Abstract
A/J and 129P3/J mouse strains have different susceptibilities to dental fluorosis due to their genetic backgrounds. They also differ with respect to several features of fluoride (F) metabolism and metabolic handling of water. This study was done to determine whether differences in F metabolism could be explained by diversities in the profile of protein expression in kidneys. Weanling, male A/J mice (susceptible to dental fluorosis, n = 18) and 129P3/J mice (resistant, n = 18) were housed in pairs and assigned to three groups given low-F food and drinking water containing 0, 10 or 50 ppm [F] for 7 weeks. Renal proteome profiles were examined using 2D-PAGE and LC-MS/MS. Quantitative intensity analysis detected between A/J and 129P3/J strains 122, 126 and 134 spots differentially expressed in the groups receiving 0, 10 and 50 ppmF, respectively. From these, 25, 30 and 32, respectively, were successfully identified. Most of the proteins were related to metabolic and cellular processes, followed by response to stimuli, development and regulation of cellular processes. In F-treated groups, PDZK-1, a protein involved in the regulation of renal tubular reabsorption capacity was down-modulated in the kidney of 129P3/J mice. A/J and 129P3/J mice exhibited 11 and 3 exclusive proteins, respectively, regardless of F exposure. In conclusion, proteomic analysis was able to identify proteins potentially involved in metabolic handling of F and water that are differentially expressed or even not expressed in the strains evaluated. This can contribute to understanding the molecular mechanisms underlying genetic susceptibility to dental fluorosis, by indicating key-proteins that should be better addressed in future studies.
Introduction
The widespread use of F has contributed to the caries decline, but excessive intake may affect both bone metabolism and enamel development, causing skeletal and dental fluorosis, respectively. There are many sources of F intake, such as drinking water, dental products, dietary supplements and infant formulas [1]. There is evidence that the prevalence of dental fluorosis (DF) is increasing worldwide both in fluoridated and non-fluoridated communities [2]. In the US, 23% of 6- to 39-yr-old subjects present enamel fluorosis, ranging from very low to relatively high in severity [3]. However, the exact mechanisms by which F affects biomineralization are not completely understood [4], [5]. It has been proposed that genetic determinants influence the susceptibility to DF in humans [6] and mice [7]. Two strains of mice have been identified with distinct responses to the effects of F in the mineralized tissues. The A/J strain is “susceptible”, with a rapid onset and severe development of DF, while the 129P3/J is “resistant”, with minimum development of DF [7]. These strains also differ regarding their susceptibilities to the effects of F in bone [8], [9].
To determine whether such differences were due to differences in F metabolism, we conducted a metabolic study in which total F intake and excretion were measured. Our results showed that, compared to A/J mice, 129P3/J mice ingested less water, excreted less urine, had lower urinary F excretion and consequently had higher F retention and plasma and femur F levels [10]. However, these findings were not able to explain the mechanisms underlying the differences in the metabolic handling of F.
Kidneys represent the major route of removal of F from the body [11]. After F enters the renal tubules, a variable amount is reabsorbed, depending on the urinary pH because transmembrane migration occurs by diffusion of HF [12]. Thus, any factor that affects urinary pH will have an impact on the amount of F that is excreted in urine [11]. Urinary F excretion is also influenced by glomerular filtration rate since its reduction, as occurs in chronic renal dysfunction as well as in the last decades of life, results in lower excretion and increased plasma F levels [13]. Considering that kidney is a key organ in the metabolism of F, we then sought to investigate the molecular mechanisms underlying the renal F metabolism in A/J and 129P3/J mice that may account for their differential metabolic handling of F. To address this, proteomic analyses were performed on kidneys of A/J and 129P3/J mice receiving both low and high level of F-containing water.
Materials and Methods
Animals and Treatment
Male mice from the A/J and 129P3/J inbred strains (3-week-old) were randomly distributed into three groups (n = 6/strain) based on the F concentrations in the drinking water. All animals were housed in pairs in metabolic cages with ad libitum access to low-F food (AIN76A, PMI Nutrition, Richmond, IN, USA, 0.95 mg/Kg F) and water, to allow analysis of water and food consumption [10]. The temperature and humidity in the climate-controlled room, which had a 12-h light/dark cycle, were 23±1°C and 40%–80%, respectively. All experimental protocols were approved by the Ethics Committee for Animal Experiments of Bauru Dental School, University of Sao Paulo (Protocol # 026/2007). Experimental groups received drinking water containing 10 (low) or 50 (high) ppm F ion (as NaF), for 60 days. Control group received deionized water for the same period. At the end of the study, the mice were anesthetized with ketamine/xylazine and kidneys were collected. The left kidney was washed with cold buffer containing Tris 100 mM, EDTA 1 mM, PMSF 1 mM, pH 7.4, frozen at liquid nitrogen and kept at -80°C until proteomic analysis. The right kidney was collected for F analysis.
F analysis in kidney
Kidneys were homogenized in deionized water for 2 min using a homogenizer (Marconi, Model MA 102). Kidney F concentrations were determined in duplicate (100 mg of kidney tissue) after overnight hexamethyldisiloxane (HMDS)-facilitated diffusion [14], [15] using the ion-specific electrode (Orion Research, Model 9409) and a miniature calomel electrode (Accumet, #13-620-79) both coupled to a potentiometer (Orion Research, Model EA 940). F standards (0.00475 to 0.19 µgF) were prepared in triplicate and diffused in the same manner as the samples. In addition, nondiffused standards were prepared to have exactly the same F concentrations as the diffused standards. Comparison of the mV readings demonstrated that the F in the diffused standards had been completely trapped and analyzed (recovery>95%). The mV potentials were converted to µg F using a standard curve with a coefficient correlation of r≥0.99.
Sample Preparation for 2DE
Kidney samples were homogenized using mortar and pestle in liquid nitrogen. Denaturation buffer (7 M urea, 2 M de thioureia, 4% CHAPS, 1% DTT and 0.5% IPG pH 3–10, GE Healthcare, Uppsala, Sweden) was added. After 1 h vortexing at 4°C, samples were centrifuged at 25000×g for 30 min at 4°C for supernatants collection. The proteins were precipitated by using the kit PlusOne 2D Cleanup (GE Healthcare, Uppsala, Sweden), as recommended by the manufacturer. The pellets were resuspended in rehydration buffer (8 M urea, 0.5% CHAPS, 10% glycerol, 0.5% IPG buffer pH 3–10, 7 mg/2.5 mL DTT, 0.002% bromophenol blue). Protein concentration was measured in each sample by Bradford protein assay. After quantification, 1000 µg of kidney proteins from each animal of the same group and strain were combined to constitute a pool [16] that was submitted to proteomic analysis in triplicate, as described below.
2-DE Separation
Renal proteins (1000 µg) were taken from each pooled sample and mixed in rehydration buffer to a volume of 400 µL which was then loaded onto 24-cm IPG strips (linear pH 3–10). Rehydration and first-dimensional IEF were performed on IPGphor IEF system at 20°C with the following parameters: 50V for 12 h, 500V for 1 h, 1000V gradient for 1 h, then 10000 V for a total 40,000 V. Ettan DALTsix (GE Healthcare) (GE Healthcare, Uppsala, Sweden) with homemade12.5% acrylamide gels was used for the second dimension separation. Electrophoresis was performed at 15 mA/gel (80V) for 1 h and at 60 mA/gel (500V) until bromophenol blue line had reached the bottom of the gels. The resolved protein spots were stained with Colloidal Coomassie Brilliant Blue G-250 [17]. Gels were scanned with an Imagemaster scanner, and all images were analyzed by ImageMASTER 2D Platinum 7.0 software (GE Healthcare, Uppsala, Sweden). Parameters used for spot detection were minimal area = 5 pixels; smooth factor = 4; and saliency = 100. The gel chosen as the reference had the highest number of spots. The reference gel was then used for matching of corresponding protein spots between gels. Following average mode of background subtraction, individual spot intensity volume was normalized with total intensity volume (summation of the intensity volumes obtained from all spots in the same 2-DE gel). The normalized intensity volume values of individual protein spots were then used to determine differential protein expression between control and experimental groups. 2D spots that exhibited a twofold or more decrease or increase were tested for statistical significance. Analysis of 2D-gel variability among the replicates of each experimental condition was taken by using the relative volume (% vol). The correlation coefficients among the triplicates are shown to vary from 0.9385 to 0.9821 (Figure S4 - Supplementary information).
LC-MS/MS Analysis
Protein spots of interest were excised from the gel and destained three times with 25 mM Ambic/Acetonitrile (50∶50 v/v) for 30 min. The destained gel pieces were dehydrated twice with acetonitrile (ACN) for 10 min and dried in a vacuum concentrator (Eppendorf, Hamburg, Germany). The dried gel pieces were rehydrated with 20 mM DTT in 50 mM Ambic for 40 min at 56°C. Excess of reagent was removed and 55 mM iodoacetamide (IAA) in Ambic 50 mM was added for 30 min RT at dark. The remaining liquid was removed and washed out with 25 mM Ambic, followed by dehydration with ACN. After its removal, the gels were dried again. For digestion, dried gels were incubated with 10 ng/µL trypsin in 25 mM Ambic for 15 min (Trypsin Gold Mass spectrometry, Promega, Madison, USA). Peptides were sequentially extracted from the gels initially in 50% ACN (v/v) with 5% formic acid for 14 h at 37°C, then in 50% ACN (v/v) with 1% formic acid for 15 min, followed by 60% methanol (v/v) with 1% formic acid for 15 min and twice with 100% ACN at 45°C under sonication (40kHz/30W, Branson, Danbury, USA). Extracts were dried using a vacuum concentrator (Eppendorf, Hamburg, Germany) and kept at -20°C. Prior to MS identification, dried peptides were dissolved in 12 µL 0.1% formic acid. The peptides were identified and quantified by LC-ESI-Q-TOF MS (Liquid Chromatography Electrospray Ionization Quadrupole Time of Flight Mass Spectrometry) (Waters, Mildord, USA). MassLynx 4.1 SCN662 software (Waters, Mildord, USA) was used to submit the combined MS and MS/MS data to MASCOT database search engine (http://www.matrixscience.com) (version 2007.12.04) based on IPI (International Protein Index) protein database restricted to taxonomies Mus musculus (Mouse). The search was limited with a mass tolerance of 100 ppm and only one missed cleavage per peptide was allowed. For modification of peptides, cysteine carbamido-methylation (fixed) and methionine oxidation (variable) were considered. Significant matching protein required score of >60. Accuracy between the theoretical and experimental obtained mass and pI were also considered. When 2 or more proteins with high scores were identified in the same spot, they are excluded from analysis. Identified proteins were classified into 6 different categories according to their primary function [18].
Statistical Analysis
For kidney F concentration, the software GraphPad InStat version 4.0 for Windows (GraphPad software Inc., La Jolla, USA) was used. Data were analysed by 2-way ANOVA and Bonferroni test for individual comparisons (p<0.05).
For proteomic data, statistical analysis was performed using ANOVA available at ImageMaster 2D Platinum 7.0 software (GE Healthcare, Uppsala, Sweden). Only proteins with significantly altered levels were excised for identification by LC-MS/MS (p<0.05).
Results
Renal F Concentration
Mean kidney F (±se) concentrations for A/J mice for control, 10 ppmF and 50 ppmF groups were: 0.126±0.008, 0.174±0.007 and 0.296±0.026 µg/g. The corresponding values for 129P3/J mice were 0.139±0.015, 0.163±0.010 and 0.198±0.046 µg/g, respectively. Two-way ANOVA revealed a significant difference among the treatments (F = 35.13, p<0.0001), but not between the strains (F = 0.099, p = 0.756) without significant interaction between these criteria (F = 0.124, p = 0.884). For both strains, significantly higher kidney F concentrations were found for the 50 ppmF group, when compared with control and 10 ppmF groups that did not significantly differ from each other.
Identification of Differentially Expressed Proteins
For the statistical analysis, comparisons were performed between the strains as follows: Control groups (A/J vs 129P3/J mice), 10 ppmF groups (A/J vs 129P3/J mice), and 50 ppmF groups (A/J vs 129P3/J mice). Tables 1–3 show the proteins that were differentially expressed (p<0.05) in each comparison. Representative 2D map of each comparison is also shown in the Supplementary Information (Figures S1-S3). Quantitative intensity analysis showed 26 changed spots between control groups (Table 1). Among them, 14 spots were up-modulated, while were 12 down-regulated in control 129P3/J mice, when compared to control A/J mice. In general, the kidney proteome dataset was found to be significantly related with several metabolic and cellular processes pathways. Most of the 14 proteins up-modulated in the kidney of 129P3/J mice are related with metabolism (57.2%), while 28.6% are involved in cell processes and the remainder in information pathways (7.1%) and transport (7.1%). A similar pattern was observed for the proteins that were down-regulated in kidney 129P3/J mice. The respective percentages were 50.0, 25.0, 16.7 and 8.3 (Table 1). From the differentially expressed proteins in control groups, 10 were exclusively expressed in this comparison whereas 2, 6, and 8 proteins were also present in either 10 ppmF or 50 ppmF or both F-treated groups, respectively (Figure 1).
Table 1. Expression of differentially significant kidney proteins between control A/J vs control 129P3/J mice.
| Spot n°. | Protein | aMw (kDa)/pI Expt. | bTheor. | cNumber of peptides/Score | dDifference(P value) | eUniprot ID | fBiological Process |
| 2 | Aconitate hydratase, mitochondrial | 91/4.71 | 82.5/7.4 | 6/99 | ↑129 (0.046) | Q99KI0 | Metabolism |
| 119/280 | ATP synthase subunit beta, mitochondrial | 33/5.155 | 51.8/5.0 | 16/1129 | ↑129(0.038) | P56480 | Metabolism |
| 151 | Pyruvate dehydrogenase E1 component subunit beta, mitochondrial | 36.5/5.1 | 35.8/5.4 | 8/434 | ↑129(0.011) | Q9D051 | Metabolism |
| 175 | Hydroxyacid oxidase 2 | 38.5/7.94 | 38.7/7.6 | 11/529 | ↑129(0.012) | Q9NYQ2 | Metabolism |
| 217/221 | Medium-chain specific acyl-CoA dehydrogenase, mitochondrial | 42.5/8.055 | 43.6/7.69 | 15/715 | ↑129(0.001) | P45952 | Metabolism |
| 269 | Homogentisate 1,2-dioxygenase | 50/7.2 | 50/6.85 | 6/105 | ↑129(0.011) | O09173 | Metabolism |
| 384 | Sarcosine dehydrogenase, mitochondrial | 95/6.14 | - | 18/583 | ↑129(0.041) | Q99LB7 | Metabolism |
| 116 | Beta-lactamase-like protein 2 | 32.5/8.885 | 32.8/5.9 | 14/198 | ↑129(0.043) | Q99KR3 | Metabolism |
| 7 | Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | 38.5/5.675 | 37.4/5.9 | 9/122 | ↑129 (0.018) | P62137 | Processes/Information pathways |
| 317/321 | Catalase | 58/7.37 | 59.7/7.7 | 6/103 | ↑129(0.032) | P24270 | Processes |
| 396 | Aldehyde dehydrogenase family 1 member L1 | 100/5.445 | 98.7/5.6 | 24/517 | ↑129(0.013) | Q8R0Y6 | Processes |
| 285 | Alpha-aminoadipic semialdehyde dehydrogenase | 52/6.22 | 55.9/6.0 | 13/374 | ↑129(0.001) | Q9DBF1 | Processes |
| 324/583 | 60 kDa heat shock protein, mitochondrial | 59.5/4.875 | 58/5.35 | 22/992 | ↑129(0.022) | P63038 | Information Pathways |
| 413 | Coatomer subunit delta | 63/6.2 | 57.2/5.9 | 7/206 | ↑129(0.009) | Q5XJY5 | Transport |
| 3 | ATP synthase subunit delta, mitochondrial | 15.5/2.82 | 51.7/5.0 | 4/188 | ↓129(0.022) | P56480 | Metabolism |
| 171 | Alcohol dehydrogenase [NADP+] | 38/7.51 | 36.5/6.9 | 17/775 | ↓129(0.049) | Q9JII6 | Metabolism |
| 189 | Sorbitol dehydrogenase | 40.5/6.885 | 38.2/6.6 | 13/853 | ↓129(0.041) | Q64442 | Metabolism |
| 209 | Isovaleryl-CoA dehydrogenase, mitochondrial | 42.5/6.6 | 43/6.3 | 12/535 | ↓129(0.000) | Q9JHI5 | Metabolism |
| 233 | Ornithine aminotransferase, mitochondrial | 45/5.72 | 45.8/5.7 | 10/255 | ↓129(0.024) | P29758 | Metabolism |
| 237 | Creatine kinase U-type, mitochondrial | 45/8.05 | 43.2/7.3 | 9/133 | ↓129(0.033) | P30275 | Metabolism |
| 60 | Lactoylglutathione lyase | 27/4.48 | 20.7/5.25 | 37/365 | ↓129(0.001) | Q9CPU0 | Processes |
| 190 | Phosphotriesterase-related protein | 41/6.58 | 39.2/6.2 | 7/293 | ↓129(0.029) | Q60866 | Processes |
| 205 | 40S ribosomal protein SA | 42.5/3.96 | 32.7/4.8 | 7/825 | ↓129(0.044) | P14206 | Processes |
| 99 | Proteasome subunit beta type-7 | 30/5.885 | 25.3/5.8 | 7/85 | ↓129(0.041) | P70195 | Information pathways |
| 385/386/520 | Meprin A subunit alpha | 93/5.43 | 77.2/5.9 | 11/187 | ↓129(0.020) | P28825 | Information pathways |
| 355 | Serum albumin | 69.5/5.33 | 65.9/5.53 | 15/635 | ↓129(0.022) | P07724 | Transport |
Experimental molecular weight (kDa)/pI of protein spot in the gel (Mean of min. and max.) based on the coordinates of landmark proteins. bTheoretical molecular weight (kDa)/pI of theoretical protein. cNumber of peptides identified and score. dDifferences in expression in relation to 129P3/J mice (↓ down-modulation; ↑ up-modulation); Individual P value after ANOVA. eIdentification is based on protein ID from IPI (international protein index) protein database (http://www.uniprot.org/). fCategory of protein based on its primary biological function according to Rison (2000) [18].
Table 3. Expression of differentially significant kidney proteins between control A/J vs control 129P3/J mice.
| Spot n°. | Protein | aMw (kDa)/pI Expt. | bTheor. | cNumber of peptides/Score | dDifference (P value) | eUniprot ID | fBiological Process |
| 2 | Aconitate hydratase, mitochondrial | 90/4.77 | 82.5/7.4 | 6/99 | ↑129(0.029) | Q99KI0 | Metabolism |
| 35 | ATP synthase subunit d, mitochondrial | 24/5.16 | 18.6/5.53 | 8/402 | ↑129(0.036) | Q9DCX2 | Metabolism |
| 116 | Beta-lactamase-like protein 2 | 32.5/6.04 | 32.7/5.9 | 7/198 | ↑129(0.045) | Q99KR3 | Metabolism |
| 133 | Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial | 33.5/9.28 | 32995.07/8.3 | 8/255 | ↑129(0.035) | Q61425 | Metabolism |
| 175 | Hydroxyacid oxidase 2 | 37/8.1 | 38.7/7.6 | 11/529 | ↑129(0.003) | Q9NYQ2 | Metabolism |
| 184 | Sorbitol dehydrogenase | 40.5/6.805 | 38.2/6.56 | 10/586 | ↑129(0.003) | Q64442 | Metabolism |
| 210 | Aminoacylase-1 | 42.5/5.92 | 45.8/5.9 | 18/831 | ↑129(0.006) | Q99JW2 | Metabolism |
| 217/221 | Medium-chain specific acyl-CoA dehydrogenase, mitochondrial | 41.5/8.22 | 43.6/7.69 | 15/715 | ↑129(0.009) | P45952 | Metabolism |
| 269 | Homogentisate 1,2-dioxygenase | 49/7.41 | 50/6.85 | 6/105 | ↑129(0.002) | O09173 | Metabolism |
| 280/119 | ATP synthase subunit beta, mitochondrial | 51/4.345 | 51.7/5.0 | 16/1129 | ↑129(0.001) | P56480 | Metabolism |
| 360 | Propionyl-CoA carboxylase alpha chain, mitochondrial | 77/6.07 | 74.4/6.0 | 13/315 | ↑129(0.033) | Q91ZA3 | Metabolism |
| 378 | Acylamino-acid-releasing enzyme | 86/5.075 | 81.5/5.3 | 10/256 | ↑129(0.001) | Q8R146 | Metabolism |
| 491 | Argininosuccinate synthase | 44/9.41 | 46.6/8.4 | 14/321 | ↑129(0.001) | P16460 | Metabolism |
| 142/146 | Aspartoacylase-2 | 36.5/4.91 | 35.3/5.3 | 9/488 | ↑129(0.003) | Q91XE4 | Metabolism |
| 285 | Alpha-aminoadipic semialdehyde dehydrogenase | 53/6.355 | 55.9/6.0 | 13/374 | ↑129(0.024) | Q9DBF1 | Processes |
| 321 | Catalase | 56/8.055 | 59.7/7.7 | 6/103 | ↑129(0.008) | P24270 | Processes |
| 298 | ATP synthase subunit alpha, mitochondrial | 51/8.62 | 55.3/8.28 | 15/552 | ↑129(0.026) | Q03265 | Information Pathways |
| 372 | Radixin | 84/6.305 | 68.5/5.91 | 9/125 | ↑129(0.015) | P26043 | Structure |
| 90 | Phosphoglycerate mutase 1 | 28.5/7.235 | 28.7/6.75 | 23/324 | ↓129(0.003) | Q9DBJ1 | Metabolism |
| 157 | L-lactate dehydrogenase B chain | 37.5/5.49 | 36.4/5.7 | 10/705 | ↓129(0.009) | P16125 | Metabolism |
| 209 | Isovaleryl-CoA dehydrogenase, mitochondrial | 43/6.76 | 43/6.3 | 12/535 | ↓129(0.000) | Q9JHI5 | Metabolism |
| 233 | Ornithine aminotransferase, mitochondrial | 46/5.855 | 45.8/5.7 | 10/255 | ↓129(0.006) | P29758 | Metabolism |
| 244 | Glycine amidinotransferase, mitochondrial | 45/7.07 | 44.2/6.4 | 15/234 | ↓129(0.003) | Q9D964 | Metabolism |
| 518 | Nucleoside diphosphate-linked moiety X motif 19, mitochondrial | 41.5/5.99 | 39.3/6.0 | 7/245 | ↓129(0.018) | P11930 | Metabolism |
| 60 | Lactoylglutathione lyase | 27/4.525 | 20.7/5.25 | 10/365 | ↓129(0.003) | Q9CPU0 | Processes |
| 190 | Phosphotriesterase-related protein | 40.5/6.74 | 39.2/6.2 | 7/293 | ↓129(0.044) | Q60866 | Processes |
| 512 | Na(+)/H(+) exchange regulatory cofactor NHE-RF3 | 70/4.665 | 56.5/5.3 | 13/403 | ↓129(0.009) | Q9JIL4 | Processes |
| 267 | Actin-related protein 3 | 49.5/5.44 | 47.2/5.6 | 13/554 | ↓129(0.015) | Q99JY9 | Structure |
| 355 | Serum albumin | 70/5.42 | 65.9/5.53 | 15/635 | ↓129(0.026) | P07724 | Transport |
| 376 | Serotransferrin | 82/7.26 | 74.9/6.8 | 12/326 | ↓129(0.019) | Q921I1 | Transport |
| 385/386/520 | Meprin A subunit alpha | 96.5/5.545 | 77.2/5.9 | 11/187 | ↓129(0.014) | P28825 | Information pathways |
Experimental molecular weight (kDa)/pI of protein spot in the gel (Mean of min. and max.) based on the coordinates of landmark proteins. bTheoretical molecular weight (kDa)/pI of theoretical protein. cNumber of peptides identified and score. dDifferences in expression in relation to 129P3/J mice (↓ down-modulation; ↑ up-modulation); individual P value after ANOVA. eIdentification is based on protein ID from IPI (international protein index) protein database (http://www.uniprot.org/). fCategory of protein based on its primary biological function according to Rison (2000) [18].
Figure 1. Venn diagram showing distribution of total kidney proteins identified with differences in expression from the 2D-PAGE and LC-MS/MS-based proteome.
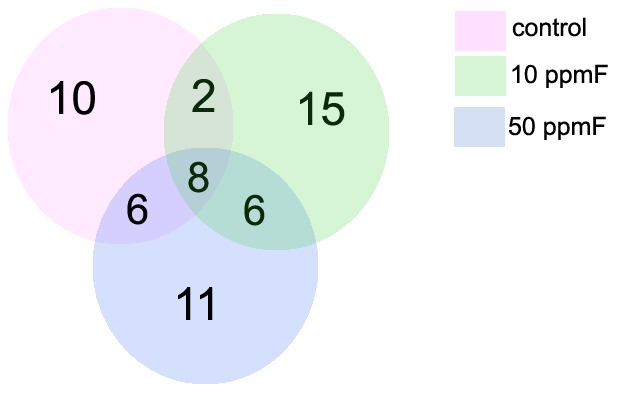
The numbers indicate the total protein identified from each comparison (control, 10 and 50 ppmF A/J and 129P3/J) and the number of proteins commonly identified between them.
For the comparison between the A/J and 129P3/J mice treated with 10 ppmF, 14 proteins were increased and 17 diminished in kidney of 129P3/J. Among the increased proteins, 64.3% are related with metabolism, while 35.7% are associated with cell processes. Most of the decreased proteins are also related to metabolism (41.1%), followed by information pathways (23.6%), cell processes (17.6%), transport (11.8%) and structure (5.9%) (Table 2). From the differentially expressed proteins in 10 ppmF group, 15 were exclusively expressed in this comparison whereas 2, 6 and 8 proteins were also present in either control or 50 ppmF or in both groups, respectively (Figure 1).
Table 2. Expression of differentially significant kidney proteins between 10 ppmF A/J vs 10 ppmF 129P3/J mice.
| Spot n°. | Protein | aMw (kDa)/pI Expt. | bTheor. | cNumber of peptides/Score | dDifference (P value) | eUniprot ID | fBiological Process | ||
| 3 | ATP synthase subunit delta, mitochondrial | 15.5/2.82 | 51.7/5.0 | 4/188 | ↑129(0.040) | P56480 | Metabolism | ||
| 182 | Aldose 1-epimerase | 40/6.625 | 37.8/6.3 | 4/146 | ↑129(0.016) | Q8K157 | Metabolism | ||
| 210 | Aminoacylase-1 | 42.5/5.94 | 45.8/5.9 | 18/831 | ↑129(0.013) | Q99JW2 | Metabolism | ||
| 221/217 | Medium-chain specific acyl-CoA dehydrogenase, mitochondrial | 43/8.25 | 43.6/7.7 | 15/541 | ↑129(0.001) | P45952 | Metabolism | ||
| 223 | Adenosylhomocysteinase | 42.5/6.6 | 43/6.3 | 15/559 | ↑129(0.047) | P50247 | Metabolism | ||
| 261 | Fumarate hydratase, mitochondrial | 45/5.72 | 50/7.9 | 8/142 | ↑129(0.015) | P97807 | Metabolism | ||
| 462 | Short/branched chain specific acyl-CoA dehydrogenase, mitochondrial | 42/6.11 | 44/6.0 | 2/66 | ↑129(0.047) | Q9DBL1 | Metabolism | ||
| 467 | 3-hydroxyanthranilate 3,4-dioxygenase | 34/6.415 | 32.8/6.1 | 8/181 | ↑129(0.039) | Q78JT3 | Metabolism | ||
| 489 | Citrate synthase, mitochondrial | 43/9.1 | 49/8.2 | 3/107 | ↑129(0.035) | Q9CZU6 | Metabolism | ||
| 142/146 | Aspartoacylase-2 | 35.5/4.915 | 35.3/5.3 | 9/488 | ↑129(0.003) | Q91XE4 | Processes | ||
| 7 | Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | 38.5/5.675 | 37.4/5.9 | 9/122 | ↑129 (0.018) | P62137 | Processes/Information pathways | ||
| 285 | Alpha-aminoadipic semialdehyde dehydrogenase | 52.5/6.34 | 55.9/6.0 | 13/374 | ↑129(0.005) | Q9DBF1 | Processes | ||
| 317/321 | Catalase | 58/7.445 | 59.7/7.7 | 6/103 | ↑129(0.032) | P24270 | Processes | ||
| 289 | Selenium-binding protein 1 | 53.5/5.68 | 52.5/5.9 | 13/374 | ↑129(0.037) | P17563 | Processes | ||
| 5 | Ester hydrolase C11orf54 homolog | 37/5.905 | 51.7/5.0 | 4/188 | ↓129(0.022) | Q91V76 | Metabolism | ||
| 87 | Enoyl-CoA hydratase, mitochondrial | 28/8.615 | 28.5/7.78 | 13/353 | ↓129(0.017) | Q8BH95 | Metabolism | ||
| 157 | L-lactate dehydrogenase B chain | 37/5.505 | 36.4/5.7 | 10/705 | ↓129(0.002) | P16125 | Metabolism | ||
| 185 | 3′(2′),5′-bisphosphate nucleotidase 1 | 41/5.535 | 33.1/5.54 | 7/216 | ↓129(0.016) | Q9Z0S1 | Metabolism | ||
| 209 | Isovaleryl-CoA dehydrogenase, mitochondrial | 43/6.715 | 43/6.3 | 12/535 | ↓129(0.021) | Q9JHI5 | Metabolism | ||
| 233 | Ornithine aminotransferase, mitochondrial | 45/5.72 | 45.8/5.7 | 10/255 | ↓129(0.024) | P29758 | Metabolism | ||
| 518 | Nucleoside diphosphate-linked moiety X motif 19, mitochondrial | 42/6.015 | 39.3/6.0 | 7/245 | ↓129(0.041) | P11930 | Metabolism | ||
| 60 | Lactoylglutathione lyase | 27/4.48 | 20.7/5.25 | 37/365 | ↓129(0.001) | Q9CPU0 | Processes | ||
| 85 | Rho GDP-dissociation inhibitor 1 | 27/4.55 | 23.3/5.1 | ↓129(0.005) | Q99PT1 | Processes | |||
| 512 | Na(+)/H(+) exchange regulatory cofactor NHE-RF3 | 69.5/4.645 | 56.5/5.3 | 13/403 | ↓129(0.041) | Q9JIL4 | Processes | ||
| 75 | GTP:AMP phosphotransferase, mitochondrial | 26.5/10.135 | 25.3/8.88 | 7/85 | ↓129(0.050) | Q9WTP7 | Information pathways | ||
| 320 | Protein disulfide-isomerase A3 | 59.5/5.73 | 54.3/5.7 | 16/362 | ↓129(0.043) | P27773 | Information pathways | ||
| 385/386/520 | Meprin A subunit alpha | 89.5/5.5 | 77.2/5.9 | 11/187 | ↓129(0.002) | P28825 | Information pathways | ||
| 400 | Heat shock protein 90, beta (Grp94), member 1 | 99.5/3.99 | 92.5/4.74 | 24/515 | ↓129(0.011) | Q91V38 | Information Pathways | ||
| 267 | Actin-related protein 3 | 49.5/5.445 | 47.2/5.6 | 13/554 | ↓129(0.016) | Q99JY9 | Structure | ||
| 311 | Vitamin D-binding protein | 57.5/4.76 | 51.9/5.2 | 9/109 | ↓129(0.028) | P21614 | Transport | ||
| 355 | Serum albumin | 69.5/5.42 | 65.9/5.53 | 15/635 | ↓129(0.020) | P07724 | Transport | ||
Experimental molecular weight (kDa)/pI of protein spot in the gel (Mean of min. and max.) based on the coordinates of landmark proteins. bTheoretical molecular weight (kDa)/pI of theoretical protein. cNumber of peptides identified and score. dDifferences in expression in relation to 129P3/J mice (↓ down-modulation; ↑ up-modulation); individual P value after ANOVA. eIdentification is based on protein ID from IPI (international protein index) protein database (http://www.uniprot.org/). fCategory of protein based on its primary biological function according to Rison (2000) [18].
Regarding the comparison between the groups treated with 50 ppmF, 18 proteins were significantly up-regulated and 13 down-modulated in kidney of 129P3/J mice when compared with A/J mice. Fourteen of eighteen enhanced proteins are associated with metabolism (77.8%), followed by processes (11.1%), information pathways (5.6%) and processes pathways (5.6%). Among the down-modulated proteins, most are also related to metabolism (46.2%), followed by cell processes (23.0%), transport (15.4%), information pathways (7.7%) and structure (7.7%) (Table 3). Among the differentially expressed proteins in kidney of animals treated with 50 ppmF, 11 proteins are exclusively expressed in this group while 6, 6 and 8 proteins are also present in either control or 10 ppmF or both groups, respectively (Figure 1).
Among the 8 proteins differentially expressed between the mice strains, regardless of the treatment with F, catalase, medium-chain specific acyl-CoA dehydrogenase and alpha-aminoadipic semialdehyde dehydrogenase were up-regulated, while isovaleryl-CoA dehydrogenase, ornithine aminotransferase, lactoylglutathione lyase, meprin A subunit alpha and albumin were down-regulated in the kidney of 129P3/J mice.
Identification of Unique Proteins
A/J and 129P3/J mice exhibited 11 and 3 exclusive proteins, respectively. From these, 9 (64.3%) are related to metabolism, followed by cell processes (4 or 28.6%) and information pathways (1 or 7.1%). This profile was not altered upon exposure to F (Table 4).
Table 4. Expression of unique kidney proteins between A/J and 129P3/J mice.
| Spot n°. | Protein | aMw (kDa)/pI Expt. | bTheor. | cNumber of peptides/Score | dUniprot ID | eBiological Process |
| Kidney A/J mice | ||||||
| 563 | Transaldolase | 37.5/5.975 | 37.4/6.6 | 5/151 | Q93092 | Metabolism |
| 564 | Isobutyryl-CoA dehydrogenase, mitochondrial | 40.5/6.905 | 42.8/7.2 | 6/202 | Q9D7B6 | Metabolism |
| 565 | Short-chain specific acyl-CoA dehydrogenase, mitochondrial | 41/6.805 | 42.2/6.3 | 9/396 | Q07417 | Metabolism |
| 567 | Cystathionine gamma-lyase | 43.5/7.99 | 43.6/7.6 | 5/163 | Q8VCN5 | Metabolism |
| 568 | Hydroxymethylglutaryl-CoA synthase, cytoplasmic | 56/5.7 | 57.6/5.65 | 2/62 | Q8JZK9 | Metabolism |
| 586 | Probable D-lactate dehydrogenase, mitochondrial | 37.5/8.23 | 19.1/6.2 | 2/84 | Q7TNG8 | Metabolism |
| 562 | Thiomorpholine-carboxylate dehydrogenase | 37.5/5.09 | 33.5/5.44 | 8/395 | O54983 | Metabolism |
| 560 | Phenazine biosynthesis-like domain-containing protein 2 | 34/4.795 | 32/5.2 | 4/130 | Q9CXN7 | Process |
| 561 | Biliverdin reductase A | 37.5/6.755 | 33.3/6.5 | 3/86 | Q9CY64 | Process |
| 576 | Sorting nexin-5 | 49.5/6.165 | 46.7/6.2 | 4/97 | Q9D8U8 | Process |
| 558 | Serum amyloid P-component | 30/5.3 | 23.9/6.4 | 3/52 | P12246 | Information Pathways |
| Kidney 129P3/J mice | ||||||
| 534/535 | Peroxisomal acyl-coenzyme A oxidase 1 | 20.5/9.99 | 74.6/8.6 | 7/227 | Q9R0H0 | Metabolism |
| 552 | Nicotinate-nucleotide pyrophosphorylase [carboxylating] | 36/6.49 | 31.5/6.2 | 4/61 | Q91X91 | Metabolism |
| 539 | Arsenite methyltransferase | 43/5.38 | 41.8/5.6 | 8/217 | Q91WU5 | Process |
Experimental molecular weight (kDa)/pI of protein spot in the gel (Mean of min. and max.) based on the coordinates of landmark proteins. bTheoretical molecular weight (kDa)/pI of theoretical protein. cNumber of peptides identified and score. dIdentification is based on protein ID from IPI (international protein index) protein database (http://www.uniprot.org/). eCategory of protein based on its primary biological function according to Rison (2000) [18].
Discussion
In the present study, we identified proteins potentially involved in renal F metabolism that are either exclusively or differentially expressed in A/J and 129P3/J mice. This highlights the molecular mechanisms underlying the differential metabolic handling of F by these two strains of mice. Exclusive proteins expressed in A/J or 129P3/J mice exhibited the same profile, regardless exposure to F. This suggests that the genetic background per se accounts for such differences between these two strains of mice. We have focused on identified proteins that may be associated with metabolic handling of F and water and renal functions. Unique metabolic proteins in kidney from A/J mice are involved in carbohydrate (probable D-lactate dehydrogenase), carbon (transaldolase), aminoacid (isobutyryl-CoA dehydrogenase, hydroxymethylglutaryl-CoA synthase (HMGCS2), cystathionine gamma-lyase (CSE), thiomorpholine-carboxylate dehydrogenase), and fatty acid [short-chain specific acyl-CoA dehydrogenase (SCAD)] metabolism. CSE is an enzyme that breaks down cystathione into cysteine and α-ketobutyrate and catalyses elimination of L-homoserine, L-cystine and L-cysteine producing ammonia and hydrogen sulfide (H2S) [19], [20]. Considering that the urine is the main excretion route for ingested F [1], the presence of CSE in A/J mice might contribute to increase the urinary pH, which can help to explain the higher urinary F excretion observed for this strain, when compared with the 129P3/J mice [10]. The pH-dependency found for urinary F excretion is due to the fact that F can cross cell membranes in general, including the walls of the renal tubules in the form of HF. Thus, the higher the urinary pH, the higher the concentration of F− that remains in the tubule to be excreted in urine [12]. Recently, it was shown that the expression of HMGCS2 was increased fourfold in diabetic kidneys, which leads to increased renal ketogenesis and plays an important role in the pathogenesis of diabetic nephropathy in type 2 diabetes [21]. In our data, the presence of HMGCS2 in kidney of A/J mice might turn these animals more prone to nephropathy, which could impair F reabsorption in kidneys [10].
Besides presenting unique proteins involved in metabolism, A/J mice also expresses exclusive proteins involved in cell processes (phenazine biosynthesis-like domain containing protein 2 (PBLD), biliverdin reductase A (BVR) and sorting nexin-5) and information pathways [serum amyloid P-component (SAP)]. Among these, SAP constitutes amyloid deposits characterized by the ordered aggregation of normal globular proteins and peptides into insoluble fibrils, which disrupt tissue architecture and are associated with cell death [22]. The presence of SAP only in A/J mice might increase the probability of kidney damage that could account for their diminished capacity to reabsorb various solutes including F, helping to explain the higher urinary F excretion seen in this strain previously [10].
From those proteins found exclusively in kidneys of 129P3/J mice, the peroxisomal acyl-coenzymeA oxidase 1 (AOX), a fatty acid metabolic protein, is shown to be expressed in proximal tubules and enhancement of its activity is associated with the preservation of kidney function during ischemia [23]. Another exclusive protein called arsenite-methyltransferase, presented only in 129P3/J mice, is a detoxifying protein involved in the arsenic biotransformation and elimination in proximal tubule epithelial cells [24]. The presence of these proteins in 129P3/J but not in A/J mice suggest that the former might have a higher capacity to reduce renal damage caused by different hostile conditions, such as exposure to F. Thus, the 129P3/J mice would be able to maintain F reabsorption in kidneys even under exposure to high F doses [10].
As mentioned above, F exposure did not alter the profile of unique proteins in either strain of mice. However, among the proteins differentially expressed in the comparisons between the two strains, only 8 were present in the control, 10 and 50 ppmF groups (catalase, medium-chain specific acyl-CoA dehydrogenase and alpha-aminoadipic semialdehyde dehydrogenase (α-AASA), isovaleryl-CoA dehydrogenase, ornithine aminotransferase, lactoylglutathione lyase, meprin A subunit alpha and albumin). Some of these significantly altered proteins with potential roles to contribute for the intrinsic differences in F and water handling by A/J and 129P3/J mice are highlighted below. Meprin A, an information pathways related protein, is an enzyme that hydrolyzes protein and peptide substrates including components of the extracellular matrix [25]. It is highly expressed at the brush border membrane of proximal tubule cells of the kidney. Inbred strains of mice subjected to ischemia reperfusion that express low levels of meprin A in kidney have markedly less kidney damage [26]. Our data show that meprin A is consistently reduced in 129P3/J kidney in all experimental conditions. This suggests that this protein could act in concert with SAP to decrease renal damage caused by F in 129P3/J mice. Among the proteins related to cellular processes, it is important to highlight α-AASA dehydrogenase and catalase. α-AASA dehydrogenase metabolyzes irreversibly betaine aldehyde to betaine, which is the most effective osmoprotectant accumulated by eukariotic organisms to cope with osmotic stress [27]. This enzyme was increased in the 129P3/J kidney, regardless F exposure. This can explain the lower volume of water consistently ingested by the 129P3/J mice throughout the study, which led us to adjust water F concentrations throughout the experiment in order that both strains had the same amount of F intake from the water [10]. The increased expression of the antioxidant enzyme catalase might indicate a higher capacity of the 129P3/J mice to deal with oxidative stress [28].
Two and 6 proteins with differential expression between the two strains in the control group were also identified upon exposure to 10 and 50 ppmF, respectively. Low F level increased the expression of serine/threonine-protein phosphatase PP1 and ATP synthase subunit delta. High F level kidney up-expressed aconitate hydratase, ATP synthase subunit beta, hydroxyacid oxidase 2, homogentisate 1,2-dioxygenase and beta-lactamase-like protein 2 and down-expressed phosphotriesterase-related protein. Besides, 6 proteins presented altered expression only in F-treated groups. Aminoacylase-1 and aspartoacylase-2 were increased, whereas L-lactate dehydrogenase B chain, nucleoside diphosphate-linked moiety X motif 19, Na(+)/H(+) exchange regulatory cofactor NHE-RF3 (PDZK1) and actin-related protein 3 were diminished in 129P3/J kidney. These proteins may act as molecular targets for the differential F metabolism between these strains induced by the treatment. Protein phosphatase 1 (PP1) is a serine/threonine protein phosphatase involved in diverse cellular processes, such as transcription, replication, pre-mRNA splicing, protein synthesis, carbohydrate metabolism, neuronal signaling, cell survival, and cell cycle progression [29], [30]. Phosphatases typically function antagonistically with kinases to achieve fine control over the phosphorylation state of proteins. Phosphatases are widely expressed enzymes that mediate the functional regulation of many proteins, including some renal channels and transporters such as the inwardly rectifying K+ channel, Na+-K+-Cl− cotransporter (NKCC1), CFTR, epithelial Na+ channel (ENaC), aquaporin-2 (AQP2) and Na+/H+ exchanger 3 (NHE3) [30], [31], [32], [33], [34], [35], [36]. In general, these ions and water channels are responsible to maintain the urine normal volume and acid-base status under varying physiological conditions and are under direct or indirect phosphorylation state control [37], [38]. It was shown that the prevention of phosphorylation of specific sites in AQP2 increases localization of AQP2 vesicles to the apical plasma membrane leading to water reabsorption and urine concentration [38]. Thus, we could speculate that the fact that 129P3/J mice excrete less urine could be possibly explained by the PP1-mediated enhancement of AQP2 vesicles trafficking, which should be confirmed in future studies.
PDZK1 is a scaffold protein that connects plasma membrane proteins and regulatory components, regulating their surface expression in epithelial cells apical domains. Within the kidney, PDZK1 is localized exclusively in the brush border of the proximal tubule and interacts with several renal proteins including NHE3, a Na-H exchanger, and CFEX, a Cl-anion exchanger [39]. These exchanger transporters play principal roles in the reabsorption of Na+ and Cl− in the proximal tubule of the mammalian kidney. Besides regulating reabsorption of filtered solutes, PDZK1 also plays a direct and essential role in maintaining normal brush border expression and function of CFEX in the proximal tubule in vivo [39]. The diminished expression of PDZK1 in kidney of 129P3/J mice may indicate an undisclosed impaired ability of ion reabsorption by this strain, which is consistent with the lower volume of urine excreted by these mice.
We conclude that the renal proteome indicates several specific target proteins, both strain and F-induced, which possibly regulate the water and F metabolism in kidney of mice with distinct susceptibilities to F. In addition, although we did not focus in the correlation between target kidney proteins and DF, we found that some of those changed proteins are also codified by chromosomes 2 (13 proteins: sarcosine dehydrogenase, catalase, sorbitol dehydrogenase, isovaleryl-CoA dehydrogenase, creatine kinase U-type, phosphotriesterase-related protein, proteasome subunit beta type-7, adenoxylhomocysteinase, protein disulfide-isomerase A3, argininosuccinate synthase, glycine amidinotransferase, biliverdin reductase A and sorting nexin-5) and 11 (3 proteins: peroxisomal acyl-coenzyme A oxidase 1, ATP synthase subunit d and Rho GDP-dissociation inhibitor 1), previously characterized to determine susceptibility and resistance to DF in A/J and 129P3/J mice, respectively [40], [41]. This correlation may provide a database for future hypothesis-driven researches.
Supporting Information
2D gel analysis of renal proteome. Representative 2D maps of control kidneys. Selected spots in green represent those with differential expression in the comparison between control A/J (A) vs control 129P3/J mice (B). In Figure B, spot identification numbers in boundaries or not represents increases or decreases in protein expression when compared to A/J, respectively (Figure A). Dashed lines represent unique spots in the AJ group (A) and 129P3/J group (B), regardless exposure to F.
(TIF)
2D gel analysis of renal proteome. Representative 2D maps of 10 ppmF treated-groups. Selected spots in green represent those with differential expression in the comparison between 10 ppmF treated- A/J (A) vs 10 ppmF treated- 129P3/J mice (B). In Figure B, spot identification numbers in boundaries or not represents increases or decreases in protein expression when compared to A/J, respectively (Figure A).
(TIF)
2D gel analysis of renal proteome. Representative 2D maps of 50 ppmF treated-groups. Selected spots in green represent those with differential expression in the comparison between 50 ppmF treated- A/J (A) vs 50 ppmF treated- 129P3/J mice (B). In Figure B, spot identification numbers in boundaries or not represents increases or decreases in protein expression when compared to A/J, respectively (Figure A).
(TIF)
2D gel variability analysis. Scatter plot of binary comparisons among the ratios of relative spots volumes detected in the representative gel (replicate 1) and the respective replicates (replicates 2 and 3). (A) Control A/J mice. (B) 10 ppmF treated-A/J mice. (C) 50 ppmF treated-A/J. (D) Control 129P3/J mice. (E) 10 ppmF treated-129P3/J mice. (F) 50 ppmF treated-129P3/J.
(TIF)
Acknowledgments
The authors thank the Laboratório Max Feffer de Genética de Plantas, Departamento de Genética da Escola Superior de Agricultura 'Luiz de Queiroz' ESALQ/USP, Piracicaba, São Paulo, Brazil and Adriana Franco Paes Leme from Associação Brasileira de Tecnologia de Luz Sincrotron, Campinas, São Paulo, Brazil for their scientific and technical assistances.
Funding Statement
This work was supported by São Paulo State Research Support Foundation (FAPESP) (grant 2005/04671-0) and CNPq (grant 471485/2007-1). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Buzalaf MA, Levy SM (2011) Fluoride intake of children: considerations for dental caries and dental fluorosis. In: Lussi, A.; Huysmans, M.C.D.N.J.M.; Weber, H.P, editors. Fluoride and the oral enviroment. Monographs in Oral Science. Basel: Karger. 1–19. [DOI] [PubMed]
- 2. Khan A, Moola MH, Cleaton-Jones P (2005) Global trends in dental fluorosis from 1980 to 2000: a systematic review. SADJ 60: 418–421. [PubMed] [Google Scholar]
- 3. Beltran-Aguilar ED, Barker LK, Canto MT, Dye BA, Gooch BF, et al. (2005) Surveillance for dental caries, dental sealants, tooth retention, edentulism, and enamel fluorosis–United States, 1988–1994 and 1999–2002. MMWR Surveill Summ 54: 1–43. [PubMed] [Google Scholar]
- 4. Bronckers AL, Lyaruu DM, DenBesten PK (2009) The impact of fluoride on ameloblasts and the mechanisms of enamel fluorosis. J Dent Res 88: 877–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Everett ET (2011) Fluoride's effects on the formation of teeth and bones, and the influence of genetics. J Dent Res 90: 552–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huang H, Ba Y, Cui L, Cheng X, Zhu J, et al. (2008) COL1A2 gene polymorphisms (Pvu II and Rsa I), serum calciotropic hormone levels, and dental fluorosis. Community Dent Oral Epidemiol 36: 517–522. [DOI] [PubMed] [Google Scholar]
- 7. Everett ET, McHenry MA, Reynolds N, Eggertsson H, Sullivan J, et al. (2002) Dental fluorosis: variability among different inbred mouse strains. J Dent Res 81: 794–798. [DOI] [PubMed] [Google Scholar]
- 8. Mousny M, Banse X, Wise L, Everett ET, Hancock R, et al. (2006) The genetic influence on bone susceptibility to fluoride. Bone 39: 1283–1289. [DOI] [PubMed] [Google Scholar]
- 9. Mousny M, Omelon S, Wise L, Everett ET, Dumitriu M, et al. (2008) Fluoride effects on bone formation and mineralization are influenced by genetics. Bone 43: 1067–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carvalho JG, Leite AL, Yan D, Everett ET, Whitford GM, et al. (2009) Influence of genetic background on fluoride metabolism in mice. J Dent Res 88: 1054–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buzalaf MAR, Whitford GM (2011) Fluoride metabolism. In: Lussi, A.; Huysmans, M.C.D.N.J.M.; Weber, H.P, editors. Fluoride and the oral environment Monographs in Oral Science. Basel: Karger. 20–36. [DOI] [PubMed]
- 12. Whitford GM, Pashley DH, Stringer GI (1976) Fluoride renal clearance: a pH-dependent event. Am J Physiol 230: 527–532. [DOI] [PubMed] [Google Scholar]
- 13. Schiffl HH, Binswanger U (1980) Human urinary fluoride excretion as influenced by renal functional impairment. Nephron 26: 69–72. [DOI] [PubMed] [Google Scholar]
- 14. Taves DR (1968) Determination of submicromolar concentrations of fluoride in biological samples. Talanta 15: 1015–1023. [DOI] [PubMed] [Google Scholar]
- 15. Whitford GM (1996) The metabolism and toxicity of fluoride. Monogr Oral Sci 16 Rev 2: 1–153. [PubMed] [Google Scholar]
- 16. Tilton RG, Haidacher SJ, Lejeune WS, Zhang X, Zhao Y, et al. (2007) Diabetes-induced changes in the renal cortical proteome assessed with two-dimensional gel electrophoresis and mass spectrometry. Proteomics 7: 1729–1742. [DOI] [PubMed] [Google Scholar]
- 17. Candiano G, Bruschi M, Musante L, Santucci L, Ghiggeri GM, et al. (2004) Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 25: 1327–1333. [DOI] [PubMed] [Google Scholar]
- 18. Rison SC, Hodgman TC, Thornton JM (2000) Comparison of functional annotation schemes for genomes. Funct Integr Genomics 1: 56–69. [DOI] [PubMed] [Google Scholar]
- 19. Levonen AL, Lapatto R, Saksela M, Raivio KO (2000) Human cystathionine gamma-lyase: developmental and in vitro expression of two isoforms. Biochem J 347 Pt 1: 291–295. [PMC free article] [PubMed] [Google Scholar]
- 20. Yang G, Wu L, Bryan S, Khaper N, Mani S, et al. (2010) Cystathionine gamma-lyase deficiency and overproliferation of smooth muscle cells. Cardiovasc Res 86: 487–495. [DOI] [PubMed] [Google Scholar]
- 21. Zhang D, Yang H, Kong X, Wang K, Mao X, et al. (2011) Proteomics analysis reveals diabetic kidney as a ketogenic organ in type 2 diabetes. Am J Physiol Endocrinol Metab 300: E287–295. [DOI] [PubMed] [Google Scholar]
- 22. Gillmore JD, Hawkins PN (1999) Amyloidosis and the respiratory tract. Thorax 54: 444–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Portilla D, Dai G, Peters JM, Gonzalez FJ, Crew MD, et al. (2000) Etomoxir-induced PPARalpha-modulated enzymes protect during acute renal failure. Am J Physiol Renal Physiol 278: F667–675. [DOI] [PubMed] [Google Scholar]
- 24. Peraza MA, Carter DE, Gandolfi AJ (2003) Toxicity and metabolism of subcytotoxic inorganic arsenic in human renal proximal tubule epithelial cells (HK-2). Cell Biol Toxicol 19: 253–264. [DOI] [PubMed] [Google Scholar]
- 25. Kronenberg D, Bruns BC, Moali C, Vadon-Le Goff S, Sterchi EE, et al. (2010) Processing of procollagen III by meprins: new players in extracellular matrix assembly? J Invest Dermatol 130: 2727–2735. [DOI] [PubMed] [Google Scholar]
- 26. Bond JS, Matters GL, Banerjee S, Dusheck RE (2005) Meprin metalloprotease expression and regulation in kidney, intestine, urinary tract infections and cancer. FEBS Lett 579: 3317–3322. [DOI] [PubMed] [Google Scholar]
- 27. Roberts MF (2000) Osmoadaptation and osmoregulation in archaea. Front Biosci 5: D796–812. [DOI] [PubMed] [Google Scholar]
- 28. Blaszczyk I, Grucka-Mamczar E, Kasperczyk S, Birkner E (2010) Influence of methionine upon the activity of antioxidative enzymes in the kidney of rats exposed to sodium fluoride. Biol Trace Elem Res 133: 60–70. [DOI] [PubMed] [Google Scholar]
- 29. Aggen JB, Nairn AC, Chamberlin R (2000) Regulation of protein phosphatase-1. Chem Biol 7: R13–23. [DOI] [PubMed] [Google Scholar]
- 30. Ceulemans H, Bollen M (2004) Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol Rev 84: 1–39. [DOI] [PubMed] [Google Scholar]
- 31. Becchetti A, Malik B, Yue G, Duchatelle P, Al-Khalili O, et al. (2002) Phosphatase inhibitors increase the open probability of ENaC in A6 cells. Am J Physiol Renal Physiol 283: F1030–1045. [DOI] [PubMed] [Google Scholar]
- 32. Darman RB, Flemmer A, Forbush B (2001) Modulation of ion transport by direct targeting of protein phosphatase type 1 to the Na-K-Cl cotransporter. J Biol Chem 276: 34359–34362. [DOI] [PubMed] [Google Scholar]
- 33. Li D, Aperia A, Celsi G, da Cruz e Silva EF, Greengard P, et al. (1995) Protein phosphatase-1 in the kidney: evidence for a role in the regulation of medullary Na(+)-K(+)-ATPase. Am J Physiol 269: F673–680. [DOI] [PubMed] [Google Scholar]
- 34. Mori Y, Kawasaki A, Takamaki A, Kitano I, Yoshida R, et al. (2001) Ca(2+)-dependent inhibition of inwardly rectifying K(+) channel in opossum kidney cells. Jpn J Physiol 51: 371–380. [DOI] [PubMed] [Google Scholar]
- 35. Valenti G, Procino G, Carmosino M, Frigeri A, Mannucci R, et al. (2000) The phosphatase inhibitor okadaic acid induces AQP2 translocation independently from AQP2 phosphorylation in renal collecting duct cells. J Cell Sci 113 (Pt 11): 1985–1992. [DOI] [PubMed] [Google Scholar]
- 36. Dynia DW, Steinmetz AG, Kocinsky HS (2010) NHE3 function and phosphorylation are regulated by a calyculin A-sensitive phosphatase. Am J Physiol Renal Physiol 298: F745–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moeller HB, Knepper MA, Fenton RA (2009) Serine 269 phosphorylated aquaporin-2 is targeted to the apical membrane of collecting duct principal cells. Kidney Int 75: 295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Moeller HB, Praetorius J, Rutzler MR, Fenton RA (2010) Phosphorylation of aquaporin-2 regulates its endocytosis and protein-protein interactions. Proc Natl Acad Sci U S A 107: 424–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thomson RB, Wang T, Thomson BR, Tarrats L, Girardi A, et al. (2005) Role of PDZK1 in membrane expression of renal brush border ion exchangers. Proc Natl Acad Sci U S A 102: 13331–13336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Everett ET, Yan D, Weaver M, Liu L, Foroud T, et al. (2009) Detection of dental fluorosis-associated quantitative trait Loci on mouse chromosomes 2 and 11. Cells Tissues Organs 189: 212–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Everett ET, Yin Z, Yan D, Zou F (2011) Fine mapping of dental fluorosis quantitative trait loci in mice. Eur J Oral Sci 119 Suppl 18–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
2D gel analysis of renal proteome. Representative 2D maps of control kidneys. Selected spots in green represent those with differential expression in the comparison between control A/J (A) vs control 129P3/J mice (B). In Figure B, spot identification numbers in boundaries or not represents increases or decreases in protein expression when compared to A/J, respectively (Figure A). Dashed lines represent unique spots in the AJ group (A) and 129P3/J group (B), regardless exposure to F.
(TIF)
2D gel analysis of renal proteome. Representative 2D maps of 10 ppmF treated-groups. Selected spots in green represent those with differential expression in the comparison between 10 ppmF treated- A/J (A) vs 10 ppmF treated- 129P3/J mice (B). In Figure B, spot identification numbers in boundaries or not represents increases or decreases in protein expression when compared to A/J, respectively (Figure A).
(TIF)
2D gel analysis of renal proteome. Representative 2D maps of 50 ppmF treated-groups. Selected spots in green represent those with differential expression in the comparison between 50 ppmF treated- A/J (A) vs 50 ppmF treated- 129P3/J mice (B). In Figure B, spot identification numbers in boundaries or not represents increases or decreases in protein expression when compared to A/J, respectively (Figure A).
(TIF)
2D gel variability analysis. Scatter plot of binary comparisons among the ratios of relative spots volumes detected in the representative gel (replicate 1) and the respective replicates (replicates 2 and 3). (A) Control A/J mice. (B) 10 ppmF treated-A/J mice. (C) 50 ppmF treated-A/J. (D) Control 129P3/J mice. (E) 10 ppmF treated-129P3/J mice. (F) 50 ppmF treated-129P3/J.
(TIF)


