Abstract
Background
Oculocutaneous albinism (OCA) is caused by a group of genetically heterogeneous inherited defects that result in the loss of pigmentation in the eyes, skin and hair. Mutations in the TYR, OCA2, TYRP1 and SLC45A2 genes have been shown to cause isolated OCA. No comprehensive analysis has been conducted to study the spectrum of OCA alleles prevailing in Pakistani albino populations.
Methods
We enrolled 40 large Pakistani families and screened them for OCA genes and a candidate gene, SLC24A5. Protein function effects were evaluated using in silico prediction algorithms and ex vivo studies in human melanocytes. The effects of splice-site mutations were determined using an exon-trapping assay.
Results
Screening of the TYR gene revealed four known (p.Arg299His, p.Pro406Leu, p.Gly419Arg, p.Arg278*) and three novel mutations (p.Pro21Leu, p.Cys35Arg, p.Tyr411His) in ten families. Ex vivo studies revealed the retention of an EGFP-tagged mutant (p.Pro21Leu, p.Cys35Arg or p.Tyr411His) tyrosinase in the endoplasmic reticulum (ER) at 37°C, but a significant fraction of p.Cys35Arg and p.Tyr411His left the ER in cells grown at a permissive temperature (31°C). Three novel (p.Asp486Tyr, p.Leu527Arg, c.1045-15 T > G) and two known mutations (p.Pro743Leu, p.Ala787Thr) of OCA2 were found in fourteen families. Exon-trapping assays with a construct containing a novel c.1045-15 T > G mutation revealed an error in splicing. No mutation in TYRP1, SLC45A2, and SLC24A5 was found in the remaining 16 families. Clinical evaluation of the families segregating either TYR or OCA2 mutations showed nystagmus, photophobia, and loss of pigmentation in the skin or hair follicles. Most of the affected individuals had grayish-blue colored eyes.
Conclusions
Our results show that ten and fourteen families harbored mutations in the TYR and OCA2 genes, respectively. Our findings, along with the results of previous studies, indicate that the p.Cys35Arg, p.Arg278* and p.Gly419Arg alleles of TYR and the p.Asp486Tyr and c.1045-15 T > G alleles of OCA2 are the most common causes of OCA in Pakistani families. To the best of our knowledge, this study represents the first documentation of OCA2 alleles in the Pakistani population. A significant proportion of our cohort did not have mutations in known OCA genes. Overall, our study contributes to the development of genetic testing protocols and genetic counseling for OCA in Pakistani families.
Keywords: TYR, OCA2, TYRP1, SLC45A2, SLC24A5, Pakistan, Exon-trapping, Oculocutaneous Albinism, Melanocytes, Hypopigmentation
Introduction
Among the most visible phenotypic traits in humans is skin color. Loss of skin, hair and iris pigmentation, a condition known as oculocutaneous albinism (OCA), represents a significant load of human genetic diseases. OCA can manifest itself in syndromic and nonsyndromic forms under a variety of inheritance models [1]. At present, mutations in at least 16 loci have been causally linked with OCA [1,2]. More genes in humans are likely to be identified as implicated by new OCA syndromes [3,4]. Mutations at four loci, OCA1 (TYR), OCA2 (OCA2), OCA3 (TYRP1) and OCA4 (SLC45A2), have been shown to be necessary and sufficient to cause isolated OCA [1]. Approximately 450 different pathogenic alleles of these four genes have been documented (Human Gene Mutation Database (HGMD), http://www.hgmd.org/), and most of these sequence variations are located in the TYR gene.
Human chromosome 11q14.3 harbors the TYR gene (MIM# 606933), which encodes tyrosinase [5]. Tyrosinase is expressed in melanocytes and controls the biosynthesis of melanin from tyrosine at three levels [5]. To date, 291 pathogenic variants of the TYR gene have been identified in individuals with the OCA1 phenotype (HGMD). There is a presumptive genotype-phenotype correlation in which the severe pathogenic or null alleles of the TYR gene result in the OCA1A (MIM# 203100) phenotype, characterized by the loss of pigmentation in the skin, hair and eyes with translucent irises [6]. Hypomorphic alleles produce a spectrum of clinical phenotypes, known as OCA1B (MIM# 606952), which range from low to nearly normal levels of skin and hair pigmentation in adults.
OCA2 (MIM# 203200) is located on human chromosome 15q11-q13 and has two non-coding and 23 coding exons. OCA2 encodes a polypeptide of ~110 kDa with 12 putative transmembrane helices. As a member of the Na+/H+ antiporter family, the OCA2 protein is thought to play an essential role in maintaining the pH of the melanosomes, which regulates tyrosinase activity [7-10]. The OCA2 protein also participates in the sorting and transport of tyrosinase and tyrosinase-related protein 1 (TYRP1) to the plasma membrane [11-13]. OCA2 mutations are the most common causes of OCA in Africa, with a prevalence rate as high as 1:3,900 being observed [14].
Human chromosome 9q23 harbors the TYRP1 gene (MIM# 115501), which is known to cause OCA type 3 (MIM 203290; a.k.a Rufous OCA). The seven known coding exons of TYRP1 (GenBank NM_000550) encode a tyrosinase-related protein of ~61 kDa with 41% sequence identity and 58% similarity to tyrosinase [15]. TYRP1 has partial tyrosinase hydroxylase activity and catalyzes the oxidation of 5,6-dihydroxyindole-2-carboxylic acid in the melanin biosynthesis pathway [16,17]. As of November 2012, only nine TYRP1 mutations have been reported in the HGMD.
The OCA4 phenotype (MIM# 606574) is caused by mutations in the SLC45A2 gene (MIM# 606202, a.k.a. MATP), which is located on human chromosome 5p13.3. In the HGMD, 76 pathogenic alleles of SLC45A2 have been reported to date. In humans, seven known coding exons of SLC45A2 transcribe four alternatively spliced variants. The longest spliced isoform (GenBank NM_016180) encodes a solute carrier family 45, member 2 (SLC45A2) protein composed of 530 amino acids and has a molecular weight of ~58 kDa. Although its precise function has not been elucidated, SLC45A2 probably acts as a melanosomal protein and substance transporter [18,19].
To the best of our knowledge, no comprehensive molecular analysis of these four known OCA genes has been conducted in Pakistani families segregating OCA. However, nine pathogenic variants of the TYR gene, including c.344delGA, p.Arg278*, p.Ser315_A316del, p.Gln328Glu, p.Glu376*, p.Gly419Arg, p.Pro431Thr, p.Pro431Leu and p.Glu453*, have been identified in mostly sporadic cases from Pakistan [20-25]. In addition to TYR alleles, only a single point mutation (c.1117 C > T, p.Arg373*) in TYRP1 gene has been reported in a large consanguineous Pakistani family [25]. As a corollary of the inimitable socio-cultural customs in the population of Pakistan, approximately 60% of marriages are consanguineous, of which more than 80% are between first cousins [26]. These large consanguineous families are a powerful resource for genetic studies of recessively inherited disorders like OCA. In the present study, we analyzed the four OCA genes in 40 large Pakistani families to characterize the genetic lesion and to establish a mutational profile of the Pakistani albino population. In addition, we screened SLC24A5 (MIM# 609802), which is responsible for ocular albinism and hypopigmentation in Slc24a5 knockout mice and is known to regulate melanogenesis in humans [27,28]. The results of this study will be important for future diagnosis, genetic counseling, and molecular epidemiology of OCA.
Materials and methods
Family participation and clinical evaluation
This study was approved by the IRB Committees at the Children’s Hospital Research Foundation, USA (2010–0452) and the Institute of Molecular Biology & Biotechnology, Pakistan. Informed written consent was obtained from the adult subjects and the parents of minor subjects. Detailed clinical histories were obtained from participating family members and affected individuals were examined by an Ophthalmologist and a physician to rule out any obvious syndromic forms of OCA. Clinical features of OCA, such as hypopigmentation of the hair and skin and the presence of eye aberrations, including nystagmus, strabismus, photophobia and poor vision, were evaluated. Peripheral blood samples were collected from each participating individual for genomic DNA extraction.
Mutational analysis
The primers used for PCR amplification and sequencing of the TYR, OCA2, TYRP1, SLC45A2 and SLC24A5 genes were designed using the Primer3 web site. The sequencing method for the PCR products has been described previously [29]. For specific amplification of exons 4 and 5 of TYR, we used the primers and PCR conditions described previously [30]. Briefly, the coding and noncoding exons of TYROCA2TYRP1SLC45A2, SLC24A5 were PCR amplified from 50 ng genomic DNA, using ABI Veriti thermocyclers (Applied Biosystems, Austin, TX). PCR reactions (final volume 20 μl) were performed with genomic DNA in the presence of 5 pmol each of forward and reverse primers, 200 mM each dNTP, 1 x PCR buffer (GenSrcipt), 1.5–2.5 mM MgCl2 (GenScript), and 0.5 U of a thermostable DNA polymerase. For sequencing reaction, we added 3.2 pmol of primer, 0.2 μl of Big Dye Terminator Ready Reaction Mix (ABI Biosystems), and 2 μl of 5 x dilution buffer (400 mM Tris–HCl pH9 and 10 mM MgCl2). An ABI 3730xl DNA capillary sequencer was used to resolve the products, and Lasergene DNAstar software was used to analyze the results. Co-segregation of the mutations with OCA in each family was confirmed by sequencing. Control DNA samples from an ethnically matched Pakistani population were sequenced for mutant alleles of TYR and OCA2. Three prediction programs, Polyphen-2 [31], SNPs3D [32], and MutationTaster [33], were used to determine the effect of novel missense mutations. Effects of missense mutations on the structure of tyrosinase and OCA2 were also analyzed using the Project HOPE web server [34].
Fluorescently tagged TYR expression constructs
The EGFP-tagged, full-length human TYR cDNA construct was generated using PCR primers located in exons 1 and 5. A retinal cDNA library (Clontech, Mountain View, CA) was used as the template source. The sequence-verified PCR product was inserted into the pEGFP-C2 vector (Clontech, Mountain View, CA). Constructs encoding the p.Pro21Leu, p.Cys35Arg and p.Tyr411His mutant forms of tyrosinase were prepared by site-directed mutagenesis (Agilent Technologies, Santa Clara, CA).
Cell culture conditions, transfection and immunostaining
Human melanocyte cells were transiently transfected using Fugene-6 (Promega, Madison, WI) with 1.5 μg of the desired construct per well in a 6-well dish. After transfection, cells were incubated for 48 hours at either 37°C or 31°C followed by fixation with 4% paraformaldehyde. For visualization of the endoplasmic reticulum and early endosomes, anti-calregulin (Santa Cruz Biotechnology, Santa Cruz, CA) and anti-EEA1 (Abcam, Cambridge, MA) antibodies were used, respectively. A Zeiss LSM700 confocal microscope was used for imaging.
Exon-trapping assay
To determine the effect of splice site mutation (c.1045-15 T > G) found in five OCA families, the wild-type and mutant exon 10 along with 200 bp flanking introns of OCA2 gene were PCR-amplified, cloned into the pSPL3 vector (Invitrogen, Carlsbad, CA), and sequence-verified. Purified, cloned DNA of the experimental, wild-type, and empty vector control constructs were separately transfected into COS-7 cells using the Fugene-6 reagent. Forty-eight hours after transfection of the pSPL3 constructs, RNA was extracted from the COS-7 cells using TRIzol reagent (Invitrogen, Carlsbad, CA), and single-stranded cDNA was synthesized (Clontech, Mountain View, CA). Primary PCR amplification of the cDNA was performed with vector primers. Ten microliters of each amplimer was analyzed on a 2% agarose gel. DNA bands were extracted and sequenced with the vector primers.
Results
Genetic and clinical analyses of TYR (OCA1)
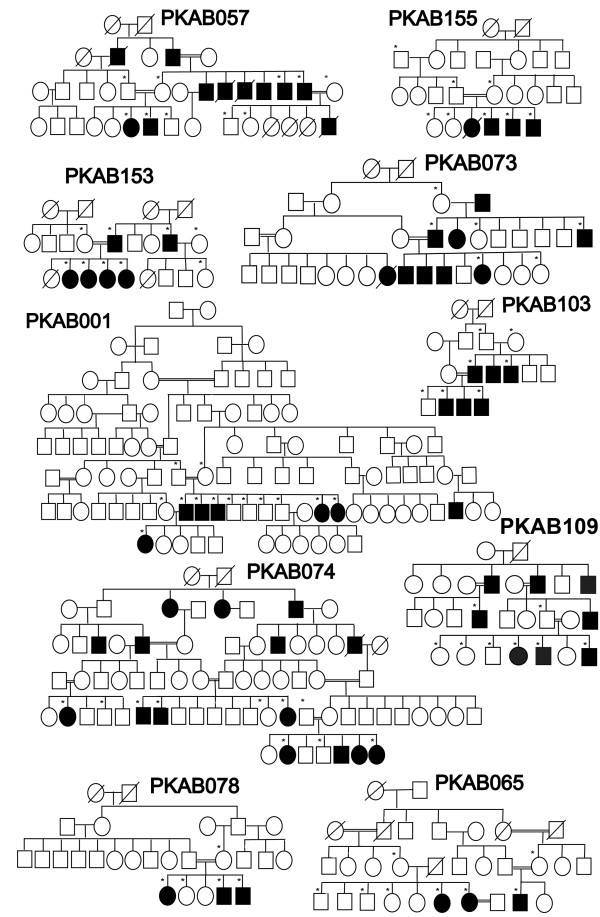
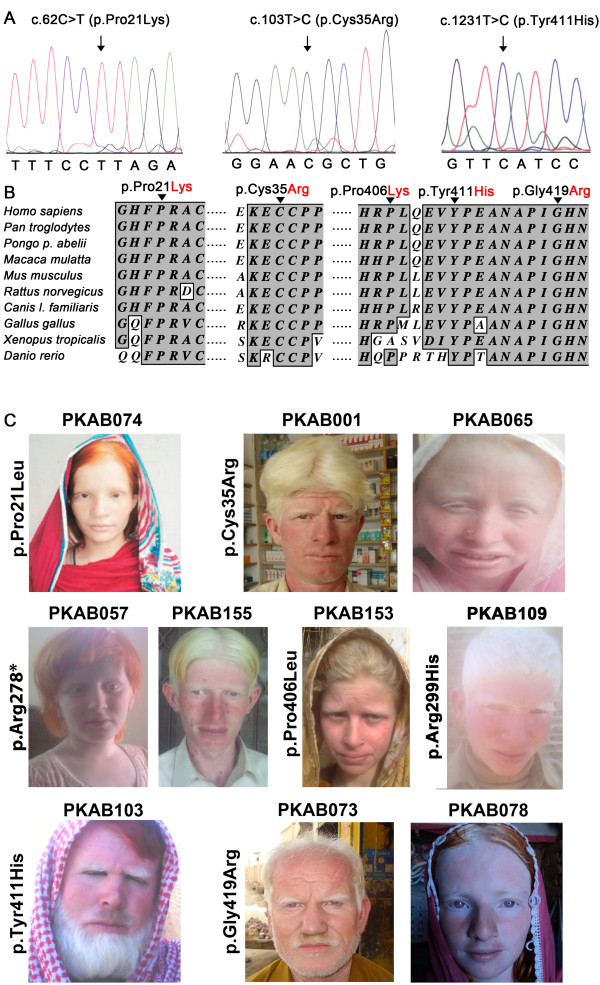
Study subjects from forty families, segregating congenital onset, nonsyndromic, recessive OCA, were enrolled from different cities in the Punjab province of Pakistan. Sequence analysis of the TYR gene revealed seven probable pathogenic variants in ten of these families (Figure 1, Tables 12). Of these seven mutant alleles, four have been previously reported and include c.832 C > T (p.Arg278*), c.896A > G (p.Arg299His), c.1217 C > T (p.Pro406Leu) and c.1255 G > A (p.Gly419Arg) [5,20,24,35]. Three novel missense substitutions were identified including p.Pro21Leu (c.62 C > T), p.Cys35Arg (c.103 T > C) and p.Tyr411His (c.1231 T > C) (Figure 2A). All of these missense mutations affected amino-acid residues that are conserved among the tyrosinase orthologs (Figure 2B). We performed a haplotype analysis using eight closely linked single nucleotide polymorphisms (SNPs) in TYR to identify potential founder effects for the recurrent variants (p.Arg278*, p.Gly419Arg and p.Cys35Arg). Selection of these eight SNPs for haplotype analysis in Pakistani OCA families was based on the observance of high heterozygosity (> 0.3) in 32 control samples randomly collected from the Pakistani population. The results were consistent with common ancestors for each of the recurrent alleles in the Pakistani families studied here (data not shown).
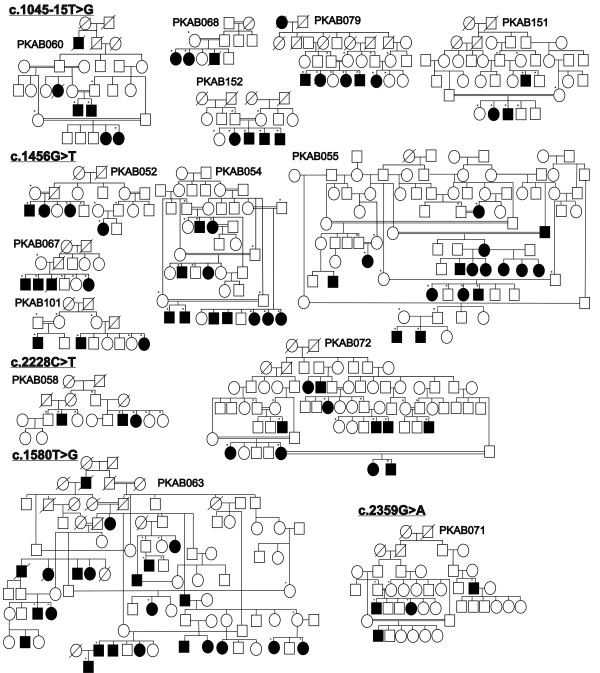
Figure 1 .
Pedigrees of Pakistani families carrying TYR mutations. Pedigrees of ten multi-generational families segregating recessive nonsyndromic OCA due to mutations in the TYR gene. Filled and empty symbols represent affected and unaffected individuals, respectively. Double lines indicate consanguineous marriages. Asterisks indicate subjects enrolled in the protocol that contributed DNA samples.
Table 1.
Mutant alleles of TYR found in ten Pakistani families
| Nucleotide change# | Exon | Effect on protein | Frequencies in control samples* | Family | Ethnicity | Polyphen2 | SNPs3D | Mutation Taster | Allele frequency in our OCA1 families | Known frequencies in other populations | References |
|---|---|---|---|---|---|---|---|---|---|---|---|
|
Missense | |||||||||||
|
c.62 C > T |
1 |
p.Pro21Leu |
0/380 |
PKAB074 |
Sayyed |
Damaging |
Damaging |
Pathogenic |
10% |
N/A |
This study |
|
c.103 T > C |
1 |
p.Cys35Arg |
0/380 |
PKAB001 PKAB065 |
Malik Malik Jutt |
Damaging |
Damaging |
Pathogenic |
20% |
N/A |
This study |
| c.896A > G |
2 |
p.Arg299His |
0/372 |
PKAB109 |
Warraich |
Damaging |
Damaging |
Pathogenic |
10% |
Caucasian 12.5%; Arab-Christian 1.6%, 2.6% and 3.3%; Chinese 18.75%; Indian 4.34%. |
[10,36-40] |
| c.1217 C > T |
4 |
p.Pro406Leu |
0/372 |
PKAB153 |
Malik Jutt |
Damaging |
Damaging |
Pathogenic |
10% |
Caucasian 2.94% and 25%; German 14.28%. |
[37,41,42] |
|
c.1231 T > C |
4 |
p.Tyr411His |
0/372 |
PKAB103 |
Arian |
Damaging |
Damaging |
Pathogenic |
10% |
N/A |
This study |
| c.1255 G > A |
4 |
p.Gly419Arg |
0/372 |
PKAB073 PKAB078 |
BhatiJutt |
Damaging |
Damaging |
Pathogenic |
20% |
Caucasian 0.83%; Indo-Pakistan 25%; Pakistan 0.83%; Indian 4.34% and 20%; South-Indian 16.6%. |
[20,36,37,43,44] |
|
Nonsense |
|
||||||||||
| c.832 C > T | 2 | p.Arg278* | 0/372 | PKAB057 PKAB155 | Shaikh Rajpoot | 20% | Guayanan 12.5%; Jewish 2.6%; Japanese 12.5%, 22.2% and 100%; European 2.5%; Mexican 0.83%; Indian 0.83% and 4.34%; Eastern Indian 8.3%, 25% and 100%; Syrian 0.83%; Chinese 18.75%. | [9,10,20,36,37,39,43-48] | |||
#Novel mutations are in bold. *Frequencies were determined by sequencing at least 372 chromosomes from geographically and ethnically-matched subjects without any history of ocular disease. N/A: not applicable.
Table 2.
Clinical assessment of the affected individuals with mutations in TYR and OCA2
| Gene/Family | Mutation (protein) | Sex | Age (yrs) | Hair color | Skin color | Iris color | Visual Acuity Right Left | Type of refraction error | Fundus | Foveal hypoplasia | Photophobia# | Nystagmus | Con | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
TYR | ||||||||||||||
| PKAB074 |
p.Pro21Leu |
M |
19 |
Brown |
Pinkish white |
Grey/Brown |
4/60 |
4/60 |
Myopic |
Albinotic |
Yes |
Present |
Yes |
Yes |
| PKAB001 |
p.Cys35Arg |
M |
40 |
White |
Pinkish white |
Grey/Blue |
6/38 |
6/38 |
Compound myopic hypermetropic astigmatism |
Albinotic |
Yes |
Present |
Yes |
Yes |
| PKAB065 |
p.Cys35Arg |
F |
28 |
White |
Whitea |
Grey/Blue |
N/A |
N/A |
N/A |
N/A |
N/A |
Present |
Yes |
No |
| PKAB057 |
p.Arg278* |
F |
7 |
White |
Whitea |
Light Brown |
N/A |
N/A |
N/A |
N/A |
N/A |
Present |
Yes |
Yes |
| PKAB155 |
p.Arg278* |
M |
16 |
White |
Whitea,b |
Grey/Blue |
N/A |
N/A |
N/A |
N/A |
N/A |
Present |
Yes |
Yes |
| PKAB109 |
p.Arg278* |
M |
24 |
N/A |
N/A |
N/A |
N/A |
N/A |
N/A |
N/A |
N/A |
Present |
Yes |
Yes |
| PKAB153 |
p.Pro406Leu |
F |
18 |
Yellow |
Whitea |
Light Grey |
N/A |
N/A |
N/A |
N/A |
N/A |
Present |
Yes |
Yes |
| PKAB103 |
p.Tyr411His |
M |
30 |
White |
Pinkish White |
Grey/Blue |
6/60 |
6/60 |
Compound hypermetropic |
Albinotic |
Yes |
Present |
Yes |
No |
| PKAB073 |
p.Gly419Arg |
M |
45 |
White |
Whitea |
Grey/Blue |
N/A |
N/A |
N/A |
N/A |
N/A |
Present |
Yes |
No |
| PKAB078 |
p.Gly419Arg |
F |
12 |
White |
Whitea,b |
Grey/Blue |
N/A |
N/A |
N/A |
N/A |
N/A |
Present |
Yes |
Yes |
|
OCA2 | ||||||||||||||
| PKAB052 |
p.Asp486Tyr |
F |
1.5 |
White |
White |
Grey/Blue |
N/A |
N/A |
N/A |
N/A |
N/A |
Present |
Present |
Yes |
| PKAB054 |
p.Asp486Tyr |
F |
13 |
Yellowish- White |
Whitea,b |
Blue |
6/60 |
6/60 |
Compound Myopic |
Albinotic |
Hypoplasia |
Present |
Present |
Yes |
| PKAB055 |
p.Asp486Tyr |
F |
1.5 |
White |
White |
Light Grey |
N/A |
N/A |
N/A |
N/A |
N/A |
Present |
Present |
Yes |
| PKAB067 |
p.Asp486Tyr |
M |
35 |
White |
Whitea |
Grey/Blue |
N/A |
N/A |
N/A |
N/A |
N/A |
Present |
Present |
No |
| PKAB101 |
p.Asp486Tyr |
M |
25 |
White |
Whitea |
Grey/Blue |
6/60 |
6/60 |
Mixed Astigmatism |
Albinotic |
Hyoplasia |
Present |
Present |
Yes |
| PKAB063 |
p.Met318Ile p.Leu527Arg |
F |
5 |
White |
Whitea |
Grey/Blue |
N/A |
N/A |
N/A |
N/A |
N/A |
Present |
Present |
Yes |
| PKAB058 |
p.Pro743Leu |
M |
30 |
White |
Whitea |
Grey/Blue |
1/60 |
4/60 |
Hypomyopic |
Albinotic |
N/A |
Present |
Present |
No |
| PKAB072 |
p.Pro743Leu |
M |
6 |
White |
Whitea |
Grey/Blue |
N/A |
N/A |
N/A |
N/A |
N/A |
Present |
Present |
Yes |
| PKAB071 |
p.Ala787Thr |
M |
10 |
Yellowish-White |
Whitea,b |
Blue |
Fc3 |
Fc3 |
Compound Hypermetropic |
Albinotic |
Hypoplasia |
Present |
Present |
Yes |
| PKAB60 |
c.1045-15 T > G |
F |
6 |
White |
Whitea,b |
Grey/Brown |
6/36 |
6/36 |
Hypermetropic Astigmatism |
Albinotic |
Hypoplasia |
Present |
Present |
Yes |
| PKAB068 |
c.1045-15 T > G |
F |
7 |
Yellowish-White |
Whitea |
Blue/Brown |
N/A |
N/A |
N/A |
N/A |
N/A |
Present |
Present |
Yes |
| PKAB079 |
c.1045-15 T > G |
M |
19 |
Yellowish-White |
Reddish |
Grey/Brown |
N/A |
N/A |
N/A |
N/A |
N/A |
Present |
Present |
Yes |
| PKAB151 |
c.1045-15 T > G |
F |
6 |
White |
Whitea |
Grey/Blue |
N/A |
N/A |
N/A |
N/A |
N/A |
Present |
Present |
Yes |
| PKAB152 | c.1045-15 T > G | F | 12 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | Present | Present | Yes |
#All individuals show squinting in normal sunlight. aReddish spots throughout the skin and lips appeared sun damaged. bShow blistering on exposed skin and generalized sunburn redness. N/A not available, Cons consanguineous union.
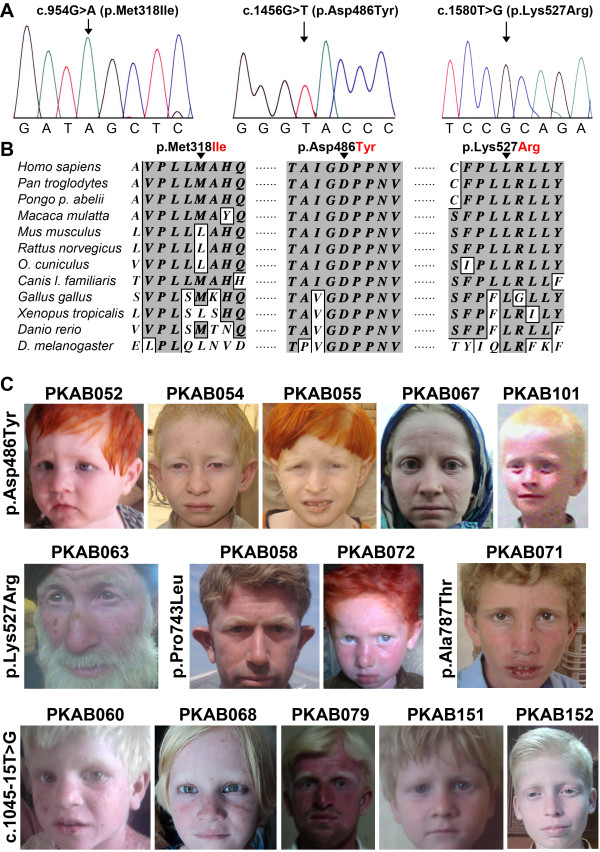
Figure 2 .
Novel TYR mutations and resulting OCA1 phenotypes. A. Electropherograms of amplimers from genomic DNA templates illustrating homozygosity for the substitution mutations found in the affected individuals of the families. Arrows indicate the site of the mutations. All of the mutations described here are numbered from the ATG start codon (GenBank NM_000372). B. Clustal W alignment of tyrosinase proteins from various species that shows the conservation of residues at positions 21, 35 and 411 among ten species. The conserved amino acids are shown with a dark gray background, and the nonconserved amino acids are shown with a white background. C. Photographs of ten OCA1 probands. The family number and the mutation identified in the TYR gene are given for each proband; some of the probands have used hair dyes.
All of the affected individuals from the ten families had nystagmus and photophobia, regardless of their sex and age (Figure 2C, Table 2 and Additional file 1: Table S1). Inter-familial variation of hair color was noted among individuals, ranging from white to honey blonde or brown (Figure 2C, Table 2 and Additional file 1: Table S1). Notably, reddish spots and marked sun-damage on the skin and grossly enlarged veins in the cheeks and lips were observed (Figure 2C).
Functional analysis of novel missense alleles of tyrosinase
Three prediction programs, specifically, Polyphen-2 [31], SNPs3D [32], and MutationTaster [33], suggested that each of the three new missense mutations were deleterious (Table 1). We also used the HOPE prediction program [34] to assess further the effect of the missense mutations on the secondary structure of the encoded protein. Both the p.Pro21 and p.Cys35 residues are located in the amino terminal region. Due to their charge, size, and hydrophilic properties, the amino acids lysine and arginine at positions 21 and 35, respectively, were predicted to disrupt protein topology, which could result in protein misfolding. Also, p.Tyr411 is located close to a defined copper-binding site within the luminal domain; inserting histidine at this position was predicted to cause an empty space in the core of the protein and the loss of hydrophobic interactions, because of the smaller size and hydrophilicity of histidine.
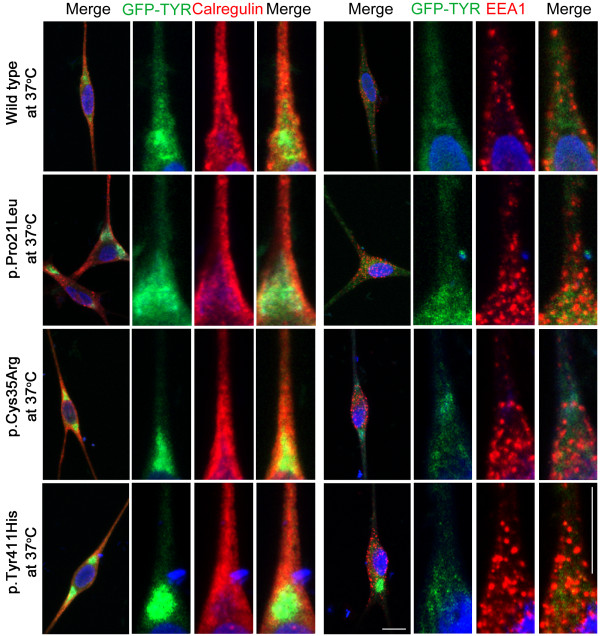
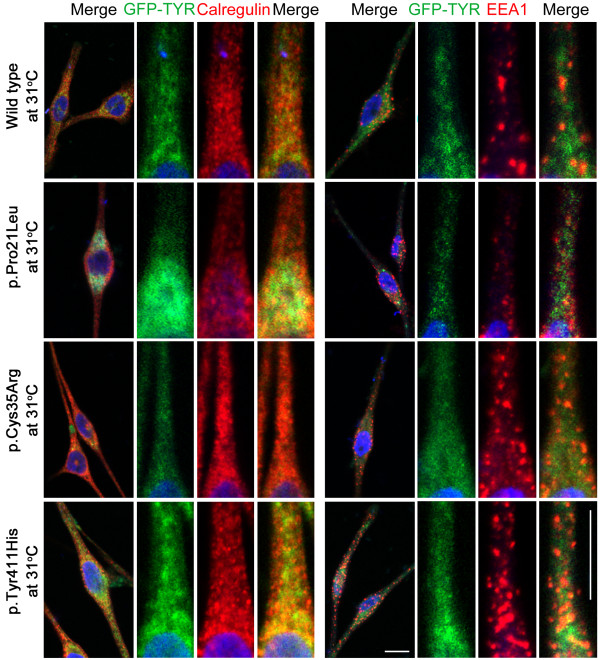
Generally, missense alleles of the TYR gene result in the retention of the encoded mutant protein in the endoplasmic reticulum (ER) [49]. To determine the effect of the three novel missense mutations (p.Pro21Leu, p.Cys35Arg and p.Tyr411His) on the localization of tyrosinase, we transiently transfected human melanocytes with GFP-tagged, full-length wild-type and mutant TYR cDNA constructs (Figure 3). Wild-type tyrosinase was localized predominantly throughout the cytoplasm of melanocytes with some expression in the ER (Figure 3). The low expression of wild-type tyrosinase in the ER might be due to newly synthesized polypeptides that are retained in the ER by chaperones until they are properly folded and assembled [50]. Immunofluorescence studies with calregulin (an ER marker) and EEA1 (an early endosome marker) demonstrated that the mutant proteins predominantly co-localized with calregulin, indicating retention in the ER (Figure 3). A portion of the known human and mouse TYR mutations, especially those present in the copper-binding region, have shown temperature-sensitive behavior [5,35,51-54]. Therefore, we also tested the effect of temperature on the localization of wild-type and mutant tyrosinase proteins by growing transfected melanocytes at 37°C and 31°C (Figures 3 and 4). Interestingly, a decrease in temperature resulted in an increase in the cytoplasmic vesicular co-localization of the p.Cys35Arg and p.Tyr411His mutant protein with EEA1 (Figure 4). Melanocytes that were transfected with wild-type and p.Pro21Leu constructs and grown at 31°C, did not show any significant change in the localization pattern relative to cells grown at 37°C (Figure 4).
Figure 3 .
Subcellular distribution of wild-type and mutant tyrosinase in human melanocytes grown at 37°C. Subcellular distribution of wild-type and mutant (p.Pro21Leu, p.Cys35Arg and p.Tyr411His) tyrosinase proteins (green) in transiently transfected human melanocytes grown at 37°C. Calregulin and EEA1 were used as markers for the endoplasmic reticulum (red) and the early endosome (red), respectively. For each construct, boxed regions were magnified in the adjacent panels. Merged images show the co-localization of the p.Pro21Leu, p.Cys35Arg and p.Tyr411His tyrosinase mutations with calregulin, which indicates ER retention. The scale bar represents 10 μm for all panels.
Figure 4 .
Subcellular distribution of wild-type and mutant tyrosinase in human melanocytes grown at 31°C. The subcellular localization of GFP-tyrosinase in wild-type and p.Pro21Leu-transfected cells was not significantly different when cells were incubated at 31°C or at 37°C. After transfection with either p.Cys35Arg or p.Tyr411His mutant constructs, the melanocytes grown at 31°C showed an increase in the cytoplasmic vesicular co-localization of the mutant protein with EEA1. For each construct, boxed regions were magnified in the adjacent panels. The scale bar represents 10 um for all panels.
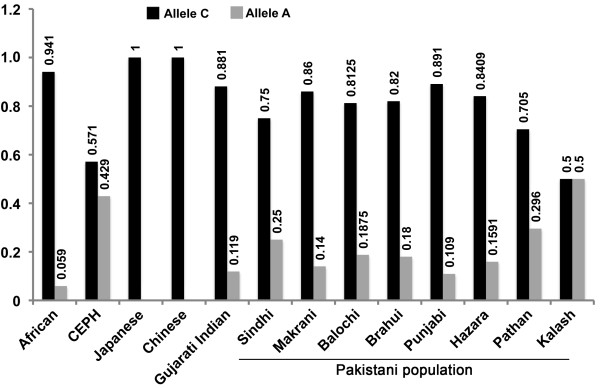
Frequency of the rs1042602 cSNP in the Pakistani population
Previous studies have shown a biased distribution of the TYR cSNP, p.Ser192Tyr (rs1042602) among the various populations studied in the International HapMap Project (http://hapmap.ncbi.nlm.nih.gov/). To determine the minor allele frequency (MAF) of the rs1042602 cSNP in the Punjab province of Pakistan, we genotyped 200 unrelated, normal individuals from different ethnic groups (Figure 5). Significant variation in the frequency of c.575 C > A in different regions of Pakistan was observed (Figure 5 and Additional File 2: Figure S1).
Figure 5 .
Allele frequency of the rs1042602 cSNP in the Pakistani population. The distribution of an ancestral C and derived A allele of TYR among Pakistani population. DNA samples from 200 individuals belonging to various ethnic groups within province of Punjab, Pakistan were genotyped for rs1042602. Also shown is the Human Genome Diversity Project data for comparison [55]. Details are available at the HGDP website (http://hgdp.uchicago.edu/).
All of the families in our study were enrolled from the Punjab province of Pakistan, and five of the forty families that were screened for TYR had a c.575 C > A (p.Ser192Tyr) polymorphism. In two of these five families, we did not find any other mutation in TYR, except for the heterozygous c.575 C > A. We found that c.575 C > A in these two families did not co-segregate with the OCA phenotype. Thus, we did not consider p.Ser192Tyr to be a pathogenic variant.
Genetic and clinical analyses of OCA2
Sequence analysis of the OCA2 gene revealed six variants in fourteen families (Figures 6 and 7, Table 3), including c.954 G > A, p.Met318Ile; c.1045-15 T > G; c.1456 G > T, p.Asp486Tyr; c.1580 T > G, p.Lys527Arg; c.2228 C > T, p.Pro743Lys; and c.2359 G > A, p.Ala787Thr. Three different prediction algorithms were used to determine the effects of the missense mutations identified in our cohort. Three of the four missense alleles were predicted to be damaging by all three programs (Table 3). The fourth change, c.954 G > A (p.Met318Ile), was predicted to be a benign polymorphism (Table 3). Furthermore, ClustalW alignment of the OCA2 proteins from eleven different species showed that the methionine residue at position 318 is not conserved and in fact, mouse, rat, rabbit, frog, and Drosophila have a leucine at this position, which is an amino acid closely related to isoleucine (Figure 7). In contrast, the remaining two residues (p.Asp486 and p.Lys527) mutated in other families are highly conserved in the eleven species analyzed here (Figure 7). Although not found in the DNA samples from 200 ethnically matched control individuals, p.Met318Ile segregated in cis with another missense mutation (p.Lys527Arg) of OCA2 in the PKAB063 family, thereby preventing us from exploring its functional role in OCA2 pathogenesis.
Figure 6 .
Pedigrees of Pakistani families segregating OCA2 mutations. Pedigrees of fourteen multi-generational families with mutations identified in the OCA2 gene. Filled and empty symbols represent affected and unaffected individuals, respectively. Double lines indicate consanguineous marriages. Asterisks indicate subjects enrolled in the protocol that contributed DNA samples.
Figure 7 .
Novel OCA2 mutations and resulting OCA2 phenotypes. A.Electropherograms of amplimers from genomic DNA templates illustrating homozygosity for the substitution mutations found in the affected individuals of the families. Arrows indicate the site of the mutations. All mutations described here are numbered from the ATG start codon (GenBank NM_000275). B. Clustal W alignment of OCA2 proteins from various species shows conservation of the residues at positions 318, 486 and 527 among twelve species. The conserved amino acids are shown with a dark gray background, and the nonconserved amino acids are shown with a white background. C. Photographs of fourteen OCA2 probands. The family number and the mutation identified in the OCA2 gene are given for each proband; a number of the probands shown have used hair dyes.
Table 3.
Mutations of OCA2 segregating in Pakistani families
| Nucleotide change# | Frequency in control samples | Effect on protein | Location | Family | Ethinicity | Haplotype* S1-S2-S3-S4-S5-S6-S7-S8 | Polyphen 2 | SNPs3D | Mutation Taster | Allele frequency this study | Regulatory region sequence¥ |
|---|---|---|---|---|---|---|---|---|---|---|---|
|
Missense | |||||||||||
|
c.954 G > A |
0/200 |
p.Met318Ile |
loop TM2-3 |
PKAB063 |
Warraich |
C-C-C-G-A-T-G-A |
Benign |
Benign |
Polymorphism |
7.14% |
TAAATG |
|
c.1456 G > T |
0/344 |
p.Asp486Tyr |
within TM7 |
PKAB052 |
Lanjay |
T-C-T-A-A-C-A-G |
Damaging |
Damaging |
Pathogenic |
35.71% |
TAAGTG |
| |
|
|
|
PKAB054 |
Mehay |
T-C-T-A-A-C-A-G |
|
|
|
|
TAAGTG |
| |
|
|
|
PKAB055 |
Mehay |
T-C-T-A-A-C-A-G |
|
|
|
|
TAAGTG |
| |
|
|
|
PKAB067 |
Ghallu |
T-C-T-A-A-C-A-G |
|
|
|
|
TAAGTG |
| |
|
|
|
PKAB101 |
Chaaki |
T-C-T-A-A-C-A-G |
|
|
|
|
TAAATG |
|
c.1580 T > G |
0/298 |
p.Leu527Arg |
within TM8 |
PKAB063 |
Warraich |
C-C-C-G-A-T-G-A |
|
|
|
7.14% |
TAAATG |
| c.2228 C > T |
|
p.Pro743Leu |
loop TM12-13 |
PKAB058 |
Arain |
C-C-T-G-A-C-A-A |
Damaging |
Damaging |
Pathogenic |
14.30% |
TAAATG |
| |
|
|
|
PKAB072 |
Joyia |
C-C-T-G-A-C-A-A |
|
|
|
|
TAAATG |
| c.2359 G > A |
|
p.Ala787Thr |
within TM13 |
PKAB071 |
Chohan |
C-C-C-G-A-C-G-A |
Damaging |
Damaging |
Pathogenic |
7.14% |
TAAATG |
| |
|
|
|
|
|
|
|
|
|
|
|
|
Splice site | |||||||||||
|
c.1045-15 T > G |
0/364 |
splicing |
within TM3 |
PKAB060 |
Bubar |
C-C-T-G-A-T-G-A |
Damaging |
Damaging |
Pathogenic |
35.71% |
TAAATG |
| |
|
error§ |
|
PKAB068 |
Sindhu Jutt |
C-C-T-G-A-T-G-A |
|
|
|
|
TAAATG |
| |
|
|
|
PKAB079 |
Abbasi |
C-C-T-G-A-T-G-A |
|
|
|
|
TAAATG |
| |
|
|
|
PKAB151 |
Ansari |
T-C-T-G-A-T-G-A |
|
|
|
|
TAAATG |
| PKAB152 | Ansari | C-C-T-G-A-T-G-A | TAAATG | ||||||||
#Given in bold are the novel variants found in this study. *SNPs used for haplotyping: S1, rs17565841; S2, rs12592307 (p.S788S); S3, rs1800411 (p.C517C); S4, rs1900758; S5, rs1800410; S6, rs10852218; S7, rs1800404; S8, rs12913832. According to Feb. 2009 UCSC Human Genome browser assembly (GRCh37/hg19), the S2 to S6 are present within the coding (S2 and S3) or intronic regions of OCA2. §Human Splicing Finder program (http://www.umd.be/HSF/) predicted a cryptic splice donor site 12 nucleotide upstream of known splice acceptor site for exon 9 due to the mutation. ¥Conserved regulatory element sequence present within intron 86 of the HERC2 gene. N/A not applicable. TM transmembrane domain. TMpred-Prediction of transmembrane regions and orientation (http://www.ch.embnet.org/software/TMPRED_form.html) was used for OCA2 domain prediction.
Five of the OCA2 families segregated a c.1045-15 T > G change, and five other families had a c.1456 G > T (p.Asp486Tyr) mutation (Figure 6 and Table 3). We performed haplotype analyses of eight closely linked short tandem repeats and single nucleotide polymorphisms to identify potential founder effects for recurrent variants of the OCA2 gene. The results are consistent with common ancestors for each of the two alleles (Table 3).
There was no obvious genotype-phenotype correlation observed in the affected individuals harboring different OCA2 gene alleles (Table 2 and Additional file 1: Table S1). The affected individuals from the fourteen families had nystagmus and photophobia, regardless of their sex and age (Figure 7C, Table 2 and Additional file 1: Table S1). Most of the affected individuals had white or yellowish white hair color (Figure 7C, Table 2 and Additional file 1: Table S1). Inter-familial variation in iris color was noted, with tones ranging from light grey to blue-brown (Table 2). Similar to OCA1 families, the presence of reddish spots and marked sun-damage on the skin with grossly enlarged veins in the cheeks and lips was observed. Some of the affected individuals were observed to use hair dyes to color their hair (Figure 7C, Table 2 and Additional file 1: Table S1).
Although there were no pathogenic mutations in the OCA2 promoter or in the cis-regulating element present in intron 86 of the HERC gene regulatory region [56], we found the blue eye color-associated allele (TAAGTG) of SNP rs12913832 in five of the OCA2 families (Table 3). In these five families, the rs12913832 “G” allele was found to be in linkage disequilibrium with the c.1456 G > T (p.Asp486Tyr) mutation in the OCA2 gene (Table 3). Affected individuals of these five families had grayish-blue or blue eye color (Table 2 and Additional file 1: Table S1). The presence of the blue eye color allele of SNP rs12913832 might contribute to the OCA phenotype by reducing the expression of the mutated OCA2 protein in these affected individuals [56].
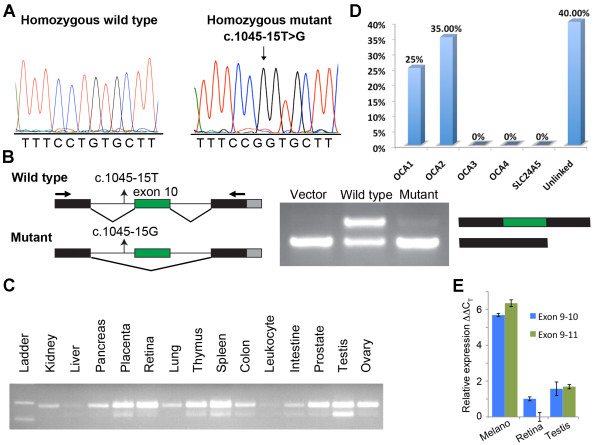
Effect of the c.1045-15 T > G mutation on splicing
To determine if c.1045-15 T > G (Figure 8A) alters the normal splicing of OCA2 mRNA, we made two constructs of genomic DNA for exon trapping. One construct was for the wild-type genomic sequence spanning intron 10 through intron 11, and the other had the splice site mutation c.1045-15 T > G (Figure 8B). The transfected empty pSPL3 vector produced the expected product of 177 bp, whereas the wild-type exon 10 splice site produced two bands, one with (249 bp) and one without (177 bp) exon 10 splicing, when amplified with vector primers (Figure 8B), which might indicate the presence of weak splice junctions around exon 10. When transfected in the exon trapping system, the construct with the mutated 5’ splice site (c.1045-15 T > G) produced a band of 177 bp that had exon 10 spliced out (Figure 8B). Although a weak band of ~249 bp was occasionally observed, sequencing revealed an aberrant spliced product. The results from the exon-trapping assay demonstrate that the c.1045-15 T > G mutation results in an mRNA that skips exon 10. If only exon 10 is skipped, then there is a deletion of twenty-four amino acids, thereby resulting in the loss of the third transmembrane domain of the full-length protein. However, analysis of known OCA2 transcripts indicated that exon 10 is alternatively spliced in most human tissues (Figure 8C). Quantitative real-time PCR analysis indicated that the OCA2 transcript including exon 10 is more abundant in human retina compared to transcripts without exon 10 (Figure 8E).
Figure 8 .
Functional analysis of c.1045-15 T > G mutation. A. Electropherograms of amplimers from genomic DNA templates illustrating homozygosity for the wild-type and c.1045-15 T > G substitution mutation found in the affected individuals of the five OCA2 families. The arrow indicates the site of the mutation. B. To determine the effect of the c.1045-15 T > G mutation on splicing, exon 10 with 200 bp of the flanking intron of OCA2 was introduced into the pSPL3 vector and analyzed through an in vitro splicing assay. The transfected empty pSPL3 vector produced the expected product of 177 bp, whereas the wild-type exon 10 splice site produced two bands with (249 bp) and without (177 bp) exon-10 splicing when amplified with vector primers, which might indicate the presence of weak splice junctions around exon 10. The construct with the c.1045-15 T > G mutation produced a band of 177 bp, which upon sequencing, revealed the skipping of exon 10. With the mutant construct, a weak band of ~249 bp was occasionally observed, but sequencing revealed an aberrant splice product. C. Human OCA2 isoforms with and without exon 10 are expressed in many tissues. D. Molecular genetic analysis of known OCA genes in a cohort of forty Pakistani families indicates that (a) OCA2 mutations are the most common cause of OCA, and (b) a significant number of families do not have mutations in the known OCA genes. E. Real-time quantitative RT–PCR analysis of TYR mRNAs level in human melanocytes, retina and testis cDNA libraries. CT is the observed threshold number of PCR cycles required for detection of the amplification product; ΔCT is the calculated difference in CT between the TYR gene and an internal control standard (GAPDH) measured in the same sample. ΔΔCT is the calculated difference in ΔCT between the experimental and exon 9–11 isoform in retina. Compared to the retina and testis, melanocytes have a relatively high expression of both exon 9–10 and exon 9–11 isoforms of TYR.
Genetic analysis of TYRP1, SLC45A2 and SLC24A5 genes
Although we did not screen the unknown regulatory regions, sequencing of all the non-coding and coding exons of the TYRP1, SLC45A2 and SLC24A5 genes in the 40 families segregating nonsyndromic OCA did not reveal any obvious pathogenic mutations (Figure 8D).
Discussion
In this study, we identified seven different pathogenic variants, three of which are novel, as the cause of OCA1 in ten Pakistani families. Two of the novel missense mutations, p.Pro21Leu and p.Cys35Arg, replaced highly evolutionarily conserved amino acid residues. The p.Cys35Arg mutation was found in two families, and SNP analysis revealed a common haplotype harboring this allele, which indicates a founder effect. The other common alleles found in the OCA1 families were p.Gly419Arg and p.Arg278* (Table 2). Both of these variants have been previously found in other Pakistani families [23,25,35]. Although no genotype data are available from the previous studies, our SNP analysis revealed common haplotypes in families sharing the same mutations. Our findings, along with the results of previous studies, indicate that p.Cys35Arg, p.Arg278* and p.Gly419Arg are the three most common mutations causing OCA1 in Pakistani families [23,25,35].
Interestingly, two of the novel variants, p.Cys35Arg and p.Tyr411His, might have temperature-sensitive behavior that could be due to a subtle conformational defect or gross protein misfolding [57]. A phenotypic evaluation of individuals homozygous for this allele did not reveal a temperature-sensitive phenotype. The loss of pigmentation in exposed skin areas was not strikingly different than that in less-exposed skin areas (e.g., legs, chest or abdomen); this finding could be due to the effect of the hot local climate (37°C - 52°C) on melanocyte growth and melanogenesis in the skin [58].
The association of the rs1042602 cSNP of TYR with squamous cell carcinoma of the skin in Caucasians and with pigmentation variation in the south Asian population has been documented [59,60]. This cSNP results in the substitution of serine with tyrosine at position 192 (p.Ser192Tyr) within the first copper-binding site (CuA) of tyrosinase [61]. Enzymatic analyses have revealed an approximately 40% reduction in the catalytic activity of tyrosinase, due to the p.Ser192Tyr mutation [61]. Interestingly, the distribution of the rs1042602 cSNP alleles varies significantly among different individuals from different geographical origins within Pakistan and thus provides a useful marker for epidemiological studies (Additional file 2: Figure S1).
In the cohort of Pakistani families segregating OCA studied here, fourteen out of forty families have mutations in OCA2. These results indicate that OCA2 is more prevalent than OCA1 in Pakistan in contrast to the Indian population [36,62]. Of the six distinct mutations in the OCA2 gene, three (p.Asp486Tyr, p.Leu527Arg, c.1045-15 T > G) have not been found in any of the various ethnic populations analyzed to date; therefore, they may be specific to Pakistani albino individuals. Of the two known mutations, p.Pro743Leu was previously identified in individuals of Caucasian, African-American and European ancestry [37,63-65], whereas the second mutation, p.Ala787Thr, was initially identified in the Chinese population [66]. Another missense mutation, p.Ala787Val, affecting the same codon of the OCA2 gene as p.Ala787Thr, was also reported to cause oculocutaneous albinism [67], further confirming the necessity of the alanine residue at this position for proper OCA2 protein function.
Affected individuals of five OCA2 families were homozygous for the c.1045-15 T > G mutation. Although the in vivo effects of c.1045-15 T > G are not known, this mutation is expected to produce only the OCA2 isoform without exon 10, which is predicted to encode a protein with no third transmembrane domain. Therefore, the level of normal OCA2 full-length protein required for the transport of tyrosinase to the plasma membrane might be affected by c.1045-15 T > G and thus cause OCA.
We found inter-familial variation in the clinical phenotype among the families segregating the same alleles of TYR or OCA2 (Table 2 and Additional file 1: Table S1). However, no obvious genotype-phenotype correlation was observed. A significant overlap in the range of phenotypes in individuals with TYR (OCA1) and OCA2 mutations was found, which makes genetic screening obligatory for the diagnosis of the type of albinism of the affected individuals. We compared the frequencies of all mutant TYR and OCA2 alleles among our forty OCA families. Mutations in exons 1, 2 and 4 combined accounted for all of the mutant alleles of the TYR gene in our cohort (Table 1). For the OCA2 gene, mutations in exons 10 and 14 collectively accounted for ~67% (10/15) of mutant alleles. Taken together, hierarchical mutation screening of these five exons of OCA genes in the nonsyndromic albino Pakistani population might reveal pathogenic alleles in approximately 43% (95% confidence interval: 28.5 - 57.9%) of cases and would be a cost-effective approach for molecular diagnosis.
Mutations in the protein coding exons or in the splice junctions of TYROCA2TYRP1 and SLC45A2 and SLC24A5 were not found in 16 families. There are at least three possible reasons to explain these findings. First, cryptic mutations might be present in the regulatory or splicing elements of these genes. Presently, we do not know the locations of the regulatory elements of these genes. Secondly, although family clinical histories and evaluation suggested no other clinical phenotype besides OCA, some of these families might have syndromic OCA. Currently, mutations in at least 12 loci have been causally linked with syndromic OCA [1,2]. There may also be an additional gene in which mutant alleles cause nonsyndromic OCA. Several previous studies of the four known OCA genes revealed no mutations in some of the affected individuals screened [36,68,69]. A need clearly exists for further genetic examination of this disease, as does the opportunity to understand this complex disorder more clearly.
Conclusions
Our results show that twenty-four families harbor twelve mutations (six previously reported and six novel mutations), but 40% of the ascertained OCA families had no apparent pathological mutations in the known OCA genes. Although our sample size was not large enough, based on our results, it is tempting to speculate that OCA2 mutations are more prevalent than OCA1 mutations in the Pakistani population. Nevertheless, this information would be useful for future diagnosis, genetic counseling and molecular epidemiology of OCA in the Pakistani population.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Z.M.A. and R.S.S conceived and designed the study; T.J.J. performed RT-PCR, mutational analyses, cloned isoforms, performed transfection studies and provided bioinformatic evaluations; T.K., N.T., M.I.M., A.S., F.I. and Sh.R. enrolled families and obtained clinical data; S.M.B. performed mutational screening; R.S.S and M.A. supervised the work at the Institute of Molecular Biology and Biotechnology, Multan; Z.M.A. and S.R. supervised the work at CCHMC. T.J.J. and Z.M.A. wrote the manuscript; All authors read and approved the final manuscript.
Supplementary Material
Table S1. Clinical assessment of the affected individuals with mutations in TYR and OCA2.
Figure S1. Schematic and geographical representation of allele frequency of the rs1042602 cSNP in the Pakistan. The distribution of an ancestral C (black) and derived A allele (gray) of TYR among Pakistani population.#All individuals show squinting in normal sunlight. aReddish spots throughout the skin and lips appeared sun damaged. bShow blistering on exposed skin and generalized sunburn redness. Cons: consanguineous union.
Contributor Information
Thomas J Jaworek, Email: thomas.jaworek@cchmc.org.
Tasleem Kausar, Email: tasleem.kausar@cchmc.org.
Shannon M Bell, Email: shannon.bell810@gmail.com.
Nabeela Tariq, Email: nabeela_trq@yahoo.com.
Muhammad Imran Maqsood, Email: itsmani85@yahoo.com.
Asma Sohail, Email: sohail82@gmail.com.
Muhmmmad Ali, Email: mohammad_03ali@yahoo.com.
Furhan Iqbal, Email: furhan.iqbal@bzu.edu.pk.
Shafqat Rasool, Email: rehansadiq80@yahoo.com.
Saima Riazuddin, Email: saima.riazuddin@cchmc.org.
Rehan S Shaikh, Email: rehansadiq80@bzu.edu.pk.
Zubair M Ahmed, Email: zubair.ahmed@cchmc.org.
Acknowledgments
We thank the families for their participation and cooperation. We also thank Faheem Ali and M. Asim for technical assistance and Dr. Ali Hussnain for critique of the manuscript. The work in Pakistan was supported by a research grant (HEC-1262) to RSS from the Higher Education Commission, Islamabad. This work was supported by the Cincinnati Children’s Hospital Research Foundation (CCHMC) intramural research funds provided to SR and ZA and the National Institute on Deafness and Other Communication Disorders (NIDCD/NIH) research grant R00-DC009287-03. ZA is also a recipient of an RPB Career Development Award.
References
- Tomita Y, Suzuki T. Genetics of pigmentary disorders. Am J Med Genet C Semin Med Genet. 2004;131C:75–81. doi: 10.1002/ajmg.c.30036. [DOI] [PubMed] [Google Scholar]
- Li W, He M, Zhou H, Bourne JW, Liang P. Mutational data integration in gene-oriented files of the Hermansky-Pudlak Syndrome database. Hum Mutat. 2006;27:402–407. doi: 10.1002/humu.20309. [DOI] [PubMed] [Google Scholar]
- Seven M, Yosunkaya E, Yilmaz SB, Karaca E, Guven G, Yuksel A. A new syndrome presenting with dysmorphic facies, oculocutaneous albinism, glaucoma, cryptorchidism and mental retardation. Genet Couns. 2011;22:25–34. [PubMed] [Google Scholar]
- Budisteanu M, Arghir A, Chirieac SM, Cardos G, Lungeanu A. Oculocutaneous albinism associated with multiple malformations and psychomotor retardation. Pediatr Dermatol. 2010;27:212–214. doi: 10.1111/j.1525-1470.2010.01115.x. [DOI] [PubMed] [Google Scholar]
- Tripathi RK, Hearing VJ, Urabe K, Aroca P, Spritz RA. Mutational mapping of the catalytic activities of human tyrosinase. J Biol Chem. 1992;267:23707–23712. [PubMed] [Google Scholar]
- Schnur RE, Sellinger BT, Holmes SA, Wick PA, Tatsumura YO, Spritz RA. Type I oculocutaneous albinism associated with a full-length deletion of the tyrosinase gene. J Invest Dermatol. 1996;106:1137–1140. doi: 10.1111/1523-1747.ep12340185. [DOI] [PubMed] [Google Scholar]
- Puri N, Gardner JM, Brilliant MH. Aberrant pH of melanosomes in pink-eyed dilution (p) mutant melanocytes. J Invest Dermatol. 2000;115:607–613. doi: 10.1046/j.1523-1747.2000.00108.x. [DOI] [PubMed] [Google Scholar]
- Preising MN, Forster H, Tan H, Lorenz B, de Jong PT, Plomp AS. Mutation analysis in a family with oculocutaneous albinism manifesting in the same generation of three branches. Mol Vis. 2007;13:1851–1855. [PubMed] [Google Scholar]
- Matsunaga J, Dakeishi-Hara M, Miyamura Y, Nakamura E, Tanita M, Satomura K, Tomita Y. Sequence-based diagnosis of tyrosinase-related oculocutaneous albinism: successful sequence analysis of the tyrosinase gene from blood spots dried on filter paper. Dermatology. 1998;196:189–193. doi: 10.1159/000017897. [DOI] [PubMed] [Google Scholar]
- Gershoni-Baruch R, Rosenmann A, Droetto S, Holmes S, Tripathi RK, Spritz RA. Mutations of the tyrosinase gene in patients with oculocutaneous albinism from various ethnic groups in Israel. Am J Hum Genet. 1994;54:586–594. [PMC free article] [PubMed] [Google Scholar]
- Rosemblat S, Sviderskaya EV, Easty DJ, Wilson A, Kwon BS, Bennett DC, Orlow SJ. Melanosomal defects in melanocytes from mice lacking expression of the pink-eyed dilution gene: correction by culture in the presence of excess tyrosine. Exp Cell Res. 1998;239:344–352. doi: 10.1006/excr.1997.3901. [DOI] [PubMed] [Google Scholar]
- Orlow SJ, Brilliant MH. The pink-eyed dilution locus controls the biogenesis of melanosomes and levels of melanosomal proteins in the eye. Exp Eye Res. 1999;68:147–154. doi: 10.1006/exer.1998.0599. [DOI] [PubMed] [Google Scholar]
- Chen K, Manga P, Orlow SJ. Pink-eyed dilution protein controls the processing of tyrosinase. Mol Biol Cell. 2002;13:1953–1964. doi: 10.1091/mbc.02-02-0022.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Inigo AE, Ladizinski B, Sethi A. Albinism in Africa: stigma, slaughter and awareness campaigns. Dermatol Clin. 2011;29:79–87. doi: 10.1016/j.det.2010.08.015. [DOI] [PubMed] [Google Scholar]
- Cohen T, Muller RM, Tomita Y, Shibahara S. Nucleotide sequence of the cDNA encoding human tyrosinase-related protein. Nucleic Acids Res. 1990;18:2807–2808. doi: 10.1093/nar/18.9.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Urabe K, Winder A, Jimenez-Cervantes C, Imokawa G, Brewington T, Solano F, Garcia-Borron JC, Hearing VJ. Tyrosinase related protein 1 (TRP1) functions as a DHICA oxidase in melanin biosynthesis. EMBO J. 1994;13:5818–5825. doi: 10.1002/j.1460-2075.1994.tb06925.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Cervantes C, Solano F, Kobayashi T, Urabe K, Hearing VJ, Lozano JA, Garcia-Borron JC. A new enzymatic function in the melanogenic pathway. The 5,6-dihydroxyindole-2-carboxylic acid oxidase activity of tyrosinase-related protein-1 (TRP1) J Biol Chem. 1994;269:17993–18000. [PubMed] [Google Scholar]
- Fernandez LP, Milne RL, Pita G, Aviles JA, Lazaro P, Benitez J, Ribas G. SLC45A2: a novel malignant melanoma-associated gene. Hum Mutat. 2008;29:1161–1167. doi: 10.1002/humu.20804. [DOI] [PubMed] [Google Scholar]
- Fukamachi S, Shimada A, Shima A. Mutations in the gene encoding B, a novel transporter protein, reduce melanin content in medaka. Nat Genet. 2001;28:381–385. doi: 10.1038/ng584. [DOI] [PubMed] [Google Scholar]
- Tripathi RK, Bundey S, Musarella MA, Droetto S, Strunk KM, Holmes SA, Spritz RA. Mutations of the tyrosinase gene in Indo-Pakistani patients with type I (tyrosinase-deficient) oculocutaneous albinism (OCA) Am J Hum Genet. 1993;53:1173–1179. [PMC free article] [PubMed] [Google Scholar]
- Spritz RA. Molecular genetics of oculocutaneous albinism. Semin Dermatol. 1993;12:167–172. [PubMed] [Google Scholar]
- Oetting WS, King RA. Molecular analysis of type I-A (tyrosinase negative) oculocutaneous albinism. Hum Genet. 1992;90:258–262. doi: 10.1007/BF00220074. [DOI] [PubMed] [Google Scholar]
- Oetting WS, Fryer JP, King RA. A dinucleotide deletion (−delta GA115) in the tyrosinase gene responsible for type I-A (tyrosinase negative) oculocutaneous albinism in a Pakistani individual. Hum Mol Genet. 1993;2:1047–1048. doi: 10.1093/hmg/2.7.1047. [DOI] [PubMed] [Google Scholar]
- Giebel LB, Tripathi RK, King RA, Spritz RA. A tyrosinase gene missense mutation in temperature-sensitive type I oculocutaneous albinism. A human homologue to the Siamese cat and the Himalayan mouse. J Clin Invest. 1991;87:1119–1122. doi: 10.1172/JCI115075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forshew T, Khaliq S, Tee L, Smith U, Johnson CA, Mehdi SQ, Maher ER. Identification of novel TYR and TYRP1 mutations in oculocutaneous albinism. Clin Genet. 2005;68:182–184. doi: 10.1111/j.1399-0004.2005.00460.x. [DOI] [PubMed] [Google Scholar]
- Hussain R, Bittles AH. The prevalence and demographic characteristics of consanguineous marriages in Pakistan. J Biosoc Sci. 1998;30:261–275. doi: 10.1017/S0021932098002612. [DOI] [PubMed] [Google Scholar]
- Ginger RS, Askew SE, Ogborne RM, Wilson S, Ferdinando D, Dadd T, Smith AM, Kazi S, Szerencsei RT, Winkfein RJ. et al. SLC24A5 encodes a trans-Golgi network protein with potassium-dependent sodium-calcium exchange activity that regulates human epidermal melanogenesis. J Biol Chem. 2008;283:5486–5495. doi: 10.1074/jbc.M707521200. [DOI] [PubMed] [Google Scholar]
- Vogel P, Read RW, Vance RB, Platt KA, Troughton K, Rice DS. Ocular albinism and hypopigmentation defects in Slc24a5−/− mice. Vet Pathol. 2008;45:264–279. doi: 10.1354/vp.45-2-264. [DOI] [PubMed] [Google Scholar]
- Ahmed ZM, Riazuddin S, Bernstein SL, Ahmed Z, Khan S, Griffith AJ, Morell RJ, Friedman TB, Wilcox ER. Mutations of the protocadherin gene PCDH15 cause Usher syndrome type 1 F. Am J Hum Genet. 2001;69:25–34. doi: 10.1086/321277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaki M, Mukhopadhyay A, Ray K. Determination of variants in the 3'-region of the tyrosinase gene requires locus specific amplification. Hum Mutat. 2005;26:53–58. doi: 10.1002/humu.20171. [DOI] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue P, Melamud E, Moult J. SNPs3D: candidate gene and SNP selection for association studies. BMC Bioinformatics. 2006;7:166. doi: 10.1186/1471-2105-7-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- Venselaar H, Te Beek TA, Kuipers RK, Hekkelman ML, Vriend G. Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinformatics. 2010;11:548. doi: 10.1186/1471-2105-11-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RA, Mentink MM, Oetting WS. Non-random distribution of missense mutations within the human tyrosinase gene in type I (tyrosinase-related) oculocutaneous albinism. Mol Biol Med. 1991;8:19–29. [PubMed] [Google Scholar]
- Renugadevi K, Sil AK, Perumalsamy V, Sundaresan P. Spectrum of candidate gene mutations associated with Indian familial oculocutaneous and ocular albinism. Mol Vis. 2010;16:1514–1524. [PMC free article] [PubMed] [Google Scholar]
- King RA, Pietsch J, Fryer JP, Savage S, Brott MJ, Russell-Eggitt I, Summers CG, Oetting WS. Tyrosinase gene mutations in oculocutaneous albinism 1 (OCA1): definition of the phenotype. Hum Genet. 2003;113:502–513. doi: 10.1007/s00439-003-0998-1. [DOI] [PubMed] [Google Scholar]
- Zahed L, Zahreddine H, Noureddine B, Rebeiz N, Shakar N, Zalloua P, Haddad F. Molecular basis of oculocutaneous albinism type 1 in Lebanese patients. J Hum Genet. 2005;50:317–319. doi: 10.1007/s10038-005-0257-5. [DOI] [PubMed] [Google Scholar]
- Wang Y, Guo X, Li W, Lian S. Four novel mutations of TYR gene in Chinese OCA1 patients. J Dermatol Sci. 2009;53:80–81. doi: 10.1016/j.jdermsci.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Tripathi RK, Strunk KM, Giebel LB, Weleber RG, Spritz RA. Tyrosinase gene mutations in type I (tyrosinase-deficient) oculocutaneous albinism define two clusters of missense substitutions. Am J Med Genet. 1992;43:865–871. doi: 10.1002/ajmg.1320430523. [DOI] [PubMed] [Google Scholar]
- Passmore LA, Kaesmann-Kellner B, Weber BH. Novel and recurrent mutations in the tyrosinase gene and the P gene in the German albino population. Hum Genet. 1999;105:200–210. doi: 10.1007/s004390051090. [DOI] [PubMed] [Google Scholar]
- Giebel LB, Tripathi RK, Strunk KM, Hanifin JM, Jackson CE, King RA, Spritz RA. Tyrosinase gene mutations associated with type IB ("yellow") oculocutaneous albinism. Am J Hum Genet. 1991;48:1159–1167. [PMC free article] [PubMed] [Google Scholar]
- Miyamura Y, Verma IC, Saxena R, Hoshi M, Murase A, Nakamura E, Kono M, Suzuki T, Yasue S, Shibata S. et al. Five novel mutations in tyrosinase gene of Japanese and Indian patients with oculocutaneous albinism type I (OCA1) J Invest Dermatol. 2005;125:397–398. doi: 10.1111/j.0022-202X.2005.23815.x. [DOI] [PubMed] [Google Scholar]
- Chaki M, Sengupta M, Mukhopadhyay A, Subba Rao I, Majumder PP, Das M, Samanta S, Ray K. OCA1 in different ethnic groups of india is primarily due to founder mutations in the tyrosinase gene. Ann Hum Genet. 2006;70:623–630. doi: 10.1111/j.1469-1809.2006.00247.x. [DOI] [PubMed] [Google Scholar]
- Tanita M, Matsunaga J, Miyamura Y, Dakeishi M, Nakamura E, Kono M, Shimizu H, Tagami H, Tomita Y. Polymorphic sequences of the tyrosinase gene: allele analysis on 16 OCA1 patients in Japan indicate that three polymorphic sequences in the tyrosinase gene promoter could be powerful markers for indirect gene diagnosis. J Hum Genet. 2002;47:1–6. doi: 10.1007/s10038-002-8648-3. [DOI] [PubMed] [Google Scholar]
- Sundaresan P, Sil AK, Philp AR, Randolph MA, Natchiar G, Namperumalsamy P. Genetic analysis of oculocutaneous albinism type 1 (OCA1) in Indian families: two novel frameshift mutations in the TYR Gene. Mol Vis. 2004;10:1005–1010. [PubMed] [Google Scholar]
- Goto M, Sato-Matsumura KC, Sawamura D, Yokota K, Nakamura H, Shimizu H. Tyrosinase gene analysis in Japanese patients with oculocutaneous albinism. J Dermatol Sci. 2004;35:215–220. doi: 10.1016/j.jdermsci.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Chaki M, Mukhopadhyay A, Chatterjee S, Das M, Samanta S, Ray K. Higher prevalence of OCA1 in an ethnic group of eastern India is due to a founder mutation in the tyrosinase gene. Mol Vis. 2005;11:531–534. [PubMed] [Google Scholar]
- Halaban R, Svedine S, Cheng E, Smicun Y, Aron R, Hebert DN. Endoplasmic reticulum retention is a common defect associated with tyrosinase-negative albinism. Proc Natl Acad Sci USA. 2000;97:5889–5894. doi: 10.1073/pnas.97.11.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausner RD, Sitia R. Protein degradation in the endoplasmic reticulum. Cell. 1990;62:611–614. doi: 10.1016/0092-8674(90)90104-M. [DOI] [PubMed] [Google Scholar]
- Toyofuku K, Wada I, Spritz RA, Hearing VJ. The molecular basis of oculocutaneous albinism type 1 (OCA1): sorting failure and degradation of mutant tyrosinases results in a lack of pigmentation. Biochem J. 2001;355:259–269. doi: 10.1042/0264-6021:3550259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon BS, Halaban R, Chintamaneni C. Molecular basis of mouse Himalayan mutation. Biochem Biophys Res Commun. 1989;161:252–260. doi: 10.1016/0006-291X(89)91588-X. [DOI] [PubMed] [Google Scholar]
- King RA, Townsend D, Oetting W, Summers CG, Olds DP, White JG, Spritz RA. Temperature-sensitive tyrosinase associated with peripheral pigmentation in oculocutaneous albinism. J Clin Invest. 1991;87:1046–1053. doi: 10.1172/JCI115064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidson SH, Fabian BC. The effect of temperature on tyrosinase activity in Himalayan mouse skin. J Exp Zool. 1981;215:91–97. doi: 10.1002/jez.1402150111. [DOI] [PubMed] [Google Scholar]
- Pickrell JK, Coop G, Novembre J, Kudaravalli S, Li JZ, Absher D, Srinivasan BS, Barsh GS, Myers RM, Feldman MW, Pritchard JK. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009;19:826–837. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiberg H, Troelsen J, Nielsen M, Mikkelsen A, Mengel-From J, Kjaer KW, Hansen L. Blue eye color in humans may be caused by a perfectly associated founder mutation in a regulatory element located within the HERC2 gene inhibiting OCA2 expression. Hum Genet. 2008;123:177–187. doi: 10.1007/s00439-007-0460-x. [DOI] [PubMed] [Google Scholar]
- Berson JF, Frank DW, Calvo PA, Bieler BM, Marks MS. A common temperature-sensitive allelic form of human tyrosinase is retained in the endoplasmic reticulum at the nonpermissive temperature. J Biol Chem. 2000;275:12281–12289. doi: 10.1074/jbc.275.16.12281. [DOI] [PubMed] [Google Scholar]
- Nakazawa K, Sahuc F, Damour O, Collombel C, Nakazawa H. Regulatory effects of heat on normal human melanocyte growth and melanogenesis: comparative study with UVB. J Invest Dermatol. 1998;110:972–977. doi: 10.1046/j.1523-1747.1998.00204.x. [DOI] [PubMed] [Google Scholar]
- Stokowski RP, Pant PV, Dadd T, Fereday A, Hinds DA, Jarman C, Filsell W, Ginger RS, Green MR, van der Ouderaa FJ, Cox DR. A genomewide association study of skin pigmentation in a South Asian population. Am J Hum Genet. 2007;81:1119–1132. doi: 10.1086/522235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan H, Kraft P, Hunter DJ, Han J. Genetic variants in pigmentation genes, pigmentary phenotypes, and risk of skin cancer in Caucasians. Int J Cancer. 2009;125:909–917. doi: 10.1002/ijc.24327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaki M, Sengupta M, Mondal M, Bhattacharya A, Mallick S, Bhadra R, Ray K. Indian Genome Variation C. Molecular and functional studies of tyrosinase variants among Indian oculocutaneous albinism type 1 patients. J Invest Dermatol. 2010;131:260–262. doi: 10.1038/jid.2010.274. [DOI] [PubMed] [Google Scholar]
- Sengupta M, Mondal M, Jaiswal P, Sinha S, Chaki M, Samanta S, Ray K. Comprehensive analysis of the molecular basis of oculocutaneous albinism in Indian patients lacking a mutation in the tyrosinase gene. Br J Dermatol. 2010;163:487–494. doi: 10.1111/j.1365-2133.2010.09830.x. [DOI] [PubMed] [Google Scholar]
- Lee ST, Nicholls RD, Schnur RE, Guida LC, Lu-Kuo J, Spinner NB, Zackai EH, Spritz RA. Diverse mutations of the P gene among African-Americans with type II (tyrosinase-positive) oculocutaneous albinism (OCA2) Hum Mol Genet. 1994;3:2047–2051. [PubMed] [Google Scholar]
- Lee ST, Nicholls RD, Bundey S, Laxova R, Musarella M, Spritz RA. Mutations of the P gene in oculocutaneous albinism, ocular albinism, and Prader-Willi syndrome plus albinism. N Engl J Med. 1994;330:529–534. doi: 10.1056/NEJM199402243300803. [DOI] [PubMed] [Google Scholar]
- Hutton SM, Spritz RA. Comprehensive analysis of oculocutaneous albinism among non-Hispanic caucasians shows that OCA1 is the most prevalent OCA type. J Invest Dermatol. 2008;128:2442–2450. doi: 10.1038/jid.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan HL, Li HY, Wu WQ, Zheng H, Chen Z. A novel P gene mutation in a Chinese family with oculocutaneous albinism. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2006;23:614–617. [PubMed] [Google Scholar]
- Oetting WS, Gardner JM, Fryer JP, Ching A, Durham-Pierre D, King RA, Brilliant MH. Mutations of the human P gene associated with Type II oculocutaneous albinism (OCA2). Mutations in brief no. 205. Online. Hum Mutat. 1998;12:434. doi: 10.1002/(SICI)1098-1004(1998)12:6<434::AID-HUMU16>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Gronskov K, Ek J, Sand A, Scheller R, Bygum A, Brixen K, Brondum-Nielsen K, Rosenberg T. Birth prevalence and mutation spectrum in danish patients with autosomal recessive albinism. Invest Ophthalmol Vis Sci. 2009;50:1058–1064. doi: 10.1167/iovs.08-2639. [DOI] [PubMed] [Google Scholar]
- Gargiulo A, Testa F, Rossi S, Di Iorio V, Fecarotta S, de Berardinis T, Iovine A, Magli A, Signorini S, Fazzi E. et al. Molecular and clinical characterization of albinism in a large cohort of Italian patients. Invest Ophthalmol Vis Sci. 2011;52:1281–1289. doi: 10.1167/iovs.10-6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Clinical assessment of the affected individuals with mutations in TYR and OCA2.
Figure S1. Schematic and geographical representation of allele frequency of the rs1042602 cSNP in the Pakistan. The distribution of an ancestral C (black) and derived A allele (gray) of TYR among Pakistani population.#All individuals show squinting in normal sunlight. aReddish spots throughout the skin and lips appeared sun damaged. bShow blistering on exposed skin and generalized sunburn redness. Cons: consanguineous union.