Abstract
The purified T-antigen origin binding domain binds site specifically to site II, the central region of the simian virus 40 core origin. However, in the context of full-length T antigen, the origin binding domain interacts poorly with DNA molecules containing just site II. Here we investigate the contributions of additional core origin regions, termed the flanking sequences, to origin recognition and the assembly of T-antigen hexamers and double hexamers. Results from these studies indicate that in addition to site-specific binding of the T-antigen origin binding domain to site II, T-antigen assembly requires non-sequence-specific interactions between a basic finger in the helicase domain and particular flanking sequences. Related studies demonstrate that the assembly of individual hexamers is coupled to the distortions in the proximal flanking sequence. In addition, the point in the double-hexamer assembly process that is regulated by phosphorylation of threonine 124, the sole posttranslational modification required for initiation of DNA replication, was further analyzed. Finally, T-antigen structural information is used to model various stages of T-antigen assembly on the core origin and the regulation of this process.
Recent studies have established many of the factors required for initiation of DNA replication in eukaryotic organisms (reviewed in references 3 and 75). However, at the molecular level, this process is still poorly understood. In higher eukaryotic organisms this situation reflects, in part, the failure to identify sequences that define origins of replication (26). As a result, little is known about the protein-DNA interactions that are essential for initiation of replication. A further limitation is that structural information is available for only a few of the proteins involved in initiation of eukaryotic DNA replication.
Therefore, basic issues related to initiation of eukaryotic DNA replication have been addressed by using viral model systems. One of the more useful viral model systems for studying DNA replication is simian virus 40 (SV40) (reviewed in references 8, 25, and 61). The SV40 origin of replication is well defined (reviewed in references 5 and 8), as is the viral initiator protein, termed T antigen (T-ag), a 708-amino-acid phosphoprotein containing several domains (8, 25, 61). Moreover, the interaction of T-ag with the core origin has been extensively studied (reviewed in references 8, 25, and 61). A further advantage afforded by this system is that considerable information about the structure of T-ag has been obtained. For instance, the structure of the T-ag domain that site-specifically binds to the SV40 origin, the origin binding domain (T-ag-obd) (residues 131 to 260), was determined by nuclear magnetic resonance methods (41). In addition, crystallographic techniques were used to determine the structures of the J domain-containing (11, 67) N terminus of T-ag (residues 7 to 117) (35) and the C-terminal helicase domain (residues 251 to 627) (39). That this structural information is of general interest to the replication field was demonstrated by recent studies, for example, those that established that the structure of the T-ag-obd is nearly identical to those of the papillomavirus E1 DNA binding domain (21), the DNA binding domain from adeno-associated virus (28), and the catalytic domain of Rep, the initiator protein of tomato yellow leaf curl virus (12). These observations have raised the possibility that viral initiators interact with their origins via conserved mechanisms. Moreover, structural homology between viral and cellular J domains have been reported (see references 11 and 35 and references therein). In addition, the recent determination of the structure of the C-terminal domain of T-ag has provided a wealth of information regarding the organization and functioning of a eukaryotic helicase (39).
When T-ag monomers assemble on the core origin, they oligomerize into hexamers and double hexamers (15, 31, 44, 49); preformed hexamers may also bind to origin-containing DNA (72). When assembled into a double hexamer, T-ag is able to function as a helicase (1, 14, 64, 68). Electron microscopy studies have provided additional information regarding the structures of T-ag hexamers (43, 44, 56, 74) and double hexamers (73). For example, they revealed that T-ag hexamers are planar rings containing a central hole through which DNA was proposed to pass (56, 73, 74), a conclusion supported by more recent studies (39). They also established the relative positions of T-ag domains within the hexamers; for instance, the N-terminal face of the hexamer contains the J/T-ag-obds, and the C-terminal side contains domains required for ATP hydrolysis and helicase activity (39, 73, 74). Less is known about the structure of T-ag double hexamers; nevertheless, double hexamers assembled on the core origin have been imaged by negative-staining techniques (73). Among the findings of these studies is that the J/T-ag-obds are situated at the hexamer-hexamer interface (73), a finding consistent with previous biochemical studies (76).
Regarding the protein-DNA interactions that take place during assembly of T-ag hexamers on the core origin, the A1 and B2 loops (41, 62, 83) within the T-ag-obd are required for site-specific binding to individual GAGGC pentanucleotides within site II (34, 41, 70; reviewed in reference 8). Consistent with these reports, the purified T-ag-obd can site-specifically bind to oligonucleotides containing just site II (36). In contrast, site II-based oligonucleotides are poor substrates for T-ag assembly (36, 70). When assayed via electrophoretic mobility shift assay (EMSA), T-ag assembly into hexamers requires site II and the presence of either of the flanking sequences (36, 66). Based on these studies, it was concluded that efficient hexamer formation on the core origin requires at least two T-ag-origin contacts: site-specific binding of the T-ag-obd to site II and an additional contact(s) between the flanking sequences and residues in the helicase domain of T-ag (36, 51).
The protein-DNA interactions required for double-hexamer formation are complex, since certain of them appear to be regulated by cell cycle-dependent phosphorylation events (2, 47, 76). For example, T-ag's interactions with subfragments of the core origin are regulated by phosphorylation of Thr124 (2), an essential posttranslational modification required for initiation of DNA replication (45, 47, 57). Moreover, peptides containing the CDK/NLS region of T-ag interact with DNA in a manner that is regulated by phosphorylation of Thr124 (37), which is additional evidence that phosphorylation regulates certain T-ag interactions with the origin.
To further characterize the protein-DNA interactions necessary for T-ag and T124A assembly on the core origin, we have analyzed the amount of flanking sequence required for hexamer and double-hexamer formation. Molecular modeling techniques have also been used to predict the region of the T-ag helicase domain that interacts with the flanking sequences during assembly on the viral origin. Moreover, to further explore the mechanism of DNA unwinding, we have examined whether the structural distortions in the core origin (7, 51) are coupled to double-hexamer formation or to earlier events during T-ag assembly. Results from these studies are presented here.
MATERIALS AND METHODS
Commercial supplies of enzymes, DNA, reagents, and oligonucleotides.
T4 polynucleotide kinase was purchased from Gibco-BRL. Plasmid pBR322 DNA, used as competitor DNA, was purified by standard procedures (54) and digested with HaeIII purchased from New England Biolabs.
Oligonucleotides were synthesized on an Applied Biosystems 394 DNA synthesizer, purified by electrophoresis through urea-10% polyacrylamide gels, and isolated as described previously (54, 65). Double-stranded oligonucleotides, 32P labeled at their 5′ termini, were prepared by standard procedures (54, 65).
Purification of wild-type T-ag and T-ag containing the T124A mutation.
SV40 T-ag and the T124A mutant were expressed in insect (Sf9) cells by using baculovirus expression vectors. The wild-type vector was previously described (50), while the T124A expression vector was developed by L. Chen, R. Upson, and D. Simmons (unpublished data). Proteins were purified by using immunoaffinity techniques with the PAb 419 monoclonal antibody (19, 60). Purified proteins were stored in T-ag storage buffer (20 mM Tris-HCl [pH 8.0], 50 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol [DTT] [29], 0.1 mM phenylmethylsulfonyl fluoride, 0.2 μg of leupeptin per ml, 0.2 μg of antipain per ml, 10% glycerol) (2, 79) and frozen at −80°C until use.
EMSAs.
T-ag-and T124A-based EMSAs were conducted under SV40 in vitro replication conditions (79). The reaction mixtures (20 μl) contained 7 mM MgCl2, 0.5 mM DTT, 4 mM adenylyl imidodiphosphate (AMP-PNP), 40 mM creatine phosphate (di-Tris salt [pH 7.6]), 0.48 μg of creatine phosphate kinase, 5 μg of bovine serum albumin, 0.8 μg of HaeIII-digested pBR322 (6 pmol) (used as a nonspecific competitor), 25 fmol of double-stranded oligonucleotide (∼106 cpm/pmol), and 0.5 μg of T-ag or T124A (6 pmol) as indicated. After a 20-min incubation at 37°C, glutaraldehyde was added (0.1% final concentration), and the reaction products were further incubated for 5 min. The reactions were stopped by the addition of 5 μl of 6× loading dye II (15% Ficoll, 0.25% bromophenol blue, and 0.25% xylene cyanol) (54) to the samples. The reaction products were applied to 4 to 12% gradient polyacrylamide gels and electrophoresed in 0.5% Tris-borate-EDTA (pH 8.4) for ∼1.5 h (10 W). The gels were dried on Whatman 3MM paper and subjected to autoradiography.
KMnO4 footprinting.
The KMnO4 footprinting technique (7), performed in 30-μl reaction mixtures, was conducted under replication conditions (79) as previously described (33). DNA substrates included the core origin containing plasmid pSV01ΔEP and four derivatives of this plasmid in which the core origin was replaced with mutant origins, each containing a single pentanucleotide. For any given mutant origin, the other three pentanucleotides were replaced with transition mutations. As in previous studies (33, 36), oligonucleotide 1 (5′ TGAGCGGATACATATTTG 3′), 5′ end labeled with [γ-32P]ATP and T4 polynucleotide kinase (54), was used in the primer extension reactions. Upon completion of the reactions, the samples were ethanol precipitated, washed with 80% ethanol, and electrophoresed for ∼2.5 h at 1,500 V and 40 mA on a 7% polyacrylamide gel containing 8 M urea. The locations of the modified residues were determined by using a dideoxy sequencing ladder (55), with oligonucleotide 1 as the primer.
Molecular modeling of T-ag assembly on core origin subfragments.
The molecular model of the T-ag monomer bound to DNA was generated by using the computer program InsightII by MSI. Standard B-form DNA was assembled by using the sequence of the 64-bp SV40 core origin (see reference 16 and references therein). The T-ag-obd (41) (Protein Data Base [PDB] accession code 1TBD) was bound to pentanucleotide 1 by using the A1 and B2 loops (41, 83). Docking was based on recent studies indicating that B2 residue His203 contacts the middle GC base pair in the GAGGC sequence (E. M. Bradshaw et al., submitted for publication) and an alignment suggested by the E1-obd-DNA costructure (22). The J domain (35) (PDB accession code 1GH6) was docked to the T-ag-obd by using constraints previously described by VanLoock et al. (74). The T-ag helicase domain (39) (PDB accession code 1N25) was juxtaposed next to the C terminus of the T-ag-obd. The orientation of the helicase domain was established (i) by aligning residues known to be in the inner channel with flanking sequence DNA (39) and (ii) by using spatial constraints generated by previous electron microscopy studies of T-ag hexamers (74).
RESULTS
Analyses of the helicase domain-flanking sequence interactions required for hexamer formation.
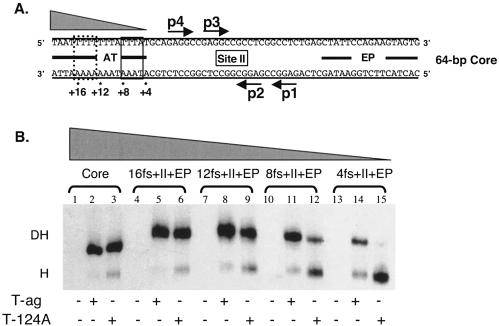
Efficient assembly of monomers of T-ag, or T124A, into hexamers requires a pentanucleotide (p) and either of the adjacent flanking sequences in the origin (36, 66, 70): the early palindrome (EP) or the AT-rich region (AT). The experiments with the results shown in Fig. 1 and 2 were conducted to establish how much of the proximal flanking sequence (fs) is required for hexamer formation on any given pentanucleotide. (In the following sections the abbreviation fs is used to refer to sequences derived from the EP, AT regions, and, in certain instances, site II.)
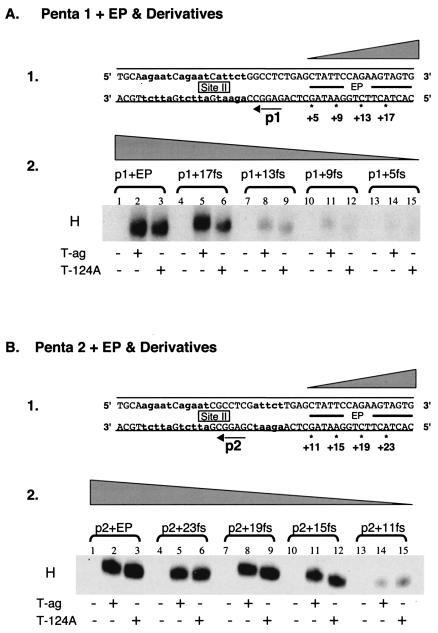
FIG. 1.
Experiments used to establish the sequences within the EP region required for hexamer formation on pentanucleotides 1 and 2. (A) Studies of the EP requirements for hexamer assembly on pentanucleotide 1. Panel 1, oligonucleotides used in these studies. In the DNA sequence, lowercase boldface letters are used to symbolize the transition mutations used to replace pentanucleotides 2, 3, and 4. The 48-bp p1 + EP oligonucleotide is the only core origin subfragment presented; however, the numbers with asterisks indicate the ends of additional oligonucleotides generated as a result of progressive truncations of the EP. The wedge symbolizes the progressive truncation of these sequences in this and subsequent figures. Panel 2, band shift reactions conducted with the indicated oligonucleotides. The reactions in lanes 2, 5, 8, 11, and 14 were conducted with T-ag (6 pmol); those in lanes 3, 6, 9, 12, and 15 were conducted with T124A (6 pmol); and those in lanes 1, 4, 7, 10, and 13 served as protein-free controls. The position of T-ag hexamers is indicated by H in this and subsequent figures. (B) Studies of the EP requirements for assembly on pentanucleotide 2. Panel 1, oligonucleotides used in this set of studies. Lowercase boldface letters indicate the transition mutations used to replace pentanucleotides 1, 3, and 4. The 48-bp p2 + EP oligonucleotide is the only core origin subfragment presented; however, the numbers with asterisks indicate the ends of additional oligonucleotides generated as a result of progressive truncations of the EP. Panel 2, band shift reactions conducted with the indicated oligonucleotides. The reactions in lanes 2, 5, 8, 11, and 14 were conducted with T-ag (6 pmol); those in lanes 3, 6, 9, 12, and 15 were conducted with T124A (6 pmol); and those in lanes 1, 4, 7, 10, and 13 served as protein-free controls.
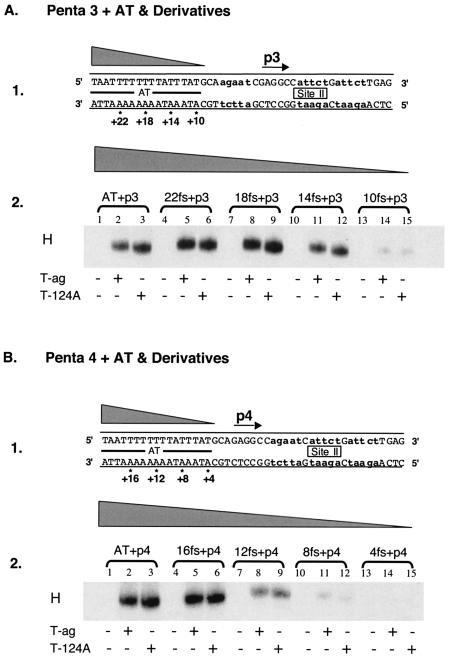
FIG. 2.
Experiments used to establish the sequences within the AT-rich region required for hexamer formation on pentanucleotides 3 and 4. (A) Studies of the AT requirements for hexamer assembly on pentanucleotide 3. Panel 1, oligonucleotides used in these experiments. Lowercase boldface letters indicate the transition mutations used to replace pentanucleotides 1, 2, and 4. The 47-bp p 3 + AT oligonucleotide is the only core origin subfragment presented; however, the numbers with asterisks indicate the ends of additional oligonucleotides that were generated as a result of progressive truncations of the AT. Panel 2, band shift reactions conducted with the indicated oligonucleotides. The reactions in lanes 2, 5, 8, 11, and 14 were conducted with T-ag (6 pmol); those in lanes 3, 6, 9, 12, and 15 were conducted with T124A (6 pmol); and those in lanes 1, 4, 7, 10, and 13 served as protein-free controls. (B) Studies of the AT requirements for assembly on pentanucleotide 4. Panel 1, oligonucleotides used in these studies. Lowercase boldface letters indicate the transition mutations used to replace pentanucleotides 1, 2, and 3. While the 47-bp p4 + AT oligonucleotide is the only core origin subfragment presented, the numbers with asterisks indicate the ends of additional oligonucleotides generated as a result of progressive truncations of the AT. Panel 2, band shift reactions conducted with the indicated oligonucleotides. The reactions in lanes 2, 5, 8, 11, and 14 were conducted with T-ag (6 pmol); those in lanes 3, 6, 9, 12, and 15 were conducted with T124A (6 pmol); and those in lanes 1, 4, 7, 10, and 13 served as protein-free controls.
(i) Hexamer assembly on substrates containing truncations of the flanking sequences located 5′ of the pentanucleotides.
In an initial set of studies, the flanking sequence requirements for hexamer formation on pentanucleotide 1 were determined by using a series of truncated forms of the 48-bp p1 + EP oligonucleotide (Fig. 1A, panel 1) and AMP-PNP (a nonhydrolyzable analog of ATP used in all of the experiments presented in Fig. 1 to 3). Inspection of Fig. 1A, panel 2 (lanes 1 to 3), confirms that the 48 bp p1 + EP oligonucleotide readily assembles T-ag and T124A hexamers (66). The experiments for lanes 4 to 15 were conducted with oligonucleotides containing progressively larger truncations in the EP region. It is apparent from lanes 4 to 6 that for either T-ag or T124A, 4 bp of the EP can be removed without a significant reduction in hexamer formation. However, inspection of lanes 7 to 15 reveals that DNA substrates smaller than the 44-bp p1 + 17fs oligonucleotide supported T-ag or T124A assembly at greatly reduced levels. Based on these studies, it is concluded that T-ag assembly on pentanucleotide 1 requires the GAGGC and sequences extending into the EP for an additional 14 to 17 bp. In a second series of experiments, the flanking sequence requirements for hexamer formation on pentanucleotide 2 were studied by using truncated derivatives of the 48-bp p2 + EP oligonucleotide (Fig. 1B, panel 1). Examination of Fig. 1B, panel 2 (lanes 1 to 3), demonstrates that the 48-bp p2 + EP oligonucleotide is a substrate for hexamer formation. The experiments for lanes 4 to 15 were conducted with oligonucleotides containing progressively larger truncations in the EP region. Inspection of lanes 4 to 12 reveals that a considerable amount of the EP can be deleted without disrupting hexamer formation on pentanucleotide 2. In contrast, very little hexamer was formed on the 32-bp p2 + 11fs oligonucleotide. It is concluded that T-ag assembly on pentanucleotide 2 requires the pentanucleotide and at least 12 to 15 bp of DNA proximal to this site.
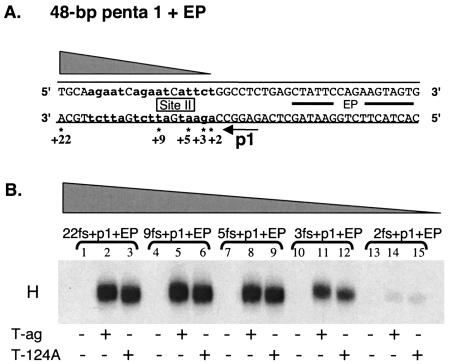
FIG. 3.
Establishment of the extent to which sequences 3′ to a given GAGGC pentanucleotide are important for hexamer formation. (A) Core origin-based oligonucleotides, containing pentanucleotide 1, the EP, and various amounts of site II, were used to establish the minimum flanking sequence, 3′ of pentanucleotide 1, needed for hexamer formation. The 48-bp p1 + EP is the only oligonucleotide presented; however, the numbers with asterisks indicate the ends of additional oligonucleotides used in these studies. Lowercase boldface letters indicate the transition mutations used to replace pentanucleotides 2, 3, and 4. (B) A representative band shift experiment, conducted with the indicated oligonucleotides in the presence of AMP-PNP. The reactions in lanes 2, 5, 8, 11, and 14 were conducted with wild-type T-ag (6 pmol), while those in lanes 3, 6, 9, 12, and 15 were conducted with the T124A mutant protein (6 pmol). As a control, the reactions in lanes 1, 4, 7, 10, and 13 were performed without protein.
The contribution of the AT-rich region to hexamer formation on pentanucleotide 3 was examined in the next series of reactions. It is apparent from Fig. 2A, panel 2 (lanes 1 to 3), that hexamers readily form on the single-pentanucleotide-containing 47-bp AT + p3 oligonucleotide (Fig. 2A, panel 1). To determine how much of the AT-rich region is required for assembly on pentanucleotide 3, additional experiments were conducted with the truncated derivatives of the 47-bp AT + p3 oligonucleotide (Fig. 2A, panel 1). Inspection of Fig. 2A, panel 2, reveals that hexamers efficiently form on truncated forms of the AT + p3 oligonucleotide (lanes 4 to 12) (e.g., the 43-bp 22fs + p3, 39-bp 18fs + p3, and 35-bp 14fs + p3 oligonucleotides). In contrast, assembly is greatly reduced on the 31-bp 10fs + p3 oligonucleotide (lanes 13 to 15). It is concluded that T-ag assembly on pentanucleotide 3 requires the pentanucleotide and at least 11 to 14 bp of the AT region proximal to this site. Finally, the flanking sequence requirements for hexamer formation on pentanucleotide 4 were studied by using truncated derivatives of the 47-bp AT + p4 oligonucleotides (Fig. 2B, panel 1). The data in Fig. 2B, panel 2 (lanes 1 to 3), confirm previous experiments (2, 66) that demonstrated that T-ag and T124A hexamers readily form on the 47-bp AT + p4 oligonucleotide. Further inspection of lanes 4 to 6 reveals that the 43-bp 16fs + p4 oligonucleotide supports T-ag and T124A hexamers. In contrast, hexamers assemble at reduced levels on smaller subfragments of the origin (e.g., the 39-bp 12 fs + p4 [lanes 7 to 9], 35-bp 8 fs + p4 [lanes 10 to 12], and 31-bp 4fs + p4 [lanes 13 to 15] oligonucleotides). These results demonstrate that efficient T-ag assembly on pentanucleotide 4 requires the GAGGC and at least 13 to 16 bp of the AT-rich region proximal to the pentanucleotide, a length requirement similar to that for hexamer formation at the other three pentanucleotides.
Finally, T-ag assembles in an ATP-dependent manner (15, 17). Therefore, to establish whether the interactions with the flanking sequences are altered by nucleotide hydrolysis, the experiments in Fig. 1 and 2, which were conducted in the presence of AMP-PNP, were repeated in the presence of ADP, and very similar results were obtained. They were also repeated in the presence of ATP and in the absence of nucleotides (data not shown). However, as previously reported (2, 66), greatly reduced levels of hexamers are formed in the in the presence of ATP (a reflection of T-ag's helicase activity [14, 27, 68]) or in the absence of nucleotide (a cofactor for hexamer formation [39]).
(ii) Sequences 3′ to a given GAGGC pentanucleotide are not required for hexamer formation.
When it is bound to the core origin, it is unlikely that T-ag makes extensive contact with origin sequences 3′ to a given pentanucleotide. This hypothesis is based, in part, on the observation that double hexamers preferentially form on pentanucleotides 1 and 3 (33, 73), two sites that are separated by only 7 bp. Therefore, the arrangement of pentanucleotide pairs (e.g., 1 and 3) provides limited room for protein-DNA contacts 3′ of any given pentanucleotide.
To test this prediction, a series of oligonucleotides that had progressively larger truncations of site II DNA located 3′ of pentanucleotide 1 were synthesized (Fig. 3A). After radiolabeling (see Materials and Methods), the oligonucleotides were used in an additional series of band shift experiments (Fig. 3B). Inspection of this Fig. 3B (lanes 1 to 9) reveals that hexamer formation on pentanucleotide 1 requires very little DNA 3′ of the GAGGC sequence. Furthermore, the phosphorylation status of Thr124 did not change the sequence requirements for T-ag assembly. However, hexamers formed at reduced levels on a substrate (i.e., the 28-bp 3fs + p1 + EP oligonucleotide) containing three base pairs 3′ of pentanucleotide 1 (lanes 10 to 12). In addition, an oligonucleotide containing two residues 3′ of pentanucleotide 1 (i.e., the 27-bp 2fs + p1 + EP oligonucleotide) was a very poor substrate for hexamer formation. Whether the 2- to 3-bp 3′ flanking region is required for protein-DNA interactions or to prevent fraying the ends of the oligonucleotide has not been determined. Nevertheless, these experiments confirm that hexamer formation on a given pentanucleotide does not require extensive protein-DNA contacts with sequences 3′ of the GAGGC sequence.
(iii) Summary of sequence requirements for hexamer formation.
To illustrate the common DNA requirements for hexamer formation on the core origin, the sequences that support hexamer assembly (Fig. 1 to 3) were aligned relative to the individual pentanucleotides (Fig. 4A). The boundary regions are shown; sequences between a given pentanucleotide and those within its associated boundary region are necessary for hexamer formation. Relative to the pentanucleotides, the AT-rich boundary regions have been mapped to residues located 11 to 17 bp from the 5′ ends of the GAGGC sequences. Nevertheless, it is noted that a possible common feature of the pentanucleotide-boundary region arrangement is a separation of 14 bp (corresponding to ∼47.6 Å). The relatively exact arrangement of the pentanucleotides and their associated boundary regions suggests that an equally distinct spatial arrangement exists in T-ag between the A1 and B2 loops in the T-ag-obd and the DNA binding residues in the helicase domain (see below). In addition, the experiments conducted with the T124A mutant demonstrated that the phosphorylation status of Thr124 does not alter the sequence requirements for hexamer formation, an observation consistent with previous studies (2, 47).
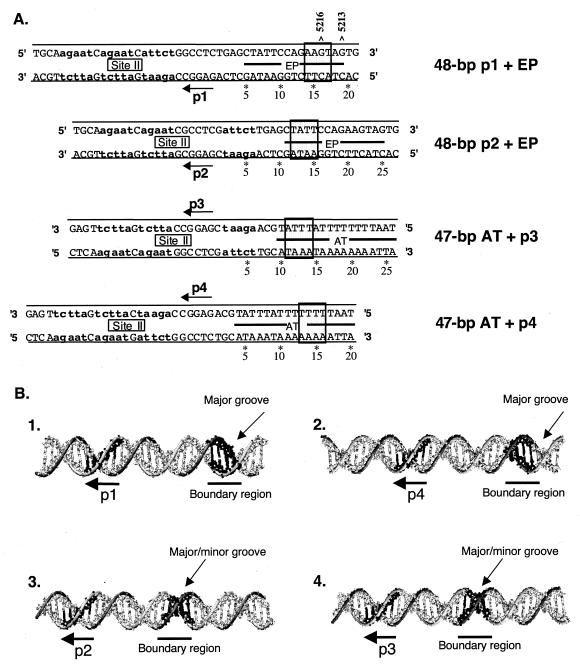
FIG. 4.
Conserved sequence requirements for hexamer formation on individual pentanucleotides. (A) The four parental oligonucleotides (i.e., the largest origin subfragments used in the truncation studies) were aligned via their individual pentanucleotides. The boxes indicate the locations of the boundary regions. As noted in the text, a common feature of the boundary regions may be a distance of 14 bp from the end of a given pentanucleotide. Finally, the positions of origin residues 5216 and 5213 are indicated. As mentioned in Discussion, SenGupta and Borowiec (58) reported that ethylation of these residues inhibited T-ag binding. (B) Positions of pentanucleotides 1, 2, 3, and 4 relative to their associated boundary regions (underlined), projected onto B-form DNA. In these models, the pentanucleotides are projecting out of the page; the locations of the major and minor grooves, relative to the boundary regions, are indicated.
To gain further insights into the spatial relationship between the pentanucleotides and boundary regions, their relative positions were mapped on standard B-form DNA (Fig. 4B). To construct these models, the individual pentanucleotides were oriented such that they face out of the plane of the paper. Inspection of Fig. 4B, panel 1, reveals that in this orientation, the boundary region associated with pentanucleotide 1 is within a major groove that also faces out of the plane of the paper. Likewise, the boundary region associated with pentanucleotide 4 is within a major groove that is coplanar with the pentanucleotide (Fig. 4B, panel 2). In contrast, inspection of Fig. 4B, panels 3 and 4, reveals that the boundary regions associated with pentanucleotides 2 and 3 have a different topology; they are situated within both major and minor grooves. This arrangement is of interest given that it was previously demonstrated that hexamers preferentially form on pentanucleotides 1 and 4, while levels of hexamer formation on pentanucleotides 2 and 3 were relatively low (33). These observations suggest that hexamers preferentially form on pentanucleotides associated with boundary regions situated primarily in the major groove.
(iv) Modeling the binding of a T-ag monomer to the SV40 origin of replication.
Given recent advances in determining the structure of T-ag (see e.g., references 35, 39, 41, 73, and 74), the formation of a T-ag monomer on pentanucleotide 1 was modeled (Fig. 5). Steps taken to assemble a T-ag monomer on core origin DNA, using the J, T-ag-obd, and helicase domains, are described in Materials and Methods. The length of a single hexamer has been reported to be ∼100 (74) to 110 (73) Å (∼29 to 32 bp). In reasonable agreement, the monomer depicted in Fig. 5 covers ∼115 Å (∼34 bp). However, Li et al. solved the structure of a fragment of the helicase domain extending between residues 251 and 627 (39). Given that T-ag contains 708 residues, the model is lacking 81 residues from the C terminus. Furthermore, based on a number of observations, including those presented in Fig. 3, the N-terminal J domain was positioned such that it does not make significant contacts with core origin sequences 3′ of a given pentanucleotide (a feature of the model that is not readily apparent from Fig. 5). The model also indicates that the interaction of the T-ag-obd with the 5′ end of pentanucleotide 1 terminates at the junction between site II and the EP, a characteristic of the model that is supported by previous phenanthroline-copper footprinting of T-ag-obd-origin complexes (33).
FIG. 5.
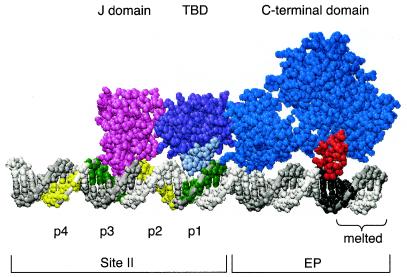
A model of a T-ag monomer docked to the 48-bp p1 + EP oligonucleotide. The positions of two SV40 core origin domains, site II and the EP, are indicated. Pentanucleotides 1 and 3 are shown in green, while pentanucleotides 2 and 4 are shown in yellow. The boundary region associated with pentanucleotide 1 is shown in black. Also depicted is the region of the EP that is melted upon T-ag binding (Fig. 9) (reviewed in reference 5). The N terminus of T-ag (residues 7 to 117) is represented in pink. The T-ag-obd (TBD) (residues 139 to 247) is in purple. The A1 and B2 binding loops, shown in light blue (reviewed in reference 8), were docked to pentanucleotide 1. The C-terminal domain (residues 266 to 627) is in dark blue. The basic finger (residues 503 to 523), depicted interacting with the boundary region associated with pentanucleotide 1, is in red. Finally, since the structures of the J domain, T-ag-obd, and C-terminal domain were solved separately, there is some uncertainty regarding the relative orientations of the individual domains. Moreover, there is no evidence that the J-domain-containing N terminus is actually in contact with origin DNA (see, e.g., Fig. 3); therefore, this model should not be interpreted as suggesting DNA binding.
The location of the boundary region associated with pentanucleotide 1 is based on the studies presented in Fig. 1A. Regarding the location of the residues within the helicase domain that contact the boundary region, it is noted that a finger, extending between residues 503 and 523, is situated opposite the boundary region. Residues at the apex of the loop are highly basic (i.e., 511KKHLNKR517), a useful feature for DNA binding; a similar cluster of basic residues is not present in any other region of the helicase domain. Furthermore, rotation of the inner channel of the helicase domain around duplex DNA, using the program Insight II, failed to suggest additional DNA binding motifs (data not shown). Finally, the distance between Arg154 in the A1 loop and the apex of the basic finger (Lys512) is ∼51 Å, a distance that is similar to the 47.6-Å separation between the 5′ ends of the pentanucleotides and the nucleotides located 14 bp away within the boundary regions.
Studies of the flanking sequence requirements for double-hexamer formation on the core origin.
Having analyzed the flanking sequence requirements for hexamer formation on individual pentanucleotides (e.g., pentanucleotide 1), we performed a similar analysis of the flanking sequence requirements for double-hexamer formation. Reasons for conducting these experiments included a desire to establish whether the initially bound hexamer, presumably bound to the pentanucleotides proximal to the flanking sequences (e.g., 1 and 4) (66), modulated the flanking sequence requirements for the assembly of the second hexamer (e.g., 3 and 2). In addition, these studies were performed both with wild-type T-ag, phosphorylated at Thr124 (30), and with the T124A mutant (45, 47, 57). The use of both proteins was designed to provide further insights into how phosphorylation of Thr124 regulates T-ag-DNA interactions and the assembly of double hexamers.
(i) Establishment of the length of the AT-rich region required for assembly of the second hexamer.
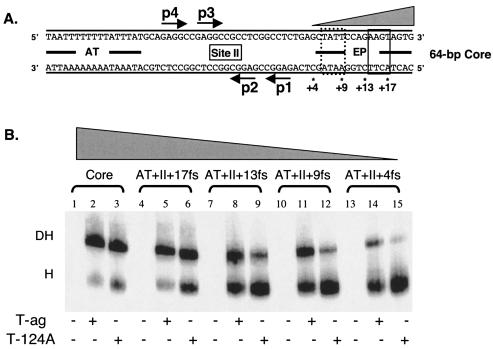
An initial series of experiments was conducted with the 64-bp core origin oligonucleotide and derivatives containing truncations in the AT-rich region (Fig. 6A). The positions of the boundary regions required for hexamer formation on pentanucleotides 3 and 4 (Fig. 2) are indicated. As a positive control, T-ag and T124A double hexamers were formed on oligonucleotides containing the 64-bp core origin (Fig. 6B, lanes 1 to 3). Double hexamers also formed on origin subfragments containing limited truncations in the AT-rich region (lanes 4 to 9) (i.e., the 60-bp 16fs + site II + EP and 56-bp 12 fs + site II + EP oligonucleotides). Since the 12 fs + site II + EP oligonucleotide lacks the pentanucleotide 4 boundary region, it is concluded that this boundary region is not needed for assembly of the second hexamer (compare lanes 4 to 6 and 7 to 9), a finding consistent with preferential assembly of T-ag double hexamers on pentanucleotides 1 and 3 (33, 66). On origin subfragments containing progressively larger truncations (e.g., the 52-bp 8fs + site II + EP and 48-bp 4fs + site II + EP oligonucleotides), T-ag continued to form double hexamers (lanes 11 and 14). In contrast, on the same substrates, T124A molecules preferentially formed hexamers (lanes 12 and 15). Thus, the second T-ag hexamer to assemble is less sensitive to truncations in the AT-rich region than the second T124A hexamer (compare lanes 11 and 14 with lanes 12 and 15). Moreover, these experiments indicate that the boundary region associated with pentanucleotide 3 is required for assembly of the second T124A hexamer but not for assembly of the second T-ag hexamer. Finally, it was previously demonstrated that double hexamers readily form on the 64-bp AT + p(1,3) + EP oligonucleotide (33). The experiments in Fig. 6B were repeated with similar truncated forms of the 64-bp AT + p(1,3) + EP oligonucleotide, and nearly identical results were obtained (data not shown).
FIG. 6.
Determination of how much of the AT-rich region is required for efficient double-hexamer formation on derivatives of the 64-bp core origin. (A) DNA substrates used in this set of experiments. The 64-bp core origin oligonucleotide is the only molecule depicted; however, the numbers with asterisks indicate the ends of additional oligonucleotides generated as a result of progressive truncations of the AT. The boundary regions associated with pentanucleotide 3 (solid rectangle) and pentanucleotide 4 (dotted rectangle) are indicated. (B) Band shift reactions conducted with the indicated oligonucleotides in the presence of AMP-PNP. The reactions in lanes 2, 5, 8, 11, and 14 were conducted with T-ag (6 pmol); those in lanes 3, 6, 9, 12, and 15 were conducted with T124A (6 pmol); and those in lanes 1, 4, 7, 10, and 13 served as protein-free controls. The positions of T-ag hexamers (H) and double hexamers (DH) are indicated.
(ii) Establishment of the length of the EP region required for assembly of the second hexamer.
Additional studies were conducted with oligonucleotides containing the 64-bp core origin and smaller derivatives possessing truncations in the EP region (Fig. 7A); the boundary regions required for hexamer formation on pentanucleotides 1 and 2 are depicted. As previously demonstrated (see, e.g., Fig. 6B), T-ag and T124A double hexamers readily form on oligonucleotides containing the 64-bp core origin (Fig. 7B, lanes 1 to 3). Similar levels of double hexamers form on the 60-bp AT + site II + 17 fs oligonucleotide, although a slight increase in T124A hexamer assembly was observed (compare lanes 5 and 6). T-ag continues to form relatively high levels of double hexamers on oligonucleotides that do not contain the pentanucleotide 1 boundary region (e.g., the 56-bp AT + site II + 13 fs and 52-bp AT + site II + 9 fs oligonucleotides [lanes 7 to 12]). In contrast, these same oligonucleotides are progressively poorer substrates for T124A double-hexamer assembly (compare lanes 8 and 11 with lanes 9 and 12). Indeed, the 47-bp AT + site II + 4fs (also termed the AT + site II) oligonucleotide, lacking both boundary regions in the EP, supported reduced levels of T-ag double hexamers (lane 14) and essentially no T124A double hexamers (lane 15). Based on the data in Fig. 6 and 7, it is concluded that the second T124A hexamer will form at normal levels only in the presence of a complete second assembly unit, i.e., the pentanucleotide and its associated boundary region. In contrast, the assembly of the second T-ag hexamer, phosphorylated on Thr124 (30), does not have an absolute requirement for the boundary region associated with the second pentanucleotide.
FIG. 7.
Determination of how much of the EP-rich region is required for efficient double-hexamer formation on derivatives of the 64-bp core origin. (A) DNA substrates used in this set of experiments. The 64-bp core origin oligonucleotide is the only molecule presented; however, the numbers with asterisks indicate the ends of additional oligonucleotides generated as a result of progressive truncations of the EP. The boundary regions associated with pentanucleotide 1 (solid rectangle) and pentanucleotide 2 (dotted rectangle) are indicated. (B) Band shift reactions conducted with the indicated oligonucleotides in the presence of AMP-PNP. The reactions in lanes 2, 5, 8, 11, and 14 were conducted in the presence of T-ag (6 pmol), and those in lanes 3, 6, 9, 12, and 15 were conducted with T124A (6 pmol). The reactions in lanes 1, 4, 7, 10, and 13 served as protein-free controls. The positions of T-ag hexamers (H) and double hexamers (DH) are indicated.
(iii) Modeling of the protein-DNA interactions required for formation of T-ag and T124A dimers.
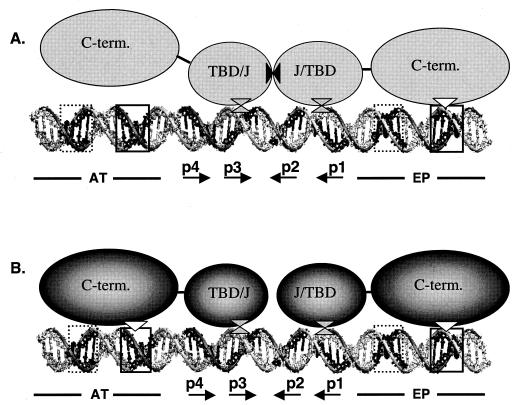
The results from the double-hexamer assembly experiments presented in Fig. 6 and 7 are summarized in Fig. 8. Regarding the assembly of the two T-ag monomers that initiate double-hexamer formation (Fig. 8A), the T-ag-obd is depicted as a dimer that protects pentanucleotides 1 to 3, a conclusion supported by phenanthroline-copper footprinting studies (33, 34). Furthermore, based on the experiments presented here (Fig. 6B), and earlier studies of wild-type T-ag (2, 66), the model indicates that formation of the initial hexamer requires flanking sequence contacts with the EP, while the second hexamer assembles in a manner that is less dependent on these interactions. A summary of the data presented in Fig. 7B would depict the C terminus of the initial hexamer contacting the AT-rich region, while the second hexamer assembles in a manner that is relatively independent of its contacts with the EP region. These contrasting observations suggest that the interactions between the C termini and the flanking sequences are dynamic.
FIG. 8.
Contrasting of the protein-DNA interactions required for T-ag and T124A double-hexamer formation. (A) A model of the interactions that take place during the docking of two T-ag monomers, phosphorylated on Thr124, to pentanucleotides 1 and 3 (66, 73). The A1 and B2 loops within the two origin binding domains (TBD) are bound to the pentanucleotides (double triangles). The helicase domain associated with T-ag bound to pentanucleotide 1 is shown making contact with its boundary region via its basic finger (rectangle-triangle interaction). In contrast, the second helicase domain, associated with T-ag assembled on pentanucleotide 3, has a lower affinity for its associated boundary region. This may stem from the orientation of the pentanucleotide 3 boundary region in a manner that prevents interactions with the major groove (Fig. 4B). Protein-protein interactions between the origin binding domains are symbolized by the solid triangles. (B) A model of the protein-DNA interactions that take place when two monomers of the T124A mutant dock to pentanucleotides 1 and 3 (66, 73). In contrast to T-ag, two basic finger-boundary region interactions are necessary for assembly (the symbols for the protein-DNA interactions are the same as in panel A). Furthermore, relative to T-ag, it is proposed that fewer protein-protein interactions take place between the two T-ag-obds and J domains, a proposal in keeping with previous studies (47, 76). Finally, T-ag double hexamers were reported to span ∼230 Å (73), corresponding to ∼68 bp of B DNA. Consistent with this estimate, previous DNase I footprinting studies indicated that double hexamers protect a region slightly larger than the 64-bp core origin (6). Moreover, these estimates are in reasonable agreement with predictions of the size of the double hexamer (∼230 Å [∼68 bp]) based on doubling the dimensions of the hexamer (Fig. 5) (∼115 Å [∼34 bp]).
The T124A mutant is a model for T-ag prior to cell cycle dependent phosphorylation (2, 23, 45-47, 57, 76). The present studies demonstrate that the flanking sequence requirements for the T124A mutant are essentially those needed for formation of two independent hexamers (Fig. 8B). Therefore, assembly of both T124A hexamers requires all of the previously described T-ag-origin interactions. This observation is consistent with previous studies indicating that when two T124A hexamers assemble on the core origin, they act as independent species (2, 47) and make weak or inappropriate protein-protein interactions (47, 76).
Flanking sequence distortions can be detected on substrates containing single pentanucleotides.
The interaction between the boundary regions and the basic finger is of interest from the point of view of the mechanism of DNA unwinding. It has been suggested that the structural distortions in the core origin (7, 51) are generated by the opposing movement of the two hexamers within a given double hexamer (24, 39). However, if at least two DNA binding sites exist within a given hexamer (the A1 and B2 loops and the basic finger within the helicase domain), then opposing movement of T-ag domains within a single hexamer could contribute torque necessary for the origin structural distortions. Therefore, to examine the stage during T-ag assembly at which the structural distortions take place, we tested whether hexamers assembled on single pentanucleotides (33, 66) can distort the proximal flanking sequence.
Standard KMnO4 assays were used for these experiments, using plasmids containing mutant core origins that have a single pentanucleotide (see Materials and Methods). After the addition of T-ag, KMnO4 assays were conducted as previously described (7). Results from a representative study, performed in the presence of AMP-PNP, are presented in Fig. 9. As a positive control, a reaction was conducted with a plasmid containing the intact core origin (lane 2); the previously described alterations in the AT and EP regions (7, 51) are indicated. As additional controls, the reactions in the odd-numbered lanes were conducted with the indicated plasmids in the absence of T-ag. Reactions performed with T-ag and plasmids containing either pentanucleotide 4, 3, 2 and 1 are displayed in lanes 4, 6, 8, and 10, respectively. It is apparent that T-ag hexamers formed on individual pentanucleotides are able to distort the proximal flanking sequence at or near wild-type levels.
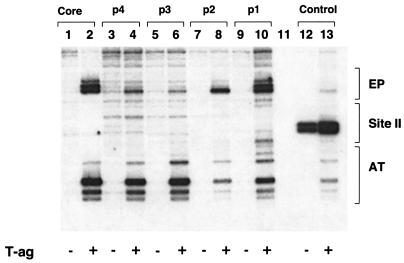
FIG. 9.
pSV01ΔEP mutants, containing single pentanucleotides, are able to catalyze T-ag-dependent structural changes in the core origin. T-ag (1 μg) was incubated under replication conditions with pSV01ΔEP(core) (lane 2), pSV01ΔEP(p 4) (lane 4), pSV01ΔEP(p 3) (lane 6), pSV01ΔEP(p 2) (lane 8), pSV01ΔEP(p 1) (lane 10), and vector alone (control). Reactions in the odd-numbered lanes were conducted with the indicated plasmids in the absence of T-ag. After treatment with KMnO4, the sites of oxidation were probed by primer extension reactions with a 32P-labeled oligonucleotide (see Materials and Methods). The primer extension products were analyzed by electrophoresis on a 7% polyacrylamide gel containing 8 M urea. The locations of the EP, site II, and AT-rich regions are shown on the right. It is noted that the control reactions conducted with the pSV01ΔEP control mutant, containing transition mutations in all four pentanucleotides (lanes 12 and 13), lack significant structural distortions in the AT-rich and EP regions. However, a T-ag-independent distortion was detected over the mutant pentanucleotides. The basis for the structural distortion over this region is not currently understood. Finally, we are also uncertain about the cause(s) of the relatively low levels of distortions detected in the distal flanking sequences (lanes 4, 6, and 8) or the more substantial distortions in the distal flanking sequences associated with the pentanucleotide 1-containing substrate (lane 10).
DISCUSSION
While scanning duplex DNA for the SV40 core origin, T-ag interacts with the surrounding sequences via nonspecific contacts, a process that requires the A1 and B2 loops in the T-ag-obd (41, 83) and a region previously mapped between residues 269 and 522 (40). When the origin is encountered, T-ag undergoes a transition between nonspecific and site-specific binding. The protein-DNA interactions required for site-specific binding to the origin include contacts between the A1 and B2 loops and the GAGGC pentanucleotides (reviewed in reference 8) and additional interactions between residues in the helicase domain and the flanking sequences (36, 52, 58, 66, 70). In addition to nonspecific DNA binding and origin recognition, the C terminus is also involved in subsequent helicase activity and single-stranded DNA binding (81, 82). In view of the importance of the C terminus-DNA interactions for viral replication, we elected to further characterize the sequence requirements for binding of the helicase domain to the SV40 origin. An additional reason for undertaking these experiments is the possibility that they might further our understanding of related interactions between the helicase domain and DNA.
Regarding the flanking sequence requirements for assembly of T-ag on the viral origin, EMSA experiments were used to identify the boundary regions necessary for efficient hexamer assembly. Subsequent molecular modeling studies indicate that a loop in T-ag centered on the highly basic (511KKHLNKR517) motif is situated opposite the boundary regions. That this motif might play an important role in DNA replication is supported by previous mutagenesis studies (53), for instance, those that characterized the Lys516→Arg mutation (42). While this conservative T-ag mutant bound to the core origin, it was defective in replication. Moreover, T-ag molecules containing a deletion of residues 507 to 510 did not support DNA replication in vivo (71). In addition, Pro 522 is situated at the distal end of the loop; the C11A T-ag mutation, Pro522→Ser, was markedly reduced in its ability to bind single-stranded DNA and partial duplex helicase substrates (48). Also of interest is that this basic finger is in close proximity with the P-loop, the ATP binding site (39). Thus, the conformation of the basic finger could be regulated by ATP hydrolysis. A similar juxtaposition of the ATP and DNA binding motifs has been found in prokaryotic helicases (see e.g., references 18, 38, and 63). These and related observations (see, e.g., the data summarized in Fig. 5) support the hypothesis that the basic finger in T-ag is critical for binding to the boundary region and for DNA replication. The relatively small size of the basic finger, and thus its limited contact with the flanking sequences, may help to explain why footprints were not detected in previous in situ footprinting assays using phenanthroline-copper ion (33). Finally, sequence alignment using the program DIALIGN 2.2 revealed that the residues centered on the 511KKHLNKR517 motif are conserved in many other initiators from the Papovaviridae viral family (e.g., polyomavirus large T-ag and human Bk and JC [data not shown]). Furthermore, a nonspecific DNA binding activity is present in the helicase domain of papillomavirus E1 (references 69 and 78 and references therein). Therefore, a number of viral initiators may rely on basic fingers, or similar motifs, for origin recognition.
It is of interest to consider possible features of the boundary region-basic finger interactions. It was previously established that nonspecific contacts with the sugar-phosphate backbone are important for the T-ag-flanking sequence association (2, 58, 59, 66). Of considerable additional interest is that the four boundary regions overlap four patches of phosphate residues previously shown to be required for T-ag binding (two in each flanking sequence, with each pair arranged on opposite strands) (58). Related experiments demonstrated, however, that the levels of hexamer and double hexamer are significantly decreased on derivatives of the core origin containing transition mutations in the flanking sequences (references 2 and 66 and data not shown). Therefore, in addition to contacts with the sugar-phosphate backbone, T-ag assembly depends upon other properties of the flanking sequences. One likely property is suggested by the observation that diethyl sulfate, an agent that ethylates guanines at the N-7 (major groove), interfered with T-ag's interactions with the EP (58). The two main sites of ethylation were at guanines situated at positions 5213 and 5216; similar conclusions were derived from methylation protection studies (7, 51). As shown in Fig. 4A (top model depicting the 48-bp p1 + EP oligonucleotide), guanine 5216 lies within the boundary region associated with pentanucleotide 1, while guanine 5213 lies just outside this region. These observations provide additional evidence that major groove contacts with the boundary regions play an important role in T-ag assembly. In addition, the finding that the flanking sequences in the SV40 core origin undergo conformational changes upon T-ag binding (reviewed in references 5, 9, 25, and 61) suggests an additional property. Indeed, the site in the EP that is melted by T-ag (7, 51) overlaps the boundary region associated with pentanucleotide 1 (Fig. 5), and it was previously proposed that the primary function of the AT-rich and EP regions is not to promote binding but to undergo structural changes required for the initiation of DNA replication (4). As previously noted, the purified T-ag-obd site specifically binds to the core origin (34, 36) but does not catalyze structural distortions in the flanking sequences (36), further evidence that the structural distortions in the flanking sequence require interactions with the helicase domain. In light of these observations, it is proposed that basic finger-boundary region interactions play a critical role in the induction of the conformational changes in the flanking sequences. Consistent with this proposal, studies with many other DNA binding proteins have demonstrated that variations in their target flanking sequences influence protein-DNA interactions by affecting DNA conformation rather than by altering particular protein-base or protein-phosphate contacts (reviewed in reference 32).
T-ag assembly and the induction of the structural distortions in the flanking sequences are coupled processes. However, the exact stage in the assembly process at which the distortions take place is not known. Our experiments demonstrate that the flanking sequence distortions can be detected on plasmid substrates that support only hexamer formation. Therefore, the structural distortions in the flanking sequences are early events that do not require double-hexamer formation. Consistent with this proposal, Parsons et al. reported that T-ag assembled on the early (e.g., pentanucleotides 1, 2 + EP) and late (e.g., pentanucleotides 3, 4 and AT) halves of the core origin was able to catalyze the distortions of the proximal flanking sequences (52). Whether the conformational changes in the flanking sequences occur as a result of monomer binding, as a result of hexamer assembly, or at some intermediate stage is presently not known. Nevertheless, at some stage in the assembly process, the basic finger-boundary region recognition interface (32) is likely to play a role in the distortions in the flanking sequences. Furthermore, recognition of the structurally distortable sequences is probably coupled to the recognition of the GAGGC sequences by the A1 and B2 loops in the T-ag-obd.
Relative to hexamer formation, the sequence requirements for double-hexamer assembly are complex. When phosphorylated on Thr124 (30), the second hexamer oligomerizes in a manner that is relatively independent of the presence of the second flanking sequence (Fig. 6 and 7) (2, 66); thus, it has reduced affinity for one of its associated flanking sequences (modeled in Fig. 8A). It is proposed that much of the binding energy necessary for formation of the second hexamer is derived from protein-protein interactions with the initially formed hexamer, a hypothesis supported by previous studies (47, 76). In contrast, our studies with the T124A mutant indicate that prior to phosphorylation of Thr124, the interactions between the helicase domains and the EP and AT-rich regions are maintained in each of the hexamers, a conclusion supported by previous experiments (2, 47). It follows that at non-S phases of the cell cycle, two hexamers may assemble on the core origin in a manner that preserves their independent status. However, upon phosphorylation of Thr124, possibly by a cyclin-dependent kinase (25), one of the hexamers releases its nonspecific interactions with the flanking sequence and initiates critical protein-protein interactions with the second hexamer. Finally, since double hexamers prefer to assemble on pentanucleotides 1 and 3 (33, 66), it is of interest that the pentanucleotide 1 boundary region is associated with a major groove while the pentanucleotide 3 boundary region is associated with both major and minor grooves (Fig. 4B). Therefore, relatively low affinity of the basic finger for the pentanucleotide 3 boundary region may help to explain the results summarized in Fig. 8A.
How the double hexamer assembled on the core origin is able to catalyze the replication protein A- and topoisomerase I-dependent formation of the unwound species termed form U (10, 14, 20, 80) has yet to be determined. It was previously reported that while double-hexamer formation requires only two pentanucleotides (e.g., pentanucleotides 1 and 3), DNA unwinding requires all four (13, 33, 66). Therefore, at some stage in the assembly process, the double hexamer must engage the second pair of pentanucleotides (e.g., pentanucleotides 2 and 4) and their associated flanking sequences. One possibility is that initiation of DNA unwinding requires oscillating contacts between pairs of pentanucleotides and flanking sequences. It is also possible that retrograde movement of domains within a single hexamer (this study) or between double hexamers (24, 39) generates torque necessary for propagation of the initial structural distortions. Upon completion of assembly and ATP hydrolysis, basic fingers not bound to DNA (e.g., those in the second hexamer) could engage DNA in a non-sequence-specific manner. Indeed, given the orientation of T-ag double hexamers on the core origin (Fig. 8) (73) and evidence that the DNA is actively spooled through the double-hexamer helicase (see, e.g., references 1, 64, and 77), it is possible that residues in the basic finger play a role in moving DNA towards the center of the complex. This component of the T-ag helicase cycle would depend upon non-sequence-specific interactions with DNA, a known feature of residues located in the helicase domain (40).
Acknowledgments
We thank David Cullinan, Gavin Schnitzler, Julie Kerner, Anuradha Kumar, and Margaret VanLoock for useful discussions and A. J. Bullock for comments on the manuscript.
This study was funded by a grant from the National Institutes of Health (9R01GM55397).
REFERENCES
- 1.Alexandrov, A. I., M. R. Botchan, and N. R. Cozzarelli. 2002. Characterization of simian virus 40 T-antigen double hexamers bound to a replication fork. J. Biol. Chem. 277:44886-44897. [DOI] [PubMed] [Google Scholar]
- 2.Barbaro, B. A., K. R. Sreekumar, D. R. Winters, A. E. Prack, and P. A. Bullock. 2000. Phosphorylation of simian virus 40 T-antigen on Thr 124 selectively promotes double-hexamer formation on subfragments of the viral core origin. J. Virol. 74:8601-8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell, S. P., and A. Dutta. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71:333-374. [DOI] [PubMed] [Google Scholar]
- 4.Borowiec, J. A. 1992. Inhibition of structural changes in the simian virus 40 core origin of replication by mutation of essential origin sequences. J. Virol. 66:5248-5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borowiec, J. A., F. B. Dean, P. A. Bullock, and J. Hurwitz. 1990. Binding and unwinding—how T antigen engages the SV40 origin of DNA replication. Cell 60:181-184. [DOI] [PubMed] [Google Scholar]
- 6.Borowiec, J. A., and J. Hurwitz. 1988. ATP stimulates the binding of the simian virus 40 (SV40) large tumor antigen to the SV40 origin of replication. Proc. Natl. Acad. Sci. USA 85:64-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borowiec, J. A., and J. Hurwitz. 1988. Localized melting and structural changes in the SV40 origin of replication induced by T-antigen. EMBO J. 7:3149-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bullock, P. A. 1997. The initiation of simian virus 40 DNA replication in vitro. Crit. Rev. Biochem. Mol. Biol. 32:503-568. [DOI] [PubMed] [Google Scholar]
- 9.Bullock, P. A., W. S. Joo, K. R. Sreekumar, and C. Mello. 1997. Initiation of SV40 DNA replication in vitro: analysis of the role played by sequences flanking the core origin on initial synthesis events. Virology 227:460-473. [DOI] [PubMed] [Google Scholar]
- 10.Bullock, P. A., Y. S. Seo, and J. Hurwitz. 1989. Initiation of simian virus 40 DNA replication in vitro: pulse-chase experiments identify the first labeled species as topologically unwound. Proc. Natl. Acad. Sci. USA 86:3944-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell, K. S., K. P. Mullane, I. A. Aksoy, H. Stubdal, J. M. Pipas, P. A. Silver, T. M. Roberts, B. S. Schaffhausen, and J. A. DeCaprio. 1997. DnaJ/hsp40 chaperone domain of SV40 large T antigen promotes efficient viral DNA replication. Genes Dev. 11:1098-1110. [DOI] [PubMed] [Google Scholar]
- 12.Campos-Olivas, R., J. M. Louis, D. Clerot, B. Gronenborn, and A. M. Gronenborn. 2002. The structure of a replication initiator unites diverse aspects of nucleic acid metabolism. Proc. Natl. Acad. Sci. USA 99:10310-10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dean, F. B., J. A. Borowiec, Y. Ishimi, S. Deb, P. Tegtmeyer, and J. Hurwitz. 1987. Simian virus 40 large tumor antigen requires three core replication origin domains for DNA unwinding and replication in vitro. Proc. Natl. Acad. Sci. USA 84:8267-8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dean, F. B., P. Bullock, Y. Murakami, C. R. Wobbe, L. Weissbach, and J. Hurwitz. 1987. Simian virus 40 (SV40) DNA replication: SV40 large T antigen unwinds DNA containing the SV40 origin of replication. Proc. Natl. Acad. Sci. USA 84:16-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dean, F. B., M. Dodson, H. Echols, and J. Hurwitz. 1987. ATP-dependent formation of a specialized nucleoprotein structure by simian virus 40 (SV40) large tumor antigen at the SV40 replication origin. Proc. Natl. Acad. Sci. USA 84:8981-8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deb, S., A. L. DeLucia, C.-P. Baur, A. Koff, and P. Tegtmeyer. 1986. Domain structure of the simian virus 40 core origin of replication. Mol. Cell. Biol. 6:1663-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deb, S. P., and P. Tegtmeyer. 1987. ATP enhances the binding of simian virus 40 large T antigen to the origin of replication. J. Virol. 61:3649-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dillingham, M. S., P. Soultanas, and D. B. Wigley. 1999. Site-directed mutagenesis of motif III in PcrA helicase reveals a role in coupling ATP hydrolysis to strand separation. Nucleic Acids Res. 27:3310-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dixon, R. A. F., and D. Nathans. 1985. Purification of simian virus 40 large T antigen by immunoaffinity chromatography. J. Virol. 53:1001-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dodson, M., F. B. Dean, P. Bullock, H. Echols, and J. Hurwitz. 1987. Unwinding of duplex DNA from the SV40 origin of replication by T antigen. Science 238:964-967. [DOI] [PubMed] [Google Scholar]
- 21.Enemark, E. J., G. Chen, D. E. Vaughn, A. Stenlund, and L. Joshua-Tor. 2000. Crystal structure of the DNA binding domain of the replication initiation protein E1 from papillomavirus. Mol. Cell 6:149-158. [PubMed] [Google Scholar]
- 22.Enemark, E. J., A. Stenlund, and L. Joshua-Tor. 2002. Crystal structures of two intermediates in the assembly of the papillomavirus replication initiation complex. EMBO J. 21:1487-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Erdile, L. F., K. L. Collins, A. Russo, P. Simancek, D. Small, C. Umbricht, D. Virshup, L. Cheng, S. Randall, D. Weinberg, I. Moarefi, E. Fanning, and T. Kelly. 1991. Initiation of SV40 DNA replication: mechanism and control. Cold Spring Harbor Symp. Quant. Biol. 56:303-313. [DOI] [PubMed] [Google Scholar]
- 24.Fanning, E. 1994. Control of SV40 DNA replication by protein phosphorylation: a model for cellular DNA replication? Trends Cell Biol. 4:250-255. [DOI] [PubMed] [Google Scholar]
- 25.Fanning, E., and R. Knippers. 1992. Structure and function of simian virus 40 large tumor antigen. Annu. Rev. Biochem. 61:55-85. [DOI] [PubMed] [Google Scholar]
- 26.Gilbert, D. M. 2001. Making sense of eukaryotic DNA replication origins. Science 294:96-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goetz, G. S., F. B. Dean, J. Hurwitz, and S. W. Matson. 1988. The unwinding of duplex regions in DNA by the simian virus 40 large tumor antigen-associated DNA helicase activity. J. Biol. Chem. 263:383-392. [PubMed] [Google Scholar]
- 28.Hickman, A. B., R. D. Ronning, R. M. Kotin, and R. Dyda. 2002. Structural unity among viral origin binding proteins: crystal structure of the nuclease domain of adeno-associated virus Rep. Mol. Cell 10:327-337. [DOI] [PubMed] [Google Scholar]
- 29.Hingorani, M. M., M. T. Washington, K. C. Moore, and S. S. Patel. 1997. The dTTPase mechanism of T7 DNA helicase resembles the binding change mechanism of the F1-ATPase. Proc. Natl. Acad. Sci. USA 94:5012-5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Höss, A., I. Moarefi, K. H. Scheidtmann, L. J. Cisek, J. L. Corden, I. Dornreiter, A. K. Arthur, and E. Fanning. 1990. Altered phosphorylation pattern of simian virus 40 T antigen expressed in insect cells by using a baculovirus vector. J. Virol. 64:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang, S. G., K. Weisshart, I. Gilbert, and E. Fanning. 1998. Stoichiometry and mechanisms of assembly of SV40 T antigen complexes with the viral origin of DNA replication and DNA polymerase α-primase. Biochemistry 37:15345-15352. [DOI] [PubMed] [Google Scholar]
- 32.Jen-Jacobson, L. 1997. Protein-DNA recognition complexes: conservation of structure and binding energy in the transition state. Biopolymers 44:153-180. [DOI] [PubMed] [Google Scholar]
- 33.Joo, W. S., H. Y. Kim, J. D. Purviance, K. R. Sreekumar, and P. A. Bullock. 1998. Assembly of T-antigen double hexamers on the simian virus 40 core origin requires only a subset of the available binding sites. Mol. Cell. Biol. 18:2677-2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joo, W. S., X. Luo, D. Denis, H. Y. Kim, G. J. Rainey, C. Jones, K. R. Sreekumar, and P. A. Bullock. 1997. Purification of the SV40 T-antigen DNA binding domain and characterization of its interactions with the SV40 origin. J. Virol. 71:3972-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim, H.-Y., B. Y. Ahn, and Y. Cho. 2001. Structural basis for the inactivation of retinoblastoma tumor suppressor by SV40 large T antigen. EMBO J. 20:295-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim, H. Y., B. A. Barbaro, W. S. Joo, A. Prack, K. R. Sreekumar, and P. A. Bullock. 1999. Sequence requirements for the assembly of simian virus 40 T-antigen and T-antigen origin binding domain on the viral core origin of replication. J. Virol. 73:7543-7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim, R. J., S. Moine, D. K. Reese, and P. A. Bullock. 2002. Peptides containing cyclin/Cdk-nuclear localization signal motifs derived from viral initiator proteins bind to DNA when unphosphorylated. J. Virol. 76:11785-11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korolev, S., J. Hsieh, G. H. Gauss, T. M. Lohman, and G. Waksman. 1997. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell 90:635-647. [DOI] [PubMed] [Google Scholar]
- 39.Li, D., R. Zhao, W. Lilyestrom, D. Gai, R. Zhang, J. A. DeCaprio, E. Fanning, A. Jochimiak, G. Szakonyi, and X. S. Chen. 2003. The structure of the replicative helicase of the transforming protein SV40 large T-antigen. Nature 423:512-518. [DOI] [PubMed] [Google Scholar]
- 40.Lin, H. J. L., R. H. Upson, and D. T. Simmons. 1992. Nonspecific DNA binding activity of simian virus 40 large T antigen: evidence for the cooperation of two regions for full activity. J. Virol. 66:5443-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo, X., D. G. Sanford, P. A. Bullock, and W. W. Bachovchin. 1996. Structure of the origin specific DNA binding domain from simian virus 40 T-antigen. Nat. Struct. Biol. 3:1034-1039. [DOI] [PubMed] [Google Scholar]
- 42.Manos, M. M., and Y. Gluzman. 1984. Simian virus 40 large T-antigen point mutants that are defective in viral DNA replication but competent in oncogenic transformation. Mol. Cell. Biol. 4:1125-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mastrangelo, I. A., M. Bezanilla, P. K. Hansma, P. V. C. Hough, and H. G. Hansma. 1994. Structures of large T antigen at the origin of SV40 DNA replication by atomic force microscopy. Biophys. J. 66:293-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mastrangelo, I. A., P. V. C. Hough, J. S. Wall, M. Dodson, F. B. Dean, and J. Hurwitz. 1989. ATP-dependent assembly of double hexamers of SV40 T antigen at the viral origin of DNA replication. Nature 338:658-662. [DOI] [PubMed] [Google Scholar]
- 45.McVey, D., L. Brizuela, I. Mohr, D. R. Marshak, Y. Gluzman, and D. Beach. 1989. Phosphorylation of large tumor antigen by cdc2 stimulates SV40 DNA replication. Nature 341:503-507. [DOI] [PubMed] [Google Scholar]
- 46.McVey, D., S. Ray, Y. Gluzman, L. Berger, A. G. Wildeman, D. R. Marshak, and P. Tegtmeyer. 1993. cdc2 phosphorylation of threonine 124 activates the origin-unwinding functions of simian virus 40 T antigen. J. Virol. 67:5206-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moarefi, I. F., D. Small, I. Gilbert, M. Hopfner, S. K. Randall, C. Schneider, A. A. R. Russo, U. Ramsperger, A. K. Arthur, H. Stahl, T. J. Kelly, and E. Fanning. 1993. Mutation of the cyclin-dependent kinase phosphorylation site in simian virus 40 (SV40) large T antigen specifically blocks SV40 origin DNA unwinding. J. Virol. 67:4992-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mohr, I. J., M. P. Fairman, B. Stillman, and Y. Gluzman. 1989. Large T-antigen mutants define multiple steps in the initiation of simian virus 40 DNA replication. J. Virol. 63:4181-4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Myers, R. M., R. C. Williams, and R. Tjian. 1981. Oligomeric structure of a simian virus 40 T antigen in free form and bound to DNA. J. Mol. Biol. 148:347-353. [DOI] [PubMed] [Google Scholar]
- 50.O'Reilly, D. R., and L. K. Miller. 1988. Expression and complex formation of simian virus 40 large T antigen and mouse p53 in insect cells. J. Virol. 62:3109-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parsons, R., M. E. Anderson, and P. Tegtmeyer. 1990. Three domains in the simian virus 40 core origin orchestrate the binding, melting, and DNA helicase activities of T antigen. J. Virol. 64:509-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parsons, R. E., J. E. Stenger, S. Ray, R. Welker, M. E. Anderson, and P. Tegtmeyer. 1991. Cooperative assembly of simian virus 40 T-antigen hexamers on functional halves of the replication origin. J. Virol. 65:2798-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson, C. G., and J. M. Pipas. 1998. SV40 large tumor antigen (T antigen): database of mutants. Nucleic Acids Res. 26:295-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 55.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.San Martin, M. C., C. Gruss, and J. M. Carazo. 1997. Six molecules of SV40 large T antigen assemble in a propeller-shaped particle around a channel. J. Mol. Biol. 268:15-20. [DOI] [PubMed] [Google Scholar]
- 57.Schneider, J., and E. Fanning. 1988. Mutations in the phosphorylation sites of simian virus 40 (SV40) T antigen alter its origin DNA-binding specificity for sites I or II and affect SV40 DNA replication activity. J. Virol. 62:1598-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.SenGupta, D. J., and J. A. Borowiec. 1994. Strand and face: the topography of interactions between the SV40 origin of replication and T-antigen during the initiation of replication. EMBO J. 13:982-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.SenGupta, D. J., and J. A. Borowiec. 1992. Strand-specific recognition of a synthetic DNA replication fork by the SV40 large tumor antigen. Science 256:1656-1661. [DOI] [PubMed] [Google Scholar]
- 60.Simanis, V., and D. P. Lane. 1985. An immunoaffinity purification procedure for SV40 large T antigen. Virology 144:88-100. [DOI] [PubMed] [Google Scholar]
- 61.Simmons, D. T. 2000. SV40 large T antigen functions in DNA replication and transformation. Adv. Virus Res. 55:75-134. [DOI] [PubMed] [Google Scholar]
- 62.Simmons, D. T., G. Loeber, and P. Tegtmeyer. 1990. Four major sequence elements of simian virus 40 large T antigen coordinate its specific and nonspecific DNA binding. J. Virol. 64:1973-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Singleton, M. R., M. R. Sawaya, T. Ellenberger, and D. B. Wigley. 2000. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell 101:589-600. [DOI] [PubMed] [Google Scholar]
- 64.Smelkova, N. V., and J. A. Borowiec. 1997. Dimerization of simian virus 40 T-antigen hexamers activates T-antigen DNA helicase activity. J. Virol. 71:8766-8773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sreekumar, K. R., B. A. Barbaro, A. Prack, and P. A. Bullock. 2001. Methods for studying interactions between simian virus 40 T-antigen and the viral origin of replication. Methods Mol. Biol. 165:49-67. [DOI] [PubMed]
- 66.Sreekumar, K. R., A. E. Prack, D. R. Winters, B. A. Barbaro, and P. A. Bullock. 2000. The simian virus 40 core origin contains two separate sequence modules that support T-antigen double-hexamer assembly. J. Virol. 74:8589-8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Srinivasan, A., A. J. McClellan, J. Vartikar, I. Marks, P. Cantalupo, Y. Li, P. Whyte, K. Rundell, J. L. Brodsky, and J. M. Pipas. 1997. The amino-terminal transforming region of simian virus 40 large T and small t antigens functions as a J domain. Mol. Cell. Biol. 17:4761-4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stahl, H., P. Droge, and R. Knippers. 1986. DNA helicase activity of SV40 large tumor antigen. EMBO J. 5:1939-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stenlund, A. 2003. E1 initiator DNA binding specificity is unmasked by selective inhibition of non-specific DNA binding. EMBO J. 22:954-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Titolo, S., E. Welchner, P. W. White, and J. Archambault. 2003. Characterization of the DNA-binding properties of the origin-binding domain of simian virus 40 large T antigen by fluorescence anisotrophy. J. Virol. 77:5512-5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tornow, J., and C. N. Cole. 1983. Intracistronic complementation in the simian virus 40 A gene. Proc. Natl. Acad. Sci. USA 80:6312-6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Uhlmann-Schiffler, H., S. Seinsoth, and H. Stahl. 2002. Preformed hexamers of SV40 T antigen are active in RNA and origin-DNA unwinding. Nucleic Acids Res. 30:3192-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Valle, M., C. Gruss, L. Halmer, J. M. Carazo, and L. E. Donate. 2000. Large T-antigen double hexamers imaged at the simian virus 40 origin of replication. Mol. Cell. Biol. 20:34-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.VanLoock, M. S., A. Alexandrov, X. Yu, N. R. Cozzarelli, and E. H. Egelman. 2002. SV40 large T antigen hexamer structure: domain organization and DNA-induced conformational changes. Curr. Biol. 12:472-476. [DOI] [PubMed] [Google Scholar]
- 75.Waga, S., and B. Stillman. 1998. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67:721-751. [DOI] [PubMed] [Google Scholar]
- 76.Weisshart, K., P. Taneja, A. Jenne, U. Herbig, D. T. Simmons, and E. Fanning. 1999. Two regions of simian virus 40 T antigen determine cooperativity of double-hexamer assembly on the viral origin of DNA replication and promote hexamer interactions during bidirectional-origin DNA unwinding. J. Virol. 73:2201-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wessel, R., J. Schweizer, and H. Stahl. 1992. Simian virus 40 T-antigen DNA helicase is a hexamer which forms a binary complex during bidirectional unwinding from the viral origin of DNA replication. J. Virol. 66:804-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.White, P. W., A. Pelletier, K. Brault, S. Titolo, E. Welchner, L. Thauvette, M. Fazekas, M. G. Cordingley, and J. Archambault. 2001. Characterization of recombinant HPV6 and 11 E1 helicases: effect of ATP on the interaction of E1 with E2 and mapping of a minimal helicase domain. J. Biol. Chem. 276:22426-22438. [DOI] [PubMed] [Google Scholar]
- 79.Wobbe, C. R., F. Dean, L. Weissbach, and J. Hurwitz. 1985. In vitro replication of duplex circular DNA containing the simian virus 40 DNA origin site. Proc. Natl. Acad. Sci. USA 82:5710-5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wold, M. S., J. J. Li, and T. J. Kelly. 1987. Initiation of simian virus 40 DNA replication in vitro: large-tumor-antigen- and origin-dependent unwinding of the template. Proc. Natl. Acad. Sci. USA 84:3643-3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu, C., D. Edgil, and D. T. Simmons. 1998. The origin DNA-binding and single-stranded DNA-binding domains of simian virus large T antigen are distinct. J. Virol. 72:10256-10259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu, C., R. Roy, and D. T. Simmons. 2001. Role of single-stranded DNA binding activity of T antigen in simian virus 40 DNA replication. J. Virol. 75:2839-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wun-Kim, K., R. Upson, W. Young, T. Melendy, B. Stillman, and D. T. Simmons. 1993. The DNA-binding domain of simian virus 40 tumor antigen has multiple functions. J. Virol. 67:7608-7611. [DOI] [PMC free article] [PubMed] [Google Scholar]