Abstract
Individuals infected with human immunodeficiency virus type 1 (HIV-1) subtype C infrequently harbour X4 viruses. We studied R5 and X4 biological clones generated from HIV-1 subtype C-infected individuals. All subtype C R5 viruses demonstrated slower profiles of replication on CD4+ lymphocytes in comparison to subtype B viruses, whereas subtype C X4 viruses replicated with comparable efficiency to subtype B X4 viruses. No differences were identified in CC or CXC chemokine inhibitions (RANTES and SDF-1α, respectively) between subtype C and subtype B viruses. Immature dendritic cells were shown in coculture experiments to similarly enhance the infection of subtype C and subtype B R5 as well as X4 viruses. By amino acid sequence analysis, we showed that the R5 and X4 subtype C gp120 envelope gene alterations were similar to those for a switching subtype B virus, specifically with respect to the V3 charge and envelope N-linked glycosylation patterns. By phylogenetic analysis, we showed that one patient was infected with HIV-1 C′ and the other was infected with HIV-1 C" and that one of the patients harbored a virus that was a recombinant in the gp120 env gene between an R5 and an X4 virus, with the resultant virus being R5. No differences were identified between the long terminal repeat regions of the subtype C R5 and X4 biological clones. These results indicate that even though R5 subtype C viruses are restrictive for virus replication, the R5-to-X4 phenotype switch can occur and does so in a manner similar to that of subtype B viruses.
The human immunodeficiency virus type 1 (HIV-1) pandemic is characterized by a large number of viral subtypes and their recombinant forms that are present in variant frequencies throughout the world (43, 44). One of the most striking statistics concerning the spread of HIV-1 has been the emergence and expansion of subtype C virus infections in Africa, China, and India (37, 38). HIV-1 subtype C is currently responsible for the majority of the estimated 45 million HIV-1 infections worldwide and accounts for almost half of all HIV-1 infections in sub-Saharan Africa, with the countries around the horn of Africa and Southern Africa experiencing extremely high prevalence (38). Ethiopia can be considered a specific example of an epidemic dominated by subtype C viruses, while in the surrounding countries subtype A and D viruses are the more prevalent. Two variant genetic genotypes have been identified among subtype C viruses, and these have been termed C′ and C" (2). The other most prevalent HIV-1 strain circulating in Africa is the circulating recombinant form CRF02_AG, which is taking over in the western countries of Africa (11, 31, 55). Many studies are under way to identify whether biological differences exist among the different HIV-1 subtypes that can help explain their altered emergence patterns in different geographical regions.
HIV-1 enters the cell types that it infects through an interaction between the gp120 envelope protein of the virus and the CD4 molecule on the cell surface and a subsequent interaction with a specific CC or CXC chemokine coreceptor, thereby mediating membrane fusion and entry (5, 47). Although a multitude of coreceptors can be utilized by HIV-1, the two most significant for virus transmission and pathogenesis are the CC chemokine receptor CCR5 and the CXC chemokine receptor CXCR4 (6, 57). The preferred phenotypic designations are R5 for the non-syncytium-inducing (NSI) CCR5-using viruses and X4 for the syncytium-inducing (SI) CXCR4-using viruses (5). It is well documented that R5 viruses are those associated with viral transmission and that X4 viruses are those found later in infection, associated with CD4 decline and disease progression (15, 49-51). A number of studies with subtype B-infected individuals have determined that between 40 and 50% of AIDS patients can harbor viruses of the SI, and presumably the X4, phenotype (27, 28). Numerous studies have revealed that the frequency of SI emergence among subtype C-infected individuals is far lower than that identified for the other subtypes (1, 7, 12, 35, 39), although a number of recent studies have found a higher frequency of the X4 phenotype (13, 26).
The molecular alterations associated with the R5-to-X4 switch in vivo are not fully understood, although many of the features of the gp120 envelope viral protein involved in coreceptor usage have been revealed. The V3 region is highly associated with the coreceptor phenotype, with the overall amino acid charge being central to coreceptor usage: higher positive charges are associated with the SI phenotype and utilization of the CXCR4 coreceptor (17, 18, 41, 48). The V1V2 region has also been associated with coreceptor usage, especially in cooperation with the V3 region of the envelope (41). Cooperation between the V3 and the V1V2 regions of gp120 have been shown to influence not only the receptor usage pattern but also its replication phenotype, with a major determinant being the N-linked glycosylation site downstream of the first V3 cysteine (41). The N-linked glycosylation patterns of the gp120 protein also confer a significant effect on the HIV-1 antibody neutralization responses mounted, with alterations in N-linked glycosylation patterns being able to successfully mask effective antibody neutralization responses (3, 56). The CC chemokines RANTES, macrophage inflammatory protein 1α (MIP-1α), and MIP-1β, the natural ligands for the CCR5 chemokine receptor, and stromal cell-derived factor 1α (SDF-1α), the natural ligand for the CXCR4 coreceptor, successfully block the replication of HIV-1 in vitro (9, 14). Furthermore, the association between a large array of genetic polymorphisms within the chemokine and chemokine receptor genes and disease progression rates indicates a strong association between the chemokine network and viral replication in vivo (53). Alterations within the gp120 envelope are therefore thought to influence the extent to which HIV-1 can be controlled by antibody-mediated immune responses as well as host chemokine and chemokine expression levels.
There is evidence that specific genetic alterations within the long terminal repeat (LTR) regions of HIV-1 can be associated with the different virus subtypes. The LTR regions of HIV-1 subtype C viruses have been shown to have an additional NF-κB site inserted in comparison to the other subtypes (24, 36). It has been speculated that an additional NF-κB site may help confer a replication advantage to the subtype C viruses over the other subtypes, and a number of studies have described enhancements to tat-induced transcription as well as virus replication through an additional NF-κB site (25, 33, 34, 36), although another study has suggested that this additional site is redundant with respect to biological function (45).
Why individuals infected with one HIV-1 subtype should switch their virus phenotype from R5 to X4 more frequently than those infected with other subtypes infers either a restriction in the host favoring the expansion of one subtype over another or, alternatively, differences in the biological characteristics of these viruses. The observation that different subtypes can predominate in similar geographical locations and among individuals with similar environmental settings suggests a restriction at the level of the virus (55). In this study we have chosen to investigate the phenotype and genotype of HIV-1 biological clones generated from two individuals who were infected with HIV-1 subtype C viruses and who had been identified as harboring primary isolates with the SI phenotype (1).
MATERIALS AND METHODS
Patient selection and generation of biologically cloned HIV-1.
Two individuals were selected for study from a previously described Ethiopian cohort of HIV-1 subtype C-infected individuals (1). Both individuals had CD4 cell counts of <150/mm3 and harbored SI viruses as determined by replication on the MT-2 cell line (1). The cocirculating HIV-1 subtype C quasi-species were cloned biologically from peripheral blood mononuclear cells (PBMC) by a previously published method, and the individual biological clones were subsequently studied (28, 54). In summary, graded numbers of patient PBMCs (range, 1 × 104 to 2 × 104 cells/well) were cocultivated in 96-well plates with 1 × 105 phytohemagglutinin (PHA)-stimulated PBMC from healthy blood donor volunteers. The proportion (F) of infected cells was determined from the formula for the Poisson distribution, F = −ln (F0), where F0 is the fraction of negative cultures. Only virus clones obtained from a dilution that gave rise to progeny virus in fewer than 33% of parallel cultures were considered clonal. The homogeneity of the isolates was tested by amplification of the V1V3 region and the subsequent ligation into the TOPO-A plasmid (Invitrogen, Breda, The Netherlands). Between 10 and 20 individual bacterial colonies were picked and subsequently DNA sequenced. Only homogeneous biological clones were used in these studies. Generated biologically cloned viruses were propagated on activated CD4+ lymphocytes, and viral growth was monitored by p24 antigen production in the culture supernatants by using a standard enzyme-linked immunosorbent assay (ELISA).
Virus replication on U87.CD4 coreceptor cells, CD4+ lymphocytes, and macrophages.
The virus coreceptor utilization phenotype was determined by measuring viral replication on the U87.CD4 cell line expressing, independently, an array of variant chemokine receptors (CCR1, CCR2b, CCR3, CCR5, and CXCR4), a gift from D. Littman, Skirball Institute, New York, N.Y. These cells were maintained in Dulbecco minimal essential medium supplemented with 10% fetal calf serum (FCS) plus the antibiotics puromycin (1 μg/ml) and neomycin (300 μg/ml). Coreceptor utilization was determined by adding 200 μl of virus stock to 3.0 × 104 U87.CD4 cells (plated 20 to 24 h previously in a 96-well flat-bottom culture plate) expressing the specific coreceptor under analysis. The cells were infected for 18 h before being washed twice with phosphate-buffered saline and fed with 200 μl of fresh medium. On day 10 of culture, the cells were scored for syncytium formation and the p24 levels in the culture supernatants were determined using a standard ELISA. Coreceptor usage was also monitored by infecting CD4+ lymphocytes isolated from an individual homozygous for a wild-type CCR5 gene (CCR5wt/wt) or from an individual homozygous for the 32-bp deletion in the CCR5 gene (CCR5Δ32/Δ32) and monitoring p24 production on days 7 and 10 of culture.
CD4+ lymphocytes were prepared by the following method. PBMCs were isolated from fresh buffy coats (Sanquin, Amsterdam, The Netherlands) or fresh blood draws from laboratory workers by standard Ficoll-Hypaque density centrifugation and frozen in multiple vials at high concentrations, thawed when required, and activated with 5 μg of PHA (Sigma, Zwijndrecht, The Netherlands) and cultured in RPMI 1640 medium containing 10% FCS, penicillin (100 U/ml), and streptomycin (100 U/ml) with recombinant interleukin-2 (100 U/ml). On day 4 of culture, the cells underwent CD4+ enrichment by removing CD8+ lymphocytes with CD8 immunomagnetic beads and a magnet (Dynal, Oslo, Norway), giving rise to cultures of >90% CD4+ lymphocyte purity.
Macrophages were obtained from standard buffy coats that were prepared as described above. After Ficoll-Hypaque density centrifugation, the cells were resuspended in RPMI 1640 medium supplemented with 10% AB+ human serum, containing 20% FCS, 2 mM glutamine, penicillin (100 U/ml) and streptomycin (100 U/ml), after which they were plated at 106 cells/cm2 for 5 days at 37°C in six-well tissue culture plates. Nonadherent cells were removed by extensive washing with RPMI-20% FCS medium, and the adherent cells were infected with each virus (input, between 200 and 1,000 50% tissue culture infective doses [TCID50]). Virus replication was monitored by assaying for p24 in the culture supernatant on days 7, 10, 14, and 18 postinfection.
Replication kinetics of generated virus stocks.
All virus stocks were generated by the multiple passaging of each virus through CD4+ lymphocytes. Viral stocks were assayed to determine their TCID50 on CD4+ lymphocytes by using a previously described method (41). The viruses were tested for their ability to replicate on CD4+ lymphocytes isolated from a CCR5wt/wt individual. The replication kinetics of each virus was measured by infecting CD4+ lymphocytes from CCR5wt/wt individuals with 200 TCID50 of virus, and replication was monitored by measuring p24 antigen levels in the culture supernatants on days 4, 7, 10, and 14 of culture.
Chemokine inhibition assays.
Chemokine inhibition assays were performed in which CD4+ lymphocytes (2.0 × 105/well) were incubated with twofold limiting dilutions of the desired chemokine, RANTES or SDF-1α (ITK Diagnostics BV, Uithoorn, The Netherlands) and incubated for 30 min at 37°C. AMD3100 was obtained through the National Institutes of Health repository. Each well was inoculated with 100 TCID50 of virus and cultured for 14 days. On days 10 and 14, the amount of p24 was determined using a standard ELISA. Virus inhibition was determined by calculating the percent p24 inhibition in the presence of chemokine and in comparison to a control infected well where no chemokine was added and utilizing the data from the day where peak p24 levels were reached.
DC enhancement to viral replication.
Immature dendritic cells (iDCs) were cultured from standard buffy coats (Sanquin) processed by standard Ficoll-Hypaque centrifugation. Monocytes were selected by adhesion in culture flasks for 3 h and then cultured in the presence of 500 U of IL-4 per ml and 800 U of granulocyte-macrophage colony-stimulating factor (a gift from T. Geijtenbeek, Free University, Amsterdam, The Netherlands) per ml for 7 days. These culture conditions have previously been shown to be suitable for cells expressing high levels of DC-SIGN as measured by flow cytometry (21). These iDCs were utilized in coculture experiments where suboptimal concentrations of virus were incubated with 2.0 × 104 iDCs and 2.0 × 105 CD4+ lymphocytes isolated from an individual with the the CCR5wt/wt genotype, and the amount of p24 in the culture supernatant was determined on days 5 and 7 of culture. Chemokine inhibition assays were also performed as described above in the presence of iDCs at the same ratios described.
DNA sequencing and analysis of the envelope regions.
PHA-stimulated PBMCs from healthy blood donor volunteers were infected with each virus, and on day 5 the cellular DNA was extracted from 106 cells by using a silica-based isolation method (10). The gp140 region of the envelope gene (between the KpnI and BamHI cloning sites) was amplified using the primers 5′-KpnENV (ACTTGTGGGTCACAGTCTATTATGGGGTACC) and 3′-BamENV (GCTCCGCAGGTCGTCCCAGGCAAGTGCTAAGGATCCG). The DNA was sequenced using a set of primers spanning the length of the DNA, with an average distance of 200 nucleotides between primers. Both DNA strands were sequenced, with the sequencing being performed on an automated sequencer with the Thermo Sequenase BigDye Terminator cycle-sequencing kit (Applied Biosystems, Foster City, Calif.) as specified by the manufacturer. The primer sequence and conditions for PCR amplification of the U3 region of the LTR (U3LTR) have been described in detail previously (16).
The sequences were aligned manually based on the alignment of the Los Alamos database reference sequences for subtyping. Phylogenetic analysis of the aligned sequences was performed using the neighbor-joining MEGA method (29) and confirmed by the DNADIST, NEIGHBOR, and DRAWTREE options of the PHYLIP software package (http://evolution.Genetics.Washington.edu/phylip.html) (20). The distance matrix was generated by Kimura's two-parameter estimation (29). Based on 100 replications, a bootstrap value equal to or greater than 70% is considered significant (22, 23). Other sequences obtained from the Los Alamos database were included as reference sequences. The boot-scanning method as implemented in the SIMPLOT program was used to detect and analyze the recombinant viruses (46; S. C. Ray, 1999, http://www.med.jhu.edu/deptmed/scray/download/simplot). Our analysis was performed by calculating the distances for a sliding window of 200 nucleotides of the test sequences, moving along the alignment of a panel of reference sequences by increments of 20 bp. One hundred replications were generated by the bootstrap method for each window, and the percent bootstrap values were plotted against the nucleotide position of the sequence of the reference panel.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the gp120 envelope sequences are AY452640 to AY452650, and those for the LTR sequences are AY452651 to AY452661.
RESULTS
Generation of subtype C R5 and X4 biologically cloned viruses and their ability to replicate on variant cell types.
From the study of 48 patients with CD4 counts of <150 cell/mm3, who had at least one AIDS-defining illness, 3 (6.3%) were identified as harboring viruses with the capacity to induce syncytia on MT-2 cells (1). Biological clones were generated from two of these individuals (PHD74 and PHD79), and the resultant viruses were studied for replication on various cell types, namely, U87.CD4 cells expressing variant coreceptors, CD4+ lymphocytes, and macrophages; a summary of the results is shown in Table 1. Four virus clones were studied from each donor, with both R5 and X4 viruses being generated from each individual. One virus from donor PHD74 was of the R3/X4 phenotype (C4), while all the other viruses used a single coreceptor. Five other viruses were used in this study to allow for further comparisons to be made: one X4 molecular cloned subtype C virus (SE12808), two subtype B primary isolates (one R5 and one X4 [NSI-18 and SI-19, respectively]), and two subtype B molecular cloned viruses (one R5 and one X4 [SF-162 and SF-2, respectively]). Replication of the subtype B and C viruses on MT-2 cells and on CD4+ lymphocytes isolated from either CCR5wt/wt or CCR5Δ32/Δ32 individuals correlated with the replication profile on U87.CD4 cells expressing either CCR5 or CXCR4. Only one of the R5 subtype C clones demonstrated replication on macrophages in comparison to both the subtype B viruses, suggesting that the CCR5-utilizing subtype C virus isolates replicate weakly on this cell type relative to subtype B viruses.
TABLE 1.
Characteristics of HIV-1 subtype C biological clones
| Donor | Virus name | Subtype | Replication on MT-2 cells | Replication on CD4+a:
|
Macrophageb | Receptorc | TCID50/mld | |
|---|---|---|---|---|---|---|---|---|
| CCR5wt/wt | CCR5Δ32/Δ32 | |||||||
| PHD74 | H4 | C | − | + | − | 0 | R5 | 103.09 |
| PHD74 | D3 | C | − | + | − | 0 | R5 | 102.22 |
| PHD74 | F3 | C | + | + | + | 0 | X4 | 102.56 |
| PHD74 | C4 | C | + | + | + | 0 | X4 | 105.72 |
| PHD79 | C1 | C | − | + | − | 7,260 | R5 | 103.72 |
| PHD79 | C12 | C | − | + | − | 0 | R5 | 104.15 |
| PHD79 | H8 | C | + | + | + | 0 | X4 | 103.97 |
| PHD79 | B8 | C | + | + | + | 0 | X4 | 104.85 |
| None | NSI-18 | B | − | + | − | 9,440 | R5 | 104.76 |
| None | SF-162 | B | − | + | − | >10,000 | R5 | 105.43 |
| None | SI-19 | B | + | + | + | 0 | X4 | 105.54 |
| None | SF-2 | B | + | + | + | 0 | X4 | 105.54 |
| None | SE12808 | C | + | + | + | 0 | X4 | 103.09 |
Replication on CD4+ lymphocytes isolated from an individual homozygous for a wild-type CCR5 (CCR5wt/wt) gene or homozygous for the 32-bp deletion in the CCR5 gene (CCR5Δ32/Δ32).
Replication on macrophages [culture supernatant (picrograms/milliliter)].
Replication on U87.CD4 cells expressing the coreceptor of choice.
Determined on CCR5wt/wt CD4+ lymphocytes.
Reduced replication kinetics of subtype C R5 but not X4 viruses.
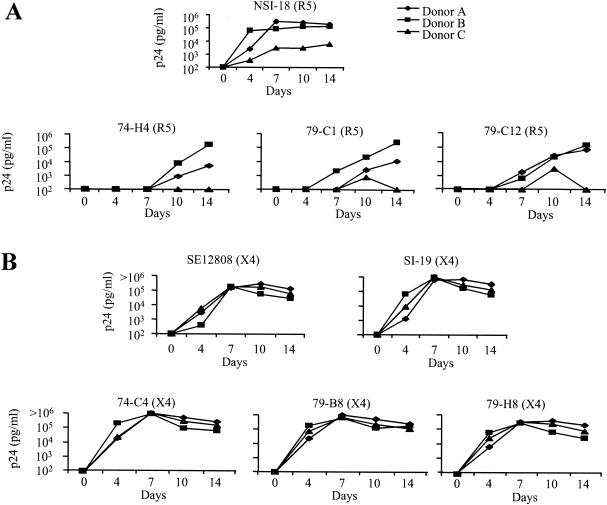
We sought to identify whether replication of the subtype C viruses on PBMCs was comparable to what is commonly found for subtype B viruses. For this purpose, we analyzed the replication of each subtype C biological clone on CD4+ lymphocytes isolated from three different individuals, wild-type for the CCR5 gene, who demonstrated variation in the replication profile of the NSI-18 subtype B R5 virus (Fig. 1A). All infections were performed with the same viral input (200 TCID50/ml) with the same activated batch of CD4+ lymphocytes. We observed low-level replication of the NSI-18 virus on CD4+ lymphocytes from donor 3, in comparison to the results for lymphocytes from the other two donors, which correlated with low cell surface expression of the CCR5 coreceptor (data not shown). All the subtype C R5 viruses demonstrated the same pattern of restricted replication on CD4+ lymphocytes from donor 3 in comparison to either donor 1 or donor 2, and the subtype C R5 viruses demonstrated slower kinetics of replication on lymphocytes from all three donors in comparison to the subtype B control virus (Fig. 1A). When comparisons were made between p24 values on day 7, the subtype B virus had reached its peak p24 value on CD4+ lymphocytes in all three donors while the subtype C viruses were only at the beginning of the logarithmic phase of their replication cycle. The replication curve we observed for the NSI-18 subtype B virus was representative of the curves for other subtype B viruses tested at the same TCID50 of infection (data not shown). In contrast, all the subtype C X4 viruses replicated to the same degree on the same CD4+-enriched lymphocyte batches as the subtype B viruses, with peak virus production in culture supernatant reached by day 7 (Fig. 1B). The similarity in replication of the X4 viruses on CD4+ lymphocytes isolated from donor 3 indicates that these cells can replicate HIV-1 as efficiently as cells from the other two donors, suggesting that the restriction observed with the CCR5-using viruses is an envelope-restricted phenomenon.
FIG. 1.
Replication kinetics of HIV-1 isolates on CD4+ lymphocytes. (A) Replication of a single subtype B R5 virus and three subtype C biologically cloned R5 viruses generated from donors PHD74 and PHD79 were monitored on PHA-activated CD4+ lymphocytes. (B) Replication of a single subtype B X4 virus, a subtype C biologically cloned X4 virus from the AIDS repository, and three biologically cloned viruses from donors PHD74 and PHD79 were monitored on PHA-activated CD4+ enriched lymphocytes. In both panels, virus replication was monitored by measuring p24 in the culture supernatants on days 4, 7, 10, and 14 of culture.
No differences between CC or CXC inhibition of the subtype C or B viruses.
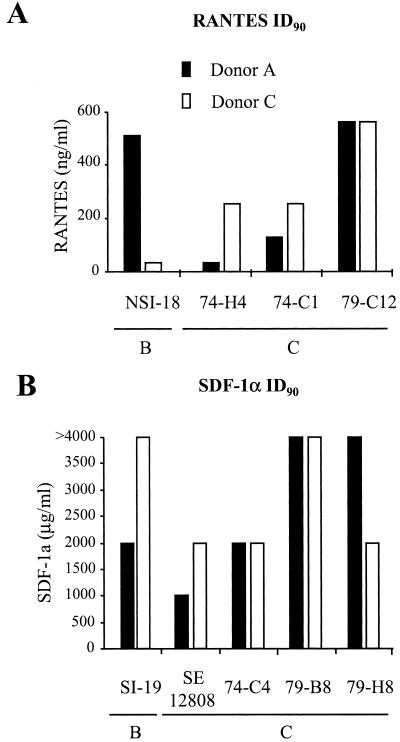
To identify the sensitivities of our viruses to the inhibitory effects of either CC or CXC chemokines, we performed in vitro inhibition experiments with limiting dilutions of either RANTES or SDF-1α for the R5 and X4 viruses, respectively (Fig. 2). The inhibitions were performed with CD4+ lymphocytes from two donors (donors A and B), and similar concentrations of CC or CXC chemokine were required to inhibit virus replication by >90% in lymphocytes from both donors (Fig. 2). We also observed a wide variation in the required concentration of CC or CXC chemokine required to inhibit replication, which was neither HIV subtype nor R5 or X4 phenotype restricted.
FIG. 2.
Chemokine inhibitions of subtype B and C viruses. (A) Inhibition of subtype B and C R5 viruses with the CC chemokine RANTES, with the results expressed as the highest concentration of RANTES required to provide >90% inhibition of virus production. (B) Inhibition of subtype B and C X4 viruses with the CXC chemokine SDF-1α, with the results expressed as the highest concentration of SDF-1α required to provide a >90% inhibition of virus production.
Efficient DC enhancement of subtype C virus infectivity.
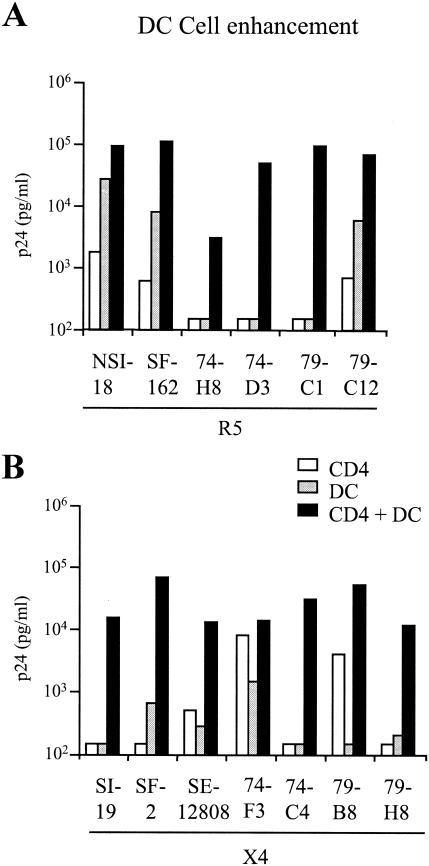
DCs enhance the replication of HIV on CD4+ lymphocytes through the interaction of the HIV gp120 envelope with the DC-SIGN molecule on the surface of the DCs. We wished to determine whether we could observe differences in DC enhancement of subtype B and C viruses and with respect to the R5 or X4 virus phenotype. We therefore performed DC enhancement experiments with each of our viruses in an iDC/CD4+ coculture experiment using a low to suboptimal level of virus input. In this experiment, we included two extra subtype B viruses, the SF-162 (R5) and SF-2 (X4) molecularly cloned viruses, to allow for extra comparisons. All viruses replicated to low levels on CD4+ cells alone and showed enhanced levels of replication in the presence of iDCs (Fig. 3). We found the enhancement to be between 10- and 100-fold, irrespective of either the virus subtype or the virus coreceptor-using phenotype. Interestingly, the two subtype B R5 viruses demonstrated the highest levels of replication on DCs alone while only one of the four subtype C R5 viruses demonstrated such high levels, probably a reflection on the poor growth of subtype C R5 viruses on CD4+ lymphocytes in general (Fig. 3A). Although we cannot rule out additional DC cytokine enhancement of viral replication, previous studies have demonstrated the necessity of the DC-SIGN interaction with the virus for increased viral transfer via the ability of a DC-SIGN-specific monoclonal antibody to inhibit enhanced viral replication (21).
FIG. 3.
iDC cell enhancement of virus infectivity. A low dose input of virus was used (<100 TCID50/ml) to infect CD4+ cells alone, iDCs alone, or CD4+ cells in the presence of iDCs (1:10 ratio) for the R5 viruses (A) and the X4 viruses (B). Virus production was determined by measuring p24 in the culture supernatants on day 7 of culture.
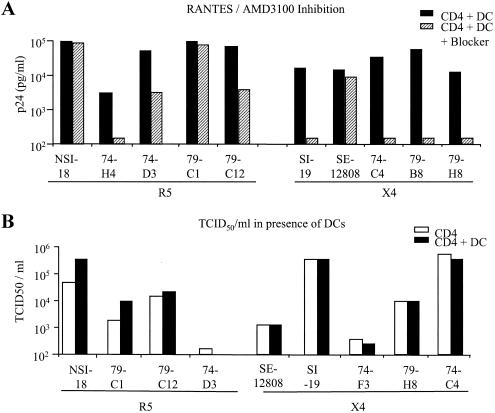
We wished to determine how sensitive the subtype C R5 viruses were to the blocking effects of the CC chemokine RANTES through interaction with iDCs. When we performed iDC/CD4+ lymphocyte transfer experiments in the presence of high concentrations of CC chemokine (100 ng/ml), sufficient to fully block CD4+ lymphocyte infection at a multiplicity of infection of 0.03, we still found a high efficiency of virus transfer (Fig. 4A). This result would suggest that CC chemokines are less efficient at blocking HIV infection of CD4+ lymphocytes when the virus is bound to iDCs. Interestingly, three of the subtype C viruses showed higher levels of inhibition in the presence of RANTES than did the R5 subtype B virus. All the X4 subtype C viruses could be fully inhibited for viral transfer in the presence of the AMD3100 inhibitor (Fig. 4A).
FIG. 4.
Effect of iDCs on virus infectivity. (A) A low-dose input of virus (<100 TCID50/ml) was used to infect CD4+ cells which had or had not been preincubated with a blocking concentration of either RANTES or AMD3100 for R5 and X4 viruses, respectively, in the presence of iDCs. Virus production was determined by measuring p24 in the culture supernatants on day 7 of culture. (B) TCID50s per milliliter were determined on CD4+ cells for viruses that had been cultured in the presence or absence of iDCs.
Since the interaction of the subtype C viruses with iDCs could enhance p24 production, we wished to determine whether this had an effect on the virus titer that could be generated from infected CD4+ lymphocytes. We therefore cultured virus on CD4+ lymphocytes in the presence of iDCs for 1 week and harvested the virus in the culture supernatant. We then determined, on the CD4+ lymphocytes isolated from the same individual, the TCID50s of the viruses produced in the presence or absence of iDCs. The TCID50s were found to be similar irrespective of the presence of iDCs during culturing (Fig. 4B), even though the p24 levels in the virus stocks differed by almost 1 log unit (data not shown).
Similarities between subtype B and subtype C R5 and X4 gp120 envelope protein sequences.
We compared the full-length gp120 envelope amino acid sequences of the cocirculating virus quasi-species from both donors PHD74 and PHD79 in order to better understand which alterations are implicated in the R5-to-X4 coreceptor switch for HIV-1 subtype C. The inclusion of two individuals in this analysis allowed for both intra- and intersample comparisons to be made. The alignments of the consensus amino acid sequences of the R5 and X4 clones from each individual are presented in Fig. 5A and B (PHD74 and PHD79, respectively), where the two phenotypes for each individual are compared. We compared these amino acid sequences with full-length gp120 envelope sequences identified in an R5-to-X4-switching individual infected with HIV-1 subtype B (Fig. 5C). Common alterations within the subtype C virus isolates were identified which had previously been linked to the R5-to-X4 switch in subtype B viruses (41).
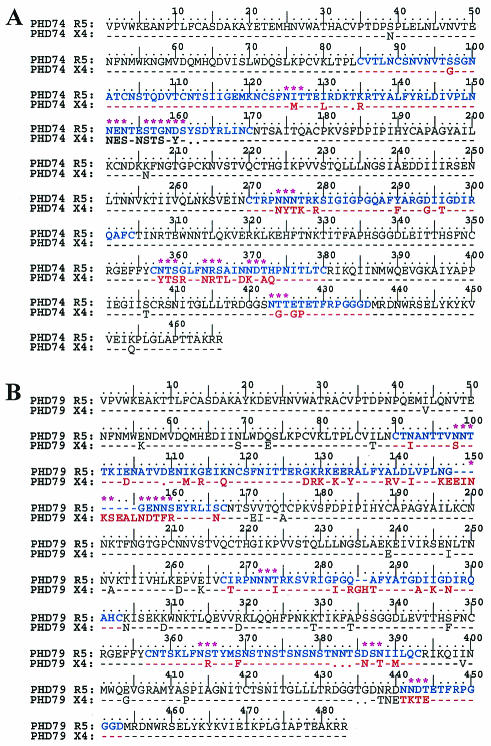
FIG. 5.
The gp120 predicted amino acid sequence comparison between the CCR5 and the CXCR4-utilizing biological clones. (A) Sequence from patient PHD74, carrying sequences of the genotype HIV-1 subtype C". (B) Sequence from patient PHD79, carrying sequences of the genotype HIV-1 subtype C′. (C) Sequence from patient ACH168, carrying sequences of the genotype HIV-1 subtype B. The sequences of the subtype C viruses are consensus sequences of the R5 and X4 biological clones isolated from the same time point, while the ACH168 sequences are the early R5 and the late X4 isolate sequences as previously described. The sequences of the variable loops of the gp120 protein, where the bulk of amino acid differences occur, are shown in color (blue for R5 and red for X4 viruses). Green asterisks indicate modifications or shifts in potential N-linked glycosylation sites between the R5 and the X4 sequences.
(i) Third-variable-loop positive charges.
CXCR4-using clones of both HIV-1 subtype B and C isolates showed alterations in their V3 loop that increased the overall positive charge compared to the CCR5-using clones, a phenomenon well described for other subtypes. The positive-charge increments were from +3/+4 in the PHD74/R5 clones to +5 and +6 in the PHD74/X4 clones while for the PHD79 isolates the increments were from +2 and +3 in the R5 clones to +5 and +7 in the X4 clones. Interestingly, the altered amino acids associated with the charge increase were not at positions 11 and 25 (in relation to the first cysteine), which has been well described for subtype B viruses (41). For PHD79, the positive charge was introduced at position 284 modifying the GPGQ motif to GPGR, the motif the most frequently observed in subtype B viruses. For PHD74 the positive charge was introduced at position 279, the other position adjacent to the positively charged motif RK at the beginning of the V3 loop. We have observed the loss of the negative charges at positions 294 and/or 298 for both patients studied, mainly with the modification of the aspartic acid (D) to asparagines (N) or glycine (G), often observed for subtype B viruses.
(ii) Third-variable-loop glycosylation modifications.
The implication of the potential N-linked glycosylation site of the NNNTR motif at the beginning of the V3 loop with the coreceptor switch has been previously documented (41). Amino acid sequence analysis of our subtype C biological clones has shown that this N-linked glycosylation site was modified in both individuals, corresponding to the R5-to-X4 switch in virus phenotype (Fig. 5A and B). We showed that in the isolates from patient PHD79 it changes from NNNTR to NNNIR, the same modification observed in donor ACH168 (Fig. 5C). In isolates from patient PHD74, the modifications were from NNNTR to NYSKR, NYIKR, or NYTKR. Although the motifs NYS and NYT are still potential N-linked glycosylation sites, the composition of the sugar chains is likely to be different.
(iii) N-linked potential glycosylations in other variable loops.
When we compared the amino acid sequences of the V1V2 regions of the two different phenotypes X4 and R5, we identified alterations in their potential N-linked glycosylation patterns. Alterations in the V1V2 regions of subtype B viruses have previously been shown to be associated with coreceptor switching and with higher virus replication rates (41). Furthermore, we have identified differences in the N-linked glycosylation patterns between the R5 and X4 viruses outside of the V1V2 and V3 variable regions, which have, interestingly, been identified in subtype B isolates that have switched in their coreceptor using phenotype (41). Of particular note, the X4 viruses of both patients lost a potential N-linked glycosylation site within the V4 region while the virus from patient PHD79 gained one N-linked glycosylation site and lost another within the V5 region.
Phylogenetic analysis of the gp120 envelope proteins of the subtype C viruses.
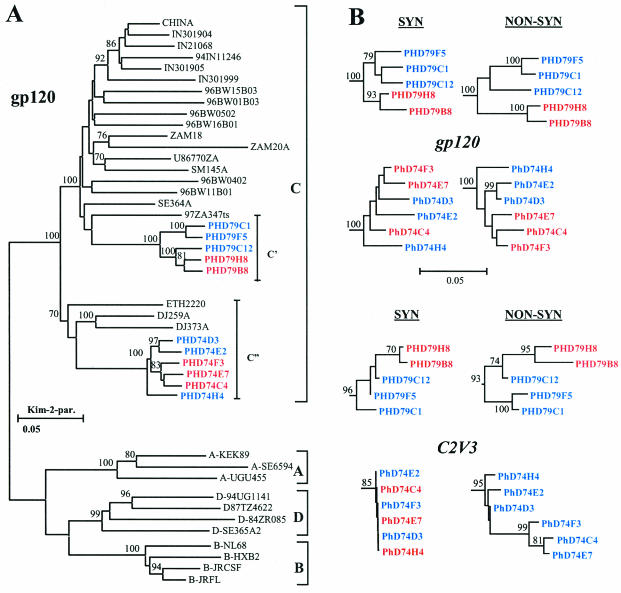
Through phylogenetic analysis of the full-length gp120 envelope sequences (1,400 nucleotides), we have shown that the viruses originating from patients PHD74 and PHD79 belong to two different cocirculating genotypes (C′ and C") (Fig. 6A). These genotype distinctions have previously been identified by analysis of the C2V3 region of the gp120 protein, and both genotypes have been shown to circulate in Ethiopia (2). The virus clones isolated from patient PHD79 belong to the HIV-1 C′ genotype, which are related to the subtype C strains dominating the epidemic in southern Africa, India, and China (40), while the clones isolated from patient PHD74 belong to the HIV-1 C" group, which includes the subtype C reference sequence ETH2220 and sequences originating in Djibouti. The similarity between the gp120 alterations between the R5 and X4 biological clones suggests that the gp120 envelope modifications leading to the coreceptor switch are similar between these two genotypes and similar to what is found for subtype B viruses.
FIG. 6.
Phylogenetic analysis of the subtype C virus biological clones. (A) Neighbor-joining/Kimura two-parameter phylogenetic analysis performed using the MEGA program. The two Ethiopian subtype C genotypes are indicated by C′ and C". The tree is composed of the biological clone gp120 sequences derived from the two AIDS patients from Ethiopia together with subtype C sequences collected in Djibouti, Somalia, as well as southern Africa, India, and China. The Ethiopian ETH2220 sequence was used as the subtype C reference, with subtype A, B, and D reference sequences from the Los Alamos database. Blue represents the CCR5-using biological clones, and red represents the CXCR4-using clones. (B) Phylogenetic branches of the C2V3 region and the gp120 full-length protein sequence of the biological clones. The analysis was performed by taking into consideration either the synonymous or the nonsynonymous nucleotide substitution differences. The numbers near the branches represent the percent bootstrap values from 1,000 replications.
Analysis of the synonymous amino acid substitutions reveal that the X4 clones, but not the R5 clones, from patient PHD79 cluster together whether the full-length protein or the C2V3 region of the gp120 protein are considered (Fig. 6B). Neither the X4 nor the R5 clones from patient PHD74 clustered together by any significant bootstrap values, indicating that coreceptor usage does not necessarily determine the phylogenetic lineage. In contrast, the nonsynonymous amino acid substitution analysis revealed that for both individuals there was a marked segregation between the X4 and R5 clones, which was particularly apparent for the C2V3 region of the gp120 protein, demonstrating the strong involvement of this region in coreceptor utilization (Fig. 6B). In addition, the synonymous-versus-nonsynonymous amino acid substitution rates (ds/dn) for the gp120 region were 1.00 for PHD74 and 0.58 and PHD79 while those for the C2V3 region were 0.02 and 0.26, respectively, again demonstrating the strong positive selection driven by coreceptor utilization.
Clone PHD79C12 is of particular interest since phylogenetic analysis (Fig. 6B) and boot-scanning analysis (data not shown) have identified it as a recombinant between the X4 and R5 quasi-species cocirculating within patient PHD79. The virus has acquired the V1V2 envelope region from the X4 clones but has remained a CCR5-using virus, indicating that for this virus the V3 region is a strong determinant of coreceptor usage. Interestingly, this virus has the highest TCID50 of all the R5 viruses (Table 1), demonstrating the highest resistance to the blocking effects of the CC chemokine RANTES on CD4+-lymphocytes (Fig. 2A).
Similarity between the LTR sequences of subtype C R5 and X4 viruses.
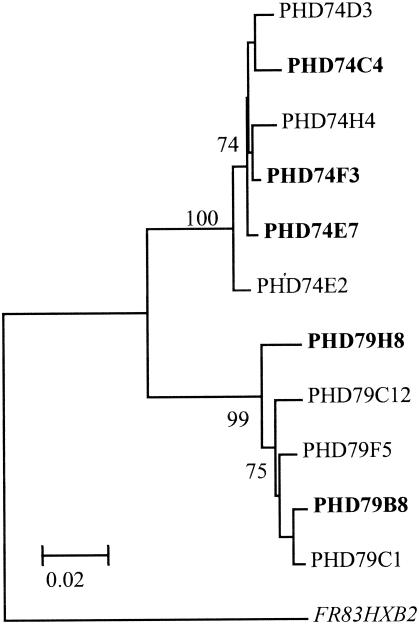
Since differences have previously been identified between the LTR regions of subtype B and C viruses (24, 36), particularly in their U3 regulatory transcription factor binding sequences, we wished to compare the LTR sequences of our generated biological clones and identify whether we could observe alterations in LTR sequences between the R5 and X4 clones, which may explain the enhanced replication capacity of the X4 viruses. We sequenced the U3R region amplified from all the biological clones generated from patients PHD74 and PHD79 and showed that they matched the subtype C consensus sequence for this region (data not shown). No differences were identified by phylogenetic analysis between clones from either individual or between the R5 and X4 viruses (Fig. 7).
FIG. 7.
Neighbor-joining phylogenetic tree of the LTR/U3R region. The distance matrix was generated by the Kimura two-parameter method, as described in the legend to Fig. 1A. The sequences from both the R5 and X4 (shown in bold type) biological clones were analyzed, and isolate FR83HXB2 (shown in italic type) was used as the outlier sequence. The numbers near the branches represent the percent bootstrap values from 1,000 replications.
Viral load and V3 positive charge comparison among subtypes.
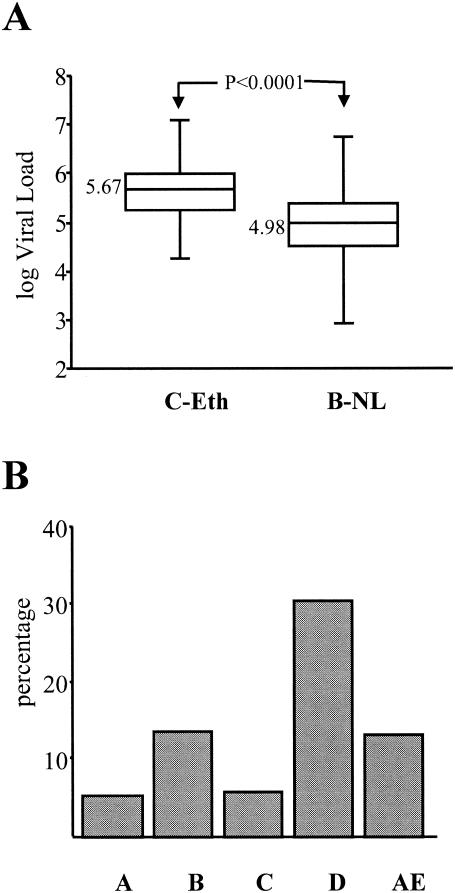
Our results demonstrating that HIV-1 subtype C R5 viruses have reduced replication capacities on CD4+ lymphocytes suggested that individuals infected with subtype C viruses may in general harbor lower viral loads than individuals infected with other subtypes. We tested this assumption by comparing the viral loads in patients infected with subtype C viruses to those in a cohort of individuals from the Netherlands infected with HIV-1 subtype B isolates. Although the cohorts are not colocalized geographically and the individuals are likely to experience different immune stimulation characteristics, we matched them by including only those individuals who were designated as AIDS patients with CD4+ cell counts below 150 cells/mm3. We showed that the subtype C-infected individuals from Ethiopia had a higher median log viral load (5.67) than the subtype B-infected individuals (4.98) from the Netherlands (Fig. 8A).
FIG. 8.
Differences between subtype B- and C-infected individuals. (A) Comparison of viral load distributions between subtype B- and C-infected individuals. The viral loads were measured in individuals who progressed to AIDS with CD4+ cell counts of less than 150 cells/mm3. The subtype C-infected individuals were from Ethiopia (n = 48), and the subtype B-infected individuals were from the Netherlands (n = 123); none had received antiretroviral therapy. The P values were found to be significant by using both the Mann-Whitney and the Wilcoxon tests. (B) Percentage of HIV-1 isolates with a V3 loop positive charge of 5 or higher. The overall positive charge was measured for the different HIV-1 subtypes analyzed: A, n = 890; B, n = 393; C, n = 531; D, n = 275; AE, n = 473. All sequences were obtained from the Los Alamos database (http://hiv-web.lanl.gov/content/index).
We analyzed the overall V3 positive charges of a large number of viruses (n = 2,562) from the Los Alamos database, compared the different subtypes A, B, C, D, and AE, and calculated the percentage of isolates with a V3 charge of +5 or higher (Fig. 8B). By doing so, we showed that subtype A and C isolates tended to have V3 regions with relatively lower overall positive V3 charges than the other subtypes and that subtype D viruses tended to have the highest charges. This observation suggests that there is either a virus or host restriction preventing the subtype A or C viruses from acquiring higher V3 charges that would lead to a switch in coreceptor usage.
DISCUSSION
Subtype C is the most prevalent HIV-1 subtype worldwide and is spreading faster than the other cocirculating forms (19, 30, 38, 42). Unlike the other HIV-1 subtypes, subtype C has been shown to infrequently switch its phenotype from NSI to SI (1, 8, 12, 35, 39). Both these points raise interesting but as yet unanswered questions. We show in this study that R5 biological clones isolated from two individuals infected with HIV-1 subtype C replicate poorly on CD4+ lymphocytes in comparison to subtype B viruses whereas X4 viruses from the same individuals replicate to comparable levels to subtype B X4 viruses. The poor replication profile of these viruses is also indicated by their low TCID50s determined on CD4+ lymphocytes and their poor replication on macrophages in comparison to subtype B viruses (Table 1). These results support a recent study showing that subtype C R5 isolates are less fit than subtype B viruses in an ex vivo competition assay (4). It will be interesting to determine in such a competition assay whether subtype B X4 viruses also prove to be fitter than subtype C viruses. The comparable replication profiles, for the subtype C R5 and X4 viruses, on different donor CD4+ lymphocytes indicates that the restriction to virus replication is at the level of virus entry via the CCR5 coreceptor. This envelope restriction to replication is further supported by the observation that there are no identifiable differences in the LTR sequences between the R5 and X4 subtype C biological clones, where alterations may have explained the observed enhanced replication profile of the X4 viruses.
When we tested our R5 and X4 subtype C viruses in iDC coculture experiments with CD4+ lymphocytes, we found that virus production was high and comparable to that seen for the subtype B viruses, demonstrating an efficient interaction between subtype C R5 gp120 envelopes with DC cells, presumably via molecules such as DC-SIGN (21). This result indicates that the efficient interaction of HIV-1 with DCs can heighten the in vitro infection of a virus that utilizes the CCR5 coreceptor relatively weakly. Although it is difficult to extrapolate in vitro data to describe what is happening in vivo and although we are comparing biological isolates generated from only two individuals, it is tempting to speculate that the interaction between HIV-1 and DCs may be significant in providing the virus with a heightened replication capacity. The interaction of HIV-1 with DCs within various tissue compartments of the infected individual may indeed contribute to higher viral loads. Interestingly, in vitro competition experiments between R5 subtype C and B viruses revealed that subtype C isolates performed relatively poorly on both CD4+ lymphocytes and macrophages but demonstrated comparable competition on skin-derived human Langerhans' cells (4). This result supports the finding that the interaction between the different virus subtypes may be modulated by the interaction between gp120 and CD4/coreceptor complexes on the cell surface and that this interaction can be differentially modulated on variant cell types. The high transmission rates of subtype C viruses demonstrating low rates of replication on CD4+ lymphocytes suggests that the interaction of HIV-1 with DCs is significant for viral transmission and initial infection, more so than the capacity of the virus to replicate on CD4+ lymphocytes.
By studying individuals from whom we have been able to isolate R5 and X4 biological clones at the same time point, we have been able to identify the genetic gp120 modifications that resulted in the R5-to-X4 coreceptor switch. From studying full-length gp120 envelopes, we have shown that the events leading to the switch are similar in both individuals as well as comparable to what is known for standard subtype B viruses. This point is pertinent considering that the viruses harbored by these individuals are genetically considerably distant from each other, representing the two cocirculating forms identified in Ethiopia (1). As predicted, the R5 viruses carry lower V3 charges than the X4 viruses and N-linked glycosylation patterns within the V1V2 and V3 region alter as expected. The V3 charges for the R5 viruses were +2/+3 for one individual and +3/+4 for the other. We showed, by analysis of a large number of isolates, that in general HIV-1 A and C subtypes include a lower proportion of isolates with high V3 positive charge while subtype D viruses have the highest. Interestingly, individuals infected with subtype D viruses reportedly have the highest incidence of X4-using viruses while those infected with subtypes A and C have the lowest incidence. This suggests that there may be some restriction at the molecular level of the gp120 envelopes of the variant subtypes. In both the subtype C-infected individuals, the R5 viruses recovered had low V3 charges, suggesting that there may be some in vivo survival advantage to these viruses over the R5 viruses with the higher V3 charges. Such a selective advantage may go some way to explaining why the incidence of X4 viruses within subtype A- and C-infected individuals is relatively low since the virus has to undergo additional mutations to arrive at the X4 phenotype. This selection of viruses with lower positive V3 charges may also be the limiting factor that results in a virus with poor replication on CD4+ lymphocytes, possibly through providing a virus with lower CCR5 affinity. Since subtype D and CRF02_AG are commonly found to be circulating in many African countries, it could be inferred that the selection is at the level of the virus rather than that of the host (55).
Our results obtained by comparing viral load measurements in individuals with T-cell counts below 150 cells/mm3 demonstrate that the low replication capacity of subtype C viruses in vitro does not necessarily correlate with lower in vivo viral load measurements for this subtype (Fig. 8A). The results also demonstrate that subtype C-infected individuals can progress to lower T-cell counts and, presumably, to disease without a switch in coreceptor usage phenotype. This finding has to be interpreted in light of the observation that Ethiopian individuals, whether HIV-1 infected or not, tend to have lower CD4 counts than other individuals (32). A number of factors, including cell type availability, coreceptor expression profiles, and HIV-1-specific immune responses, will obviously contribute to the observation that despite the low in vitro replication of the subtype C R5 viruses, these individual can develop high viral loads.
When we compared the sensitivities of the subtype C R5 viruses to the blocking effects of the CC chemokine RANTES, we found a wide variation, probably reflecting differences in the coreceptor binding domain of the gp120 envelope, which may alter the affinity for the CCR5 coreceptor. Notably, the virus with the highest resistance to RANTES neutralization (PHD79C12) was identified by phylogenetic and boot-scanning analysis to be a recombinant between an R5 virus and an X4 virus from the same patient, with the virus possessing the V3 region from an R5 virus and the V1V2 region from an X4 virus. This is not the first time that recombination between cocirculating R5 and X4 viruses has been described (52). This result would indicate that the recombinant virus either has a higher-affinity interaction with the CCR5 coreceptor or can better escape control by circulatory CC chemokines, thereby helping explain its emergence in vivo.
It is still difficult to explain why subtype C viruses do not switch coreceptors, considering that they require the same envelope alterations as subtype B viruses in order to do so. The low initial V3 charge seen in the V3 region may provide some explanation, with more time required for the mutations in the gp120 envelope to accrue, allowing the switch to occur, especially if the replication capacity of the viruses with the lower V3 charge is restricted. It has been speculated that the low X4 frequency with subtype C viruses is due to differences in cohort selection, time of monitoring, and coinfection profiles (13). Although these arguments may well be relevant, we think that they are unlikely to be the main explanations. In our Ethiopian cohort, we identified a very low incidence of X4 viruses among AIDS patients, with only 3 of 48 patients in the cohort (6.25%) harboring X4 viruses, a similar observation to those in other studies (1, 8, 39). Coinfection with other pathogens and the general immune activation profile of the host are likely to be major factors in directing viral coreceptor usage and switching through alterations in such factors as coreceptor expression levels. However, individuals from countries with similar geographical settings and infection profiles can have extremely different outcomes with respect to the HIV-1 subtype being propagated and, more significantly, the virus phenotypes, which is especially pertinent for subtype C and CRF02_AG viruses (55). Why subtype C viruses are spreading so fast is also a mystery; again, the low V3 positive charges associated with subtype C viruses may favor the presence within the infected population of viruses harboring a transmission advantage over viruses with the higher positive V3 charges and the X4 phenotype.
Acknowledgments
This work was supported by grants from the European Union (QLK2-CT-1999-01321 “EuroVac”), the American Foundation of AIDS Research (grant 02721-28-RG), and the Ethio-Netherlands AIDS Research Program (ENARP). Virus SE18208 was provided through the EU Programme EVA/MRC Centralized Facility for AIDS Reagents, NIBSC, United Kingdom (grants QLK2-CT-1999-00609 and GP828102). W.A.P. is the recipient of a research fellowship from the Royal Dutch Academy for Arts and Sciences.
We thank Patrizia Carotenuto for providing viruses NSI-18 and SI-19.
REFERENCES
- 1.Abebe, A., D. Demissie, J. Goudsmit, M. Brouwer, C. L. Kuiken, G. Pollakis, H. Schuitemaker, A. L. Fontanet, and T. F. Rinke de Wit. 1999. HIV-1 subtype C syncytium- and non-syncytium-inducing phenotypes and coreceptor usage among Ethiopian patients with AIDS. AIDS 13:1305-1311. [DOI] [PubMed] [Google Scholar]
- 2.Abebe, A., G. Pollakis, A. L. Fontanet, B. Fisseha, B. Tegbaru, A. Kliphuis, G. Tesfaye, H. Negassa, M. Cornelissen, J. Goudsmit, and T. F. Renke de Wit. 2000. Identification of a genetic subcluster of HIV type 1 subtype C (C′) widespread in Ethiopia. AIDS Res. Hum. Retrovir. 16:1909-1914. [DOI] [PubMed] [Google Scholar]
- 3.Back, N. K., L. Smit, J. J. De Jong, W. Keulen, M. Schutten, J. Goudsmit, and M. Tersmette. 1994. An N-glycan within the human immunodeficiency virus type 1 gp120 V3 loop affects virus neutralization. Virology 199:431-438. [DOI] [PubMed] [Google Scholar]
- 4.Ball, S. C., A. Abraha, K. R. Collins, A. J. Marozsan, H. Baird, M. E. Quinones-Mateu, A. Penn-Nicholson, M. Murray, N. Richard, M. Lobritz, P. A. Zimmerman, T. Kawamura, A. Blauvelt, and E. J. Arts. 2003. Comparing the ex vivo fitness of CCR5-tropic human immunodeficiency virus type 1 isolates of subtypes B and C. J. Virol. 77:1021-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger, E. A. 1997. HIV entry and tropism: the chemokine receptor connection. AIDS 11(Suppl. A):S3-S16. [PubMed] [Google Scholar]
- 6.Berger, E. A., R. W. Doms, E. M. Fenyo, B. T. Korber, D. R. Littman, J. P. Moore, Q. J. Sattentau, H. Schuitemaker, J. Sodroski, and R. A. Weiss. 1998. A new classification for HIV-1. Nature 391:240. [DOI] [PubMed] [Google Scholar]
- 7.Bjorndal, A., H. Deng, M. Jansson, J. R. Fiore, C. Colognesi, A. Karlsson, J. Albert, G. Scarlatti, D. R. Littman, and E. M. Fenyo. 1997. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J. Virol. 71:7478-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bjorndal, A., A. Sonnerborg, C. Tscherning, J. Albert, and E. M. Fenyo. 1999. Phenotypic characteristics of human immunodeficiency virus type 1 subtype C isolates of Ethiopian AIDS patients. AIDS Res. Hum. Retrovir. 15:647-653. [DOI] [PubMed] [Google Scholar]
- 9.Bleul, C. C., M. Farzan, H. Choe, C. Parolin, I. Clark-Lewis, J. Sodroski, and T. A. Springer. 1996. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature 382:829-833. [DOI] [PubMed] [Google Scholar]
- 10.Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carr, J. K., M. O. Salminen, J. Albert, E. Sanders-Buell, D. Gotte, D. L. Birx, and F. E. McCutchan. 1998. Full genome sequences of human immunodeficiency virus type 1 subtypes G and A/G intersubtype recombinants. Virology 247:22-31. [DOI] [PubMed] [Google Scholar]
- 12.Cecilia, D., S. S. Kulkarni, S. P. Tripathy, R. R. Gangakhedkar, R. S. Paranjape, and D. A. Gadkari. 2000. Absence of coreceptor switch with disease progression in human immunodeficiency virus infections in India. Virology 271:253-258. [DOI] [PubMed] [Google Scholar]
- 13.Cilliers, T., J. Nhlapo, M. Coetzer, D. Orlovic, T. Ketas, W. C. Olson, J. P. Moore, A. Trkola, and L. Morris. 2003. The CCR5 and CXCR4 coreceptors are both used by human immunodeficiency virus type 1 primary isolates from subtype C. J. Virol. 77:4449-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cocchi, F., A. L. Devico, A. Garzino-Demo, S. K. Arya, R. C. Gallo, and P. Lusso. 1995. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science 270:1811-1815. [DOI] [PubMed] [Google Scholar]
- 15.Connor, R. I., K. E. Sheridan, D. Ceradini, S. Choe, and N. R. Landau. 1997. Change in coreceptor use coreceptor use correlates with disease progression in HIV-1-infected individuals. J. Exp. Med. 185:621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Baar, M. P., A. M. van der Schoot, J. Goudsmit, F. Jacobs, R. Ehren, K. H. van der Horn, P. Oudshoorn, F. De Wolf, and A. De Ronde. 1999. Design and evaluation of a human immunodeficiency virus type 1 RNA assay using nucleic acid sequence-based amplification technology able to quantify both group M and O viruses by using the long terminal repeat as target. J. Clin. Microbiol. 37:1813-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Jong, J. J., A. De Ronde, W. Keulen, M. Tersmette, and J. Goudsmit. 1992. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J. Virol. 66:6777-6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Jong, J. J., J. Goudsmit, W. Keulen, B. Klaver, W. Krone, M. Tersmette, and A. de Ronde. 1992. Human immunodeficiency virus type 1 clones chimeric for the envelope V3 domain differ in syncytium formation and replication capacity. J. Virol. 66:757-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Martinez, A. M., E. F. Barbosa, P. C. Ferreira, F. A. Cardoso, J. Silveira, G. Sassi, C. M. da Silva, V. Mendonca-Signorini, and C. M. Antunes. 2002. Molecular epidemiology of HIV-1 in Rio Grande, RS, Brazil. Rev. Soc. Bras. Med Trop. 35:471-476. [DOI] [PubMed] [Google Scholar]
- 20.Felsenstein, J. 1996. PHYLIP: Phylogeny Inference Package, v. 3.52. University of Washington, Seattle.
- 21.Geijtenbeek, T. B., D. S. Kwon, R. Torensma, S. J. van Vliet, G. C. van Duijnhoven, J. Middel, I. L. Cornelissen, H. S. Nottet, V. N. KewalRamani, D. R. Littman, C. G. Figdor, and Y. van Kooyk. 2000. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell 100:587-597. [DOI] [PubMed] [Google Scholar]
- 22.Hillis, D. M. 1997. Phylogenetic analysis. Curr. Biol. 7:R129-R131. [DOI] [PubMed] [Google Scholar]
- 23.Hillis, D. M., and J. J. Bull. 1993. An empirical test for bootstraping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 42:182-192. [Google Scholar]
- 24.Hunt, G., and C. T. Tiemessen. 2000. Occurrence of additional NF-kappaB-binding motifs in the long terminal repeat region of South African HIV type 1 subtype C isolates. AIDS Res. Hum. Retrovir. 16:305-306. [DOI] [PubMed] [Google Scholar]
- 25.Jeeninga, R. E., M. Hoogenkamp, M. Armand-Ugon, M. de Baar, K. Verhoef, and B. Berkhout. 2000. Functional differences between the long terminal repeat transcriptional promoters of human immunodeficiency virus type 1 subtypes A through G. J. Virol. 74:3740-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnston, E. R., L. S. Zijenah, S. Mutetwa, R. Kantor, C. Kittinunvorakoon, and D. A. Katzenstein. 2003. High frequency of syncytium-inducing and CXCR4-tropic viruses among human immunodeficiency virus type 1 subtype C-infected patients receiving antiretroviral treatment. J. Virol. 77:7682-7688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karlsson, A., K. Parsmyr, K. Aperia, E. Sandstrom, E. M. Fenyo, and J. Albert. 1994. MT-2 cell tropism of human immunodeficiency virus type 1 isolates as a marker for response to treatment and development of drug resistance. J. Infect. Dis. 170:1367-1375. [DOI] [PubMed] [Google Scholar]
- 28.Koot, M., A. B. van't Wout, N. A. Kootstra, R. E. de Goede, M. Tersmette, and H. Schuitemaker. 1996. Relation between changes in cellular load, evolution of viral phenotype, and the clonal composition of virus populations in the course of human immunodeficiency virus type 1 infection. J. Infect. Dis. 173:349-354. [DOI] [PubMed] [Google Scholar]
- 29.Kumar, S., K. Tamura, and M. Nei. 1993. Molecular Evolutionary Genetics Analysis (MEGA), version 1.01. Institute of Molecular Evolutionary Genetics, Pennsylvania State University, University Park.
- 30.Long, E. M., H. L. Martin, Jr., J. K. Kreiss, S. M. Rainwater, L. Lavreys, D. J. Jackson, J. Rakwar, K. Mandaliya, and J. Overbaugh. 2000. Gender differences in HIV-1 diversity at time of infection. Nat. Med. 6:71-75. [DOI] [PubMed] [Google Scholar]
- 31.McCutchan, F. E., J. K. Carr, M. Bajani, E. Sanders-Buell, T. O. Harry, T. C. Stoeckli, K. E. Robbins, W. Gashau, A. Nasidi, W. Janssens, and M. L. Kalish. 1999. Subtype G and multiple forms of A/G intersubtype recombinant human immunodeficiency virus type 1 in Nigeria. Virology 254:226-234. [DOI] [PubMed] [Google Scholar]
- 32.Messele, T., M. Abdulkadir, A. L. Fontanet, B. Petros, D. Hamann, M. Koot, M. T. Roos, P. T. Schellekens, F. Miedema, and T. F. Rinke de Wit. 1999. Reduced naive and increased activated CD4 and CD8 cells in healthy adult Ethiopians compared with their Dutch counterparts. Clin. Exp. Immunol. 115:443-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Montano, M. A., C. P. Nixon, T. Ndung'u, H. Bussmann, V. A. Novitsky, D. Dickman, and M. Essex. 2000. Elevated tumor necrosis factor-alpha activation of human immunodeficiency virus type 1 subtype C in Southern Africa is associated with an NF-kappaB enhancer gain-of-function. J Infect. Dis. 181:76-81. [DOI] [PubMed] [Google Scholar]
- 34.Montano, M. A., V. A. Novitsky, J. T. Blackard, N. L. Cho, D. A. Katzenstein, and M. Essex. 1997. Divergent transcriptional regulation among expanding human immunodeficiency virus type 1 subtypes. J. Virol. 71:8657-8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morris, L., T. Cilliers, H. Bredell, M. Phoswa, and D. J. Martin. 2001. CCR5 is the major coreceptor used by HIV-1 subtype C isolates from patients with active tuberculosis. AIDS Res. Hum. Retrovir. 17:697-701. [DOI] [PubMed] [Google Scholar]
- 36.Novitsky, V. A., M. A. Montano, M. F. McLane, B. Renjifo, F. Vannberg, B. T. Foley, T. P. Ndung'u, M. Rahman, M. J. Makhema, R. Marlink, and M. Essex. 1999. Molecular cloning and phylogenetic analysis of human immunodeficiency virus type 1 subtype C: a set of 23 full-length clones from Botswana. J. Virol. 73:4427-4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Osmanov, S., C. Pattou, N. Walker, B. Schwardlander, and J. Esparza. 2002. Estimated global distribution and regional spread of HIV-1 genetic subtypes in the year 2000. J. Acquir. Immune Defic. Syndr. 29:184-190. [DOI] [PubMed] [Google Scholar]
- 38.Papathanasopoulos, M. A., G. M. Hunt, and C. T. Tiemessen. 2003. Evolution and diversity of HIV-1 in Africa—a review. Virus Genes 26:151-163. [DOI] [PubMed] [Google Scholar]
- 39.Peeters, M., R. Vincent, J. L. Perret, M. Lasky, D. Patrel, F. Liegeois, V. Courgnaud, R. Seng, T. Matton, S. Molinier, and E. Delaporte. 1999. Evidence for differences in MT2 cell tropism according to genetic subtypes of HIV-1: syncytium-inducing variants seem rare among subtype C HIV-1 viruses. J. Acquir. Immune Defic. Syndr. 20:115-121. [DOI] [PubMed] [Google Scholar]
- 40.Pollakis, G., A. Abebe, A. Kliphuis, T. F. Rinke de Wit, B. Fisseha, B. Tegbaru, G. Tesfaye, H. Negassa, Y. Mengistou, A. L. Fontanet, I. L. Cornelissen, and J. Goudsmit. 2003. Recombination of HIV type 1C (C′/C") in Ethiopia: possible link of EthHIV-1C′ to subtype C sequences from the high-prevalence epidemics in India and southern Africa. AIDS Res. Hum. Retrovir. 999-1008. [DOI] [PubMed]
- 41.Pollakis, G., S. Kang, A. Kliphuis, M. I. Chalaby, J. Goudsmit, and W. A. Paxton. 2001. N-linked glycosylation of the HIV type-1 gp120 envelope glycoprotein as a major determinant of CCR5 and CXCR4 coreceptor utilization. J. Biol. Chem. 276:13433-13441. [DOI] [PubMed] [Google Scholar]
- 42.Robbins, K. E., C. I. Bandea, A. Levin, J. J. Goedert, W. A. Blattner, G. Brubaker, T. M. Brown, G. Schochetman, M. L. Kalish, J. Shao, and T. R. O'Brien. 1996. Genetic variability of human immunodeficiency virus type 1 in rural northwest Tanzania. AIDS Res. Hum. Retrovir. 12:1389-1391. [DOI] [PubMed] [Google Scholar]
- 43.Robertson, D. L., J. P. Anderson, J. A. Bradac, J. K. Carr, B. Foley, R. K. Funkhouser, F. Gao, B. H. Hahn, M. L. Kalish, C. Kuiken, G. H. Learn, T. Leitner, F. McCutchan, S. Osmanov, M. Peeters, D. Pieniazek, M. Salminen, P. M. Sharp, S. Wolinsky, and B. Korber. 2000. HIV-1 nomenclature proposal. Science 288:55-56. [DOI] [PubMed] [Google Scholar]
- 44.Robertson, D. L., B. H. Hahn, and P. M. Sharp. 1995. Recombination in AIDS viruses. J. Mol. Evol. 40:249-259. [DOI] [PubMed] [Google Scholar]
- 45.Roof, P., M. Ricci, P. Genin, M. A. Montano, M. Essex, M. A. Wainberg, A. Gatignol, and J. Hiscott. 2002. Differential regulation of HIV-1 clade-specific B, C, and E long terminal repeats by NF-kappaB and the Tat transactivator. Virology 296:77-83. [DOI] [PubMed] [Google Scholar]
- 46.Salminen, M. O., J. K. Carr, D. S. Burke, and F. E. McCutchan. 1995. Identification of breakpoints in intergenotypic recombinants of HIV type 1 by bootscanning. AIDS Res. Hum. Retrovir. 11:1423-1425. [DOI] [PubMed] [Google Scholar]
- 47.Sattentau, Q. J., A. G. Dalgleish, R. A. Weiss, and P. C. Beverley. 1986. Epitopes of the CD4 antigen and HIV infection. Science 234:1120-1123. [DOI] [PubMed] [Google Scholar]
- 48.Scarlatti, G., E. Tresoldi, A. Bjorndal, R. Fredriksson, C. Colognesi, H. K. Deng, M. S. Malnati, A. Plebani, A. G. Siccardi, D. R. Littman, E. M. Fenyo, and P. Lusso. 1997. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat. Med. 3:1259-1265. [DOI] [PubMed] [Google Scholar]
- 49.Schuitemaker, H., M. Koot, N. A. Kootstra, M. W. Dercksen, R. E. de Goede, R. P. van Steenwijk, J. M. Lange, J. K. Schattenkerk, F. Miedema, and M. Tersmette. 1992. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J. Virol. 66:1354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tersmette, M., R. E. de Goede, B. J. Al, I. N. Winkel, R. A. Gruters, H. T. Cuypers, H. G. Huisman, and F. Miedema. 1988. Differential syncytium-inducing capacity of human immunodeficiency virus isolates: frequent detection of syncytium-inducing isolates in patients with acquired immunodeficiency syndrome (AIDS) and AIDS-related complex. J. Virol. 62:2026-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tersmette, M., R. A. Gruters, F. De Wolf, R. E. de Goede, J. M. Lange, P. T. Schellekens, J. Goudsmit, H. G. Huisman, and F. Miedema. 1989. Evidence for a role of virulent human immunodeficiency virus (HIV) variants in the pathogenesis of acquired immunodeficiency syndrome: studies on sequential HIV isolates. J. Virol. 63:2118-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Rij, R. P., M., Worobey, J. A. Visser, and H. Schuijtemaker. 2003. Evolution of R5 and X4 human immunodeficiency virus type 1 gag sequences in vivo: evidence for recombination. Virology 314:451-459. [DOI] [PubMed] [Google Scholar]
- 53.Van Rij, R. P., and H. Schuitemaker. 2002. Host genetic factors in the clinical course of HIV-1 infection: chemokines and chemokine receptors. Commun. Genet. 5:88-101. [DOI] [PubMed] [Google Scholar]
- 54.van't Wout, A. B., H. Blaak, L. J. Ran, M. Brouwer, C. Kuiken, and H. Schuitemaker. 1998. Evolution of syncytium-inducing and non-syncytium-inducing biological virus clones in relation to replication kinetics during the course of human immunodeficiency virus type 1 infection. J. Virol. 72:5099-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vergne, L., A. Bourgeois, E. Mpoudi-Ngole, R. Mougnutou, J. Mbuagbaw, F. Liegeois, C. Laurent, C. Butel, L. Zekeng, E. Delaporte, and M. Peeters. 2003. Biological and genetic characteristics of HIV infections in Cameroon reveals dual group M and O infections and a correlation between SI-inducing phenotype of the predominant CRF02_AG variant and disease stage. Virology 310:254-266. [DOI] [PubMed] [Google Scholar]
- 56.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, Y. J., and J. P. Moore. 1999. Will multiple coreceptors need to be targeted by inhibitors of human immunodeficiency virus type 1 entry? J. Virol. 73:3443-3448. [DOI] [PMC free article] [PubMed] [Google Scholar]