Abstract
The pseudorabies virus (PrV) proteins UL11, glycoprotein E (gE), and gM are involved in secondary envelopment of tegumented nucleocapsids in the cytoplasm. To assess the relative contributions of these proteins to the envelopment process, virus mutants with deletions of either UL11, gM, or gE as well as two newly constructed mutant viruses with simultaneous deletions of UL11 and gE or of UL11 and gM were analyzed in cell culture for their growth phenotype. We show here that simultaneous deletion of UL11 and gE reduced plaque size in an additive manner over the reduction observed by deletion of only UL11 or gE. However, one-step growth was not further impaired beyond the level of the UL11 deletion mutant. Moreover, in electron microscopic analyses PrV-ΔUL11/gE exhibited a phenotype similar to that of the UL11 mutant virus. In contrast, plaque formation was virtually abolished by the simultaneous absence of UL11 and gM, and one-step growth was significantly reduced. Electron microscopy showed the presence of huge intracytoplasmic inclusions in PrV-ΔUL11/gM-infected cells, with a size reaching 3 μm and containing nucleocapsids embedded in tegument. We hypothesize that UL11 and gM are involved in different steps during secondary envelopment and that simultaneous deletion of both interrupts both processes, resulting in the observed drastic impairment of secondary envelopment.
The herpesvirus virion is composed of four structural components: the core, which contains the double-stranded DNA genome; the icosahedral capsid; the tegument; and a lipid bilayer envelope derived from cellular cytoplasmic membranes, which contains virally encoded glycoproteins (31). The tegument and envelope are highly complex structures, with each containing more than 10 different virally encoded proteins (35). Although the major constituents of tegument and envelope have been identified, at least for the alphaherpesviruses pseudorabies virus (PrV) and herpes simplex virus type 1 (HSV-1), the molecular mechanisms that drive the assembly of the viral tegument and envelope during maturation of herpesvirions are still largely unknown. Herpesviruses acquire their final tegument and envelope in the cytoplasm by budding of nucleocapsids into trans-Golgi-derived vesicles after having traversed the nuclear membrane by a process which includes acquisition of a primary envelope by budding of intranuclear capsids at the inner leaflet and subsequent fusion at the outer leaflet of the nuclear membrane (27).
Our studies focus on the elucidation of molecular interactions that are required for tegument formation and secondary envelopment of PrV. We demonstrated that envelope glycoprotein M (gM) and gE play an important role in secondary envelopment (3, 4), which includes their interaction with the major tegument protein UL49 via their cytoplasmic tails (9). Both gM and gE are present in heterodimeric complexes with gN and gI, respectively (13, 38). In the absence of gE/gI and gM, intracytoplasmic capsids accumulated in aggregates which also contain tegument proteins. This demonstrated that in the absence of these envelope glycoproteins, capsids still acquire tegument proteins but secondary envelopment is blocked (3, 4), indicating that tegumentation of capsids can be separated from the budding process in secondary envelopment. Recently, we identified and characterized the UL11 protein of PrV as a membrane-associated tegument protein that is not necessary for virus replication (21). Analysis of a UL11 deletion mutant showed that the PrV UL11 protein is involved in secondary envelopment in virion morphogenesis. The lack of UL11 resulted in slightly reduced virus titers and in accumulation of unenveloped capsids in the cytoplasm, sometimes in association with tegument proteins in aggregations which resemble those observed in the absence of gE/gI and gM. Moreover, lack of UL11 apparently distorts the architecture of Golgi-derived membranes, i.e., of the normal site of secondary envelopment. The absence of the UL11-homologous proteins of HSV-1 (1) and human cytomegalovirus (33) has also been shown to impair virion formation in the cytoplasm.
Whereas UL11 and gM represent proteins which are conserved throughout the herpesvirus family, gE is present only in the alphaherpesviruses. To test the relative contributions of the individual proteins to virion morphogenesis of PrV, mutants with either of these proteins deleted were compared to newly isolated double mutants simultaneously lacking either UL11 and gE or UL11 and gM.
MATERIALS AND METHODS
Viruses and cells.
All PrV mutants were derived from the wild-type laboratory strain Kaplan (PrV-Ka) (14). Viruses were grown on rabbit kidney (RK13) or porcine kidney cells in Eagle's minimum essential medium supplemented with 10 or 5% fetal calf serum, respectively. Complementing UL11-expressing RK13-UL11, gM-expressing RK13-gM, and gE-expressing RK13-gE cells have been described previously (3, 21). The UL11/gM and UL11/gE double mutants (see below) are based on PrV-ΔUL11 which contains a deletion of codons 16 to 45 of the UL11 open reading frame, which were replaced by an Flp recombinase recognition target site consisting of 36 bp of foreign noncoding DNA. Expression of the N-terminal part of the protein containing the predicted N-myristoylation site was also excluded by site-specific mutagenesis of the ATG initiation codon to GTG. Moreover, at the third codon the sequence CAG was replaced by the stop codon TAG (21) (Fig. 1).
FIG. 1.
Construction of virus mutants. (A) A schematic map of the PrV genome shows the unique long (UL) and unique short (US) regions, the inverted repeat sequences (IR and TR), and the positions of BamHI restriction fragments. (B) Enlargement of the PrV UL10/11 gene region. The UL14, UL13, UL12, and UL11 genes are transcribed into 3′-coterminal mRNAs which share a common polyadenylation signal (arrow pointing down). The UL10 gene, which is transcribed in opposite direction into a monocistronic mRNA that terminates at a polyadenylation signal downstream from the UL10 open reading frame (arrow pointing up), is separated from the UL14-UL11 transcription unit by a region containing direct repeat elements (repeats). Relevant restriction sites are indicated. (C) Construction of PrV-ΔUL11/gM. The deletion and point mutations within the UL11 open reading frame are shown, as is the deletion of UL10 sequences and concomitant insertion of the kanamycin resistance (KanR) cassette. (D) Enlargement of the PrV gE gene region. (E) Construction of PrV-ΔgE.
Isolation of PrV-ΔgE.
To generate a gE-negative mutant, a recombinant plasmid comprising 5′ and 3′ flanking sequences of the gene to be deleted and a green fluorescent protein (GFP) marker gene cassette was assembled. To this end, part of the US7 gene encoding gI was amplified by PCR from cloned BamHI fragment 7 of viral DNA with primers EDELF and EDELR (Table 1), giving rise to a 1,086-bp PCR product. A second, 1,013-bp subfragment comprising part of the US8 gene encoding the C terminus of gE was amplified by using primers EDELF2 and EDELR2. Both PCR products were directly cloned into appropriately cleaved plasmid pUC19, giving rise to plasmid pUC19-EDEL1/2. In addition, a GFP marker gene cassette was inserted into the BamHI site between the two PCR fragments, giving rise to plasmid pUC19-EDEL1/2GFP. Of the US8 gene, the last 35 codons remained, to avoid interference with transcription of the following US9 gene (Fig. 1). Both plasmids, pUC19-EDEL1/2GFP and pUC19-EDEL1/2, were partially sequenced to verify correct deletion. Plasmid pUC19-EDEL1/2GFP was then used for cotransfection with PrV-Ka DNA into RK13 cells. A marker gene-expressing mutant, PrV-ΔgEGFP, was isolated on RK13 cells. To eliminate possible unwanted effects caused by insertion of the GFP cassette, a mutant without a marker gene was subsequently constructed. After cotransfection of PrV-ΔgEGFP DNA and plasmid pUC19-EDEL1/2 into RK13 cells by calcium phosphate precipitation (11), a nonfluorescing mutant virus could be isolated. One plaque isolate, designated PrV-ΔgE, was chosen for further analysis. Correct recombination was verified by Southern blot analysis of mutant virus DNA (34) (data not shown).
TABLE 1.
Primers used for construction of mutant viruses
| Primer | Sequencea | Restriction enzyme | Nucleotide position | GenBank accession no. |
|---|---|---|---|---|
| EDELF | 5′-CACAGAATTCCGGGGGCGTCCTCTTCAGGG-3′ | EcoRI | 111-129 | M14336 |
| EDELR | 5′-CACAGGATCCGCGCGGACGGGAGATAAAACGCC-3′ | BamHI | 1177-1156 | M14336 |
| EDELF2 | 5′-CACAAGGATCCCCCGCTCCGGCTTCGACG-3′ | BamHI | 2854-2871 | M14336 |
| EDELR2 | 5′-CACAAAGCTTGCTTCGAGCCGTCCGCCG-3′ | HindIII | 559-542 | D10452 |
| gM FOR | 5′-CACAGAATTCCGTCCGCCGCCATGTGC-3′ | EcoRI | 3596-3580 | X97257 |
| gM REV | 5′-CACACTCGAGCCATGTGCGATCGAAACGAACG-3′ | XhoI | 2381-2402 | X97257 |
| KAN950 NEWFW | 5′-TCCGGATCCCGATTTATTCAACAAAGCCAGG-3′ | BamHI | 1800-1819 | X06402 |
| KAN950 NEVRV | 5′-TTCGAATTCGCCAGTGTTACAACCAATTAACC-3′ | EcoRI | 2769-2747 | X06402 |
Underlining indicates the restriction enzyme site.
Isolation of PrV-ΔgM.
The previously constructed gM deletion mutant of PrV, PrV-ΔgMβ (7), contained a deletion of ca. 60% of the gM gene and insertion of a lacZ expression cassette. To generate an improved gM-negative PrV mutant based on a bacterial artificial chromosome (BAC), plasmid pcDNA-gM (3) was digested with HindIII and EcoRI to remove the vector-specific KpnI site, resulting in plasmid pcDNA-gMΔEH. Then, a 970-bp PCR-amplified kanamycin resistance gene linked with EcoRI and BamHI sites from pACYC177 (New England Biolabs, Frankfurt, Germany) by using primers KAN950NEWFW and KAN950NEVRV (Table 1) was inserted into SanDI- and KpnI-digested pcDNA-gMΔEH, resulting in deletion of a 666-bp fragment from the gM-encoding UL10 gene. The resulting 1.53-kbp insert pcDNA3-gMK fragment was amplified by PCR with primers gM FOR and gM REV (Table 1) and Pfx DNA polymerase.
The resulting product was used for mutagenesis of BAC pPrV-ΔgB (21) in Escherichia coli, utilizing the Red recombinase of bacteriophage λ. To this end, bacteria containing pPrV-ΔgB were transformed with the helper plasmid pKD46, which carries the Red recombinase under control of an arabinose-inducible promoter (6). After induction, the cells were transformed by electroshock with the 1.53-kbp pcDNA3-gMK PCR product, and recombinant clones were selected on agar plates containing 30 μg of chloramphenicol and 50 μg of kanamycin per ml. The mini-F plasmid vector and the adjacent enhanced GFP expression cassette were then removed from the gB locus after cotransfection of RK13 cells with BAC DNA and plasmid pUC-B1BclI, which contains the authentic gB gene of strain PrV-Ka within a 6,971-bp BclI fragment (21). The resulting mutant, PrV-ΔgM, was characterized by restriction analysis and Southern blot hybridization (data not shown).
Isolation of PrV-ΔUL11/gE.
A PrV-ΔUL11/gE deletion mutant was generated after cotransfection of PrV-ΔUL11 DNA and plasmid EDEL1/2 GFP into RK13-UL11 cells by calcium phosphate coprecipitation (11). Green fluorescent plaques were picked and purified. To eliminate possible unwanted effects caused by insertion of the GFP cassette, a mutant without a marker gene was also constructed. After cotransfection of PrV-ΔUL11/gEGFP DNA and pUC19-EDEL1/2 into RK13 cells, plaques lacking GFP autofluorescence were picked and purified. One virus isolate, PrV-ΔUL11/gE, was randomly chosen for further analysis.
Isolation of PrV-ΔUL11/gM.
To obtain PrV-ΔUL11/gM, the 1.53-kbp pcDNA3-gMK PCR product was introduced by electroshock into bacteria containing pPrV-ΔUL11 (21), and recombination was induced with pKD46 helper plasmid. The resulting clones were selected on agar plates containing ampicillin and kanamycin. The obtained pPrV-ΔUL11/gM BAC DNA and plasmid pUC-B1BclI were cotransfected into RK13-UL11 cells to restore gB expression (21). One plaque isolate, PrV-ΔUL11/gM, was chosen for further analysis. Correct recombination was verified by restriction analysis and Southern blot hybridization of mutant virus DNA by standard procedures (34) (data not shown).
Virus purification and immunoblotting.
For virus purification, porcine kidney (PSEK) cells were infected with PrV-Ka, PrV-ΔUL11, PrV-ΔgE, PrV-ΔgM, PrV-ΔUL11/gM, and PrV-ΔUL11/gE and incubated until a complete cytopathic effect developed. The remaining intact cells were lysed by freezing (−70°C) and thawing (37°C), and cellular debris was removed by low-speed centrifugation. For reduction of volume, the virus-containing supernatant was centrifuged for 1 h at 64,000 × g. The pellet was resuspended in TBSal (200 mM NaCl, 2.6 mM KCl, 10 mM Tris-HCl [pH 7.5], 20 mM MgCl2, 1.8 mM CaCl2), layered onto a discontinuous sucrose gradient (30, 40, and 50% sucrose), and centrifuged for 2 h at 20,000 rpm in a TST-28 rotor. Virions accumulated at the boundary between 40 and 50% sucrose. They were harvested by aspiration, pelleted, and resuspended in TBSal. Purified virions (3 μg per lane) were separated by electrophoresis in sodium dodecyl sulfate (SDS)-10 or 15% polyacrylamide gels (23), electrotransferred onto nitrocellulose membranes (36), and reacted for 1 h at room temperature with monospecific antisera against the UL11 (21) (dilution, 1:20,000), UL37 (16) (dilution, 1:100,000), UL49 (4) (dilution, 1:100,000), and US3 (18) (dilution, 1:100,000) tegument proteins as well as glycoprotein gM (21) (dilution, 1:10,000) and with monoclonal antibodies against glycoproteins gB (b43-b5; dilution, 1:500) (29), gC (B16-c8; dilution, 1:100) (18), and gE (A9-b15; dilution, 1:100) (29). After incubation with peroxidase-conjugated secondary antibody (Dianova, Hamburg, Germany), bound antibody was detected by enhanced chemiluminescence (Supersignal; Pierce, Bonn, Germany) recorded on X-ray film.
Determination of plaque size.
Plaque sizes were measured after titration of virus mutants on various cell lines after 2 days of incubation at 37°C under a methylcellulose overlay. Thereafter, cells were fixed with 5% formaldehyde and stained with crystal violet. For each combination of virus and cells, 50 plaques were measured microscopically, and the average plaque size was determined. Values were calculated compared to those of PrV-Ka, which were set at 100%. Average percentages as well as standard deviations were determined from three independent experiments.
One-step growth analysis.
To monitor one-step growth, normal, gM-, gE-, or UL11-expressing cells were infected with virus mutants at a multiplicity of infection (MOI) of 5 for 1 h at 4°C. Thereafter, the inoculum was replaced with prewarmed medium, and virus was allowed to penetrate for 60 min at 37°C. The remaining extracellular virus was then inactivated by low-pH treatment (26). Cells and supernatants were harvested immediately thereafter (0 h) and after 4, 8, 12, 24, and 36 h of incubation at 37°C. The virus progeny was titrated on RK13 and RK13-UL11 cells. Average values and standard deviations from three independent experiments were calculated.
Electron microscopy.
For ultrathin sectioning, normal, gM-, gE-, or UL11-expressing cells were infected at an MOI of 1 and fixed at 12 h postinfection. Fixation, dehydration, and embedding for routine microscopy and for intracellular immunolabeling of viral proteins were performed as described previously (12, 12a). Reactivity of monospecific antisera against the UL36 (19), UL37 (16), UL46 (20), UL47 (20), UL49 (4), and US3 (18) proteins was visualized with 10-nm-diameter-gold-tagged secondary anti-rabbit antibodies (GAR 10; British Biocell International). The counterstained ultrathin sections were analyzed with an electron microscope (EM 400T, Tecnai 12; Philips, Eindhoven, The Netherlands).
RESULTS
Immunoelectron microscopy of PrV-ΔUL11.
In PrV-ΔUL11-infected rabbit kidney cells, intracytoplasmic membranes were distorted and appeared to form tightly packed stacks that were not present in cells infected by wild-type virus (Fig. 2A) (21). Moreover, unenveloped capsids accumulated in the cytoplasm associated with electron-dense material which resembled tegument. For further analyses of these intracytoplasmic aggregates, immunoelectron microscopy was performed. The electron-dense material within the clusters was heavily labeled with the UL46 antiserum (Fig. 2B) as well as with antisera against the UL47 (Fig. 2C), UL48 (Fig. 2D), and UL49 proteins (Fig. 2E). Thus, this material apparently represents bona fide tegument. Interestingly, antisera against the UL36 and UL37 proteins exclusively labeled the nucleocapsids associated with these aggregates and not the electron-dense material in the middle (Fig. 2F and G). This further indicates that UL36 and UL37 are indeed capsid-associated tegument proteins that are acquired during tegumentation before addition of the other tegument components, such as the UL46, UL47, UL48, or UL49 protein. A two-step nucleation of tegumentation has recently been proposed (27). In contrast, the US3 protein could be detected within the electron-dense material as well as at the capsids (Fig. 2H). US3 has recently been demonstrated to be a tegument protein of mature virus particles that is also part of primary enveloped virions and is either retained during nuclear egress or reacquired very early during tegumentation (12a). These data show that primary tegumentation of capsids with UL36 and UL37 occurs independent of UL11 and that the addition of the other tegument proteins to produce mature virions is impaired in the absence of UL11.
FIG. 2.
Immunoelectron microscopy of inclusions in PrV-ΔUL11-infected cells. RK13 cells infected with PrV-ΔUL11 were analyzed by conventional (A) or immunoelectron (B to H) microscopy. Sections were labeled with monospecific rabbit antisera against the UL46 (B), UL47 (C), UL48 (D), UL49 (E), UL36 (F), UL37 (G), or US3 (H) tegument protein. Reactivity of the antisera was visualized after incubation with 10-nm-diameter-gold-tagged secondary anti-rabbit antibodies. Bars, 250 nm.
Isolation and characterization of UL11/gE and UL11/gM deletion mutants.
In previous experiments we demonstrated a role for gE and gM in secondary envelopment (3, 4). To assay for a possible functional interplay with UL11, PrV mutants that simultaneously lack either UL11 and gE or UL11 and gM were constructed. To investigate whether efficient incorporation of different structural proteins into the virion depends on the presence of UL11, gM, and gE, virions of PrV-Ka, PrV-ΔUL11, PrV-ΔgE, and PrV-ΔgM as well as PrV-ΔUL11/gE and PrV-ΔUL11/gM were purified from the supernatants of infected cells by sucrose gradient centrifugation. Equal amounts of protein were separated by SDS-polyacrylamide gel electrophoresis and analyzed in Western blots (Fig. 3). Whereas the tegument proteins UL37, US3, and UL49, as well as glycoproteins gB and gC, were present in all preparations, gE was absent in the gE deletion mutants and gM was not detectable in the gM deletion mutants, verifying the correct phenotype as expected from the introduced mutations. Also, as expected, the UL11 protein was absent from PrV-ΔUL11, PrV-ΔUL11/gE, and PrV-ΔUL11/gM. Thus, the UL11, gE, and gM gene products, either singly or in combination, are not required for efficient virion localization of any of the investigated viral structure proteins.
FIG. 3.
Characterization of mutant virions. Virions of wild-type PrV-Ka or deletion mutants PrV-ΔUL11, PrV-ΔgE, PrV-ΔgM, PrV-ΔUL11/gE, and PrV-ΔUL11/gM were purified from the supernatant of infected cells by discontinuous sucrose gradient centrifugation and lysed, and viral proteins were separated by SDS-polyacrylamide gel electrophoresis. After transfer to nitrocellulose filters, blots were probed with monospecific antisera or monoclonal antibodies against the UL37, US3, UL49, and UL11 tegument proteins and the envelope proteins gB, gE, gM, and gC. Locations of molecular mass markers are indicated on the left.
In vitro growth properties of PrV-ΔUL11/gE.
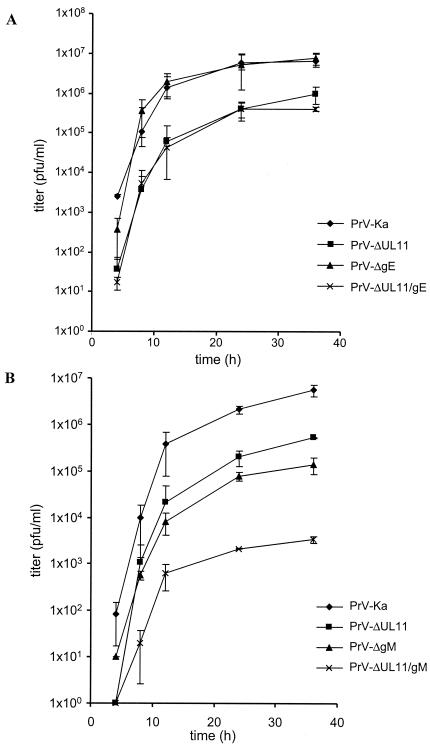
Initial characterization of PrV-ΔUL11/gE on noncomplementing cells had already indicated that simultaneous deletion of these two nonessential proteins did not drastically impair viral replication. This is reflected by the results of one-step growth analyses (Fig. 4A). On normal rabbit kidney cells PrV-ΔUL11/gE exhibited growth properties similar to those of PrV-ΔUL11. Both virus mutants exhibited decreased titers at all time points, with final titers reduced ca. 10-fold compared to those of PrV-Ka. PrV-ΔgE exhibited growth properties similar to those of PrV-Ka, with no apparent defect in one-step replication in vitro.
FIG. 4.
One-step growth analyses. RK13 cells were infected with the indicated virus mutants at an MOI of 5 and harvested at the indicated times after infection. Average values and standard deviations from three independent experiments are shown.
In contrast, plaque sizes of PrV-ΔUL11/gE (Fig. 5A) were reduced by ca. 90% compared to those of PrV-Ka, whereas deletion of gE resulted in only ca. 30% and that of UL11 resulted in ca. 60% reduced plaque sizes. Thus, there appears to be an additive effect of the gene deletions on plaque size, indicating that UL11 and gE are involved in distinct steps of plaque formation. On gE-expressing cells, PrV-ΔUL11/gE was complemented to approximately the level of PrV-ΔUL11. Although cell-to-cell spread of PrV-ΔUL11/gE was clearly improved on RK13-UL11 cells, for unknown reasons it did not reach the level of the PrV-ΔgE mutant. In contrast, complementation of the single mutants to wild-type levels occurred on UL11- or gE-expressing rabbit kidney cells. Thus, the deletion of UL11/gE from PrV-Ka severely impaired its capacity for plaque formation, i.e., direct viral cell-to-cell spread without additionally affecting viral replication beyond the level of the UL11 deletion mutant.
FIG. 5.
Determination of plaque size. Noncomplementing RK13 as well as UL11-expressing (RK13-UL11), gE-expressing (RK13-gE), or gM-expressing cells (RK13-gM) were infected with the indicated virus mutants under plaque assay conditions and incubated under a methylcellulose overlay. Plaques were microscopically analyzed 2 days after infection. Relative plaque sizes were calculated compared to those of wild-type PrV-Ka on noncomplementing cells, which was set at 100%. Average values and standard deviation from three independent experiments are shown.
In vitro growth properties of PrV-ΔUL11/gM.
In contrast to PrV-ΔUL11/gE, mutant PrV-ΔUL11/gM exhibited a drastic growth defect, indicating that simultaneous deletion of UL11 and gM severely impairs viral replication. To analyze this defect in more detail, one-step growth kinetics were analyzed on noncomplementing RK13 cells. As shown in Fig. 4B, PrV-ΔUL11 replicated to ca. 10-fold-lower titers and PrV-ΔgM replicated to ca. 50-fold-lower titers than PrV-Ka. In the simultaneous absence of UL11 and gM, titers were again ca. 100-fold lower than those of PrV-ΔUL11 and PrV-ΔgM, with final titers reaching only 103 PFU/ml. This indicated a synergistic effect of the UL11 and gM deletions on one-step replication. On UL11-expressing cells, PrV-ΔUL11 exhibited a growth rate similar to that of PrV-Ka, and the growth defect of PrV-ΔUL11/gM could be compensated to the level of PrV-ΔgM, indicating that, as expected, RK13-UL11 cells rescued the UL11 defect but not the gM-associated defect. The same could be observed in one-step growth kinetics on gM-expressing cells in which the gM defect of PrV-ΔgM and PrV-ΔUL11/gM was rescued (data not shown). To assay the ability of PrV-ΔUL11/gM to form plaques on normal and trans-complementing cells, RK13, RK13-UL11, and RK13-gM cells were infected with the different virus mutants. Two days after infection under plaque assay conditions, monolayers were fixed and stained with crystal violet. As shown in Fig. 5B, plaques produced by PrV-ΔUL11/gM were approximately 90 to 95% reduced compared to those produced by PrV-Ka. This defect could be corrected on UL11-expressing cells to approximately wild-type levels (PrV-ΔgM produces plaques that are only slightly reduced in size compared to PrV-Ka) and on RK13-gM cells up to levels of PrV-ΔUL11, which are ca. 70% reduced compared to wild-type PrV. Thus, simultaneous deletion of UL11 and gM caused a distinct synergistic effect on virus replication and direct viral cell-to-cell spread.
Electron microscopy.
To pinpoint the defect in cells infected with the double mutants PrV-ΔUL11/gE and PrV-ΔUL11/gM, electron microscopic analyses were performed. To this end, noncomplementing rabbit kidney cells were infected at an MOI of 1. Twelve hours after infection, cells were fixed, stained, and analyzed. As shown in Fig. 6, cells infected with PrV-ΔUL11/gE, besides showing all stages of intracellular virion maturation (i.e., capsid assembly in the nucleus, transit through the nuclear membrane with primary envelopment, and secondary envelopment in the trans-Golgi area, including extracellular mature virions), exhibited small local clusters of electron-dense tegument material surrounded by unenveloped capsids in the cytoplasm, as observed in cells infected with the single mutant PrV-ΔUL11 (21) (Fig. 2A). Furthermore, distorted membranes could also be observed in the cytoplasm in association with the tegument clusters that are not detectable in PrV-Ka infected cells. Thus, ultrastructurally the phenotype of PrV-ΔUL11/gE was indistinguishable from that of PrV-ΔUL11, which parallels the results from the one-step growth analyses.
FIG. 6.
Electron microscopy of PrV-ΔUL11/gE. RK13 cells were infected with PrV-ΔUL11/gE at an MOI of 1 and fixed 12 h after infection. (A) Overview of an infected cell, demonstrating the presence of intracytoplasmic aggregates containing nucleocapsids and tegument proteins (see also Fig. 2). (B) Higher-magnification view showing distorted membranes (arrows) as well as capsid-tegument aggregates (closed triangles) and envelopment stages (open triangles). Bars, 2 μm (A) and 1 μm (B).
In contrast, drastic effects were observed in PrV-ΔUL11/gM-infected rabbit kidney cells (Fig. 7). Ultrastructural analyses revealed that in the absence of UL11 and gM, very large intracytoplasmic aggregations were formed, which contained electron-dense tegument material associated with capsids. The size of these clusters could reach 3 μm (Fig. 7B), and most infected cells contained only two or three of them (Fig. 7A). Although these inclusions resembled in appearance those observed in cells infected with the triple mutant PrV-gEIM− (3), intracytoplasmic inclusions of this size had never been observed before in herpesvirus-infected cells. Secondary envelopment and release of enveloped virions were not observed, which correlated with the drastically impaired one-step replication of PrV-ΔUL11/gM (Fig. 4B). Figure 8A shows a possible intermediate step in formation of these inclusions, with condensed tegument material lined with nucleocapsids at the center, surrounded by capsids embedded in a less electron-dense matrix. We hypothesize that in the outer area tegument material did not yet become as closely packed as in the center but already interacted with capsids. Interestingly, nucleocapsid aggregations within densely packed areas of the inclusions seem to follow hexagonal symmetry (Fig. 8B), which correlates with the assumption of an ordered structure extending beyond the capsid (37). Despite the distinct impairment of secondary envelopment in the cytoplasm, the absence of UL11 and M did not visibly affect primary envelopment in the nucleus or nuclear egress of virions.
FIG. 7.
Electron microscopy of PrV-ΔUL11/gM. RK13 cells were infected with PrV-ΔUL11/gM at an MOI of 1 and fixed 12 h after infection. (A) Overview of an infected cell with two huge intracytoplasmic aggregations of capsids and tegument. (B) Inclusion at a higher magnification. Bars, 2 μm (A) and 1 μm (B).
FIG. 8.
Ultrastructure of inclusions in PrV-ΔUL11/gM-infected cells. Cells were infected and processed as described in the legend to Fig. 7. (A) An apparent intermediate step in the formation of the aggregates, with condensed tegument in the middle lined by associated capsids and more tegument material, presumably in the process of condensation, around the inclusions. nuc, nucleus; cyt, cytoplasm. (B) The arrangement of capsids within the aggregates may follow a hexagonal symmetry. Fivefold (arrowheads) and sixfold (arrows) symmetry levels are indicated. Bars, 500 nm.
To verify that these effects were indeed a result of the introduced deletions, complementing cells were infected with the double mutants and compared with single mutants which yielded the expected phenotypes (data not shown). Since simultaneous deletion of UL11 and gM resulted in a phenotype which is significantly more dramatic than that of the single mutants, we suggest that the two proteins participate in different steps in secondary envelopment.
DISCUSSION
Herpesvirus maturation requires two different budding steps. First, capsids that were formed in the nucleus bud at the inner leaflet of the nuclear membrane, thereby acquiring a primary envelope which is subsequently lost by fusion with the outer leaflet. In this process, a complex of the UL31 primary tegument and UL34 primary envelope protein, which are conserved throughout the herpesviruses, plays an important role (1, 5, 8, 17, 28, 30, 32). Final maturation of virions then occurs in the cytoplasm by budding of intracytoplasmic capsids into Golgi-derived vesicles (10, 12; reviewed in reference 27). During this maturation process, more than 20 different tegument and envelope proteins are assembled into the mature virion. Recent data indicate that formation of mature virions follows an intricate network of protein-protein interactions with a surprising functional redundancy (reviewed in reference 27). Although the participating tegument and envelope proteins are largely known, knowledge of their interaction required for virion incorporation is only recently beginning to emerge. Tegumentation may start at two sites, the nucleocapsid and the future envelopment site. At the nucleocapsid, the UL36 tegument protein presumably forms the innermost layer of tegument anchored at the vertices of the capsid (37). Capsid-associated UL36 protein nucleates tegumentation at the capsid by interaction with the conserved UL37 protein (19; M. E. Harmon and W. Gibson, Proc. Am. Soc. Virol., abstr. W35-4, p. 144, 1996). The cytoplasmic tails of the nonconserved gE and of the conserved gM have been shown to interact with the UL49 tegument protein, which may initiate tegument assembly at the future budding site (9). In the absence of gE and gM, secondary envelopment is blocked and intracytoplasmic capsids accumulate in the cytoplasm embedded in tegument (3). Similar but less prominent aggregations of tegument and capsids were also detected in the absence of gM alone (4) or when the UL11 protein, a membrane-apposed tegument component, was deleted (21).
Since all three proteins, i.e., UL11, gM, and gE, appeared to be involved in secondary envelopment, we investigated whether their function can be separated by constructing doubly deleted virus mutants lacking either UL11 and gM or UL11 and gE. A mutant lacking gE and gM has previously been characterized (3). We demonstrate here that the simultaneous absence of the two conserved proteins UL11 and gM drastically impairs virion maturation, resulting in the formation of large intracytoplasmic inclusions containing capsids and tegument. Whereas the overall appearance of these inclusions resembled that found in cells infected with UL11 and gM single mutants (3, 21), their size was drastically increased, reaching up to 3 μm in diameter. Most infected cells contained only two or three of them. The shape of these inclusions may vary from round (Fig. 7) to irregular (Fig. 8). Since simultaneous deletion of UL11 and gM resulted in a much more drastic impairment of viral replication than single deletion of either protein, we hypothesize that the two proteins, although both involved in secondary envelopment, mediate different steps in this process.
UL11 of HSV-1 has previously been demonstrated to contain targeting information for Golgi membranes. A hybrid protein consisting of HSV-1 UL11 fused to the human immunodeficiency virus type 1 Gag protein was relocated from the plasma membrane to the Golgi and was shown to bud into Golgi-derived vesicles (2). Therefore, it seems reasonable to hypothesize that UL11 may be involved in targeting tegument proteins or even (partially) tegumented capsids to the secondary envelopment site. A direct interaction between the UL11 and UL16 tegument proteins of HSV-1 has recently been demonstrated (25). In contrast, PrV gM has been observed to reduce cell-cell fusion in a transient expression-fusion assay (15), presumably by retention of fusogenic glycoproteins in the Golgi apparatus or efficient retrieval from the plasma membrane (C. Crump, H. Browne, B. Bruun, and A. C. Minson, 28th Int. Herpesvirus Workshop, abstr. 7.03, 2003). The gM-gN complexes of infectious laryngotracheitis virus (15), equine herpesvirus 1 (15), and human herpesvirus 8 have a similar activity (22). Thus, gM or the gM-gN complex might play an important role in collecting constituents of the future viral envelope at the budding site. In this scenario, both the UL11 and gM proteins play a role in secondary envelopment, but they do so at different steps, which could explain our experimental findings.
Immunoelectron microscopic analysis of capsid-tegument inclusion bodies which are formed in the absence of the UL11 protein demonstrated that the UL36 and UL37 proteins were tightly associated with the capsids, whereas the UL46, UL47, UL48, and UL49 tegument proteins were mostly detected dispersed in the inclusions. This finding substantiates our hypothesis that tegumentation at the capsid is initiated by capsid-bound UL36 interacting with UL37 (19). Moreover, the US3 tegument protein was observed at capsids as well as dispersed in the inclusions, which parallels recent findings that the US3 protein may be retained during nuclear egress or reacquired soon after, also indicating a close association with the capsid (12a).
Whereas concomitant deletion of UL11 and gM resulted in a drastically increased impairment of viral replication, deletion of UL11 and gE did not lead to a defect beyond that of a UL11 deletion mutant, at least not with regard to one-step growth. Thus, UL11 and gE may be involved in a similar step in secondary envelopment. We hypothesized that UL11 may direct tegument proteins to the envelopment site, and the UL16 tegument protein has been identified as one possible interaction partner for UL11 (25). It has also been shown that the cytoplasmic tail of gE is capable of interacting with the UL49 tegument protein (9). Conceivably, UL11 and gE cooperate in the accumulation of tegument proteins at the budding site by using the targeting capacity of UL11 to transport tegument proteins to this site and the interaction with the gE cytoplasmic tail for retention of tegument proteins. In contrast, gM may be involved in collecting envelope proteins. Deletion of UL11 and gM (or gE and gM [3]) would then interfere with both processes, whereas deletion of UL11 and gE only impaired collection of tegument without blocking the accumulation of glycoproteins. A certain “leakage” or redundancy in either pathway may then explain the continuous production of infectious progeny in the absence of either UL11 or gM.
The central role for UL11 and gM in the secondary envelopment process is reflected by their conservation throughout the herpesviruses. Indeed, UL11 of human cytomegalovirus has been shown to be required for virion formation in the cytoplasm (33). In Epstein-Barr virus, the absence of gM and its complex partner gN severely impaired virion formation (24). Thus, conservation of these proteins may also include conservation of function correlating with the role of these proteins in a central step of herpesvirus replication.
Acknowledgments
This study was supported by a grant from the Deutsche Forschungsgemeinschaft (Me 854/5-2).
We thank Uta Hartwig, Mandy Jörn, Petra Meyer, Diana Werner, and Elke Zorn for expert technical and photographical assistance.
REFERENCES
- 1.Baines, J. D., and B. Roizman. 1992. The UL11 gene of herpes simplex virus 1 encodes a function that facilitates nucleocapsid envelopment and egress from cells. J. Virol. 66:5168-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowzard, J. B., R. J. Visalli, C. B. Wislon, J. S. Loomis, E. M. Callahan, R. J. Courtney, and J. W. Wills. 2000. Membrane targeting properties of a herpesvirus tegument protein-retrovirus Gag chimera. J. Virol. 74:8692-8699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brack, A. R., J. M. Dijkstra, H. Granzow, B. G. Klupp, and T. C. Mettenleiter. 1999. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J. Virol. 73:5364-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brack, A. R., B. G. Klupp, H. Granzow, R. Tirabassi, L. W. Enquist, and T. C. Mettenleiter. 2000. Role of the cytoplasmic tail of pseudorabies virus glycoprotein E in virion maturation. J. Virol. 74:4004-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, Y. E., C. Van Sant, P. W. Krug, A. E. Sears, and B. Roizman. 1997. The null mutant of the UL31 gene of herpes simplex virus 1: construction and phenotype in infected cells. J. Virol. 71:8307-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dijkstra, J. M., T. C. Mettenleiter, and B. G. Klupp. 1997. Intracellular processing of pseudorabies virus glycoprotein M (gM): gM of strain Bartha lacks N-glycans. Virology 237:113-122. [DOI] [PubMed] [Google Scholar]
- 8.Fuchs, W., B. G. Klupp, H. Granzow, N. Osterrieder, and T. C. Mettenleiter. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but mature virions. J. Virol. 76:364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuchs, W., B. G. Klupp, H. Granzow, C. Hengartner, A. Brack, A. Mundt, L. W. Enquist, and T. C. Mettenleiter. 2002. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 76:8208-8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gershon, A. A., D. L. Sherman, Z. Zhu, C. A. Gabel, R. T. Ambron, and M. D. Gershon. 1994. Intracellular transport of newly synthesized varicella-zoster virus: final envelopment in the trans-Golgi network. J. Virol. 68:6372-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 12.Granzow, H., B. G. Klupp, W. Fuchs, J. Veits, N. Osterrieder, and T. C. Mettenleiter. 2001. Egress of alphaherpesviruses: comparative ultrastructural study. J. Virol. 75:3675-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12a.Granzow, H., B. G. Klupp, and T. C. Mettenleiter. 2004. The pseudorabies virus US3 protein is a component of primary and of mature virions. J. Virol. 78:1314-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jöns, A., J. M. Dijkstra, and T. C. Mettenleiter. 1998. Glycoproteins M and N of pseudorabies virus form a disulfide-linked complex. J. Virol. 72:550-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaplan, A. S., and A. E. Vatter. 1959. A comparison of herpes simplex and pseudorabies virus. Virology 7:394-407. [DOI] [PubMed] [Google Scholar]
- 15.Klupp, B. G., R. Nixdorf, and T. C. Mettenleiter. 2000. Pseudorabies virus glycoprotein M inhibits membrane fusion. J. Virol. 74:6760-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klupp, B. G., H. Granzow, E. Mundt, and T. C. Mettenleiter. 2001. Pseudorabies virus UL37 gene product is involved in secondary envelopment. J. Virol. 75:8927-8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063-10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2001. Effect of the pseudorabies virus US3 protein on nuclear membrane localization of the UL34 protein and virus egress from the nucleus. J. Gen. Virol. 82:2363-2371. [DOI] [PubMed] [Google Scholar]
- 19.Klupp, B. G., W. Fuchs, H. Granzow, R. Nixdorf, and T. C. Mettenleiter. 2002. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J. Virol. 76:3065-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kopp, M., B. G. Klupp, H. Granzow, W. Fuchs, and T. C. Mettenleiter. 2002. Identification and characterization of the pseudorabies virus tegument proteins UL46 and UL47: role for UL47 in virion morphogenesis in the cytoplasm. J. Virol. 76:8820-8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kopp, M., H. Granzow, W. Fuchs, B. G. Klupp, E. Mundt, A. Karger, and T. C. Mettenleiter. 2003. The pseudorabies virus UL11 protein is a virion component involved in secondary envelopment in the cytoplasm. J. Virol. 77:5339-5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koyano, S., E.-C. Mar, F. Stamey, and N. Inoue. 2003. Glycoproteins M and N of human herpesvirus 8 form a complex and inhibit cell fusion. J. Gen. Virol. 84:1485-1491. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 24.Lake, C. M., and L. Hutt-Fletcher. 2000. Epstein-Barr virus that lacks glycoprotein gN is impaired in assembly and infection. J. Virol. 74:11162-11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loomis, J. S., R. J. Courtney, and J. W. Wills. 2003. Binding partners for the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 77:11417-11424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mettenleiter, T. C. 1989. Glycoprotein gIII deletion mutants of pseudorabies virus are impaired in virus entry. Virology 171:623-625. [DOI] [PubMed] [Google Scholar]
- 27.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muranyi, W., J. Haas, M. Wagner, G. Krohne, and U. H. Koszinowski. 2002. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297:854-857. [DOI] [PubMed] [Google Scholar]
- 29.Nixdorf, R., B. G. Klupp, A. Karger, and T. C. Mettenleiter. 2000. Effects of truncation of the carboxy terminus of pseudorabies virus glycoprotein B on infectivity. J. Virol. 74:7137-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reynolds, A. E., B. J. Ryckman, J. D. Baines, Y. Zhou, L. Liang, and R. J. Roller. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 75:8803-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roizman, B., and P. Pellet. 2001. The family Herpesviridae: a brief introduction, p. 2381-2397. In D. M. Knipe and P. M. Howley (ed.), Virology, 4th ed. Lippincott-Raven, Philadelphia, Pa.
- 32.Roller, R. J., Y. Zhou, R. Schnetzer, J. Ferguson, and D. DeSalvo. 2000. Herpes simplex virus type 1 UL34 gene product is required for viral envelopment. J. Virol. 74:117-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanchez, V., E. Sztul, and W. J. Britt. 2000. Human cytomegalovirus pp28 (UL99) localizes to a cytoplasmic compartment which overlaps the endoplasmic reticulum-Golgi-intermediate compartment. J. Virol. 74:3842-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 35.Steven, A. C., and P. G. Spear. 1997. Herpesvirus capsid assembly and envelopment, p. 312-351. In W. Chiu, R. M. Burnett, and R. Garcea (ed.), Structural biology of viruses. Oxford University Press, New York, N.Y.
- 36.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou, Z., D. Chen, J. Jakana, F. J. Rixon, and W. Chiu. 1999. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J. Virol. 73:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zuckermann, F., T. C. Mettenleiter, C. Schreurs, N. Sugg, and T. Ben-Porat. 1988. Complex between glycoproteins gI and gp63 of pseudorabies virus: its effect on virus replication. J. Virol. 62:4622-4626. [DOI] [PMC free article] [PubMed] [Google Scholar]