an extended hydrophobic face characteristic of fusion peptides is evident in both the alpha-helical and beta strand conformations of the p10 HP (Fig. 7A), computer-based structural analysis of the p10 HP (12) suggested a buried beta strand as the most favorable structure. Furthermore, the evolutionary conservation and essential role of the cysteines flanking the fusion peptide suggested the presence of disulfide bonds. Electrophoretic analysis of p10 under reducing and nonreducing conditions clearly revealed that ectodomain cysteines are not involved in intermolecular disulfide bonds (Fig. 7B). This does not preclude the possible participation of cys9 and cys21 in an intramolecular disulfide bond, as small loops rarely affect the mobility of proteins sufficiently for detection by electrophoresis (24). Therefore, the p10 HP may exist as a disulfide-bonded loop, depicted as a loop of antiparallel beta strands in Fig. 7A. We undertook biophysical analysis to better predict the secondary structure of the p10 fusion peptide.
FIG. 7.
Structural characteristics of the p10 HP. (A) Three diagrams depict the amino acid arrangement within the p10 HP when depicted as an α-helix, a β-sheet, or a disulfide-bonded loop composed of antiparallel β-sheets. The residues most commonly in contact with bilayer lipids (V, I, L, F, A, and G) are in boldface and circled and the residues that can form hydrogen or covalent bonds to favor their presence within membranes (T, S, and C) are circled, while the residues that do not prefer direct contact with lipids (Q, N, H, D, E, R, and K) are neither boldface nor circled. (B) SDS-PAGE analysis of immunoprecipitated HA-p10 from transfected cells under reducing (2% β-mercaptoethanol [+ β-Me]) or nonreducing (− β-Me) conditions shows the absence of a gel mobility shift, indicating that p10 does not form intermolecular disulfide bonds. (C) CD data, expressed as the mean residue ellipticity, was obtained for the p10ehp (top) and p10hp (bottom) peptides. The top CD spectra were obtained using 100 μM extended p10 HP peptide (p10ehp) in LPC at a lipid-to-peptide molar ratio of 10, in 50% TFE, or in 100% TFE, as indicated. The spectra under oxidized and reduced conditions were superimposable. The bottom CD spectra were obtained using 100 μM p10hp peptide in LPC at a lipid-to-peptide molar ratio of 10 or in 33, 66, or 100% TFE, as indicated. (D) The secondary-structure predictions of p10ehp (top) and p10hp (bottom) were estimated with the program Selcon3 under the indicated conditions. The percents α-helix (%α), β-strand (%β), turn (%T), and random coil (%R) are indicated.
CD revealed the absence of minima at 208 and 222 nm, suggesting an almost complete lack of alpha-helical secondary structure in the p10hp and p10ehp peptides (Fig. 7C). Bilayer induction of helical secondary structure was not significant, as the absence of helical structure was maintained in the presence of the micelle-forming lipid lysophosphatidylcholine (LPC). Even in the helix-promoting solvent trifluoroethane (TFE), the p10ehp peptide failed to display significant helical structure (Fig. 7C, top). The increase in the helicity of the p10hp peptide in the presence of 66 to 100% TFE (Fig. 7C, bottom) indicated that the peptide can assume a helical structure under very favorable conditions. In addition, secondary-structure assessment by CD spectrum analysis demonstrated preservation of the lack of helical structure for both the p10ehp and p10hp peptides under reducing conditions (Fig. 7D), suggesting little helix propensity in the presence or absence of the predicted loop structure. Therefore, both secondary-structure predictions and CD data are consistent with the p10 HP existing as a disulfide-bonded loop with little or no helical structure.
Further support for the presence of a disulfide-bonded loop was obtained by assessing the ability of the p10ehp peptide to induce lipid mixing under reducing versus nonreducing conditions. The extended p10ehp peptide was synthesized with cystines under conditions that favor disulfide bond formation to permit analysis of the effect of reducing conditions on liposome fusion activity. A reducing environment decreased the extent of lipid mixing, an effect most noticeable at a high lipid-to-peptide ratio, where activity was decreased to near-background levels under disulfide bond-reducing conditions (Fig. 8A). The increased liposome fusion activity of the extended fusion peptide, therefore, may reflect an increased propensity of the extended peptide to form a cystine noose. The time course for significant changes in the extent of p10-induced syncytium formation (8 h or more) and the toxic effects associated with prolonged treatment of cells with various sulfhydryl-reducing or -reactive compounds confounded attempts to obtain in vivo support for the importance of a disulfide bond in the fusogenic activity of p10 (data not shown).
Enveloped virus fusion peptides can be categorized into internal and N-terminal fusion peptides based on their locations in the ectodomain of fusion proteins. N-terminal fusion peptides are commonly alpha helical, while recent studies have found that internal fusion peptides can adopt a loop structure consisting of beta strands held together by disulfide bonds. Interestingly, extension of the N terminus of p10 by up to three HA epitopes did not eliminate the fusion activity of p10 (Fig. 8B). Rather, each additional HA epitope only delayed the onset and development of syncytium formation. The mutagenic analysis and peptide studies are consistent with the p10 HP functioning as an internal fusion peptide, likely as a disulfide-stabilized loop with little alpha-helical content.
DISCUSSION
The ARV p10 protein shows certain similarities to the fusion proteins of enveloped viruses. As we have shown, p10 is required only in the donor membrane to promote membrane fusion. Furthermore, the N-proximal HP present in the ectodomain of p10 exhibits properties associated with fusion peptides. The putative p10 fusion peptide bears a resemblance to the internal fusion peptides of enveloped viruses with an estimated length of 16 to 19 residues flanked by charged or polar residues (58). Additional similarities between the predicted p10 fusion peptide motif and those of enveloped viruses include (i) a preponderance of glycine and alanine residues, (ii) the potential to form an amphipathic secondary structure, (iii) an essential role in membrane fusion with dependence on hydrophobicity, (iv) an attraction for membranes, and (v) the ability to promote lipid bilayer mixing. Our present results provide compelling evidence that the HP in the ectodomain of p10 represents an internal fusion peptide that serves to destabilize cell membranes.
Unlike the majority of enveloped virus fusion peptides, CD spectrum analysis and computational secondary-structure predictions both predict that the p10 fusion peptide does not form alpha helices. The conservation and essential role of the cysteines that flank the fusion peptide suggest the presence of a disulfide-stabilized loop of antiparallel beta strands. This speculation is supported by the absence of any disulfide-stabilized p10 multimers, indicating that these residues do not participate in an intermolecular disulfide bond (Fig. 7B), and by the reduced fusogenic activity displayed by the p10ehp peptides under reducing conditions (Fig. 8A). Similar structural models, consisting of a loop formed by two antiparallel beta strands, have recently been proposed for the internal fusion peptides of the tick-borne encephalitis virus and avian sarcoma/leukosis virus (ASLV) (13, 43). Furthermore, the fusion peptide loop structure of ASLV is predicted to be stabilized by an intramolecular disulfide bond (5, 14, 43), similar to our proposed structure for the p10 fusion peptide. Other internal fusion peptides, including those within the fusion proteins of Ebola virus and viral hemorrhagic septicemia virus, have also been modeled as looped structures (14, 22, 23, 43). The emerging theme of “protruding” structures in viral fusion peptides also extends to the N-terminal fusion peptides of enveloped viruses. Recent structural studies of the prototypical influenza virus HA fusion peptide revealed a kinked, amphipathic helix-hinge-helix structure (26, 30, 56). The apparent conservation of a kinked or looped fusion peptide structure in viral fusion peptides, even in the predicted fusion peptide of the unusual nonenveloped reovirus p10 fusion protein, underscores what may be an essential feature of these membrane fusion motifs.
The proposed ability of loop structures to interact with lipid bilayers is not limited to viral fusion peptides. For example, various lipases and the prothrombin Gla domain have a flexible amphipathic loop postulated to create a hydrophobic surface that interacts with the lipid phase (11, 19, 21). Disulfide bonds commonly stabilize the loops of lipases and antibacterial peptides, and loss of the disulfide bond reduces the activity of these peptide motifs (27, 28). Furthermore, several antimicrobial peptides that disrupt lipid membranes have a sheet-loop-sheet or helix-loop-helix structure (2, 3). Differences in the depth and/or angle of insertion of these membrane-invasive structures into the lipid bilayer may dictate the outcome (i.e., lysis, fusion, or no effect) of the membrane interaction (35, 41, 42, 55). Mutational analysis clearly revealed that the p10 ectodomain CR influences syncytium formation, suggesting that this region might influence the folding or function of the p10 HP. Interestingly, three-dimensional structural predictions of the p10 ectodomain (http://www.sbg.bio.ic.ac.uk/∼3dpssm) identified similarities to a group of small antimicrobial peptides that exhibit a compact, disulfide-stabilized, triple-stranded beta sheet structure (22a). Therefore, the loop of antiparallel beta strands predicted for the structure of the p10 HP (Fig. 7) might just as easily include the adjacent CR in a triple-beta-strand structure.
The reduced hydrophobicity and liposome fusion activity of the p10 fusion peptide compared with the fusion peptides of enveloped viruses suggests that p10 may have evolved a weak fusion peptide. A comparison of the biological function of p10 in ARV infection to the role of enveloped virus fusion proteins provides the rationale for the evolution of a weak fusion peptide. Expression of p10 in ARV-infected cells is not temporally regulated and resembles the expression profiles of other ARV proteins (46). At the same time, extensive syncytium formation results in altered membrane integrity and cell lysis (8). The combination of a weak fusion peptide, inefficient p10 translation (46), and extensive degradation of newly synthesized p10 molecules (Fig. 5A) may serve to delay the rate of syncytium formation to allow efficient virus replication and assembly prior to cell-cell transmission of the infection and syncytial lysis.
Apart from the decreased activity of the p10 fusion peptide, significant differences exist between p10 and other fusion proteins. The fusion proteins of enveloped viruses share a transition to the active state through conformational changes within the large ectodomain induced by specific triggers (1, 10, 18, 50). These extensive conformational changes involve either coiled-coil rearrangements or multimeric reorganization and are believed to provide the energy required to force close membrane apposition and drive the fusion reaction (31, 50, 57). Aside from the fusion peptide, only 20 residues remain within the ectodomain of p10. Furthermore, recent analysis of the multimeric status of p10 failed to provide any evidence that p10 functions as a multimer (47). The small size and potentially monomeric nature of p10 make it difficult to envision how complex structural rearrangements in p10 could contribute to overcoming the energy barrier that prevents spontaneous membrane fusion.
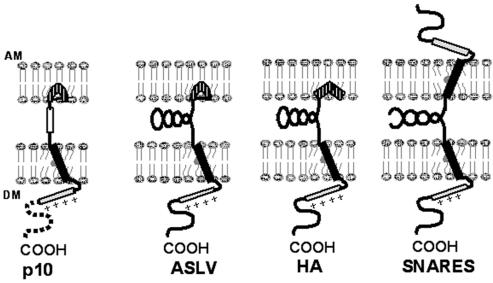
A simplified model of p10, the fusion proteins of ASLV and influenza virus, and the SNARE proteins involved in intracellular vesicle transport shows the conservation of target and donor membrane-interacting domains despite considerable variation in the intervening region (Fig. 9). Although diagrammed as interacting with the target membrane, it is also conceivable that the p10 HP may exert its influence via interactions with the donor membrane. Differences between p10 and the fusion proteins of enveloped viruses may reflect distinct limitations imposed during evolution according to their roles in the virus life cycle. Fusion proteins of enveloped viruses must traffic to the surfaces of cells in an inactive state for incorporation into newly formed virions. In order to maintain envelope integrity, the fusion proteins cannot induce significant destabilizing effects on the donor membrane. Furthermore, enveloped virus fusion proteins require a mechanism for activation following specific interactions with target membranes. Following activation, the fusion proteins must produce rapid and efficient fusion to permit prompt entry and invasion of the cell. In contrast, p10, a nonstructural protein encoded by a nonenveloped virus, need only reach the cell surface in sufficient quantity to induce slow, progressive cell-cell fusion. Additional motifs common to enveloped virus fusion proteins and absent from p10 may reflect modifications necessary for regulation, specificity, and efficiency of enveloped virus-induced membrane fusion. The activity of the fusion peptide of p10, together with domains associated with the donor membrane (47), may, therefore, represent the minimal requirements for biological membrane fusion.
FIG. 9.
Model comparison of fusion proteins. The model depicts the domains of p10 associated with the donor membrane (the TM domain; solid rectangle), the intracellular basic domain (+++), the nonessential cytoplasmic domain (dotted line), the CR (open rectangle), and the proposed kinked fusion peptide (hatched). Similar domains in the fusion protein of ASLV, the fusion protein of influenza virus (HA), and the SNARE proteins involved in intracellular vesicle fusion are depicted. Both ASLV and influenza virus fusion proteins contain complex secondary structure between the fusion peptide and TM domain. Similarly, the SNARE proteins in donor and target membranes pair through significant coiled-coil interactions. In contrast, only the CR separates the fusion peptide and the TM domain of p10.
Acknowledgments
We thank Jingyun Shou for expert technical assistance.
This research was funded by a grant from the Canadian Institutes of Health Research (CIHR). R.D. is the recipient of a CIHR-RPP Investigators Award. M.S. was funded by scholarships from the Natural Sciences and Engineering Research Council of Canada and the Killam Foundation.
REFERENCES
- 1.Allison, S. L., J. Schalich, K. Stiasny, C. W. Mandl, C. Kunz, and F. X. Heinz. 1995. Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. J. Virol. 69:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andreu, D., C. Carreno, C. Linde, H. G. Boman, and M. Andersson. 1999. Identification of an anti-mycobacterial domain in NK-lysin and granulysin. Biochem. J. 344:845-849. [PMC free article] [PubMed] [Google Scholar]
- 3.Aumelas, A., M. Mangoni, C. Roumestand, L. Chiche, E. Despaux, G. Grassy, B. Calas, and A. Chavanieu. 1996. Synthesis and solution structure of the antimicrobial peptide protegrin-1. Eur. J. Biochem. 237:575-583. [DOI] [PubMed] [Google Scholar]
- 4.Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3:309-319. [DOI] [PubMed] [Google Scholar]
- 5.Balliet, J. W., K. Gendron, and P. Bates. 2000. Mutational analysis of the subgroup A avian sarcoma and leukosis virus putative fusion peptide domain. J. Virol. 74:3731-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bentz, J. 2000. Minimal aggregate size and minimal fusion unit for the first fusion pore of influenza hemagglutinin-mediated membrane fusion. Biophys. J. 78:886-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blobel, G., and B. Dobberstein. 1975. Transfer of proteins across membranes. II. Reconstitution of functional rough microsomes from heterologous components. J. Cell Biol. 67:852-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bodelon, G., L. Labrada, J. Martinez-Costas, and J. Benavente. 2002. Modification of late membrane permeability in avian reovirus-infected cells. J. Biol. Chem. 277:17789-17796. [DOI] [PubMed] [Google Scholar]
- 9.Bordier, C. 1981. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 256:1604-1607. [PubMed] [Google Scholar]
- 10.Carr, C. M., C. Chaudhry, and P. S. Kim. 1997. Influenza hemagglutinin is spring-loaded by a metastable native conformation. Proc. Natl. Acad. Sci. USA 94:14306-14316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carriere, F., K. Thirstrup, S. Hjorth, F. Ferrato, P. F. Nielsen, C. Withers-Martinez, C. Cambillau, E. Boel, L. Thim, and R. Verger. 1997. Pancreatic lipase structure-function relationships by domain exchange. Biochemistry 36:239-248. [DOI] [PubMed] [Google Scholar]
- 12.Crofts, A. R. 1994. pSAAM (protein sequence analysis and modeling) package. University of Illinois, Urbana-Champaign.
- 13.Delos, S. E., and J. M. White. 2000. Critical role for the cysteines flanking the internal fusion peptide of avian sarcoma/leucosis virus envelope glycoprotein. J. Virol. 74:9738-9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delos, S. E., J. M. Gilbert, and J. M. White. 2000. The central proline of an internal viral fusion peptide serves two important roles. J. Virol. 74:1686-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duncan, R. 1999. Extensive sequence divergence and phylogenetic relationships between the fusogenic and nonfusogenic orthoreoviruses: a species proposal. Virology 260:316-328. [DOI] [PubMed] [Google Scholar]
- 16.Duncan, R., Z. Chen, S. Walsh, and S. Wu. 1996. Avian reovirus-induced syncytium formation is independent of infectious progeny virus production and enhances the rate, but is not essential, for virus-induced cytopathology and virus egress. Virology 224:453-464. [DOI] [PubMed] [Google Scholar]
- 17.Durell, S. R., I. Martin, J. M. Ruysschaert, Y. Shai, and R. Blumenthal. 1997. What studies of fusion peptides tell us about viral envelope glycoprotein-mediated membrane fusion. Mol. Membr. Biol. 14:97-112. [DOI] [PubMed] [Google Scholar]
- 18.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 19.Egloff, M. P., F. Marguet, G. Buono, R. Verger, C. Cambillau, and H. van Tilbeurgh. 1995. The 2.46 A resolution structure of the pancreatic lipase-colipase complex. Biochemistry 34:2751-2762. [DOI] [PubMed] [Google Scholar]
- 20.Eisenberg, D. 1984. Three-dimensional structure of membrane and surface proteins. Annu. Rev. Biochem. 53:595-623. [DOI] [PubMed] [Google Scholar]
- 21.Falls, L. A., B. C. Furie, M. Jacobs, B. Furie, and A. C. Rigby. 2001. The omega-loop region of the human prothrombin gamma-carboxyglutamic. J. Biol. Chem. 276:23895-23902. [DOI] [PubMed] [Google Scholar]
- 22.Gallaher, W. R. 1996. Similar structural models of the transmembrane proteins of Ebola and avian sarcoma viruses. Cell 17:477-478. [DOI] [PubMed] [Google Scholar]
- 22a.Gao, G. H., W. Liu, J. X. Dai, J. F. Wang, Z. Hu, Y. Zhang, and D. C. Wang. 2001. Solution structure of PAFP-S: a new knottin-type anti-fungal peptide from the seeds of Phytolacca americana. Biochemistry 40:10973-10978. [DOI] [PubMed] [Google Scholar]
- 23.Gaudin, Y., P. de Kinkelin, and A. Benmansour. 1999. Mutations in the glycoprotein of viral haemorrhagic septicaemia virus that affect virulence for fish and the pH threshold for membrane fusion. J. Gen. Virol. 80:1221-1229. [DOI] [PubMed] [Google Scholar]
- 24.Glombik, M. M., A. Kromer, T. Salm, W. B. Huttner, and H. H. Gerdes. 1999. The disulfide-bonded loop of chromogranin B mediates membrane binding and directs sorting from the trans-Golgi network to secretory granules. EMBO J. 18:1059-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gray, C., and L. K. Tamm. 1998. pH-induced conformational changes of membrane-bound influenza hemagglutinin and its effect on target lipid bilayers. Protein Sci. 7:2359-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han, X., J. H. Bushweller, D. S. Cafiso, and L. K. Tamm. 2001. Membrane structure and fusion—triggering conformational change of the fusion domain from influenza hemagglutinin. Nat. Struct. Biol. 8:715-720. [DOI] [PubMed] [Google Scholar]
- 27.Harwig, S. S., A. Waring, H. J. Yang, Y. Cho, L. Tan, and R. I. Lehrer. 1996. Intramolecular disulfide bonds enhance the antimicrobial and lytic. Eur. J. Biochem. 240:352-357. [DOI] [PubMed] [Google Scholar]
- 28.Henderson, H. E., F. Hassan, D. Marais, and M. R. Hayden. 1996. A new mutation destroying disulphide bridging in the C-terminal domain of lipoprotein lipase. Biochem. Biophys. Res. Commun. 227:189-194. [DOI] [PubMed] [Google Scholar]
- 29.Hernandez, L. D., L. R. Hoffman, T. G. Wolfsberg, and J. M. White. 1996. Virus-cell and cell-cell fusion. Annu. Rev. Cell Dev. Biol. 12:627-661. [DOI] [PubMed] [Google Scholar]
- 30.Hsu, C. H., S. H. Wu, D. K. Chang, and C. Chen. 2002. Structural characterizations of fusion peptide analogs of influenza virus hemagglutinin. Implication of the necessity of a helix-hinge-helix motif in fusion activity. J. Biol. Chem. 277:22725-22733. [DOI] [PubMed] [Google Scholar]
- 31.Kielian, M., M. R. Klimjack, S. Ghosh, and W. A. Duffus. 1996. Mechanisms of mutations inhibiting fusion and infection by Semliki Forest virus. J. Cell Biol. 134:863-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kliger, Y., A. Aharoni, D. Rapaport, P. Jones, R. Blumenthal, and Y. Shai. 1997. Fusion peptides derived from the HIV type 1 glycoprotein 41 associate within phospholipid membranes and inhibit cell-cell fusion. Structure-function study. J. Biol. Chem. 272:13496-13505. [DOI] [PubMed] [Google Scholar]
- 33.Kozlov, M., and L. Chernomordik. 1998. A mechanism of protein-mediated fusion: coupling between refolding of the influenza hemagglutinin and lipid rearrangements. Biophys. J. 75:1384-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luneberg, J., I. Martin, F. Nussler, J. M. Ruysschaert, and A. Herrmann. 1995. Structure and topology of the influenza virus fusion peptide in lipid. J. Biol. Chem. 270:27606-27614. [DOI] [PubMed] [Google Scholar]
- 35.Martin, I., M. C. Dubois, T. Saermark, R. M. Epand, and J. M. Ruysschaert. 1993. Lysophosphatidylcholine mediates the mode of insertion of the NH2-terminal SIV fusion peptide into the lipid bilayer. FEBS Lett. 333:325-330. [DOI] [PubMed] [Google Scholar]
- 36.Matlack, K. E. S., W. Mothes, and T. A. Rapoport. 1998. Protein translocation: tunnel vision. Cell 92:381-390. [DOI] [PubMed] [Google Scholar]
- 37.Mayer, A. 2001. What drives membrane fusion in eukaryotes? Trends Biochem. Sci. 26:717-723. [DOI] [PubMed] [Google Scholar]
- 38.Melikyan, G. B., R. M. Markosyan, H. Hemmati, M. K. Delmedico, D. M. Lambert, and F. S. Cohen. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151:413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ni, Y., and R. F. Ramig. 1993. Characterization of avian reovirus-induced cell fusion: the role of viral structural proteins. Virology 194:705-714. [DOI] [PubMed] [Google Scholar]
- 40.Nieva, J. L., and T. Suarez. 2000. Hydrophobic-at-interface regions in viral fusion protein ectodomains. Biosci. Rep. 20:519-533. [DOI] [PubMed] [Google Scholar]
- 41.Pritsker, M., J. Rucker, T. L. Hoffman, R. W. Doms, and Y. Shai. 1999. Effect of nonpolar substitutions of the conserved Phe11 in the fusion peptide of HIV-1 gp41 on its function, structure, and organization in membranes. Biochemistry 38:11359-11371. [DOI] [PubMed] [Google Scholar]
- 42.Rapaport, D., and Y. Shai. 1994. Interaction of fluorescently labeled analogues of the amino-terminal fusion peptide of Sendai virus with phospholipid membranes. J. Biol. Chem. 269:15124-15131. [PubMed] [Google Scholar]
- 43.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature 375:291-298. [DOI] [PubMed] [Google Scholar]
- 44.Rothman, J. E. 1994. Mechanisms of intracellular protein transport. Nature 372:55-63. [DOI] [PubMed] [Google Scholar]
- 45.Shmulevitz, M., and R. Duncan. 2000. A new class of fusion-associated small transmembrane (FAST) proteins encoded by the nonenveloped fusogenic reoviruses. EMBO J. 19:902-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shmulevitz, M., Z. Yameen, S. Dawe, J. Shou, D. O'Hara, I. Holmes, and R. Duncan. 2002. Sequential partially overlapping gene arrangement in the tricistronic S1 genome segments of avian reovirus and Nelson Bay reovirus: implications for translation initiation. J. Virol. 76:609-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shmulevitz, M., J. Salsman, and R. Duncan. 2003. Palmitoylation, membrane-proximal basic residues, and transmembrane glycine residues in the reovirus p10 protein are essential for syncytium formation. J. Virol. 77:9769-9779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siegel, D. P., and R. M. Epand. 1997. The mechanism of lamellar-to-inverted hexagonal phase transitions. Biophys. J. 73:3089-3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skehel, J. J., and D. C. Wiley. 1998. Coiled coils in both intracellular vesicle and viral membrane fusion. Cell 95:871-874. [DOI] [PubMed] [Google Scholar]
- 50.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza haemagglutinin. Annu. Rev. Biochem. 69:531-569. [DOI] [PubMed] [Google Scholar]
- 51.Sreerama, N., S. Y. Venyaminov, and R. W. Woody. 1999. Estimation of the number of alpha-helical and beta-strand segments in proteins using circular dichroism spectroscopy. Protein Sci. 8:370-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stegmann, T., R. W. Doms, and A. Helenius. 1989. Protein-mediated membrane fusion. Annu. Rev. Biophys. Biophys. Chem. 18:187-211. [DOI] [PubMed] [Google Scholar]
- 53.Struck, D. K., D. Hoekstra, and R. E. Pagano. 1981. Use of resonance energy transfer to monitor membrane fusion. Biochemistry 20:4093-4099. [DOI] [PubMed] [Google Scholar]
- 54.Suarez, T., W. R. Gallaher, A. Agirre, F. M. Goni, and J. L. Nieva. 2000. Membrane interface-interacting sequences within the ectodomain of the human immunodeficiency virus type 1 envelope glycoprotein: putative role during viral fusion. J. Virol. 74:8038-8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tamm, L. K., and X. Han. 2000. Viral fusion peptides: a tool set to disrupt and connect biological membranes. Biosci. Rep. 20:501-518. [DOI] [PubMed] [Google Scholar]
- 56.Tamm, L. K., X. Han, Y. Li, and A. L. Lai. 2002. Structure and function of membrane fusion peptides. Biopolymers 66:249-260. [DOI] [PubMed] [Google Scholar]
- 57.Weissenhorn, W., A. Dessen, L. J. Calder, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1999. Structural basis for membrane fusion by enveloped viruses. Mol. Membr. Biol. 16:3-9. [DOI] [PubMed] [Google Scholar]
- 58.White, J. M. 1990. Viral and cellular membrane fusion proteins. Annu. Rev. Physiol. 52:675-679. [DOI] [PubMed] [Google Scholar]
- 59.White, J. M. 1992. Membrane fusion. Science 258:917-924. [DOI] [PubMed] [Google Scholar]
- 60.Wimley, W. C., and S. H. White. 1996. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 3:842-848. [DOI] [PubMed] [Google Scholar]
- 61.Zimmerberg, J., and L. V. Chernomordik. 1999. Membrane fusion. Adv. Drug Deliv. Rev. 38:197-205. [DOI] [PubMed] [Google Scholar]