Abstract
Using the data set of 180,000 expressed sequence tags (ESTs) of the blood fluke Schistosoma mansoni generated recently by our group, we identified three novel long-terminal-repeat (LTR)- and one novel non-LTR-expressed retrotransposon, named Saci-1, -2, and -3 and Perere, respectively. Full-length sequences were reconstructed from ESTs and have deduced open reading frames (ORFs) with several uncorrupted features, characterizing them as possible active retrotransposons of different known transposon families. Alignment of reconstructed sequences to available preliminary genome sequence data confirmed the overall structure of the transposons. The frequency of sequenced transposon transcripts in cercariae was 14% of all transcripts from that stage, twofold higher than that in schistosomula and three- to fourfold higher than that in adults, eggs, miracidia, and germ balls. We show by Southern blot analysis, by EST annotation and tallying, and by counting transposon tags from a Social Analysis of Gene Expression library, that the four novel retrotransposons exhibit a 10- to 30-fold lower copy number in the genome and a 4- to 200-fold-higher transcriptional rate per copy than the four previously described S. mansoni retrotransposons. Such differences lead us to hypothesize that there are two different populations of retrotransposons in S. mansoni genome, occupying different niches in its ecology. Examples of retrotransposon fragment inserts were found into the 5′ and 3′ untranslated regions of four different S. mansoni target gene transcripts. The data presented here suggest a role for these elements in the dynamics of this complex human parasite genome.
Transposable elements are regarded as one of the principal forces driving the evolution of eukaryotic genomes (7), since they are associated with the generation of phenotypic diversity (8, 35) and speciation (42). Although regarded as a selfish DNA with negative impact on the host (7, 60), transposons have been shown to contribute significantly to gene evolution (16, 24, 27, 39). They can be assigned to two broad groups designated retroelements (class I) and classic transposable elements (class II). One of the classes of retroelements is retrotransposons, which can be divided into two major groups according to the presence or absence at both ends of elements of long terminal repeats (LTRs). Although both groups transpose by reverse transcription, the processes are considerably different, and there are significant discrepancies between their reverse transcriptases (RTs) at the primary sequence level.
The LTR class of retrotransposons integrates into the genome by means of an integrase with a high degree of sequence specificity (52). It has been generally accepted that LTR retrotransposons can be divided into two major groups, Ty1/copia and Gypsy/Ty3, but the existence of a third group, the BEL (or Pao-like) group, has been proposed (1, 9, 41). Usually LTR retrotransposons have one or two open reading frames (ORFs) with products that show similarities to retroviral Gag and Pol polypeptides. However, some invertebrate retrotransposons, such as Gypsy, have been shown to possess an additional ORF with properties analogous to those of retroviral env, conferring on them the ability to infect other cells (30).
The non-LTR class of retrotransposons, in contrast, integrates into the genome by using a mechanism by which an endonuclease nicks the chromosome and DNA synthesis is initiated using the 3′ hydroxyl of the broken strand of target DNA as the primer for reverse transcription (37). Non-LTR retrotransposons comprise one or two ORFs, and only the RT domain is common to all elements. Phylogenetic analysis of this domain has allowed the non-LTR transposons to be classified into 11 clades (40). Other characteristic domains present in some elements are an apurinic or apyrimidic endonuclease, an RNase H, and a putative nucleic acid binding motif.
Schistosoma mansoni, a digenetic blood fluke, is the primary causative agent of schistosomiasis in humans and an important source of morbidity on a global scale. The disease is endemic in 74 developing countries, infecting about 200 million individuals, and it is estimated that an additional 500 to 600 million are at risk (59). The Schistosoma genome has approximately 270 Mbp (54), and a considerable portion (more than 20%) is believed to be composed of retrotransposons (31). Four retroelements belonging to LTR and non-LTR classes have been previously characterized for S. mansoni (10, 14, 15). The presence of RT activity in Schistosoma extracts suggests that some of these elements are active (23).
In this work we describe the sequence and structure of full coding regions of three novel LTR retrotransposons, including a member of the BEL family, not previously described in schistosomes and one novel non-LTR retrotransposon. All have high transcriptional activity and have been reconstructed from expressed sequence tag (EST) data generated by the “Schistosoma mansoni EST Genome Project” (http://bioinfo.iq.usp.br/schisto). Acquisition and maintenance of the parasitic way of life requires the ability to evolve rapidly, which could be conferred by high retrotransposon activity. The data presented here double the number of retrotransposons described for S. mansoni, highlight the existence of two populations with distinct features regarding gene number and transcriptional activity, and show examples of retrotransposon fragment inserts in four different S. mansoni target gene transcripts, suggesting an influence of such elements in S. mansoni genome evolution.
MATERIALS AND METHODS
Construction of cDNA libraries.
A total of 179,072 ESTs from six different stages of the life cycle (cercariae, 7-day-cultured schistosomula, adults, eggs, miracidia, and germ balls) were sequenced in the Schistosoma mansoni EST Genome Project (http://bioinfo.iq.usp.br/schisto), as previously described (56). Approximately 4 μg of mRNA from each stage was obtained with the MACS kit (Miltenyi Biotec) and eluted in 200 μl of diethyl pyrocarbonate-treated water; samples were treated with Promega RQ1 RNase-free DNase (1 U/10 μl) for 30 min at 37°C. DNase was inactivated at 65°C for 10 min. mRNA purity and integrity were checked by RT-PCR using appropriate primer pairs of known genes as well as negative controls (PCR in the absence of reverse transcription). DNase-treated mRNAs obtained were used for the construction of cDNA and Social Analysis of Gene Expression (SAGE) libraries. cDNA synthesis and amplification were performed using the ORESTES low-stringency RT-PCR protocol with modifications as described elsewhere (56). A normalized poly(dT)-primed cDNA library was prepared as described elsewhere (56), by using the abundantly available mRNA from adult worms. cDNA sequencing was performed using standard fluorescence labeling dye-terminator protocols.
Reconstruction of retrotransposon sequences.
EST sequence chromatograms were stored, processed, and trimmed through a Web-based service (48); sequences with at least 100 bp with phred-15 or higher (http://www.phrap.org/) were accepted and further evaluated. S. mansoni retrotransposons were filtered by using BLASTN (http://www.ncbi.nlm.nih.gov/BLAST/) analysis with a local copy of the GenBank nucleotide database and the BlastMachine (Paracel, Inc.), and were processed with a fast parser tool (49) to select those that matched known S. mansoni retrotransposon sequences with an E value of ≤10−15 and had at least 85% identity along at least 75 nucleotides. By comparing with BLASTX against the set of transposon protein sequences from the GenBank nonredundant (NR) protein database and selecting those matching with an E value of ≤10−4 and at least 30% identity along at least 75 amino acids, we identified further potential novel S. mansoni transposon coding sequences. A total of 10,348 putative transposon EST reads were selected.
Selected reads were assembled using the Cap3 program (22) to generate the core sequences. Selected core sequences for the novel S. mansoni LTR transposons Saci 1 to -3, the novel non-LTR transposon Perere, and the previously described Boudicca retrotransposon (10) were picked by manual inspection of the longest assembled sequences and were extended by manual curation, using a local copy of BLASTN and the Bioedit program (version 5.0.6) (19) to compare each transposon consensus with the 179,072 EST reads from the project; these full-length assemblies were used as a blueprint for construction of full-length sequences by using a minimum set of EST reads selected from the pool to reconstruct an ORF for each transposon, leaving out truncated copies that generated stops. With the exception of Saci-2, all reconstructed sequences were anchored at the 3′ end by a sequence from the directional poly(dT)-primed normalized library (56), in order to avoid problems of artifacts due to incorrect, ambiguous assembly of expressed segments of LTRs. Saci-3 was anchored by a 3′-end sequence from GenBank dbEST (accession number AI018990.1). Saci-perere is a hero-trickster (50) in the native Tupi Indian mythology of South America, a very short young black boy with a red bonnet that confers his magic powers. He is hyperactive and jumps all over the place on his single leg, haunting people and playing tricks. He lives in whirlwinds and moves very fast, making loud whistling noises and scaring people when they travel alone at night in the dark forests.
Construction of phylogenetic trees.
The RT domains of novel and known S. mansoni retrotransposons were aligned with Clustal X (version 1.83) (55). The alignment of the characteristic (Y/F)XDD box was checked to ensure the quality of the alignment. Further analysis with Clustal X by using the neighbor-joining method, excluding positions with gaps, resulted in the phylogenetic trees shown in the figures. The confidence of the branches was evaluated by bootstrap analysis using 1,000 samplings. Phylogenetic trees were drawn using Treeview (version 1.6.6) (47). The GenBank sequences and accession numbers utilized for construction of alignments and phylogenetic trees are as follows: BEL, AAB03640.1; blastopia, CAA81643.1; cer-1, AAA50456.1; Copia, OFFFCP; CsRn, AAK07486.1; Dea1, T07863; Grasshopper, AAA21442.1; Gypsy, GNFFG1; HIV2, AAA76841.2; Kabuki, BAA92689.1; Kamikase, 9757434; Mag, S08405; Maggy, AAA33420.1; Micropia, CAA32198.1; MMTV, GNMVMM; Ninja, T31674; Pao, S33901; Pao (P1), BAA95569; SIV, AAA47606.1; Sushi, AAC33526.2; Ted, AAA92249; Tom, CAA80824.1; Ty1, P47100; Ty3, S53577; Ulysses, CAA39967.1; woot, AAC47271.1; Yoyo, T43046; Zam, CAA04050.1; BRG2, X60372.1; CgT1, AAA85636.1; CR1 Gallus, AAC60281.1; CR1 spixii, BAA88337.1; CRE1, M33009.2; Czar, AAA30239.1; Doc, CAA35587.1; I, AAA70222.1; Ingi, CAA29181.1; Jockey, AAA28675.1; Juan, AAA29354.1; L1Homo, AAC51279.1; L1Mouse, AAC72810.1; L1Rat, AAB41224.1; Lian, AAB65093.1; pido, AY034003.1; Q, AAA53489.1; R1Dros, CAA36227.1; R1Mori, AAC13649.1; R2Bombyx, AAB59214.1; R2dros, CAA36225.1; R2esrwig, AAC34906.1; R4, AAA97394.1; RTE1, AAC72298.1; SR1, AAC06263.1; SR2, AAC24982.2; Swimmer, AAD02928.1; T1, AAA29367.1; Tad1, AAA21781.1; Tart, AAC46494.1; Tx1, AAA49976.1.
Southern blotting.
Twenty-five micrograms of genomic DNA from S. mansoni adult worms was subjected to overnight incubation at 37°C with the EcoRI or BamHI restriction enzyme (New England Biolabs) in the appropriate buffer. Samples were divided into five aliquots of 5 μg each, which were electrophoresed in 0.8% agarose gels in Tris-acetate-EDTA (TAE) at 1 V/cm and immobilized on a Hybond-N+ nylon membrane (Amersham Biosciences) by using the Posiblot apparatus (Stratagene).
Radiolabeled probes for each retrotransposon were generated from 25 ng of fragments of approximately 700 bp labeled with [32P]dCTP by using Rediprime kits (Amersham Biosciences). After labeling, 1 μl of each probe had its radioactivity counted, and the same number of counts was used in all experiments. Overnight hybridization was performed in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.5% sodium dodecyl sulfate (SDS)-5× Denhardt's solution-25 μg of salmon testis DNA (Sigma)/ml at 68°C. Membranes were washed once with 1× SSC-0.2% SDS and twice with 0.5× SSC-0.2% SDS for 30 min each time at 68°C. Radioactive signals were detected by using a phosphor screen and a Storm apparatus (Molecular Dynamics), and images were processed by using the ImageQuant program (Molecular Dynamics).
PCR of genomic DNA for detection of full-length copies.
The following primers flanking the entire coding region of each retrotransposon were designed: Pererefw, GTTTGCGTTACGATCACACG; Perererv, ATTTCCAGTGCCAGAGCAAG; Saci-1fw, TGCCTAACAATCGTGCAAAG; Saci-1rv, GGTTCACCTAATCGCTTTCC; Saci-2fw, GAGGCTTGTGATTCCCACTG; Saci-2rv, ACTGTCCTCAGTGCCTGGTC; Saci-3fw, TTTGGAACACGCAATACAGC; Saci-3rv, CAAGCTCGAACCAAACAAGG.
These primers were used for PCR of S. mansoni genomic DNA by using the Advantage 2 polymerase mix (BD Bioscience) and a cycling program of 95°C for 3 min followed by 40 cycles of 95°C for 30 s, 60°C for 30 s, and 68°C for 5 min in a GenAmp PCR system 9700 (Applied Biosystems). The ramp of temperature transition between the annealing and extension steps was reduced to 5% of the default speed in order to increase the amount of amplified product.
Aliquots (2 μl) of each reaction product were electrophoresed in 0.8% agarose gels and further immobilized on a Hybond-N+ nylon membrane (Amersham Biosciences) by using the Posiblot apparatus (Stratagene). Hybridization with radioactive probes was performed with the same probes used for Southern blotting and with the same protocol for hybridization and washing.
Estimation of transposon copy numbers in the S. mansoni genome by use of BLASTN.
Comparative estimates of copy numbers of retrotransposons in the S. mansoni genome were obtained essentially as described by Copeland et al. (10), by using the local BLASTN program to compare each of the reconstructed full-length transposons with the database of 27,064 bacterial artificial chromosome (BAC) end sequences, obtained by filtering out the duplicated deposits of 42,017 S. mansoni genome survey sequences from GenBank. The count of hits with scores higher than 100 divided by the total length of the query transposon allowed us to calculate a gene index, an estimate of the relative abundance of the gene in the S. mansoni genome. The absolute copy number range was estimated from the gene index by using the copy number range of the Boudicca retrotransposon as a benchmark (10).
Estimation of relative transcription rate and activity.
Transcription rates for different retrotransposons were calculated by using the local BLASTN program to compare each of the full-length transposons with the database of 179,092 ESTs generated by the Schistosoma mansoni EST Genome Project; hits with matching scores higher than 100 were counted. Counts were divided by the sequence length for normalization. The resulting number was considered to reflect the overall expression level among the different stages of the S. mansoni life cycle. The results were divided by the value obtained for SR2 for normalization.
A second estimate was obtained by using a SAGE library previously generated by our group from adult worms (56). In silico restriction maps for NlaIII were produced for each retrotransposon, and the 10 bp adjacent to the 3′-most NlaIII site was recorded as the expected SAGE tag for that transposon transcript. This tag sequence was used to search the database of 68,238 SAGE tags, which had been sequenced from S. mansoni adult worm mRNAs, and the number of identical tags (exact matches) was counted.
Identification of fragments of novel transposons inserted into known genes of S. mansoni.
Sequences of retrotransposons Saci-1, -2, and -3, Perere, Boudicca, and SR2 were used as queries for BLASTN searches of our EST database. Reads with matching E values lower than 0.01 were retrieved and assembled with Cap3. A BLASTX search of the resulting contigs and singlets against the GenBank NR protein database followed by manual inspection allowed the identification of sequences containing an ORF for a known protein besides the segment of retroviral sequence. In order to exclude possible artifact chimeras generated at the vector ligation step of EST production, we considered only those transposon inserts that were confirmed by at least two different EST clones spanning the junction between the transposon and the target gene.
Genomic DNA sequences of the new transposons.
Preliminary sequence data for the S. mansoni genome was obtained from “The Schistosoma mansoni Genome Project” at The Institute for Genomic Research (TIGR) (http://www.tigr.org) and at The Sanger Institute (ftp://ftp.sanger.ac.uk/pub/databases/Trematode/S.mansoni/). We used these data for a BLASTN search, with the novel reconstructed retrotransposon sequences as queries. Results described refer to the genomic best match for each retrotransposon.
Nucleotide sequence accession numbers.
Full-length reconstructed sequences for the four new transposons were deposited in the Third Party Annotation section of the DDBJ/EMBL/GenBank databases under the following TPA accession numbers: Saci-1, BK004068; Saci-2, BK004069; Saci-3, BK004070; Perere, BK004067. The reconstructed sequence for Boudicca (10), for which no full-length sequence was available, was deposited under TPA accession number BK004066. All 7,086 ESTs matching the novel transposons and the known Boudicca and SR2 transposons were deposited in GenBank under accession numbers CF490117 to CF497202.
RESULTS
Phylogenetic trees of LTR retrotransposons containing the novel S. mansoni sequences.
Our group has recently analyzed the transcriptome of S. mansoni, for which we obtained a 92% sampling of the estimated 14,000-gene complement (56). In that analysis, transposon sequences were excluded along with rRNA and mitochondrial RNA. By performing a detailed analysis focused on transposon similarities, we have now identified four novel S. mansoni retrotransposons among the complete set of EST sequences acquired in the earlier project. Full-length sequences for each of these four transposons were reconstructed from the assembled EST reads followed by careful manual curation, as described in Materials and Methods. Three of these transposons, designated Saci-1, -2, and -3, were identified as LTR retrotransposons, and one was identified as a non-LTR retrotransposon, named Perere, as described below in detail.
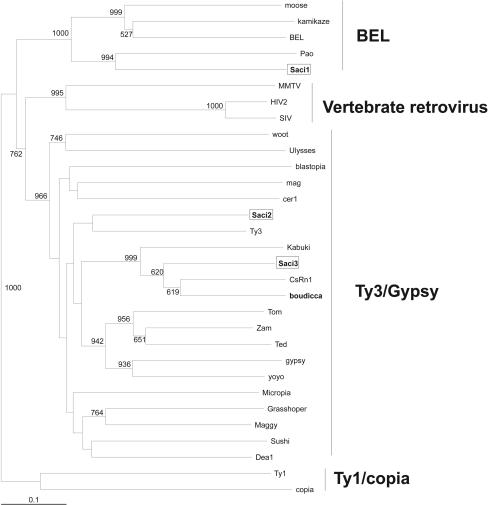
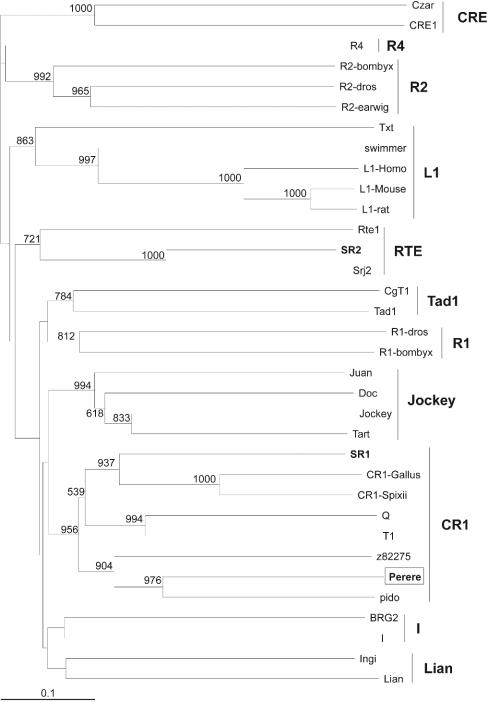
To determine the ancestral origin of these novel transposons, phylogenetic trees were constructed. Sequences from the RT domains of several members of the LTR group of retrotransposons and some retroviruses were aligned by using the Clustal X program (55), and a phylogenetic tree was constructed by using the neighbor-joining method (Fig. 1). The result clearly distinguished the three different LTR transposon families previously described and permitted classification of the three novel S. mansoni LTR retrotransposons, Saci-1, -2, and -3 (see below). The same approach was used for members of the non-LTR group of retrotransposons, and the resulting phylogenetic tree (Fig. 2) allowed the distinction of all the 11 clades previously described, permitting classification of the novel S. mansoni non-LTR transposon Perere as a member of the CR1 family (see below). The latter family includes the previously reported transposon pido of the closely related trematode Schistosoma japonicum. A set of 44,000 ESTs has recently been acquired from adult worms and eggs of S. japonicum (21). A search of that database, by using TBLASTX and sequences from the three novel S. mansoni LTR transposons as queries, showed that very few messages similar to these S. mansoni LTR transposons are expressed in S. japonicum (52 ESTs in all, with a matching cutoff E value of ≤10−5), and the four transcripts that covered the RT domains were not phylogenetically related to those of the S. mansoni transposons (data not shown).
FIG. 1.
Phylogenetic tree for the RT domains of LTR retrotransposons. The tree was constructed by the neighbor-joining method, excluding positions with gaps. Previously described S. mansoni retrotransposons are boldfaced, and the three novel S. mansoni LTR retrotransposons identified in this work are boxed. Numbers represent the confidence of the branches assigned by bootstrap analysis (in 1,000 samplings); bootstrap values lower than 500 are omitted from the figure.
FIG. 2.
Phylogenetic tree for the RT domains of non-LTR retrotransposons. The tree was constructed by the neighbor-joining method, excluding positions with gaps. Previously described S. mansoni retrotransposons are boldfaced, and the novel non-LTR S. mansoni retrotransposon identified in this work is boxed. Numbers represent the confidence of the branches assigned by bootstrap analysis (in 1,000 samplings); bootstrap values lower than 500 are omitted from the figure.
Saci-1 belongs to the BEL family of LTR retrotransposons.
The sequence of Saci-1 has one predicted ORF encoding a protein of 1,680 amino acids containing three Cys motifs, a protease, an RT, an RNase H, and an integrase domain (Fig. 3). From the phylogenetic tree obtained with the RT domains (Fig. 1), it is possible to place Saci-1 within the BEL family of retrotransposons. Phylogenetic analysis with the RNase H domain also groups Saci-1 within the BEL family, with a bootstrap value of 1,000 (data not shown). The Gag domain presents a structure similar to that previously described for the BEL family, i.e., with three Cys motifs (Fig. 3), and different from those of Gag domains of other retrotransposon families (1). The motifs of Saci-1 are similar in structure to those of Pao, with identical spacing between the conserved amino acids throughout the entire motif (Fig. 3), providing further support to their relatedness shown in the phylogenetic tree constructed from RT domains (Fig. 1). The integrase domain of Saci-1 contains a motif similar to the D(X35)E motif of most retrovirus and retrotransposon integrase proteins (28), with 49 amino acids instead of 35 between the conserved aspartic and glutamic acids (Fig. 3); the length of this interval is within the range (45 to 53 amino acids) found in other members of the BEL family (1).
FIG.3.
Multiple-sequence alignment of novel S. mansoni LTR retrotransposons. Clustal X alignments for conserved regions of Gag, protease (Pro), RT, RNase H (RH), and integrase (Int) are presented. Arrows above the Cys domain point to cysteines and histidines comprising the motifs; shaded arrows indicate that the amino acid is not present in all sequences in the alignment. In the protease domain, the D(T/S)G motif common to aspartic proteases is boxed. Arrows above RNase H and integrase alignments point to conserved motifs previously described. Regions RT1 to RT7, each containing one of the seven motifs described for RT, are indicated.
Both Saci-2 and Saci-3 belong to the Gypsy/T3 family.
Saci-3 has three predicted ORFs encoding products of 288, 1,166, and 232 amino acids; in contrast, Saci-2 has only one ORF, encoding a protein of 1,382 amino acids (Fig. 3). Alignment of the protease domain shows a conserved D(T/S)G motif and conservation of several other amino acids in this region relative to other members of the Gypsy/Ty3 family. The RTs of both Saci-2 and Saci-3 show several conserved residues among the different domains relative to other members of the Gypsy/Ty3 family (Fig. 3); in fact, the phylogenetic tree obtained with the RT domains shows that both retrotransposons fall within this family. In contrast to Saci-1, both Saci-2 and Saci-3 have a D(X35)E domain with exactly 35 amino acids between the conserved aspartic and glutamic acids, as expected for members of the Gypsy/Ty3 family (10).
Saci-2 has an unusual Cys motif.
The Cys motif of the Gag domain of Saci-2 resembles that present in other retroposons (CX2CX4HX4C) such as micropia of Drosophila melanogaster and COS41.3 of Ciona intestinalis, but a lysine replaces the conserved histidine (Fig. 3). This amino acid is confirmed by 22 out of 24 ESTs that cover that region of the retrotransposon, and neither of the other 2 EST sequences codes for a histidine. There is an additional histidine in Saci-2, 1 amino acid distant from the final cysteine of the motif. This creates a new CX2CX9CXH motif, which resembles the motif of the CCCH zinc finger (Zf) protein family (CX8CX5CX3H), shown to bind specific RNAs (32). Further experiments are warranted to determine whether this is a functional domain or simply a nonfunctional degeneration of the CX2CX4HX4C motif.
Saci-3 is closely related to Boudicca and CsRn1.
The first ORF of Saci-3 encodes a putative Gag protein presenting a Cys domain with a motif (CX2HX9CX3C) identical to that of S. mansoni Boudicca (10), Clonorchis sinensis CsRn1 (3), and Bombyx mori Kabuki (Fig. 3) and different from the usual CX2HX4CX4C motif seen in most retrotransposon and retrovirus Gags (10, 11). The second ORF has a structure typical of an ORF encoding a Pol polyprotein, with the presence of domains for protease, RT, RNase H, and integrase. The phylogenetic tree constructed from RT domains places Saci-3 in the same branch as Boudicca, CsRn1, and Kabuki, with Boudicca more closely related to CsRn1 than to Saci-3 (Fig. 1). Saci-3, like Boudicca, has a third ORF that does not exhibit identity to other known proteins and may code for an envelope protein, due to its position on the retroviral message. However, it is particularly difficult to characterize envelope proteins, since they present a low degree of similarity to each other (33).
Perere is a member of the CR1 family of non-LTR retrotransposons.
Perere has a single ORF coding for a product of 1,227 amino acids, a polyprotein with domains for endonuclease and RT (Fig. 4). The presence of the endonuclease domain suggests that the mechanism of integration of Perere involves the nicking of target DNA. The phylogenetic tree suggests that Perere belongs to the CR1 family of non-LTR retrotransposons (Fig. 2), and apparently it forms a discrete branch with S. japonicum pido and a non-LTR retrotransposon of Caenorhabditis elegans. Such a discrete branch has been described previously (31) but was weakly supported; now, with the addition of Perere, the existence of this branch gains strong bootstrap support.
FIG. 4.
Multiple-sequence alignment of the novel S. mansoni non-LTR retrotransposon Perere. Clustal X alignments for conserved regions of the endonuclease (End) and RT domains are presented. Regions RT1 to RT7, each containing one of the seven motifs described for RT, are indicated.
Evaluation of retrotransposon diversity.
We calculated the average identity (as well as the associated degree of dispersion) of all ESTs from the 180,000-sequence database of the Schistosoma mansoni EST Genome Project that aligned to the single consensus full-length sequence of each reconstructed retrotransposon with a BLASTN score higher than 100. All retrotransposon consensus sequences exhibited a high level of average identity with their matching ESTs—99.0% for Saci-1, 97.7% for Saci-3, 97.3% for Perere, 95.5% for Saci-2, and 95.2% for Boudicca—indicating that the reconstructed sequences are representative of the actual expressed sequences. The standard deviation of the average identity was calculated in order to obtain a measure of divergence among the different copies of retrotransposons. Standard deviations were ±4.9 and ±4.8% for Saci-2 and Boudicca, respectively, ±3.1% for Perere, ± 2.5% for Saci-3, and ±1.1% for Saci-1, indicating a higher divergence among the expressed copies for both Saci-2 and Boudicca.
In contrast, for SR2, a reconstruction using genomic clones (accession number AF025672), the ESTs in our data set showed a lower level of identity (91.7% ± 4.6%), suggesting that the genomic clone deposited in GenBank is highly divergent from most of its expressed copies.
Estimates of copy numbers and transcriptional activities of S. mansoni retrotransposons.
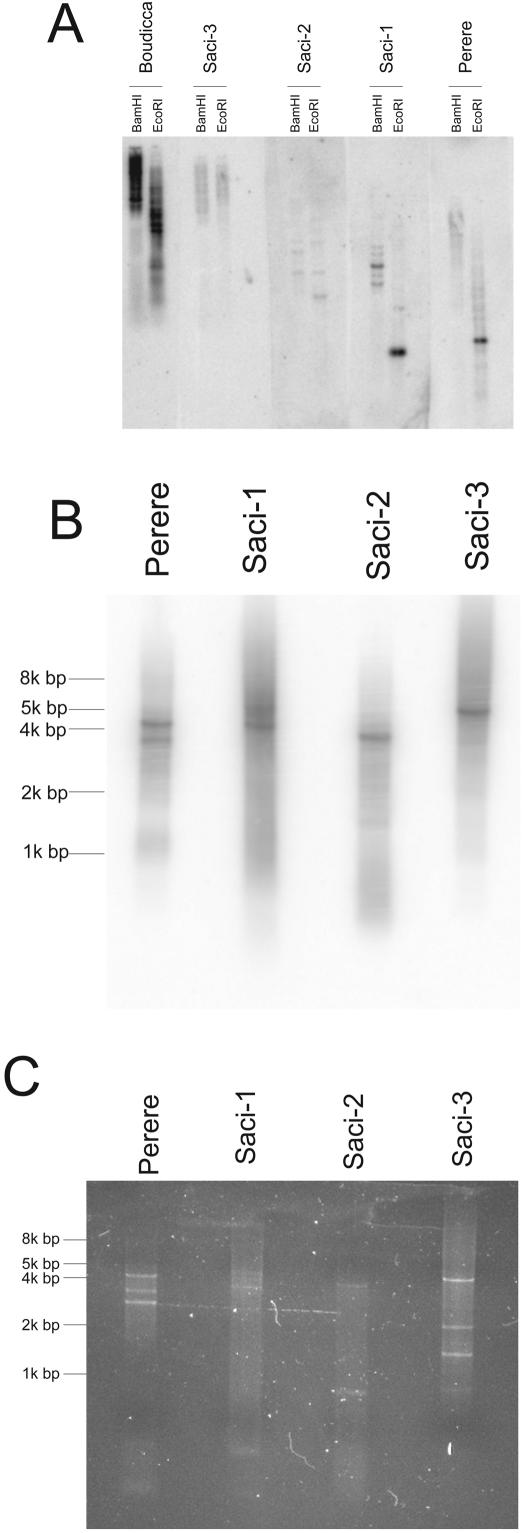
Computational estimates of copy number showed that Saci-1 and -2 display relatively low copy numbers in the genome of S. mansoni (70 to 850 copies), while Saci-3 and Perere display intermediate copy numbers (150 to 2,500 copies) compared to those of previously described S. mansoni retrotransposons (Table 1). Southern blot experiments with all four new retrotransposons, and with Boudicca as a benchmark, showed that Boudicca exhibits a significantly higher signal, indicating that it is present at a much higher copy number in the genome than the other retrotransposons (Fig. 5A).
TABLE 1.
Transcriptional activities of S. mansoni retrotransposonsa
| Gene | Transposon length (bp) | Hits in BAC endsb | Gene indexc | Estimated copy no. in the genomed | No. of ESTse | Relative transcriptional ratef | Transcriptional activity (per copy)g | Adult SAGE tagsh | Transcriptional activity in adults (per copy)i |
|---|---|---|---|---|---|---|---|---|---|
| Saci-1 | 5,980 | 48 | 0.008 | 70-700 | 1,538 | 0.61 | 23.5 | 162 | 211.4 |
| Saci-2 | 4,946 | 50 | 0.010 | 85-850 | 230 | 0.12 | 3.7 | 15 | 15.4 |
| Saci-3 | 5,217 | 89 | 0.017 | 150-1,500 | 2,261 | 1.02 | 18.5 | 29 | 18.1 |
| Perere | 4,875 | 147 | 0.030 | 250-2,500 | 1,140 | 0.55 | 5.6 | 279 | 97.5 |
| SR2 | 3,913 | 1,206 | 0.308 | 2,600-26,000 | 1,655 | 1.00 | 1.0 | 29 | 1.0 |
| Boudicca | 4,279 | 505 | 0.118 | 1,000-10,000 | 278 | 0.15 | 0.4 | 15 | 1.3 |
| Smα | 331 | 772 | 2.332 | 20,000-200,000 | 218 | 1.56 | 0.2 | ||
| SR1 | 2,337 | 568 | 0.243 | 2,000-20,000 | 84 | 0.09 | 0.1 | 0 | 0 |
The four novel transposons identified in the present work are boldfaced and are listed along with the other four known transposons of S. mansoni.
Number of BAC end sequences with matching scores higher than 100 when BLASTN was used to search with the transposon sequence as a query against the GenBank database of 27,064 S. mansoni genomic BAC end sequences.
Calculated as the number of hits in BAC ends divided by the transposon length.
Estimated by taking the range of the numbers of copies of Boudicca in the S. mansoni genome (10) as a reference and using the gene index factors to calculate the copy number ranges for the other retrotransposons.
Number of EST transcripts with matching scores higher than 100 when using BLASTN was used to search with the transposon sequence as a query against a database of 179,072 EST transcripts generated by the Schistosoma mansoni EST Genome Project.
Calculated as the number of EST transcripts divided by the transposon length and normalized in relation to SR2.
Calculated as the relative transcriptional rate divided by the gene index and normalized in relation to SR2.
Number of tags per million from an adult SAGE library sequenced by the Schistosoma mansoni EST Genome Project that matched the transposon sequence adjacent to the 3′-most NlaIII site.
Calculated as the number of SAGE tags divided by the gene index and normalized in relation to SR2.
FIG. 5.
Detection of the novel S. mansoni retrotransposons in the parasite's genome. (A) Southern blot of novel retrotransposons. S. mansoni genomic DNA was digested with one of two different restriction enzymes and analyzed by Southern blotting with radiolabeled probes specific for each of the transposons. Fragments of similar sizes and the same number of radioactive counts were used for each of the five transposon probes. The Boudicca retrotransposon (10) was included for comparison, along with the four new retrotransposons Saci-1, -2, and -3 and Perere. (B and C) Detection of full-length copies. (B) Products of PCR from genomic S. mansoni DNA with primers specific for each retrotransposon were subjected to electrophoresis in 0.8% agarose gels, transferred to nylon membranes, and hybridized with the same probes as those used for panel A. (C) A second 0.8% agarose gel with the same products was stained with ethidium bromide to visualize all products, including those not detected by the probes. The expected sizes of amplified products based on the reconstructed full-length sequences were as follows: Perere, 4,593 bp; Saci-1, 5,130 bp; Saci-2, 4,246 bp; Saci-3, 5,003 bp.
The number of redundant ESTs in a large transcript database can be used to estimate the relative transcriptional rates of the retrotransposons. BLASTN searches of each retrotransposon consensus sequence against the EST database of 180,000 reads from the Schistosoma mansoni EST Genome Project retrieved a considerable number of matching ESTs for each of the four new retrotransposons as well as for the four previously known (Table 1). Transposon ESTs were sequenced from all six stages of S. mansoni that were studied (7-day-cultured schistosomula, adults, eggs, miracidia, germ balls, and cercariae) (data not shown), indicating that transposons are expressed across all life cycle stages. A second estimate of the transcription level was obtained by using the number of SAGE tags that were sequenced for each transposon transcript from an adult SAGE library (Table 1).
The ratio of the relative transcriptional rate (or SAGE tag count) to the copy number allowed us to estimate the relative transcriptional activity, which would reflect the average level of transcription per genomic copy of each retrotransposon. It is noticeable that Saci-1, -2, and -3 and Perere have considerably higher transcriptional activities (1 to 2 orders of magnitude higher) than the other S. mansoni retrotransposons that have been described (Table 1).
The frequency of sequenced transposon transcripts for each of the life cycle stages was determined (Table 2). In cercariae, the frequency of novel transposon ESTs was 11.2% of all transcripts sequenced, and that of the known transposons Boudicca and SR2 was 3.2% (Table 2). For all types of transposons taken together, the frequency of transposon ESTs in cercariae was twofold higher than that in schistosomula and three- to fourfold higher than those in adults, eggs, miracidia, and germ balls.
TABLE 2.
Frequency of sequenced transposon transcripts in life cycle stages
| Life cycle stage | Total no. of ESTs sequenced | No. of ESTs (%)a
|
ESTs of all transposons (%)a | |
|---|---|---|---|---|
| Novel transposons | Boudicca + SR2 | |||
| Cercariae | 11,704 | 1,315 (11.2) | 375 (3.2) | 14.4 |
| Schistosomula | 29,122 | 1,671 (6.0) | 435 (1.4) | 7.4 |
| Adults | 34,237 | 747 (2.1) | 310 (0.9) | 3.0 |
| Eggs | 19,806 | 518 (2.6) | 211 (1.1) | 3.7 |
| Miracidia | 19,611 | 758 (3.8) | 215 (1.1) | 4.9 |
| Germ balls | 17,229 | 367 (2.1) | 147 (0.9) | 3.0 |
Transposon ESTs as a percentage of the total number of ESTs sequenced for that particular life cycle stage.
Genomic DNA sequences of the new transposons.
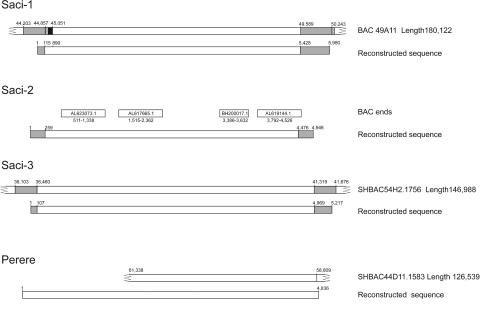
Comparison by BLASTN of the full-length sequences of the four new transposons to the database of assembled genomic BACs and BAC end-sequences from The Sanger Institute (http://www.sanger.ac.uk/Projects/S_mansoni/) and TIGR (http://www.tigr.org/tdb/e2k1/sma1/) revealed that the new elements Saci-1, Saci-3, and Perere are represented within the limited data set so far available of 13 sequenced BACs (approximately 0.5% of the S. mansoni genome). Alignment showed that the matching sequence with the highest score for Saci-1 had 97% identity and 85% coverage, that for Saci-3 had 98% identity and 100% coverage, and that for Perere had 97% identity and 52% coverage. Genomic BAC clones for both Saci-1 and Perere exhibited truncations at the 5′ ends of their sequences that resulted in partial alignment to the reconstructed transcript sequences (Fig. 6).
FIG. 6.
Genomic sequences of the novel transposons. Shown is a schematic representation of the alignment of genomic clones. Assembled BAC sequences were retrieved from the TIGR and Sanger S. mansoni genome sequencing projects, and BAC end sequences were obtained from GenBank. Numbers above the diagrams represent the position (in base pairs) in each clone. Shaded areas, LTR sequences. The hatched area in the Saci-1 alignment represents a portion of the genomic clone LTR that had no similarity to the reconstructed sequence or to any EST from our data set, and the solid area represents a gap in the sequence to allow for alignment. The GenBank accession numbers of BAC end sequences are given in open rectangles above the regions of Saci-2 to which they are aligned.
The genomic sequences of Saci-1 and Saci-3 permitted confirmation of the overall structure of the reconstructed LTR sequences and extended them by a few bases. Thus, the deduced genomic LTR from Saci-3 (Fig. 6) is 358 bp long, covering 35 bp at the 5′ end and 314 bp at the 3′ end of our reconstructed sequence. The deduced genomic LTR from Saci-1 (Fig. 6) has 848 bp; however, it aligns to the 5′ end from our reconstructed sequence only from base 544 to 655. Saci-2 was not represented in the 13 assembled BACs, but it has 53% coverage from four nonredundant genomic BAC end sequences from the public database (Fig. 6); the best matching sequence has 17% coverage.
Further confirmation of the existence of genomic full-length copies for each retrotransposon was obtained by PCR of S. mansoni genomic DNA using primers designed to flank the coding region of each retrotransposon (Fig. 5B and C). We were able to amplify a fragment of approximately the size expected for a full-length copy for each of the reconstructed retrotransposons (Fig. 5B), as detected by hybridization with the same radioactive probes used in Southern blot experiments. In addition, Perere and Saci-1 presented a few other copies with lower molecular weights, as recognized by the radioactive probes, which must correspond to truncated copies. Other amplicons of lower molecular weights, which were not recognized by the radioactive probes, were visualized by ethidium bromide staining (Fig. 5C) and possibly represent truncated copies of retrotransposons lacking the probe region.
Identification of DNA fragments of novel transposons inserted into known genes of S. mansoni.
We were able to map fragments of retrotransposon sequences to the untranslated regions (UTR) of four different S. mansoni gene transcripts (Table 3). It is noteworthy that in all cases of LTR transposon insertion, the inserted fragments were from the LTR region and were always located in the 5′ UTR of the target gene. In contrast, the insertion of SR2 was only a few bases away from the 3′ end of the sequence in both target genes. Additionally, all the retrotransposon inserts were found in the same strand as the ORFs of the target genes. Taken together, the above data may indicate that either the LTR region or the polyadenylation site of the retrotransposons may actually be used as an alternative promoter or alternative polyadenylation site for the target genes, as described for other organisms (4, 38, 43).
TABLE 3.
Mapping of transposon inserts into S. mansoni target genes
| Cluster IDa | No. of reads in cluster | Cluster length (bp) | Transposon segment (bp)b | Transposon insert cluster coordinates
|
Target ORF cluster coordinates
|
Putative target ORF product | |||
|---|---|---|---|---|---|---|---|---|---|
| bp | Strand | Identity (%) | bp | Strand | |||||
| Saci-2 (4,952 bp) | |||||||||
| SmAE601007.1 | 5 | 642 | 4836-4928 | 85-177 | + | 92 | 266-642 | + | UDP-sugar transporter sqv-7 (Squashed vulva protein 7), C. elegans |
| SmAE610579.1 | 4 | 953 | 150-260; 4644-4739 | 12-121 | + | 93 | 198-893 | + | Ribulose-5-phosphate-3-epimerase, Mus musculus |
| SmAE605584.1c | 5 | 671 | 218-260 | 12-54 | + | 97 | 406-671 | + | Phosphatidylinositol glycan, class A isoform 2, Homo sapiens |
| Saci-3 (5,210 bp) | |||||||||
| SmAE605584.1c,d | 5 | 671 | 5050-5210; 1-130 | 55-388 | + | 96 | 406-671 | + | Phosphatidylinositol glycan, class A isoform 2, H. sapiens |
| SmAE605672.1d | 5 | 723 | 5023-5210; 1-34 | 439-714 | − | 98 | 1-421 | − | Phosphatidylinositol glycan, class A isoform 2, H. sapiens |
| SR2 (3,913 bp), SmAE605208.1 + 2 singlet readse | 7 | 1423 | 3903-3817 | 22-103 | − | 88 | 275-1423 | − | Microtubule-associated protein, 215 kDa, Xenopus laevis |
Identification of the cluster in the Schistosoma mansoni EST Genome Project.
Mapping of the insert sequences in the retrotransposons.
This single cluster presented insertions from both Saci-2 and Saci-3.
These two clusters represent the same ORF of the target gene but have different patterns of Saci-3 insertion.
Complemented by two singlet EST reads (MA3-0001U-M321-D06-U.B and MA3-0001U-M334-G02-U.G).
One of the genes exhibited insertions of both Saci-2 and Saci-3 in nearby segments, an event unlikely to occur by chance. This may indicate the presence of regions particularly susceptible to retrotransposon insertions, a phenomenon that has been characterized previously (6, 58).
Curation of the S. mansoni EST database.
With the sequences used in this work, we were able to reannotate as retrotransposons 2,391 reads that have not been previously identified as such (56). These reads clustered into 452 S. mansoni assembled EST sequences (SmAEs) (56) and are accessible at the project website (http://bioinfo.iq.usp.br/schisto); they represent 1.5% of the 30,988 unique SmAE sequences previously reported (56).
DISCUSSION
This work presents the identification and full-length sequence reconstruction of four novel retrotransposons from S. mansoni, which were revealed by large-scale EST sequencing. Analysis of the coding sequences permitted the identification of the first member of the BEL family in S. mansoni, of two members of the Ty3/Gypsy family, and of one non-LTR retrotransposon of the CR-1 family. Previous work describing retrotransposons in S. mansoni (10, 14) utilized genomic clones to obtain partial or complete sequences. Such an approach has apparently created a bias toward the description of transposons with the highest numbers of copies in the genome of the parasite (Table 1). In contrast, in the present work, by using a different approach and compiling the information from a large set of ESTs to reconstruct full-length retrotransposon messages, we were able to retrieve novel transposon sequences with the highest levels of expression and lower copy numbers in the S. mansoni genome (Table 1). Although these reconstructed sequences do not represent actual genomic clones, we believe that they correspond to active copies due to consistent confirmation by hundreds of ESTs and the retention of several characteristic traits of retrotransposons. Additionally, verification of the identity of these reconstructed consensus sequences with BAC clone sequences from the S. mansoni genome, using preliminary information generated by TIGR and The Sanger Institute, confirmed that our assemblies represent the actual structures of the transposons present in the organism.
Saci-1, -2, and -3 and Perere display low to medium numbers of copies in the genome but exhibit expression levels equal to or higher than those of the other transcribed retrotransposons (Table 1). This means that in a comparison of transcript production per genome copy, the novel retrotransposons have an activity up to 2 orders of magnitude higher. These characteristics probably reflect different niches occupied by each of the retrotransposons during genome evolution. It has been shown that each retrotransposon tends to have a different ratio of distribution between euchromatin and heterochromatin (12) and that different locations in the chromosome influence the conservation and the copy number of transposable elements (26, 29). The elements tend to be more abundant in heterochromatin because of the lower density of functional genes in this region (13, 26, 29), but they are more degenerate and expressed at lower levels than elements present in the euchromatin (26). It is tempting to hypothesize that the high-copy-number retrotransposons present in the S. mansoni genome are preferentially located in heterochromatin, generating several truncated copies that would be inactive, thus accounting for their low transcriptional activity. The repetitive elements W1 and W2 are located in the heterochromatic region of the S. mansoni W sexual chromosome. Different parasite isolates are known to exhibit sex-specific polymorphisms, resulting from different numbers of copies of repetitive elements, which indicates genomic instability and suggests that replication of repetitive elements is a method of generating variability within schistosomes (18). In contrast, we propose that the low-copy number retrotransposons would have several copies located in euchromatin and would be subject to a stricter process of selection, which has been shown previously to induce the conservation of retrotransposons with active characteristics (46). It is noteworthy that retrotransposons are thought to proliferate in the sexual species, where they would propagate during sexual reproduction, as suggested by the work of Arkhipova and Meselson (2). Schistosomes are among the earliest animals in the evolutionary scale to develop sexual dimorphism and heteromorphic sex chromosomes, and a substantial fraction of the genome of this metazoan parasite is predicted to comprise repetitive sequences made up of retrotransposons (5). The fact that the non-LTR transposon Perere is phylogenetically related to the S. japonicum non-LTR transposon pido implies an ancestral acquisition.
Gene silencing caused by cosuppression, which is a mechanism that preferentially diminishes mRNA levels of high-copy-number retrotransposons (25), may provide an explanation for the differential levels of transcription of high- and low-copy-number transposons in S. mansoni. Cosuppression may trigger different pathways (25) such as methylation of DNA (36, 57), chromatin remodeling (17), and RNA silencing (20). Our group has previously identified (56) S. mansoni sequences coding for proteins with a high degree of similarity to those involved in gene silencing, such as DDM1 (SmAE 609008.1) (44), DNMAP1 (SmAE 700041.1) (51), Dicer (SmAE 604739.1), and Argonaute/Piwi (SmAEs 603705.1 and 606231.1) (20, 61). It is possible that the different levels of retrotransposon expression result from selective transposon silencing related to the copy number and triggered by cosuppression. Interestingly, retrotransposon activity as a factor in the silencing of nearby genes in the genome has been recently described (53). Thus, mapping of retrotransposons may provide clues for the silencing of some additional genes in the S. mansoni genome.
Knowledge of these four transposons should help in the assembly of the parasite's genome sequence, a task that is particularly difficult when a highly repetitive, complex genome is sequenced by the whole-genome shotgun approach (34, 45). Moreover, data from these four new elements allowed us to discern in S. mansoni two populations of retrotransposons with different copy numbers and transcriptional activities. New experiments, such as fluorescence in situ hybridization for detection of these retrotransposons in the S. mansoni chromosomes, in silico analysis of their differential distribution throughout the S. mansoni genome, and measurement of retrotransposon transcriptional activities in RNA interference experiments, should provide clues to understanding the differences between these two populations and extend our understanding of the dynamics of the S. mansoni genome and the biology of this complex human parasite.
Acknowledgments
This work was financed by the Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP) and by the Brazilian Ministry of Science and Technology, Conselho Nacional de Desenvolvimento Científico e Tecnológico (MCT, CNPq). Preliminary genome sequences used from The TIGR Schistosoma mansoni Genome Project were obtained with support from the National Institute of Allergy and Infectious Diseases (NIAID) to TIGR.
REFERENCES
- 1.Abe, H., F. Ohbayashi, T. Sugasaki, M. Kanehara, T. Terada, T. Shimada, S. Kawai, K. Mita, Y. Kanamori, M. T. Yamamoto, and T. Oshiki. 2001. Two novel Pao-like retrotransposons (Kamikaze and Yamato) from the silkworm species Bombyx mori and B. mandarina: common structural features of Pao-like elements. Mol. Genet. Genom. 265:375-385. [DOI] [PubMed] [Google Scholar]
- 2.Arkhipova, I., and M. Meselson. 2000. Transposable elements in sexual and ancient asexual taxa. Proc. Natl. Acad. Sci. USA 97:14473-14477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bae, Y. A., S. Y. Moon, Y. Kong, S. Y. Cho, and M. G. Rhyu. 2001. CsRn1, a novel active retrotransposon in a parasitic trematode, Clonorchis sinensis, discloses a new phylogenetic clade of Ty3/gypsy-like LTR retrotransposons. Mol. Biol. Evol. 18:1474-1483. [DOI] [PubMed] [Google Scholar]
- 4.Baust, C., W. Seifarth, H. Germaier, R. Hehlmann, and C. Leib-Mosch. 2000. HERV-K-T47D-related long terminal repeats mediate polyadenylation of cellular transcripts. Genomics 66:98-103. [DOI] [PubMed] [Google Scholar]
- 5.Brindley, P. J., T. Laha, D. P. McManus, and A. Loukas. 2003. Mobile genetic elements colonizing the genomes of metazoan parasites. Trends Parasitol. 19:79-87. [DOI] [PubMed] [Google Scholar]
- 6.Cantrell, M. A., B. J. Filanoski, A. R. Ingermann, K. Olsson, N. DiLuglio, Z. Lister, and H. A. Wichman. 2001. An ancient retrovirus-like element contains hot spots for SINE insertion. Genetics 158:769-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charlesworth, B., P. Sniegowski, and W. Stephan. 1994. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371:215-220. [DOI] [PubMed] [Google Scholar]
- 8.Clegg, M. T., and M. L. Durbin. 2000. Flower color variation: a model for the experimental study of evolution. Proc. Natl. Acad. Sci. USA 97:7016-7023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cook, J. M., J. Martin, A. Lewin, R. E. Sinden, and M. Tristem. 2000. Systematic screening of Anopheles mosquito genomes yields evidence for a major clade of Pao-like retrotransposons. Insect Mol. Biol. 9:109-117. [DOI] [PubMed] [Google Scholar]
- 10.Copeland, C. S., P. J. Brindley, O. Heyers, S. F. Michael, D. A. Johnston, D. L. Williams, A. C. Ivens, and B. H. Kalinna. 2003. Boudicca, a retrovirus-like long terminal repeat retrotransposon from the genome of the human blood fluke Schistosoma mansoni. J. Virol. 77:6153-6166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Covey, S. N. 1986. Amino acid sequence homology in gag region of reverse transcribing elements and the coat protein gene of cauliflower mosaic virus. Nucleic Acids Res. 14:623-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Franco, C., A. Terrinoni, P. Dimitri, and N. Junakovic. 1997. Intragenomic distribution and stability of transposable elements in euchromatin and heterochromatin of Drosophila melanogaster: elements with inverted repeats Bari 1, hobo, and pogo. J. Mol. Evol. 45:247-252. [DOI] [PubMed] [Google Scholar]
- 13.Dimitri, P., N. Junakovic, and B. Arca. 2003. Colonization of heterochromatic genes by transposable elements in Drosophila. Mol. Biol. Evol. 20:503-512. [DOI] [PubMed] [Google Scholar]
- 14.Drew, A. C., and P. J. Brindley. 1997. A retrotransposon of the non-long terminal repeat class from the human blood fluke Schistosoma mansoni. Similarities to the chicken-repeat-1-like elements of vertebrates. Mol. Biol. Evol. 14:602-610. [DOI] [PubMed] [Google Scholar]
- 15.Drew, A. C., D. J. Minchella, L. T. King, D. Rollinson, and P. J. Brindley. 1999. SR2 elements, non-long terminal repeat retrotransposons of the RTE-1 lineage from the human blood fluke Schistosoma mansoni. Mol. Biol. Evol. 16:1256-1269. [DOI] [PubMed] [Google Scholar]
- 16.Ganko, E. W., V. Bhattacharjee, P. Schliekelman, and J. F. McDonald. 2003. Evidence for the contribution of LTR retrotransposons to C. elegans gene evolution. Mol. Biol. Evol. 20:1925-1931. [DOI] [PubMed] [Google Scholar]
- 17.Gendrel, A. V., Z. Lippman, C. Yordan, V. Colot, and R. A. Martienssen. 2002. Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297:1871-1873. [DOI] [PubMed] [Google Scholar]
- 18.Grevelding, C. G. 1999. Genomic instability in Schistosoma mansoni. Mol. Biochem. Parasitol. 101:207-216. [DOI] [PubMed] [Google Scholar]
- 19.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 20.Hamilton, A., O. Voinnet, L. Chappell, and D. Baulcombe. 2002. Two classes of short interfering RNA in RNA silencing. EMBO J. 21:4671-4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu, W., Q. Yan, D. K. Shen, F. Liu, Z. D. Zhu, H. D. Song, X. R. Xu, Z. J. Wang, Y. P. Rong, L. C. Zeng, J. Wu, X. Zhang, J. J. Wang, X. N. Xu, S. Y. Wang, G. Fu, X. L. Zhang, Z. Q. Wang, P. J. Brindley, D. P. McManus, C. L. Xue, Z. Feng, Z. Chen, and Z. G. Han. 2003. Evolutionary and biomedical implications of a Schistosoma japonicum complementary DNA resource. Nat. Genet. 35:139-147. [DOI] [PubMed] [Google Scholar]
- 22.Huang, X., and A. Madan. 1999. CAP3: a DNA sequence assembly program. Genome Res. 9:868-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivanchenko, M. G., J. P. Lerner, R. S. McCormick, A. Toumadje, B. Allen, K. Fischer, O. Hedstrom, A. Helmrich, D. W. Barnes, and C. J. Bayne. 1999. Continuous in vitro propagation and differentiation of cultures of the intramolluscan stages of the human parasite Schistosoma mansoni. Proc. Natl. Acad. Sci. USA 96:4965-4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwashita, S., N. Osada, T. Itoh, M. Sezaki, K. Oshima, E. Hashimoto, Y. Kitagawa-Arita, I. Takahashi, T. Masui, K. Hashimoto, and W. Makalowski. 2003. A transposable element-mediated gene divergence that directly produces a novel type bovine Bcnt protein including the endonuclease domain of RTE-1. Mol. Biol. Evol. 20:1556-1563. [DOI] [PubMed] [Google Scholar]
- 25.Jiang, Y. W. 2002. Transcriptional cosuppression of yeast Ty1 retrotransposons. Genes Dev. 16:467-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Junakovic, N., A. Terrinoni, C. Di Franco, C. Vieira, and C. Loevenbruck. 1998. Accumulation of transposable elements in the heterochromatin and on the Y chromosome of Drosophila simulans and Drosophila melanogaster. J. Mol. Evol. 46:661-668. [DOI] [PubMed] [Google Scholar]
- 27.Kapitonov, V. V., and J. Jurka. 1999. The long terminal repeat of an endogenous retrovirus induces alternative splicing and encodes an additional carboxy-terminal sequence in the human leptin receptor. J. Mol. Evol. 48:248-251. [DOI] [PubMed] [Google Scholar]
- 28.Khan, E., J. P. Mack, R. A. Katz, J. Kulkosky, and A. M. Skalka. 1991. Retroviral integrase domains: DNA binding and the recognition of LTR sequences. Nucleic Acids Res. 19:851-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kidwell, M. G., and D. Lisch. 1997. Transposable elements as sources of variation in animals and plants. Proc. Natl. Acad. Sci. USA 94:7704-7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim, A., C. Terzian, P. Santamaria, A. Pelisson, N. Purd'homme, and A. Bucheton. 1994. Retroviruses in invertebrates: the gypsy retrotransposon is apparently an infectious retrovirus of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 91:1285-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laha, T., P. J. Brindley, C. K. Verity, D. P. McManus, and A. Loukas. 2002. pido, a non-long terminal repeat retrotransposon of the chicken repeat 1 family from the genome of the Oriental blood fluke, Schistosoma japonicum. Gene 284:149-159. [DOI] [PubMed] [Google Scholar]
- 32.Lai, W. S., E. Carballo, J. M. Thorn, E. A. Kennington, and P. J. Blackshear. 2000. Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to AU-rich elements and destabilization of mRNA. J. Biol. Chem. 275:17827-17837. [DOI] [PubMed] [Google Scholar]
- 33.Lerat, E., and P. Capy. 1999. Retrotransposons and retroviruses: analysis of the envelope gene. Mol. Biol. Evol. 16:1198-1207. [DOI] [PubMed] [Google Scholar]
- 34.Li, X., and M. S. Waterman. 2003. Estimating the repeat structure and length of DNA sequences using L-tuples. Genome Res. 13:1916-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Long, A. D., R. F. Lyman, A. H. Morgan, C. H. Langley, and T. F. Mackay. 2000. Both naturally occurring insertions of transposable elements and intermediate frequency polymorphisms at the achaete-scute complex are associated with variation in bristle number in Drosophila melanogaster. Genetics 154:1255-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lorincz, M. C., D. Schubeler, and M. Groudine. 2001. Methylation-mediated proviral silencing is associated with MeCP2 recruitment and localized histone H3 deacetylation. Mol. Cell. Biol. 21:7913-7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luan, D. D., M. H. Korman, J. L. Jakubczak, and T. H. Eickbush. 1993. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72:595-605. [DOI] [PubMed] [Google Scholar]
- 38.Mager, D. L., D. G. Hunter, M. Schertzer, and J. D. Freeman. 1999. Endogenous retroviruses provide the primary polyadenylation signal for two new human genes (HHLA2 and HHLA3). Genomics 59:255-263. [DOI] [PubMed] [Google Scholar]
- 39.Makalowski, W. 2003. Genomics. Not junk after all. Science 300:1246-1247. [DOI] [PubMed] [Google Scholar]
- 40.Malik, H. S., W. D. Burke, and T. H. Eickbush. 1999. The age and evolution of non-LTR retrotransposable elements. Mol. Biol. Evol. 16:793-805. [DOI] [PubMed] [Google Scholar]
- 41.Malik, H. S., and T. H. Eickbush. 2001. Phylogenetic analysis of ribonuclease H domains suggests a late, chimeric origin of LTR retrotransposable elements and retroviruses. Genome Res. 11:1187-1197. [DOI] [PubMed] [Google Scholar]
- 42.McFadden, J., and G. Knowles. 1997. Escape from evolutionary stasis by transposon-mediated deleterious mutations. J. Theor. Biol. 186:441-447. [DOI] [PubMed] [Google Scholar]
- 43.Medstrand, P., J. R. Landry, and D. L. Mager. 2001. Long terminal repeats are used as alternative promoters for the endothelin B receptor and apolipoprotein C-I genes in humans. J. Biol. Chem. 276:1896-1903. [DOI] [PubMed] [Google Scholar]
- 44.Miura, A., S. Yonebayashi, K. Watanabe, T. Toyama, H. Shimada, and T. Kakutani. 2001. Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 411:212-214. [DOI] [PubMed] [Google Scholar]
- 45.Mullikin, J. C., and Z. Ning. 2003. The Phusion Assembler. Genome Res. 13:81-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Navarro-Quezada, A., and D. J. Schoen. 2002. Sequence evolution and copy number of Ty1-copia retrotransposons in diverse plant genomes. Proc. Natl. Acad. Sci. USA 99:268-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 48.Paquola, A., M. Nishiyama, Jr., E. M. Reis, A. M. daSilva, and S. Verjovski-Almeida. 2003. ESTWeb: bioinformatics services for EST sequencing projects. Bioinformatics 19:1587-1588. [DOI] [PubMed] [Google Scholar]
- 49.Paquola, A. C. M., A. A. Machado, E. M. Reis, A. M. da Silva, and S. Verjovski-Almeida. 2003. Zerg: a very fast BLAST parser library. Bioinformatics 19:1035-1036. [DOI] [PubMed] [Google Scholar]
- 50.Queiroz, R. S. 1991. O herói-trapaceiro. Reflexões sobre a figura do trickster. Tempo Social Rev. Sociol. USP 3:93-107. [Google Scholar]
- 51.Rountree, M. R., K. E. Bachman, and S. B. Baylin. 2000. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 25:269-277. [DOI] [PubMed] [Google Scholar]
- 52.Sandmeyer, S. B., L. J. Hansen, and D. L. Chalker. 1990. Integration specificity of retrotransposons and retroviruses. Annu. Rev. Genet. 24:491-518. [DOI] [PubMed] [Google Scholar]
- 53.Schramke, V., and R. Allshire. 2003. Hairpin RNAs and retrotransposon LTRs effect RNAi and chromatin-based gene silencing. Science 301:1069-1074. [DOI] [PubMed] [Google Scholar]
- 54.Simpson, A. J., A. Sher, and T. F. McCutchan. 1982. The genome of Schistosoma mansoni: isolation of DNA, its size, bases and repetitive sequences. Mol. Biochem. Parasitol. 6:125-137. [DOI] [PubMed] [Google Scholar]
- 55.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Verjovski-Almeida, S., R. DeMarco, E. A. Martins, P. E. Guimaraes, E. P. Ojopi, A. C. Paquola, J. P. Piazza, M. Y. Nishiyama, J. P. Kitajima, R. E. Adamson, P. D. Ashton, M. F. Bonaldo, P. S. Coulson, G. P. Dillon, L. P. Farias, S. P. Gregorio, P. L. Ho, R. A. Leite, L. C. Malaquias, R. C. Marques, P. A. Miyasato, A. L. Nascimento, F. P. Ohlweiler, E. M. Reis, M. A. Ribeiro, R. G. Sa, G. C. Stukart, M. B. Soares, C. Gargioni, T. Kawano, V. Rodrigues, A. M. Madeira, R. A. Wilson, C. F. Menck, J. C. Setubal, L. C. Leite, and E. Dias-Neto. 2003. Transcriptome analysis of the acoelomate human parasite Schistosoma mansoni. Nat. Genet. 35:148-157. [DOI] [PubMed] [Google Scholar]
- 57.Walsh, C. P., J. R. Chaillet, and T. H. Bestor. 1998. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 20:116-117. [DOI] [PubMed] [Google Scholar]
- 58.Withers-Ward, E. S., Y. Kitamura, J. P. Barnes, and J. M. Coffin. 1994. Distribution of targets for avian retrovirus DNA integration in vivo. Genes Dev. 8:1473-1487. [DOI] [PubMed] [Google Scholar]
- 59.World Health Organization. 2002. TDR strategic direction for research: schistosomiasis. World Health Organization, Geneva, Switzerland.
- 60.Yoder, J. A., C. P. Walsh, and T. H. Bestor. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13:335-340. [DOI] [PubMed] [Google Scholar]
- 61.Zilberman, D., X. Cao, and S. E. Jacobsen. 2003. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299:716-719. [DOI] [PubMed] [Google Scholar]