Abstract
Satellite RNAs usurp the replication machinery of their helper viruses, even though they bear little or no sequence similarity to the helper virus RNA. In Cereal yellow dwarf polerovirus serotype RPV (CYDV-RPV), the 322-nucleotide satellite RNA (satRPV RNA) accumulates to high levels in the presence of the CYDV-RPV helper virus. Rolling circle replication generates multimeric satRPV RNAs that self-cleave via a double-hammerhead ribozyme structure. Alternative folding inhibits formation of a hammerhead in monomeric satRPV RNA. Here we determine helper virus requirements and the effects of mutations and deletions in satRPV RNA on its replication in oat cells. Using in vivo selection of a satRPV RNA pool randomized at specific bases, we found that disruption of the base pairing necessary to form the non-self-cleaving conformation reduced satRPV RNA accumulation. Unlike other satellite RNAs, both the plus and minus strands proved to be equally infectious. Accordingly, very similar essential replication structures were identified in each strand. A different region is required only for encapsidation. The CYDV-RPV RNA-dependent RNA polymerase (open reading frames 1 and 2), when expressed from the nonhelper Barley yellow dwarf luteovirus, was capable of replicating satRPV RNA. Thus, the helper virus's polymerase is the sole determinant of the ability of a virus to replicate a rolling circle satellite RNA. We present a framework for functional domains in satRPV RNA with three types of function: (i) conformational control elements comprising an RNA switch, (ii) self-functional elements (hammerhead ribozymes), and (iii) cis-acting elements that interact with viral proteins.
Satellite RNAs depend on helper viruses for replication, encapsidation, and dissemination among hosts (47, 48). Satellite RNAs that form circles (circular satellite RNAs) (2, 20) encode no functional open reading frames (ORFs). Thus, the helper virus and the host must provide all trans-acting factors necessary for replication. Satellite RNAs accumulate to high levels in the presence of a helper virus, despite having no sequence homology with the helper virus RNA. In contrast, viroids are autonomously replicating circular RNA pathogens that do not require a helper virus or virion (2, 14, 20). Circular satellite RNAs and viroids (4) are the smallest replicating nucleic acids, ranging from 225 to about 450 nucleotides (nt) in length. Thus, they serve as models for understanding the minimum requirements for the replication of genetic information. This can shed light on virus replication processes and lead to means of interfering with virus infection.
Numerous studies of replication intermediates and the processing of satellite RNAs support the following rolling circle mechanism for replication of circular satellite RNAs (4, 5). Upon exiting the virion in the cell, the infectious circular plus-strand RNA is copied into a linear, multimeric minus strand (4, 5). (For satellite RNAs that encode no ORFs, we define the encapsidated strand as plus sense.) The minus strand either self-cleaves and religates to form circular monomers or remains as a multimer, depending on whether the satellite uses a symmetrical or asymmetrical replication strategy (12, 45). Either form of minus strand serves as a template for production of a multimeric plus strand. The multimeric plus-strand RNA cleaves, yielding monomers. Cleavage of the plus strand of satellite RNAs and of viroids in the Avsunviroidae family occurs at a hammerhead ribozyme site to generate the linear monomeric unit (18, 46). In most cases monomeric plus strands are ligated into circular forms (45) that are encapsidated in the virions provided by the helper virus. In other examples, the encapsidated form of the satellite RNA is linear (27, 34), and it is circularized only in the cell (8, 47, 48).
Because rolling circle satellite RNAs encode no proteins, the sequence and higher-order structure of the RNA itself must confer the biological properties, as has been shown for other noncoding, infectious RNAs (reviewed in references 19 and 47). The RNA sequence and structures involved in pathogenicity and replication are well characterized for the linear satellite RNAs of Cucumber mosaic virus (19) and Turnip crinkle virus (satRNA C) (42, 44, 52). However, little is known about replication and encapsidation signals on rolling circle satellite RNAs (8).
We have been investigating the structure and function of the satellite RNA in the RPV serotype (satRPV RNA) of Cereal yellow dwarf virus. SatRPV RNA was previously called satellite Barley yellow dwarf virus (sBYDV) RNA (27, 29). However, the helper virus, formerly known as the RPV serotype of BYDV (BYDV-RPV), proved to be quite distantly related to the type isolate of BYDV (BYDV-PAV) outside of the coat protein genes and, thus, has been reclassified as Cereal yellow dwarf virus-RPV (CYDV-RPV) in the genus Polerovirus (10). BYDV-PAV is now a member of the genus Luteovirus. Both genera are in the Luteoviridae family (10). BYDV serotypes that remain classified in the genus Luteovirus do not support replication of satRPV RNA or any other known satellite RNA (41). In contrast, Beet western yellows polerovirus (BWYV) does support replication of satRPV RNA (37). We proposed that this is because the RNA-dependent RNA polymerase (RdRp) gene of CYDV-RPV is much more closely related to that of BWYV than it is to that of BYDV. It has been proposed that circular satellite RNAs are recognized and replicated by helper virus replicase, but this has not been demonstrated unequivocally. Here we show that this is indeed the case.
Previously it has been reported that two pentameric sequences called L1 and L2a that are loops in the functional hammerhead ribozyme of satRPV RNA can also form alternative base pairing that prevents formation of the hammerhead in the most stable predicted conformation of the monomer (43). Instead, self-cleavage occurs via a double-hammerhead structure that can arise only in multimers after formation of L1-L1 and L2a-L2a helices. It was suggested that the noncleaving alternative tertiary structure is essential for some other aspect of replication than cleavage and that the L1-L2a rearrangements serve as a “riboswitch” to switch satRPV RNA between self-cleavage-competent and replication-competent conformations (43). Here we provide evidence for the overall most stable structure of the satRPV RNA monomer and the putative functions of the sequences and/or structures in satRPV RNA. Our results show that L1 and L2a are important for conformational changes in satRPV RNA. We also map the putative specific packaging signal and cis-acting elements (promoters) for satRPV RNA synthesis in both strands.
MATERIALS AND METHODS
Structure probing by nuclease T1 and imidazole.
Unlabeled, dimeric satRPV RNAs were synthesized by in vitro transcription of EcoRI-linearized templates by using T7 RNA polymerase (RiboMax kit; Promega, Madison, Wis.). To induce self-cleavage, the RNA transcripts were incubated at 37°C in cleavage buffer for approximately 3 h. satRPV transcripts were 5′ end labeled with [γ-32P]ATP and T4 polynucleotide kinase (40, 43) on RNA previously treated with alkaline phosphatase. The labeled 322-nt monomer obtained by self-cleavage was eluted from a 6% polyacrylamide-7 M urea gel after electrophoresis and staining with ethidium bromide. Structural probing with imidazole was performed in 0.04 mM NaCl, 1 mM EDTA, and 10 mM MgCl2 with 0, 0.4, 0.8, or 1.6 M imidazole for 15 h at 25°C (16, 21). Partial digestion with RNase T1 was done as described by Miller and Silver (29). Reaction products were separated via denaturing 6% polyacrylamide-7 M urea gel electrophoresis. The gels were dried and exposed to phosphorimager screens for 1 to 3 days and visualized with a STORM 840 PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
Generation of L1 and L2a randomized pool for in vivo genetic selection.
RNA molecules were synthesized by in vitro transcription of plasmid pWT as described previously (43). Plasmid pWT contains a one-and-a-half-unit length (1.5-mer) of wild-type satRPV cDNA inserted in pGEM3Zf(-) (Promega). To generate L2a-randomized mutants (L2a-pool), oligonucleotides 5L2a (TA ATACG ACTCA CTATA GGGTA TTTCG TGGAT AACAG AGCGC GTACT GTCTG ACGAC GTATC NNNNN GGACT AGAAG GCTGG, where N indicates equal amounts of all four nucleotides) and SP6pro (CGATT TAGGT GACAC TATA) were used as primers in the first PCR round, with plasmid pWT as a template (43). To generate the pool containing the randomized L1 and L2a sequences, oligonucleotides 5L1 (TAA TACGA CTCAC TATAG GGTAT TTCGT GGATA ACAGA NNNNN TACTG TCTGA CGACG) and SP6pro were used as primers in the second-round PCR with L2a-pool. The cDNA products of the second PCR contained a T7 RNA polymerase promoter upstream of a satRPV 1.5-mer pool randomized at the L1 and L2a pentamers. The uncloned products of the second PCR were transcribed by using T7 polymerase. The 322-nt gel-purified monomer obtained by self-cleavage (as above) was coelectroporated into oat protoplasts with 50 ng of viral RNA purified from a mixture of CYDV-RPV and BYDV-PAV isolates (41, 44). (The CYDV-RPV mixture accumulates to a higher titer than pure CYDV-RPV in the presence of BYDV-PAV, and the mixture supports satRPV RNA as efficiently as does pure CYDV-RPV [41].) For the second-round inoculations, total RNA was extracted from infected protoplasts (48 h postinfection [hpi]) and was used to coinoculate fresh protoplasts with 50 ng of helper viral RNA. After the second-round inoculation (48 hpi), total RNA was extracted from protoplasts and selected negative-strand mutants were cloned by reverse transcription-PCR with SuperScript II (Invitrogen).
Generation of deletion mutants for in vitro transcription.
Serial deletion mutants (see Fig. 3A) of satRPV were generated by using exonuclease III (ExoIII; Erase-a-Base system; Promega) after digestion of plasmid pWT with MscI (construct M122), AvaI (A222, A241, A243) or ClaI (C321). Deletion mutants were constructed as follows. Separate pWT aliquots were digested with each of the following sets of restriction enzymes: BglII and MscI (construct BM1), MscI and AvaI (MA5), BglII and AvaI (BA9), and ClaI (CEH14) and then end filled with the Klenow fragment of DNA polymerase in a 2 mM mixture of all four deoxyribonucleoside triphosphates. These products were self-ligated. Mutant MHH, in which bases 154 to 254 are deleted, was constructed as follows: two oligonucleotides, C253SAT (TTGAT CGATT GTTTC CCAAA GCAAG TCTCC TCACT) and 104SAT (AGGTG GCCAC CACTC TTTGA AGTGA GGAGA CTTGC), were annealed at 22°C for 30 min and then end filled with Klenow fragment. The products were digested at their ends with MscI and ClaI (underlined) and cloned into the wild-type 1.5-mer cDNA clone pWT, which had been digested with the same enzymes.
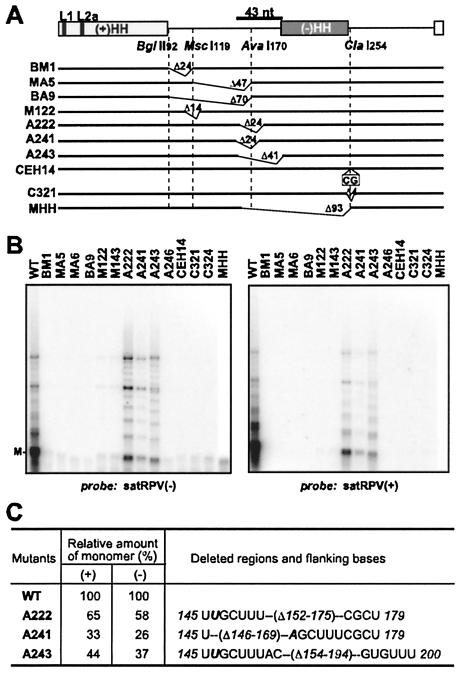
FIG. 3.
Effect of deletions and insertions on satRPV RNA accumulation. (A) Maps of satRPV RNA mutants. A diagram of satRPV RNA is shown at the top, with shaded boxes indicating the locations of hammerhead (HH) structures in plus and minus strands. Vertical bars indicate L1 and L2a. The positions of the restriction enzyme sites are shown. The thick bar at the top (43 nt) indicates a region that could be deleted while allowing significant satRPV RNA accumulation (mutant A243). Deletions in individual mutants are indicated by thin angled lines, with the number of deleted (Δ) nucleotides shown. CEH14 contains a CG insertion at the ClaI254 site. (B) Northern blot analysis of total RNA extracted from oat protoplasts (48 hpi) coelectroporated with CYDV-RPV helper virus genomic RNA and the indicated monomeric wild-type (WT) or mutant transcripts. The same blot was probed with plus strand, stripped, and then probed with minus-strand satRPV RNA. M, mobility of monomeric bands. (C) Quantification of deletion mutants that accumulated to significant levels in both strands. Deletions and flanking bases are shown at right.
Construction of BYDV expression vectors.
We converted our infectious clone of BYDV-PAV, pPAV6, into expression vector pCP393 by replacing ORF3, ORF4, and ORF5 of pPAV6 with unique restrictions sites ApaI and BssHI. Bases 2832 to 4599 were deleted from pPAV6 (13) by inverted PCR-mediated deletion with Vent DNA polymerase (New England Biolabs) and the primers PAV5END (TGGGG GCCCG CGGCA GAAAT TGAGA GAAGC CGCGA ATGC), spanning bases 4570 to 4599 with nine nonviral bases at the 5′ end (ApaI site underlined), and PAV3END (GGGGC GCGCT CACCA CCTCT CTAGT GGTGT CTGAA), complementary to bases 2832 to 2857 with nine nonviral bases at the 5′ end (BssHII site underlined). HpaI-digested pPAV6 was used as a template. The PCR product was self-ligated prior to transformation of Escherichia coli strain DH5α.
The chimeric helper virus genome CP393RPV, in which ORF3, ORF4, and ORF5 of PAV6 RNA were replaced by ORF1 and ORF2 of CYDV-RPV New York isolate (CYDV-RPV-NY), was constructed by inserting bases 324 to 3578 of CYDV-RPV-NY into the BssHI-ApaI sites of pCP393. We used a full-length clone of CYDV-RPV-NY, pRPV-NY, as the source of ORF1 and ORF2. First, a SmaI2780 site within ORF2 of pRPV-NY was removed to allow linearization of full-length pCP393RPV at the SmaI site at the genomic 3′ end. This was done by using a two-step PCR method for site-directed mutagenesis (22). For both rounds of PCR, pRPV-NY was used as the template. The first round used the mutagenic oligomer NoSma (TTGAT CAATT GGTAG CCCGT GTTTT GTTTC AAAGA CAA; the altered base is underlined), spanning bases 2763 to 2800, and the downstream primer RPV-ATG (TGGGG GCCCT GTCCG GCTAG TTTTG TGCTC AGT; the ApaI site is underlined, and the complement of the ORF2 stop codon is in italics), complementary to bases 3555 to 3578 with nine nonviral bases at the 5′ end. The PCR product was gel purified and used as a downstream primer paired with the upstream primer RPV-TGA (GGGGC GCGCA TGAAA TCGAT TTATT TTGTG; the BssHII site is underlined, and the ORF1 start codon is in italics), spanning bases 324 to 348 with nine nonviral bases at the 5′ end. The product was digested with the restriction endonucleases BssHII and ApaI, gel purified, and inserted into vector pCP393 cut with the same enzymes. The sequence across mutated regions of plasmid pCP393RPV was determined by sequencing and SmaI digestion.
Electroporation of protoplasts, extraction of total RNA, and partial purification of virus.
Oat (Avena sativa cv. Stout) protoplasts were isolated from cell suspension culture (cell line S226 obtained from Howard Rines, U.S. Department of Agriculture, Agricultural Research Service, University of Minnesota) as previously described (41). Protoplasts were electroporated with 50 ng of viral RNA purified from CYDV-RPV virions and 20 ng of gel-purified monomeric satRPV RNA transcript. At designated times, cells were collected by centrifugation, quick-frozen in liquid nitrogen, and stored at −80°C. Total RNA was isolated from protoplasts by using the RNeasy Plant mini kit (QIAGEN, Valencia, Calif.) and resuspended in diethyl pyrocarbonate-treated H2O. Virus particles were partially purified from oat protoplasts by resuspending a pellet in cold 0.1 M potassium phosphate buffer, pH 7.0, containing 0.5% Triton X-100. The homogenized, lysed cells were centrifuged at 800 × g for 10 min, and virus particles were pelleted from the supernatant by centrifugation through a 20% sucrose cushion in 0.1 M potassium phosphate buffer, pH 7.0, at 51,000 rpm in a Beckman TLA 100.3 rotor for 40 min. Pellets were resuspended overnight on ice in 50 μl of phosphate buffer.
RNA analysis.
Denaturing 1.5% agarose gel electrophoresis and Northern blot hybridization were performed according to the method of Rasochova and Miller (36). Each lane was loaded with equal amounts of total RNA as determined by spectrophotometry and confirmed by ethidium bromide staining of rRNA before Northern blot hybridization. 32P-labeled RNA probes were synthesized by in vitro transcription (Promega) by using [α-32P]CTP label. Antisense satRPV RNA probe was synthesized by SP6 RNA polymerase transcription of HindIII-cut plasmid pT7Sat (36, 39). Sense satRPV RNA probe was prepared by T7 RNA polymerase transcription of EcoRI-cut plasmid pT7Sat. Before blots were reprobed with a second probe, they were stripped by boiling in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.1% sodium dodecyl sulfate and exposed to ensure that all counts had been removed. The MFOLD program was used to predict the secondary structure of the lowest free energy state for RNAs (46, 53).
RESULTS
Secondary structure model for satRPV RNA by structure probing.
As a first step toward understanding the structure-function relationships of satRPV RNA, we determined the secondary structure of plus-sense monomeric satRPV RNA in vitro. Previously, we predicted that the most stable structure of monomeric satRPV RNA forms a series of stem-loops that does not include the hammerhead. Here we mapped single- and double-stranded regions of the entire satRPV RNA by probing with structure-sensitive agents under nondenaturing conditions. Figure 1A shows autoradiograms from monomeric satRPV RNA digested partially with RNase T1, which cuts single-stranded Gs (15), or imidazole, which cuts unstructured (mostly single-stranded) nucleotides (50). Although some nonspecific cleavages by RNase T1 occurred under native conditions, they do not affect interpretation of the data. Such nonspecific cleavages by RNase T1 were also observed in studies of Cucumber mosaic virus satRNA (3).
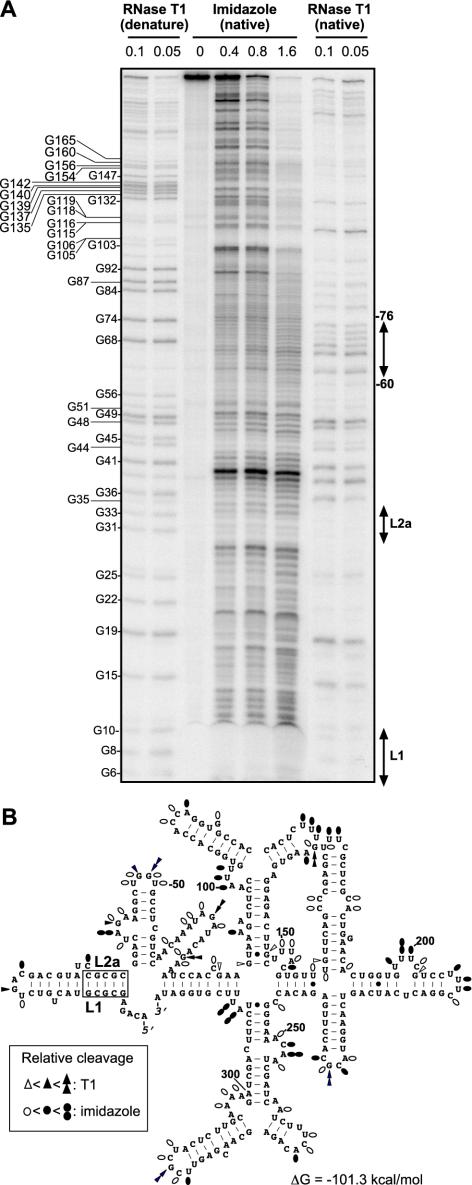
FIG. 1.
Secondary structure of satRPV RNA monomer. (A) Autoradiograph of 5′-end-labeled transcript of monomeric satRPV RNA after partial digestion with imidazole or RNase T1. Gel-purified, end-labeled RNA was incubated in three different molarities of imidazole under nondenaturing (native) conditions for 15 h (lanes 3 to 6). Indicated units of RNase T1 were used for digestion in nondenaturing (native) conditions for 5 min at 25°C. To generate the G-track sequencing ladder, indicated units of RNase T1 were incubated with RNA under denaturing conditions for 5 min at 50°C (left two lanes). The positions of G residues are indicated at left, numbered from the 5′ end. The positions of L1 and L2a sequences and nucleotides 60 to 76 are indicated at right. (B) Secondary-structure model of satRPV RNA predicted by MFOLD (53) superimposed with markers indicating intensity of cleavages in panel A. Open, filled, and double symbols indicate weak, moderate, and strong cuts, respectively.
The RNase T1 and imidazole cleavage sites are in good agreement with the predicted satRPV RNA monomer structure (Fig. 1B). The L1 and L2a bases resisted RNase T1 digestion, consistent with the prediction that they are base paired to each other. A slight discrepancy is in nucleotides 60 to 76, which are predicted to be single stranded but which were cut poorly by imidazole (Fig. 1A). This region may fold into a stable higher-order structure with other parts of satRPV RNA or it may be stabilized by non-Watson-Crick interactions. The two guanosine residues in this region (G68 and G74) were cut strongly by RNase T1, consistent with the predicted single-stranded nature of this region.
Replication of satRPV RNA containing randomized L1 and L2a sequences.
Previously, it was proposed that the L1-L2a base pairing was required for a role in replication other than self-cleavage of multimeric satRPV RNA (29, 43). To analyze the L1 and L2a sequences and structures important for satRPV RNA replication, we observed the replication of satRPV RNA mutants containing random bases in place of the five bases of L1 and the five bases of L2a. The pools of monomeric transcripts of plus-strand satRPV RNA containing these 10 randomized bases were coelectroporated with CYDV-RPV helper virus genomic RNA into oat protoplasts (first-round selection). After 48 h, total RNA was extracted from the infected protoplasts, and the isolated total RNA (which included detectable satRPV RNA) was coelectroporated again with CYDV-RPV helper virus genomic RNA into oat protoplasts (second-round selection). The progeny satRPV RNAs were cloned by using a primer complementary to the satRPV RNA minus strand for first-strand reverse transcription. This technique prevents cloning of any nonreplicating, contaminating transcripts from the original pool of plus-sense inocula. However, it may bias the pool of clones in favor of mutants that produce significant levels of minus strand. The sequences of the L1 and L2a regions of nine second-round progeny are shown in Fig. 2A. One clone, L1L2a-18, had wild-type sequences for L1 and L2a. Three mutants had wild-type L1, and one had a wild-type L2a sequence.
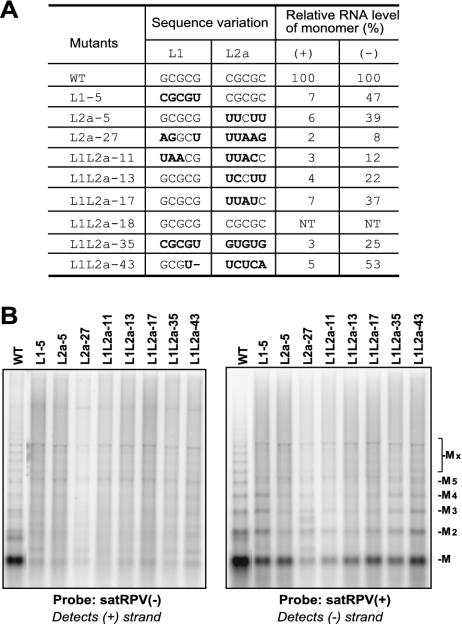
FIG. 2.
Effect of L1 and L2a sequence randomization on satRPV RNA accumulation. (A) Sequences in L1 and L2a of a sample of cloned “winners” after two rounds of replication in oat protoplasts. The nucleotides that differ from the wild type are in bold. The relative RNA levels quantified from the Northern blot hybridization in panel B are shown. The relative counts were measured with a PhosphorImager and quantified with ImageQuantNT4.0 software (Molecular Dynamics). (B) Northern blot hybridization of total RNA extracted from oat protoplasts (48 hpi) inoculated with CYDV-RPV RNA and either wild-type (WT) satRPV RNA or the indicated mutant transcripts. After being probed with plus-strand satRPV RNA, the blot was stripped, exposed to ensure that all signal was removed, and reprobed with minus-strand satRPV RNA. Mobilities (M) of monomeric and multimeric forms of satRPV RNAs are indicated at right (with subscripts indicating the number of satRPV repeat units up to X, which represents bands larger than pentamers).
To determine the replication properties of the selected replicating mutants, in separate inoculations monomeric transcript of each cloned mutant was coelectroporated with CYDV-RPV helper virus genomic RNA into oat protoplasts. After 48 h, accumulation of both strands of each mutant satRPV RNA was detected by Northern blot hybridization. All the mutants accumulated approximately 15- to 50-fold less plus strand than wild-type satRPV RNA (Fig. 2B). However, they accumulated only about 2- to 4.5-fold less negative strand than satRPV RNA except L2a-27 (12.5-fold less) and L1L2a-11 (8.4-fold less). Thus, base pairing between L1 and L2a appears to be more important for plus-strand accumulation than for minus-strand accumulation (43).
Effects of deletions and an insertion on replication of satRPV RNA.
We next sought to determine the roles of other portions of satRPV RNA by determining the effects of deletions on replication. Complete deletions between BglII91-MscI119 (construct BM1), BglII91-AvaI170 (BA9), and MscI119-AvaI170 (MA5) were constructed (subscripts indicate the bases after which the enzymes cut) (Fig. 3A). Also, three sets of the internal deletion mutants were generated by using ExoIII nuclease digestion at the MscI119 (construct M122), AvaI170 (A222, A241, A243), or ClaI254 (C321) restriction site (Fig. 3A). The only deletion mutants that replicated significantly were three similar constructs with ExoIII deletions around the AvaI170 site (A222, A241, and A243) (Fig. 3B). Mutants A222 and A241 both had 24-nt deletions that were staggered by six bases. Because five of the six bases adjacent to the deletions were the same, the net result was the same 24-nt deletion with a single base difference (U146 in A222 and A146 in A241) (Fig. 3C). The A146 in A241 reduced satRPV RNA accumulation in both strands to about half the level of that of mutant A222 (Fig. 3B and C) or about 30% of the wild-type level (Fig. 3C).
The 41-nt deletion in A243, which encompasses the 24-nt deletion in A222, reduced satRPV RNA accumulation in both strands by only about 60% compared to the level of wild-type satRPV RNA (Fig. 3). A243 accumulated about the same or slightly more progeny RNA than mutant A241. Both A222 and A243 have base U146, indicating that base U146 or its complement may participate in a sequence or secondary structure that controls accumulation of satRPV RNA (see Discussion). The computer-predicted secondary structures (MFOLD) of both strands of mutant A243 are the same as those of wild-type satRPV RNA outside of the deleted region. Thus, bases 154 to 194 are not essential for satRPV RNA replication. In contrast, the insertion of two bases (CG) between bases 256 and 257 (construct CEH14) or deletion of nucleotides 254 to 257 (C321) completely abolished replication (Fig. 3). Deletion of nucleotides 154 to 254 also abolished replication (Fig. 3, MHH). This sequence includes the nonessential region from nucleotides 154 to 194 plus the adjacent sequence that comprises the minus-strand hammerhead structure. Therefore, bases 254 to 257 and 195 to 254 (or their complements) directly or indirectly play a key role in the accumulation of satRPV RNA. Thus, all or part of the minus-strand hammerhead region is essential for accumulation of satRPV RNA, or this deletion causes improper folding elsewhere in the satRPV RNA.
Matching cis-acting elements in plus and minus strands are essential for satRPV RNA accumulation.
Previously, it was shown that, unlike most infectious single-stranded RNAs, both strands of satRPV RNA accumulate to similar levels (43). Thus, we propose that both strands have similar activities as replication templates. In this case, both strands may function equally as inocula to initiate infection. To investigate this possibility, we tested the infectivity of minus-strand satRPV RNA. As shown in Fig. 4, both strands of satRPV RNA are about equally infectious in oat protoplasts. This is a striking difference from other satellite RNAs in which only the encapsidated strand is infectious.
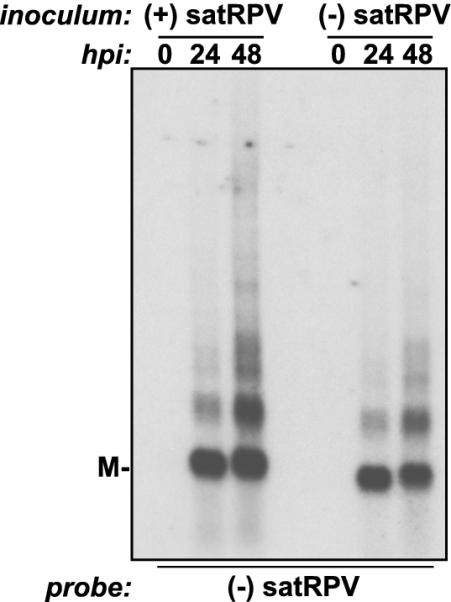
FIG. 4.
Both strands of satRPV RNA are infectious in oat protoplasts. Northern blot analysis of total RNA extracted from oat protoplasts inoculated with CYDV-RPV and plus-strand or minus-strand satRPV RNA monomer is shown. M, mobility of linear, monomeric forms of satRPV RNAs.
Because each strand indeed seems to serve equally well as a replication template, we suspected that the origins of replication and/or replicase binding sites in each strand might have similar sequences or secondary structures. The secondary structure of monomeric negative-strand satRPV RNA was predicted by MFOLD (53) (Fig. 5A). The predicted minus strand resembles the plus strand (Fig. 1B) in that the lowest free energy secondary structure does not form an active hammerhead structure. However, when the sequences complementary to L1 and L2a are forced to base pair in silico, the predicted secondary structure of monomeric minus-strand satRPV contains an active hammerhead structure at a site far from L1 and L2a (Fig. 5B).
FIG. 5.
Predicted secondary structures of satRPV RNA minus-strand monomer and putative replicase recognition sites in both strands. (A) The predicted secondary structure of the lowest free energy state for monomeric minus-strand satRPV RNA (using MFOLD) does not involve L1-L2a base pairing. (B) The most stable structure with forced base pairing between L1 and L2a (boxed) has a functional hammerhead ribozyme structure. Both strands form a branched structure proposed to be the origin of replication or a replicase binding site (indicated). (C) Similar structures in the plus and minus strands that are essential for replication may be replicase binding sites and/or origins of replication. Base positions of both strands are numbered in the plus sense. Restriction enzyme sites are shaded and in italics. Bases common to both structures are boxed.
Intriguingly, the most stable secondary structures for plus- and minus-strand monomers both harbor very similar branched stem-loop structures (Fig. 5C). Bases 245 to 310 in the plus strand and the complement of bases 87 to 153 in the negative strand are predicted to form similar bulged Y-shaped secondary structures with common primary sequences (Fig. 5C, outlined) as well. All mutations and deletions in the branched stem-loop in the plus strand, including small mutations around the ClaI site, totally eliminated satRPV RNA accumulation (Fig. 3 and 5C, MHH, CEH14 and C321). Four of the five mutations in the minus-strand branched stem-loop eliminated replication. The single transversion of A to U in mutant A241 reduced the accumulation of satRPV RNA by about twofold compared to the level in A222 (Fig. 3). This point mutation exists in the middle of a sequence (CAA) in a predicted bulge that is conserved in both strands (Fig. 5C). These results are consistent with a key role in RNA synthesis for the similar branched stem-loop structures in both strands.
The replicase proteins are the only CYDV-RPV proteins necessary for replication of satRPV RNA.
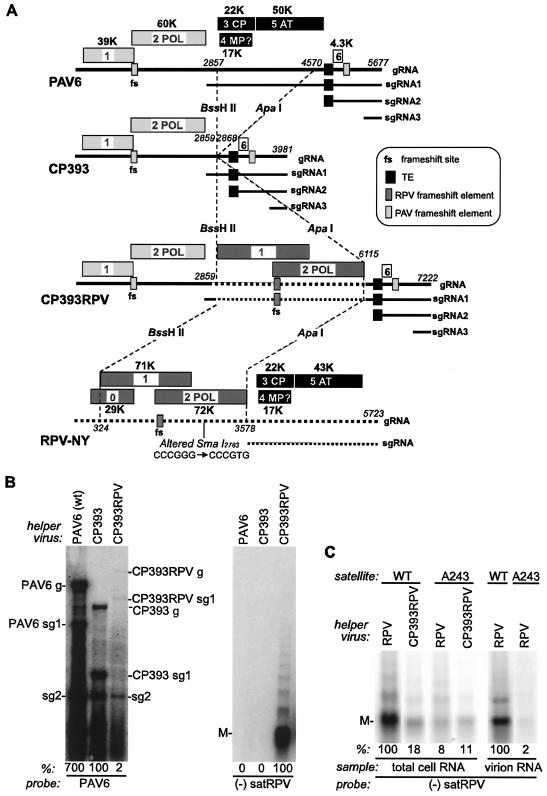
To better understand the interactions of the various cis-acting elements in satRPV RNA with functions supplied by the helper virus, we next sought to identify the helper virus proteins necessary for satRPV RNA replication. To do this, we used modified BYDV-PAV as a vector to express CYDV-RPV genes. BYDV does not support satRPV RNA replication (41). BYDV has significant sequence similarity to CYDV-RPV only in the coat protein and putative movement protein genes (ORF3, ORF4, and ORF5) (28) that are not needed for BYDV RNA replication in protoplasts (30). In contrast, BWYV has significant amino acid sequence identity to CYDV-RPV in ORF1 and ORF2 (37 and 61%, respectively) and is capable of supporting satRPV RNA replication (37). ORF1 and ORF2 (formerly known as ORF2 and ORF3 in BWYV) are fused during translation by ribosomal frameshifting to produce the viral RdRp (39). Thus, it was suggested that it is the viral RdRp that recognizes and replicates satRPV RNA (37). To test this, we subcloned the portion of the CYDV-RPV genome encompassing ORF1 and ORF2 into the vector pCP393 (a deletion version of infectious BYDV clone pPAV6, with ORF3, ORF4, and ORF5 replaced by a cloning site) to create vector pCP393RPV. This should allow expression of CYDV-RPV ORF1 and ORF2 from BYDV subgenomic RNA1 (sgRNA1), in place of the usual BYDV ORF3, ORF4, and ORF5 (Fig. 6A).
FIG. 6.
Helper proteins involved in replication and encapsidation of satRPV RNA. (A) Construction of BYDV-derived expression vector. Structural (CP, AT) and putative movement (MP?) genes of BYDV-PAV infectious clone PAV6 were replaced (dashed lines) by two unique restriction sites, BssHII and ApaI, to generate vector CP393. This was modified to express CYDV-RPV ORFs 1 and 2 from sgRNA1 by inserting bases 324 to 3578 of CYDV-RPV RNA into the BssHII-ApaI sites to generate CP393RPV. Molecular masses of encoded proteins are indicated. Bold horizontal lines indicate genomic RNAs, with CYDV-RPV-derived sequences dashed. Subgenomic RNAs (sgRNAs) are also indicated. Positions of key nucleotides are shown in italics, with the length of each infectious transcript indicated at right (3′ end). K, kilodalton; CP, coat protein; AT, aphid transmission; TE, cap-independent translation element. (B and C) Northern blot analysis of total RNA extracted from oat protoplasts (48 hpi) inoculated with satRPV RNA and the indicated helper virus transcript. The 32P-labeled probes used to detect RNAs are indicated below each blot. Relative RNA levels detected (%) were quantified with a STORM 840 PhosphorImager and Imagequant 1.2 for Macintosh (Molecular Dynamics) software. In panel B, protoplasts were coinoculated with the indicated viral transcript (left panel) and gel purified, monomeric satRPV RNA (right panel). Mobilities of genomic (g) and subgenomic (sg) RNAs and satRPV RNA monomer (M) are indicated. Panel C shows a Northern blot of total cellular RNA (left four lanes) or partially purified virion RNA (right two lanes) 48 h after inoculation with the indicated combination of helper RNA and wild-type or A243 mutant satellite RNA. Virions were partially purified as described in Materials and Methods.
Transcripts from SmaI-linearized vector pCP393 (transcript CP393) and vector pCP393RPV (transcript CP393RPV) replicated in oat protoplasts (Fig. 6B). CP393 progeny RNAs accumulated about sevenfold less than those of wild-type PAV6. CP393RPV genomic and sgRNA1 accumulated to far lower levels than CP393 RNAs. Most importantly, only this chimeric viral RNA supported replication of satRPV RNA in oat protoplasts (Fig. 6B). Thus, CYDV-RPV ORF1 and ORF2 are the only CYDV-RPV ORFs necessary to support replication of satRPV RNA, and they are expressed at sufficient levels from CP393RPV to do so.
While CYDV-RPV ORF1 and ORF2 are sufficient for replication of satRPV RNA, another CYDV-RPV protein(s) may be necessary for maximum accumulation of satRPV RNA. In side-by-side comparisons, CP393RPV helper RNA supported 5- to 10-fold less satRPV RNA accumulation than did CYDV-RPV helper virus (Fig. 6C, left two lanes). However, satRPV RNA mutant A243, which lacks nucleotides 152 to 194, accumulated to the same levels in the presence of either helper virus RNA (Fig. 6C, middle two lanes), and this level was similar to that of wild-type satRPV RNA supported by CP393RPV. Thus, CYDV-RPV ORF1 and ORF2 are sufficient for the full accumulation of mutant A243 RNA, but the deletion in A243 prevents its accumulation to higher levels in the presence of wild-type CYDV-RPV helper.
Sequence between bases 152 and 194 participates in encapsidation of satRPV RNA.
We hypothesize that satRPV RNA accumulates to higher levels in the presence of wild-type helper than CP393RPV helper because the former provides coat proteins in which satRPV RNA is encapsidated and protected from nucleases (30). In contrast, we predict that mutant A243 lacks the origin of assembly sequence (OAS) required for encapsidation, causing it to accumulate to equally low levels in the presence or absence of coat proteins. To test this, we examined levels of wild-type and mutant satRPV RNAs encapsidated in the presence of wild-type helper virus by performing Northern blots on virions isolated from infected protoplasts. As expected (37), mostly monomeric and dimeric satRPV RNAs were encapsidated. Higher multimers were greatly reduced relative to their levels in total cellular RNAs (Fig. 6C, compare total cell RNA with virion RNA). Most importantly, A243 RNA was present in virions at only 2% of the level of wild-type satRPV RNA (Fig. 6C, right two lanes). This result, combined with the lack of difference of wild-type and A243 accumulation in the presence of CP393RPV helper virus lacking coat proteins, strongly suggests that bases 152 to 194 play a key role in encapsidation function and little role in self-cleavage, ligation, or replicase recognition.
DISCUSSION
This molecular dissection of satRPV RNA reveals three kinds of functional elements (discussed below): (i) conformational change control elements (RNA switches), e.g., L1 and L2a that facilitate but are not absolutely essential for replication; (ii) self-functioning RNA (the hammerhead ribozymes); and (iii) cis-acting elements that interact with helper virus and/or host components, e.g., origins of replication and assembly.
L1 and L2a may comprise an RNA switch.
Alternative structures formed by metastable foldings are likely to play important roles in the replication of all rolling circle RNAs (20, 24, 25, 38, 43). Rolling circle satellite RNAs, the Avsunviroidae, and the circular hepatitis delta virus RNA fold into diverse structures (Fig. 1B) (6, 8, 9, 23, 31). One feature the structures share is that active ribozymes are absent in the most stable structure. Thus, the hammerhead ribozyme probably forms only transiently (see, e.g., reference 7) and, in some cases, including satRPV RNA, requires a double hammerhead (9, 17, 43). Consistent with a role for a nonfunctional hammerhead conformation, we showed that alternative base-paired regions that prevent formation of the plus-strand hammerhead are required for efficient satRPV RNA accumulation (Fig. 2 and reference 43). The replicating mutants selected from the pools randomized at L1 and L2A showed no conserved sequence, but sequences similar to those of the wild type predominate. Those that did not form either an L1-L1 or L1-L2a-like base pairing showed poor RNA accumulation in both strands (Fig. 2, L2a-27 and L1L2a-11).
The L1-L2a helix also appears to play a role in conformational change in minus-strand satRPV RNA. The predicted secondary structure of lowest free energy state for monomeric minus-strand satRPV does not form a hammerhead structure (Fig. 5A). However, the predicted most stable secondary structures of the monomeric minus strand of all the selected randomized mutants lacking L1-L2a complementarity do contain an active hammerhead structure (data not shown). The mutants all showed poor accumulation of plus-strand satRPV RNA (Fig. 2). Therefore the L1-L2a helix may play a direct role in plus-strand RNA synthesis. Alternatively, the L1-L2a helix may act indirectly simply by reducing the rate of hammerhead formation. This reduces self-cleavage of the minus strand, which must be a circle or multimer to generate multimeric plus strand, as necessitated by the rolling circle replication mechanism (45).
The most stable conformation of plus-strand satRPV RNA (Fig. 1B) may favor ligation of linear monomers. Alternative conformations for cleavage and ligation have been identified in other RNAs. In potato spindle tuber viroid, which requires host components for cleavage, the switch from cleavage to ligation is driven by a change from a tetraloop to a conformation that resembles the loop E structure found in 5S rRNA (2). Chay et al. (8) showed that linear satTRSV RNA folds into a nonhammerhead conformation to facilitate efficient ligation to form a circular molecule. This ligation structure juxtaposes the 5′ and 3′ ends rigidly in close proximity via base pairing. Unlike satTRSV, several bases remain single stranded at the 5′ end of the most stable satRPV RNA structure (Fig. 1B), and considerable flexibility would be predicted between the 5′ and 3′ ends. Alternatively, the 5′ and 3′ ends would be held in close proximity with little flexibility, in a previously predicted pseudoknotted structure containing L1-L2a base pairing (29, 43). Perhaps this structure is the optimal ligation substrate.
Previously we proposed a “sliding model” for the possible folding pathway of multimeric plus-strand satRPV RNA, in which the base pairing of L1-L2a and L1-L1 induces formation of an active double-hammerhead structure (43). We propose that L1 and L2a sequences are conformational control elements (RNA switches) for the satRPV RNA folding pathway in the replication cycle. All the L1 and L2a mutants accumulated satRPV RNAs at different levels. We predict that more passages of the satRPV RNA would select for L1-L1, L2a-L2a, and L1-L2a complementarity as in wild-type RNA. However, it is important that these interactions are not absolutely essential for replication. The results are consistent with other in vivo genetic selection experiments in which a surprisingly wide variety of sequences often can be tolerated after several rounds (7).
Putative cis-acting elements for replication of satRPV RNAs.
It is possible that there is no single specific initiation site for replication, because the satRPV RNA template is probably circular or multimeric. However, to outcompete helper virus RNA for the replicase, satRPV RNAs most likely contain cis-acting sequences or structures recognized specifically by the helper virus's replicase. In contrast to the L1-L2a conformational control elements, a replicase recognition site should be essential for replication. Because both strands of satRPV RNA are equally infectious and accumulate to similar levels, similar cis-acting sequences and structures may exist in each strand for replicase recognition. Indeed, strikingly similar structures in regions that are essential for satRPV RNA accumulation were found in nonoverlapping portions of each strand (Fig. 5C). All mutations and deletions in the minus-strand structure and most mutations in the plus-strand structure prevented or drastically reduced plus-strand accumulation and eliminated minus-sense accumulation (Fig. 3). Mutants A222 and A243, which merely reduce satRPV RNA accumulation, disrupt only the bottom two base pairs in the main stem (Fig. 3 and 7). The presence of similar replication origins in both strands was demonstrated previously in the self-cleaving, circular Avocado sunblotch viroid (ASBV) (32). However, unlike satRPV RNA, the origins of ASBV RNA are complementary to each other and ASBV plus strand accumulates in vast excess to minus strand (11).
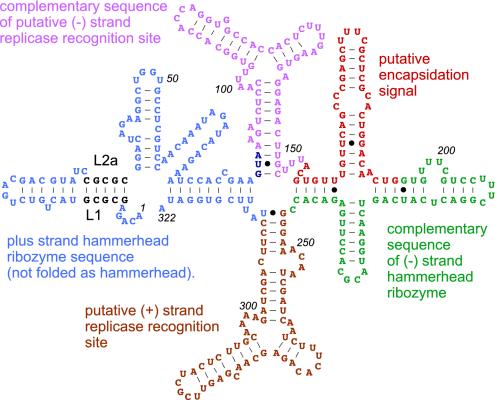
FIG. 7.
Known and putative functional domains in the most stable secondary structure of satRPV RNA. Structural domains are color-coded for each known or proposed function as indicated, with L1-L2a bases shown in black.
Sequences in the satRPV RNA branched stem-loops bear some resemblance to the likely origins of replication of the satRPV RNA helper virus RNAs. The conserved 5′-terminal five bases of the genomic and subgenomic RNAs of the known satRPV helper viruses, CYDV-RPV and BWYV, is 5′-ACPuAA (37), where Pu is purine, and the 5′ end of the minus strand of all helpers is AC. The motif ACAAA occurs far more frequently in Polerovirus genomes than expected by chance (26, 28). Note that a similar motif, PuCAA, is present in the lower stem bulge and in the bulge at the right branch point in both satRPV RNA strands in Fig. 5C. Bulge bases may participate in replicase recognition, due to their accessibility. A single point mutation in one of the bulged PuCAA motifs in the minus-strand structure (Fig. 5C, mutant A241) reduced satRPV RNA accumulation by three- to fourfold (Fig. 3). Similar Y-shaped structures were not obvious in the termini of helper virus RNAs. However, satRPV RNA replication recognition sites may not resemble the helper virus RNA, because both satRPV RNA strands are much better templates than the helper virus RNA itself (36).
In addition to the conserved branched stem-loops, other regions may participate in replication. The ACPuAA motif is also present at nucleotides 61 to 65 and in the negative strand complementary to nucleotides 199 to 203 (Fig. 5A and 7). We proposed previously that the structure and accessibility of the 61ACAAA65 sequence, which is a single-stranded region in the plus-strand hammerhead domain, may be affected by alternative conformations involving L1-L2a base pairing and that this could regulate replication (43). In keeping with the theme of symmetry between plus and minus strands, the 203ACGAA199 motif in the minus strand is in a single-stranded bulge in the minus-strand hammerhead domain (Fig. 7). A key role for this sequence may explain why deletion of the entire hammerhead domain in mutant MHH prevented satellite RNA accumulation, whereas point mutations that blocked only self-cleavage in the minus strand did not prevent replication (43).
The sequences from nucleotides 154 to 194 are necessary for specific packaging of satRPV RNA.
Only the plus strand of satRPV RNA is encapsidated in CYDV-RPV virions along with CYDV-RPV genomic RNA (27). Thus, satRPV RNA presumably contains a specific OAS. All or part of the OAS is likely to be within nucleotides 152 to 194 (Fig. 7) because the presence of this region increases accumulation of satRPV RNA only in the presence of a coat protein-expressing helper virus, and the absence of this region prevents encapsidation (Fig. 6C). Most of this region forms a bulged stem-loop (Fig. 7). While few OASs have been identified for plant viruses, bulged stem-loops serve as OAS for Tobacco mosaic virus (49) and are involved in specific packaging of the viral RNAs of Turnip crinkle virus genomic (35) and satellite (51) RNAs.
The replicase of the helper virus is the only viral component needed for satRPV RNA replication.
Support of significant satRPV RNA replication by the CP393-RPV chimeric virus strongly supports our hypothesis (37) that it is the helper virus replicase, rather than the host or other viral genes, that determines the ability of a virus to support a rolling circle satellite RNA. The data do not rule out the possibility that the CYDV-RPV RNA itself, rather than the product of ORF1 and ORF2, is necessary for satRPV replication, but it is difficult to conceive a mechanism to explain that possibility. The requirement for only the replicase protein from the helper virus was shown previously for the very different linear Satellite tobacco necrosis virus (1). However, a role for coat protein was not ruled out because Satellite tobacco necrosis virus encodes its own coat protein. Our data also support the notion that rolling circle satellite RNAs reduce helper virus levels by directly competing for the helper virus replicase. We rule out an essential role for CYDV-RPV ORF0, which is a suppressor of host posttranscriptional gene silencing in a related Polerovirus (33). This and other helper viral proteins may contribute to optimal satRPV RNA accumulation, but they are clearly not essential.
satRPV RNA levels are reduced in the presence of CP393RPV compared to wild-type helper virus, probably in large part because CP393RPV RNA accumulates to much lower levels. This difference is likely due to the very large (3.2-kb) insert of ORF1 and ORF2, resulting in a net addition of 1,487 nt to the wild-type PAV6 transcript. It is remarkable that CP393-RPV replicates at all, considering that it encodes two separate polymerases that probably compete for host components. Each polymerase must “find” the appropriate template RNA(s). Finally, this experiment reveals the sensitivity of satRPV RNA as a detector of functional CYDV-RPV replicase. While the mRNA for the replicase (sgRNA1 of CP393RPV) is so rare that it is very difficult to detect by Northern blotting, the satRPV RNA accumulates to high and readily detectable levels (Fig. 6). Thus, we show that it is possible to (i) convert a nonhelper virus into a helper virus of a satellite RNA by inserting genes from the helper into the nonhelper, (ii) express two unrelated replicases from one virus for replication of two different RNAs, and (iii) replicate a rolling circle satellite RNA supported only by the replicase of its helper virus.
Acknowledgments
We thank Randy Beckett for technical assistance.
This research was funded by a grant from the U.S. Department of Agriculture National Research Initiative.
REFERENCES
- 1.Andriessen, M., F. Meulewaeter, and M. Cornelissen. 1995. Expression of tobacco necrosis virus open reading frames 1 and 2 is sufficient for the replication of satellite tobacco necrosis virus. Virology 212:222-224. [DOI] [PubMed] [Google Scholar]
- 2.Baumstark, T., A. R. Schroder, and D. Riesner. 1997. Viroid processing: switch from cleavage to ligation is driven by a change from a tetraloop to a loop E conformation. EMBO J. 16:599-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernal, J. J., and F. Garcia-Arenal. 1997. Analysis of the in vitro secondary structure of cucumber mosaic virus satellite RNA. RNA 3:1052-1067. [PMC free article] [PubMed] [Google Scholar]
- 4.Branch, A. D., and H. D. Robertson. 1984. A replication cycle for viroids and other small infectious RNA's. Science 223:450-455. [DOI] [PubMed] [Google Scholar]
- 5.Bruening, G., B. K. Passmore, H. van Tol, J. M. Buzayan, and P. A. Feldstein. 1991. Replication of a plant virus satellite RNA: evidence favors transcription of circular templates of both polarities. Mol. Plant-Microbe Interact. 4:219-225. [DOI] [PubMed] [Google Scholar]
- 6.Bussiere, F., J. Ouellet, F. Cote, D. Levesque, and J. P. Perreault. 2000. Mapping in solution shows the peach latent mosaic viroid to possess a new pseudoknot in a complex, branched secondary structure. J. Virol. 74:2647-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carpenter, C. D., and A. E. Simon. 1998. Analysis of sequences and predicted structures required for viral satellite RNA accumulation by in vivo genetic selection. Nucleic Acids Res. 26:2426-2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chay, C. A., X. Guan, and G. Bruening. 1997. Formation of circular satellite tobacco ringspot virus RNA in protoplasts transiently expressing the linear RNA. Virology 239:413-425. [DOI] [PubMed] [Google Scholar]
- 9.Collins, R. F., D. L. Gellatly, O. P. Sehgal, and M. G. Abouhaidar. 1998. Self-cleaving circular RNA associated with rice yellow mottle virus is the smallest viroid-like RNA. Virology 241:269-275. [DOI] [PubMed] [Google Scholar]
- 10.D'Arcy, C. J., L. L. Domier, and M. A. Mayo. 2000. Family Luteoviridae, p. 775-784. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 11.Daròs, J. A., J. F. Marcos, C. Hernández, and R. Flores. 1994. Replication of avocado sunblotch viroid: evidence for a symmetric pathway with two rolling circles and hammerhead ribozyme processing. Proc. Natl. Acad. Sci. USA 91:12813-12817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies, C., J. Haseloff, and R. H. Symons. 1990. Structure, self-cleavage, and replication of two viroid-like satellite RNAs (virusoids) of subterranean clover mottle virus. Virology 177:216-224. [DOI] [PubMed] [Google Scholar]
- 13.Di, R., S. P. Dinesh-Kumar, and W. A. Miller. 1993. Translational frameshifting by barley yellow dwarf virus RNA (PAV serotype) in Escherichia coli and in eukaryotic cell-free extracts. Mol. Plant-Microbe Interact. 6:444-452. [DOI] [PubMed] [Google Scholar]
- 14.Diener, T. O. 1996. Understanding replication mechanisms in viroids and viroidlike RNAs. Trends Microbiol. 4:85-87. [DOI] [PubMed] [Google Scholar]
- 15.Ehresmann, C., F. Baudin, M. Mougel, P. Romby, J. P. Ebel, and B. Ehresmann. 1987. Probing the structure of RNAs in solution. Nucleic Acids Res. 15:9109-9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felden, B., H. Himeno, A. Muto, J. McCutcheon, J. Atkins, and R. F. Gesteland. 1997. Probing the structure of the Escherichia coli 10Sa RNA (tmRNA). RNA 3:89-103. [PMC free article] [PubMed] [Google Scholar]
- 17.Forster, A. C., C. Davies, C. C. Sheldon, A. C. Jeffries, and R. H. Symons. 1988. Self-cleaving viroid and newt RNAs may only be active as dimers. Nature 334:265-267. [DOI] [PubMed] [Google Scholar]
- 18.Forster, A. C., and R. H. Symons. 1987. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell 49:211-220. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Arenal, F., and P. Palukaitis. 1999. Structure and functional relationships of satellite RNAs of cucumber mosaic virus. Curr. Top. Microbiol. Immunol. 239:37-63. [DOI] [PubMed] [Google Scholar]
- 20.Gultyaev, A. P., F. H. van Batenburg, and C. W. Pleij. 1998. Dynamic competition between alternative structures in viroid RNAs simulated by an RNA folding algorithm. J. Mol. Biol. 276:43-55. [DOI] [PubMed] [Google Scholar]
- 21.Koev, G., B. R. Mohan, and W. A. Miller. 1999. Primary and secondary structural elements required for synthesis of barley yellow dwarf virus subgenomic RNA1. J. Virol. 73:2876-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Landt, O., H. P. Grunert, and U. Hahn. 1990. A general method for rapid site-directed mutagenesis using the polymerase chain reaction. Gene 96:125-128. [DOI] [PubMed] [Google Scholar]
- 23.Lazinski, D. W., and J. M. Taylor. 1995. Intracellular cleavage and ligation of hepatitis delta virus genomic RNA: regulation of ribozyme activity by cis-acting sequences and host factors. J. Virol. 69:1190-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lazinski, D. W., and J. M. Taylor. 1995. Regulation of the hepatitis delta virus ribozymes: to cleave or not to cleave? RNA 1:225-233. [PMC free article] [PubMed] [Google Scholar]
- 25.Loss, P., M. Schmitz, G. Steger, and D. Riesner. 1991. Formation of a thermodynamically metastable structure containing hairpin II is critical for infectivity of potato spindle tuber viroid RNA. EMBO J. 10:719-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, W. A., S. P. Dinesh-Kumar, and C. P. Paul. 1995. Luteovirus gene expression. Crit. Rev. Plant Sci. 14:179-211. [Google Scholar]
- 27.Miller, W. A., T. Hercus, P. M. Waterhouse, and W. L. Gerlach. 1991. A satellite RNA of barley yellow dwarf virus contains a novel hammerhead structure in the self-cleavage domain. Virology 183:711-720. [DOI] [PubMed] [Google Scholar]
- 28.Miller, W. A., S. Liu, and R. Beckett. 2002. Barley yellow dwarf virus: Luteoviridae or Tombusviridae? Mol. Plant Pathol. 3:177-183. [DOI] [PubMed] [Google Scholar]
- 29.Miller, W. A., and S. L. Silver. 1991. Alternative tertiary structure attenuates self-cleavage of the ribozyme in the satellite RNA of barley yellow dwarf virus. Nucleic Acids Res. 19:5313-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mohan, B. R., S. P. Dinesh-Kumar, and W. A. Miller. 1995. Genes and cis-acting sequences involved in replication of barley yellow dwarf virus-PAV RNA. Virology 212:186-195. [DOI] [PubMed] [Google Scholar]
- 31.Navarro, B., and R. Flores. 1997. Chrysanthemum chlorotic mottle viroid: unusual structural properties of a subgroup of self-cleaving viroids with hammerhead ribozymes. Proc. Natl. Acad. Sci. USA 94:11262-11267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Navarro, J. A., and R. Flores. 2000. Characterization of the initiation sites of both polarity strands of a viroid RNA reveals a motif conserved in sequence and structure. EMBO J. 19:2662-2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfeffer, S., P. Dunoyer, F. Heim, K. E. Richards, G. Jonard, and V. Ziegler-Graff. 2002. P0 of beet western yellows virus is a suppressor of posttranscriptional gene silencing. J. Virol. 76:6815-6824. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Prody, G., J. T. Bakos, J. M. Buzayan, I. R. Schneider, and G. Bruening. 1986. Autolytic processing of dimeric plant virus satellite RNA. Science 231:1577-1580. [DOI] [PubMed] [Google Scholar]
- 35.Qu, F., and T. J. Morris. 1997. Encapsidation of turnip crinkle virus is defined by a specific packaging signal and RNA size. J. Virol. 71:1428-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rasochova, L., and W. A. Miller. 1996. Satellite RNA of barley yellow dwarf-RPV virus reduces accumulation of RPV helper virus RNA and attenuates RPV symptoms in oats. Mol. Plant-Microbe Interact. 9:646-650. [Google Scholar]
- 37.Rasochova, L., B. K. Passmore, B. W. Falk, and W. A. Miller. 1997. The satellite RNA of barley yellow dwarf virus-RPV is supported by beet western yellows virus in dicotyledonous protoplasts and plants. Virology 231:182-191. [DOI] [PubMed] [Google Scholar]
- 38.Repsilber, D., S. Wiese, M. Rachen, A. W. Schroder, D. Riesner, and G. Steger. 1999. Formation of metastable RNA structures by sequential folding during transcription: time-resolved structural analysis of potato spindle tuber viroid (−)-stranded RNA by temperature-gradient gel electrophoresis. RNA 5:574-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reutenauer, A., V. Ziegler-Graff, H. Lot, D. Scheidecker, H. Guilley, K. Richards, and G. Jonard. 1993. Identification of beet western yellows luteovirus genes implicated in viral replication and particle morphogenesis. Virology 195:692-699. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Silver, S. L., L. Rasochova, S. P. Dinesh-Kumar, and W. A. Miller. 1994. Replication of barley yellow dwarf virus satellite RNA transcripts in oat protoplasts. Virology 198:331-335. [DOI] [PubMed] [Google Scholar]
- 42.Simon, A. E. 1999. Replication, recombination, and symptom-modulation properties of the satellite RNAs of turnip crinkle virus. Curr. Top. Microbiol. Immunol. 239:19-36. [DOI] [PubMed] [Google Scholar]
- 43.Song, S. I., S. L. Silver, M. A. Aulik, L. Rasochova, B. R. Mohan, and W. A. Miller. 1999. Satellite cereal yellow dwarf virus-RPV (satRPV) RNA requires a double hammerhead for self-cleavage and an alternative structure for replication. J. Mol. Biol. 293:781-793. [DOI] [PubMed] [Google Scholar]
- 44.Sun, X., and A. E. Simon. 2003. Fitness of a turnip crinkle virus satellite RNA correlates with a sequence-nonspecific hairpin and flanking sequences that enhance replication and repress the accumulation of virions. J. Virol. 77:7880-7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Symons, R. H. 1997. Plant pathogenic RNAs and RNA catalysis. Nucleic Acids Res. 25:2683-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Symons, R. H. 1992. Small catalytic RNAs. Annu. Rev. Biochem. 61:641-671. [DOI] [PubMed] [Google Scholar]
- 47.Symons, R. H., and J. W. Randles. 1999. Encapsidated circular viroid-like satellite RNAs (virusoids) of plants. Curr. Top. Microbiol. Immunol. 239:81-105. [DOI] [PubMed] [Google Scholar]
- 48.Taliansky, M. E., and P. F. Palukaitis. 1999. Satellite RNAs and satellite viruses, p. 1607-1615. In A. Granoff and R. G. Webster (ed.), Encyclopedia of virology, 2nd ed., vol. 3. Academic Press, San Diego, Calif. [Google Scholar]
- 49.Turner, D. R., L. E. Joyce, and P. J. Butler. 1988. The tobacco mosaic virus assembly origin RNA. Functional characteristics defined by directed mutagenesis. J. Mol. Biol. 203:531-547. [DOI] [PubMed] [Google Scholar]
- 50.Vlassov, V. V., G. Zuber, B. Felden, J. P. Behr, and R. Giege. 1995. Cleavage of tRNA with imidazole and spermine imidazole constructs: a new approach for probing RNA structure. Nucleic Acids Res. 23:3161-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, J., and A. E. Simon. 2000. 3′-end stem-loops of the subviral RNAs associated with turnip crinkle virus are involved in symptom modulation and coat protein binding. J. Virol. 74:6528-6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, G., and A. E. Simon. 2003. A multifunctional turnip crinkle virus replication enhancer revealed by in vivo functional SELEX. J. Mol. Biol. 326:35-48. [DOI] [PubMed] [Google Scholar]
- 53.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]