Abstract
The LIM homeodomain transcription factor Lmx1b is essential for the development of the isthmic organizer and mesodiencephalic dopaminergic neurons. The uncoupling of Pitx3 and Th expression, in the Lmx1b null mutant, suggests that Lmx1b may act as a positional activator of the mdDA domain, eventually leading to properly differentiating mdDA neurons. In this study, we aimed to elucidate how Lmx1b functions mechanistically in this developmental process, by searching for molecular interactors of Lmx1b at the protein level. Initially, affinity-purification of LMX1B-HIS overexpressed protein in MN9D dopaminergic cells followed by mass-spectrometry analysis, resulted in the identification of PSPC1 protein as a possible binding partner of LMX1B. Subsequent immunoprecipitation experiments revealed an interaction between LMX1B and PSPC1 in a larger protein complex also containing PSF. This complex was observed in vitro and in vivo, and we hypothesize that, via PSF and PSPC1, LMX1B may be part of the previously identified Nurr1 transcriptional complex wherein interaction with the co-repressor PSF and the transcription factor Pitx3 is needed to drive expression of Nurr1 target genes in specifying the dopaminergic phenotype. Furthermore, we identified GRLF1, DHX9, MYO1C, HSP70 and TMPO as potential LMX1B interactors. DHX9 and GRLF1 are highly expressed in the developing mdDA neuronal field, and GRLF1 and MYO1C have both been linked to neurite outgrowth. The identification of these proteins suggests that Lmx1b may act directly in the transcriptional activation of Nurr1 target genes and be involved in other processes like neurite outgrowth as well.
Introduction
One of the essential transcription factors involved in mesodiencephalic dopaminergic (mdDA) neuron development, is the LIM homeodomain (LIM-HD) transcription factor 1 beta (Lmx1b). The first sign of this relevance was provided through the analysis of the Lmx1b null mutant, which showed a clear midbrain defect and uncoupling of Th and Pitx3 expression in the mdDA region [1]. Lmx1b is expressed before the expression of Nurr1, Pitx3 and Th and has intrinsic properties as a developmental regulator. The (partial) loss of Pitx3 and later of Th in the Lmx1b null mutant suggest that Lmx1b may act as an upstream activator of these genes in the development of mdDA neurons [1]–[3], or Lmx1b may be involved in specifying the dopaminergic niche in the midbrain region. This possibility is underlined by the fact that Lmx1b is also involved in regulation of Fgf8 and Wnt1, and several isthmus-related transcription factors, and it is essential for inductive activity of the isthmic organizer (IsO) [4], [5]. Loss of Lmx1b likely affects mid-hindbrain (MHB) patterning, resulting in an early loss of a large part of the midbrain [4]. In a recent study, it was shown that specific inactivation of Lmx1b in mdDA progenitors, but not in the IsO, resulted in normally developing neurons, and it was suggested that Lxm1b is not required for the specification and differentiation of mdDA progenitors on its own [6].
Furthermore, it was shown that Foxa1 and Foxa2 are able to specify mdDA progenitors by positively regulating the expression of Lmx1a and Lmx1b. Foxa1/Foxa2 together with Lmx1a/Lmx1b, induce expression of Nurr1, and it was suggested that these genes cooperate in order to promote mdDA development by regulating common targets that are important for mdDA differentiation [7]–[9].
Despite the many studies of the function of Lmx1b in mdDA development and differentiation, the precise role is still not clear. Most of the studies focus on identifying genes in the molecular cascades in which Lmx1b is involved and not much is known about the functional level of Lmx1b in the proposed pathways. However, two studies identified CLIM2 (LDB1) and PAX2 as proteins that directly interact with LMX1B protein, via yeast-two-hybrid assays [10], [11]. Ldb1 is an essential LIM-HD co-factor that can, in a transcriptional complex, act as a central signaling integrator [12]–[14].
In the current study, we specifically focus on the identification of physical interactors with LMX1B protein. In an open search for binding partners, based on affinity purification, and immunoprecipitation (IP) techniques followed by mass spectrometry analysis, we identified PSPC1, GRLF1, DDX9, MYO1C, HSP70 and TMPO as possible interactors binding to LMX1B. Furthermore, via IP experiments in vitro and in vivo, a protein complex was identified containing PSPC1, PSF and LMX1B, suggesting the existence of this complex in mdDA neurons.
Materials and Methods
Animals
Experiments were carried out in C57Bl/6J wild-type mice (Charles River). Pregnant mice were decapitated or euthanized by CO2 asphyxiation and embryos were collected at E14.5 (the day on which the copulatory plug was detected was considered E0.5). Mice were maintained under standard conditions, all efforts were made to minimize suffering, and all procedures were according to and fully approved by the Dutch Ethical Committee for animal experimentation of the University Medical Center Utrecht (DEC UMC-U, The Netherlands).
PCR and Cloning
pBSK(+)-Lmx1b cDNA vector (kind gift of the lab of R. Johnson, Houston) was used to clone a 1.3 kB fragment containing the full Lmx1b coding sequence, into the expression vector pcDNA3.1(-) (Invitrogen), by using the EcoRI restriction sites. To generate myc-HIS tagged LMX1B protein, primers were designed for the pcDNA3.1(-)-LMX1B template, to introduce an EcoRI restriction site at the 5′ side, and a BamHI restriction site at the 3′ end, plus an elimination of the stop-codon that was initially incorporated in the LMX1B construct (forward 5′-CAGAATTCGGGCGCTGGAGAGG-3′, reverse 5′-AGGATCCGGAGGCAAAGTAGGAGCTC-3′). The PCR product was cloned into pcDNA3.1(-)myc-HIS_A (Invitrogen) using the EcoRI and BamHI restriction sites. The resulting vector was sequenced (Baseclear, Netherlands).
MN9D Cell Culture and Transfection
MN9D cells (a kind gift of Dr. Thomas Perlmann; for literature: [15], [16]) were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% (v/v) heat-inactivated fetal calf serum (hiFCS), 100 units/mL penicillin, 100 units/mL streptomycin and 2 mM L-Glutamine, in a standard incubator with 5% CO2 at 37°C. Cells were grown on 10 cm dishes, additionally coated with poly-L-lysine. At least 2 hours before transfection, culture medium was replaced by antibiotics free medium. Transfection was performed with Lipofectamine 2000 (Invitrogen), according to manufacturer’s protocol. Expression vector pcDNA3.1(-)-LMX1B-mycHIS was transfected in an amount of 22 ug DNA. Control (empty) vector was transfected in equimolar amounts. Cloning vector pBluescript SKII(+) was added as carrier DNA. 5–6 hours post transfection, the cells were split (1∶3) and cultured in fresh medium with antibiotics. Cells were harvested when the plates were 90–95% confluent and RNA or protein was isolated for further analysis.
Ni-NTA Magnetic Agarose Bead Purification
Transfected and harvested cells were centrifuged and resuspended in Qialysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 0.05% Tween 20, pH 8.0). Lysis was done on ice for 30–40 min. and cellpellets were homogenized by passing a 27/3–4 G syringe 5 times to ensure complete cell and nuclear lysis before centrifugation. Lysates were kept on ice before being incubated with Ni-NTA magnetic agarose beads (Qiagen; 100 uL/mL beads) on an end-over-end shaker in a cold room (4–8°C) overnight. Ni-NTA beads were washed four times (50 mM NaH2PO4, 300 mM NaCl, 40 mM imidazole, 0.05% Tween 20, pH 8.0). Beads were captured using a magnetic separator (>20 mega-oersted, Qiagen). Beads were incubated in elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, 0.05% Tween 20, pH 8.0) with frequent flicking of the tube, for 20 minutes. Eluates were stored for later analysis at −80°C. All buffers were supplemented with a complete protease inhibitor cocktail (Roche).
Immunoprecipitation
MN9D or E14.5 embryonic tissue cells were homogenized in lysis buffer (50 mM Tris-HCl pH 8, 150 mM NaCl, 5 mM MgCl2, 0.5 mM EDTA, 0,2% NP-40, 5% glycerol, and 1× Protease Inhibitor (Roche)). Per plate of MN9Ds or 5–6 dissected E14.5 midbrains, approximately 500 uL lysis buffer was used. Cells were lysed for 20 minutes on ice and lysate was homogenized by passing 5× through a 27G syringe and centrifuged. Magnetic Protein A Dynal beads (Invitrogen; 35 uL beads/mL lysate) were blocked in 0,5% BSA in 1× PBS and incubated with antibody overnight on a rotator at 4°C. The following antibodies were used for (Co-)IP: rabbit anti-PSPC1 (Santa Cruz), rabbit anti-HIS (Abcam), goat anti-LMX1B (Santa Cruz), mouse anti-PSF (Sigma), and rabbit anti-NURR1 (Santa Cruz). As a control, host normal serum was used. Lysate was incubated with antibody-bound beads overnight on a rotator at 4°C. Beads were washed in lysis buffer (five times) and captured using a magnetic separator (Qiagen). Proteins were eluted in elution buffer (50 mM TrisHCl pH 8, 10 mM EDTA, 1% (v/v) SDS, and 1× Protease Inhibitor (Roche)) at 65°C for 10 minutes and with frequent vortexing. Eluates were stored for analysis at −80°C.
Western Blot
Proteins were separated by standard SDS-PAGE on 11% acrylamide gels (Biorad), and transferred to a Hybond C extra membrane (Amersham). Membranes were blocked overnight in 5% milk powder in PBS, at 4°C. Subsequently, membranes were incubated with primary antibodies in PBS-T (0.05% v/v Tween) on a shaker for 2 hours at room temperature. Antibodies used: rabbit anti-HIS (Abcam, ab9108; 1∶20.000), rabbit anti-PSPC1 (Santa Cruz, sc-84577; 1∶10.000), goat anti-LMX1B (Santa Cruz, sc-21231; 1∶5.000), mouse anti-PSF (Sigma, B92; 1∶2.500), rabbit anti-NURR1 (Santa Cruz, sc-990; 1∶200). When using goat anti-LMX1B, blots were blocked in 5% normal donkey serum instead of milk, to reduce high background due to secondary donkey anti-goat antibody binding to bovine IgGs. Membranes were incubated with SuperSignal West Dura Extended Duration Substrate (Pierce; Thermo Scientific) and exposed to ECL films (Pierce; Thermo Scientific).
Silverstaining
Protein eluates were separated by means of SDS PAGE. Subsequently, the gel was silver-stained to detect the proteins. The gel was fixed in 50% methanol (2×15 min) and 5% methanol (10 min), rinsed 3 times with water, soaked in 10 µM DTT (20 min), incubated in 0.1% AgNO3 (20 min), and rinsed with water. Next, it was incubated in developer solution (3% NaCO3, 0.05% formaldehyde) until protein bands were visible. The reaction was stopped by adding citric acid and the gel was washed in water. Protein bands of interest were excised form the gel and subjected to nanoLC-ESI-MS Mass Spectrometry analysis and subsequent database analysis (The Proteome Factory, Berlin). Additionally, several SDS PAGE gels were silver-stained by using the FireSilver staining kit from the Proteome Factory.
NanoLC-ESI-MS/MS
Protein identification was performed by the Proteome Factory (Berlin, Germany): The MS system consisted of an Agilent 1100 nanoLC system (Agilent, Waldbronn, Germany), PicoTip emitter (New Objective, Woburn, USA) and a Qtof Ultima mass spectrometer (Micromass/Waters, Manchester, UK). Protein spots were in-gel digested by trypsin (Promega, Mannheim, Germany) and applied to nanoLC-ESI-MS/MS. Peptides were trapped and desalted on the enrichment column (Zorbax SB C18, 0.3×5 mm, Agilent) for five minutes using 1% acetonitrile/0.5% formic acid as eluent, then peptides were separated on a Zorbax 300 SB C18, 75 µm x 150 mm column (Agilent) using an acetonitrile/0.1% formic acid gradient from 5% to 40% acetonitrile within 40 minutes. MS spectra were automatically recorded by the mass spectrometer according to manufacturer’s instrument settings for nanoLC-ESI-MS/MS analysis. Proteins were identified using MS/MS ion search of the Mascot search engine (Matrix Science, London, England) and the non-redundant protein database (National Center for Biotechnology Information, Bethesda, USA). Ion charges in search parameters for ions from ESI-MS/MS data acquisition were set to 1+,2+ or 3+ according to the instrument’s and method’s common charge state distribution. Individual ions scores >30 indicate identity or extensive homology (p<0.05).
Results
Paraspeckle Protein (PSPC1) Interacts with an LMX1B-myc-HIS Fusion Protein
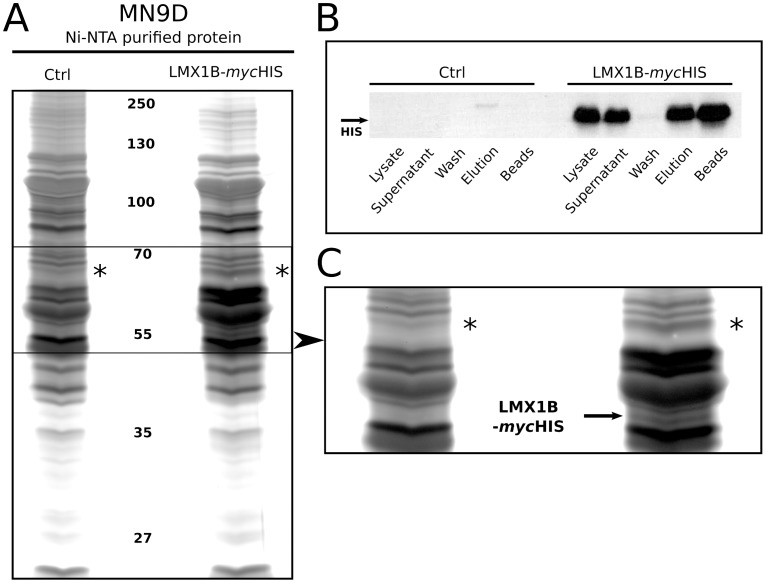
In order to determine possible interactors of LMX1B, an open search was performed by means of affinity purification of LMX1B. To achieve this, we overexpressed (OE) LMX1B protein translationally fused to a myc-HIS sequence, in MN9D dopaminergic cells. Through Ni-NTA affinity purification, we were able to purify the HIS-tagged LMX1B protein from cell lysates, and analyzed this by means of protein gel silver-staining (FIG. 1A). The set-up was validated by means of Western blot (FIG. 1B), that clearly revealed the successful purification of OE HIS-tagged LMX1B protein, compared to a similar purification experiment on MN9D cells transfected with an empty vector as control (pcDNA3.1(-)-myc-HIS). A large amount of HIS-tagged LMX1B protein was detected in the total lysate of OE cells, and after binding to Ni-NTA beads, unbound LMX1B-HIS remained in the supernatant. Bound LMX1B-HIS protein was retrieved after elution. Since almost no protein was detected in the first washing steps, the LMX1B-HIS protein was strongly bound to the Ni-NTA beads.
Figure 1. Affinity purified Lmx1b-HIS proteins.
(A) Lmx1b-HIS purified proteins from MN9D cells, by means of HIS tagged affinity purification via Ni-NTA agarose beads, followed by separation on silver-stained SDS gel. The left lane represents proteins purified from control transfected MN9D cells. The right lane shows purified proteins from LMX1B-HIS overexpressing MN9D cells. Asterisks mark an observed differential protein band. (B) Western blot validation of the LMX1B-HIS overexpression in MN9D cells, followed by successful purification of LMX1B-HIS protein. Lysate shows clear LMX1B-HIS overexpression. Supernatant reveals unbound LMX1B-HIS after Ni-NTA bead incubation. Protein is strongly bound to the beads, as shown by an extremely low amount of protein that was detected in the first washing-steps. Following successful elution, beads were heated and used for a second, thorough elution; a large amount of protein was detected that was not eluted in the first elution step. (C) Detailed image of the differential protein band in the LMX1B-HIS OE sample (asterisks). Overexpressed LMX1B-HIS protein (based on size) was detected as well (arrow). Ctrl, purified protein from control transfected MN9D cells; LMX1B-mycHIS, purified protein from MN9D cells overexpressing LMX1B-HIS.
Due to the large amount of background protein, only small differences were observed in protein composition of both samples. In the OE sample lane, a small, extra protein band was observed at approximately 55 kDA, which most likely represents the LMX1B-HIS OE protein, based on the size. In addition, a clear protein band was observed between 65 and 70 kDA (FIG. 1A and C, asterisks). This protein band was excised from gel, together with the same material (same relative mobility position) from the control lane, and sent for mass spectrometry analysis (Proteome Factory, Berlin). The resulting data analysis (MASCOT) identified several proteins (Table 1). Unfortunately, three of the four identified proteins, were also found in the control sample, and they might represent background proteins. However, one protein was identified which was not present in the control lane: Paraspeckle protein 1 (PSPC1).
Table 1. Mascot search results of proteins from the Ni-NTA purification set-up.
| Protein name | Uniprot name | Mass | Score | Peptide match | In control protein band |
| Heterogeneous nuclear ribonucleoprotein L | HnRNP L | 60085 | 193 | 6 | yes |
| Paraspeckle protein 1 | PSPc1 | 58736 | 138 | 4 | no |
| DEAD (Ala-Glu-Ala-Asp) box polypeptide 5 | Ddx5 | 69237 | 108 | 2 | yes |
| Splicing factor 1 | Sf1 | 63100 | 90 | 3 | yes |
Pspc1 is Expressed in the mdDA Neuronal Field, Overlapping with Lmx1b Expression
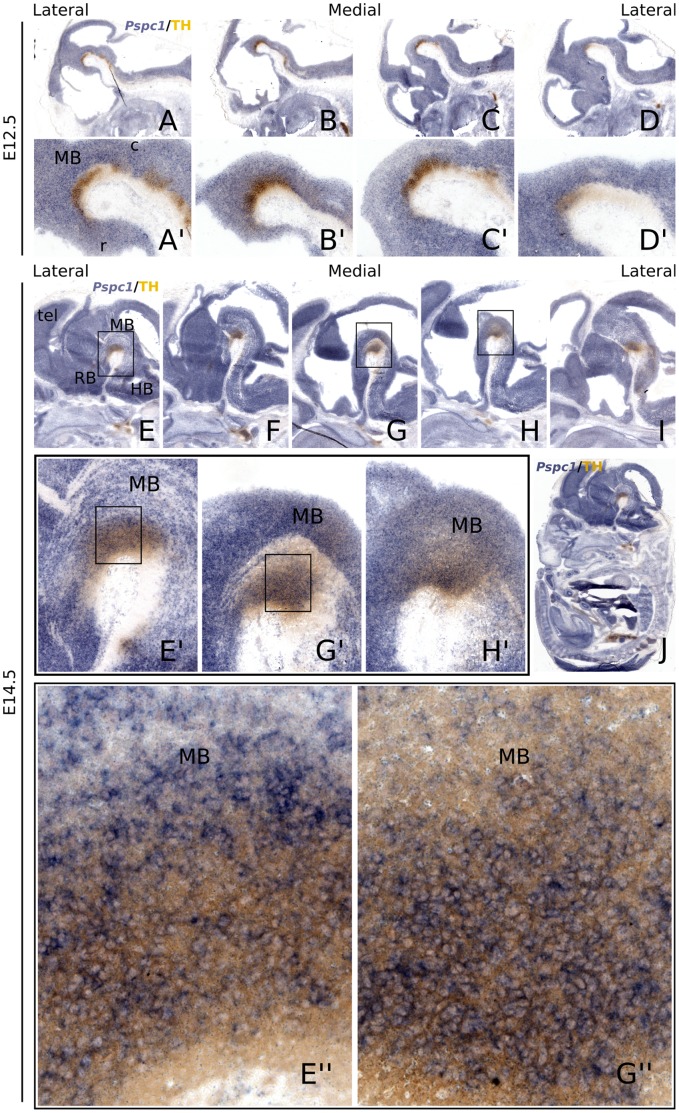
To address whether the described interaction between LMX1B-HIS and PSPC1 is relevant in mdDA neurons, we performed in situ hybridization (ISH) analysis of Pspc1 on E12.5 and E14.5 wild-type (C57Bl/6J) tissue, with TH immunohistochemistry as a reference. At E12.5, Pspc1 transcript was observed throughout the entire CNS (FIG. 2A–D), and was clearly present in the mdDA area (FIG. 2A’-D’). At E14.5, a similar expression pattern was detected; Pspc1 was broadly but specifically expressed in several embryonic areas and CNS (FIG. 2E–I, and J), thereby clearly overlapping with the TH expression domain (Fig. 2.E’–H’). When analyzing the mdDA neuronal field into more detail, Pspc1 mRNA was expressed in TH positive neurons in the mdDA neuronal field (FIG. 2E”,G”).
Figure 2. Pspc1 is expressed in the midbrain during development.
(A-D’) Sagittal analysis of the expression pattern of Pspc1 transcript (blue) in E12.5 wild-type (C57Bl/6J) mice. TH immunohistochemistry staining (brown) was taken along to mark the mdDA neuronal field. (E-I) At E14.5, Pspc1 is expressed in the mdDA area, but also in the rest of the CNS, as revealed by an overview image (J). (E’-H’) High levels of Pspc1 transcript are observed in the brain, overlapping with the TH positive domain. (E”-G”) Pspc1 is co-expressed in most TH positive neurons.
To conclude, Pspc1 transcripts were clearly present in the developing midbrain and mdDA neurons, suggesting that the found interaction may be relevant in terms of Lmx1b function in mdDA neurons.
PSPC1 is not Present in LMX1B Immunoprecipitated Complexes
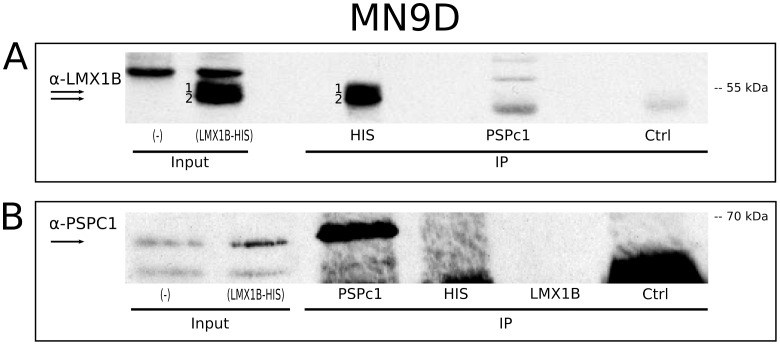
Since the outcome of the mass spectrometry analysis and the overlapping transcripts of Lmx1b and Pspc1 in the developing midbrain, suggested a potential interaction, we aimed to validate this by immunoprecipitation (IP). In order to achieve this, we transfected MN9D cells with LMX1B-HIS and control constructs and IP experiments were performed with antibodies against LMX1B, HIS, and PSPC1 (FIG. 3).
Figure 3. Pull-down of LMX1B(-HIS) and PSPC1 protein from MN9D cells.
(A) Overexpression of LMX1B-HIS reveals two specific protein bands (1,2). HIS IP shows clear protein bands (1,2). PSPC1 IP reveals three bands of which none completely overlaps with the HIS IP protein bands. (B) PSPC1 is clearly pulled down in a PSPC1 IP. No PSPC1 interaction could be detected in the HIS IP. HC, heavy chain detection; Ctrl, control IP with normal host serum; IP, immunoprecipitation; (-), MN9D cells transfected with control (empty) vector; (OE) MN9D cells transfected with LMX1B-HIS overexpression vector.
Western blot analysis with an LMX1B antibody clearly showed successful overexpression and HIS-immunoprecipitation of LMX1B-HIS protein (FIG. 3A, bands 1,2). In the PSPC1 IP, several protein bands were detected (FIG. 3A), however the observed protein of interest appeared to be located in a position that runs lower than the detected LMX1B-HIS protein bands (1 and 2). Moreover, in this experiment, the product observed in the control IP, clearly suggested partial overlap with the product in the PSPC1 IP. Furthermore, the performed PSPC1, LMX1B and HIS IP experiments were analyzed on Western blot by anti-PSPC1 antibody. We confirmed PSPC1 pull down in the PSPC1 IP, but we were not able to detect PSPC1 protein in either the HIS IP, or LMX1B IP at the expected size (FIG. 3B).
LMX1B Interacts with the Previously Identified Binding Partner of PITX3 and NURR1, PSF
The previously described interaction between PSPC1 and PSF [17]–[19], and the fact that PSF was coupled to a recently described NURR1/PITX3 transcriptional protein complex [20], made us hypothesize that LMX1B may also interact with PSF. Therefore, we aimed to investigate a possible interaction between LMX1B and PSF through co-immunoprecipitation experiments.
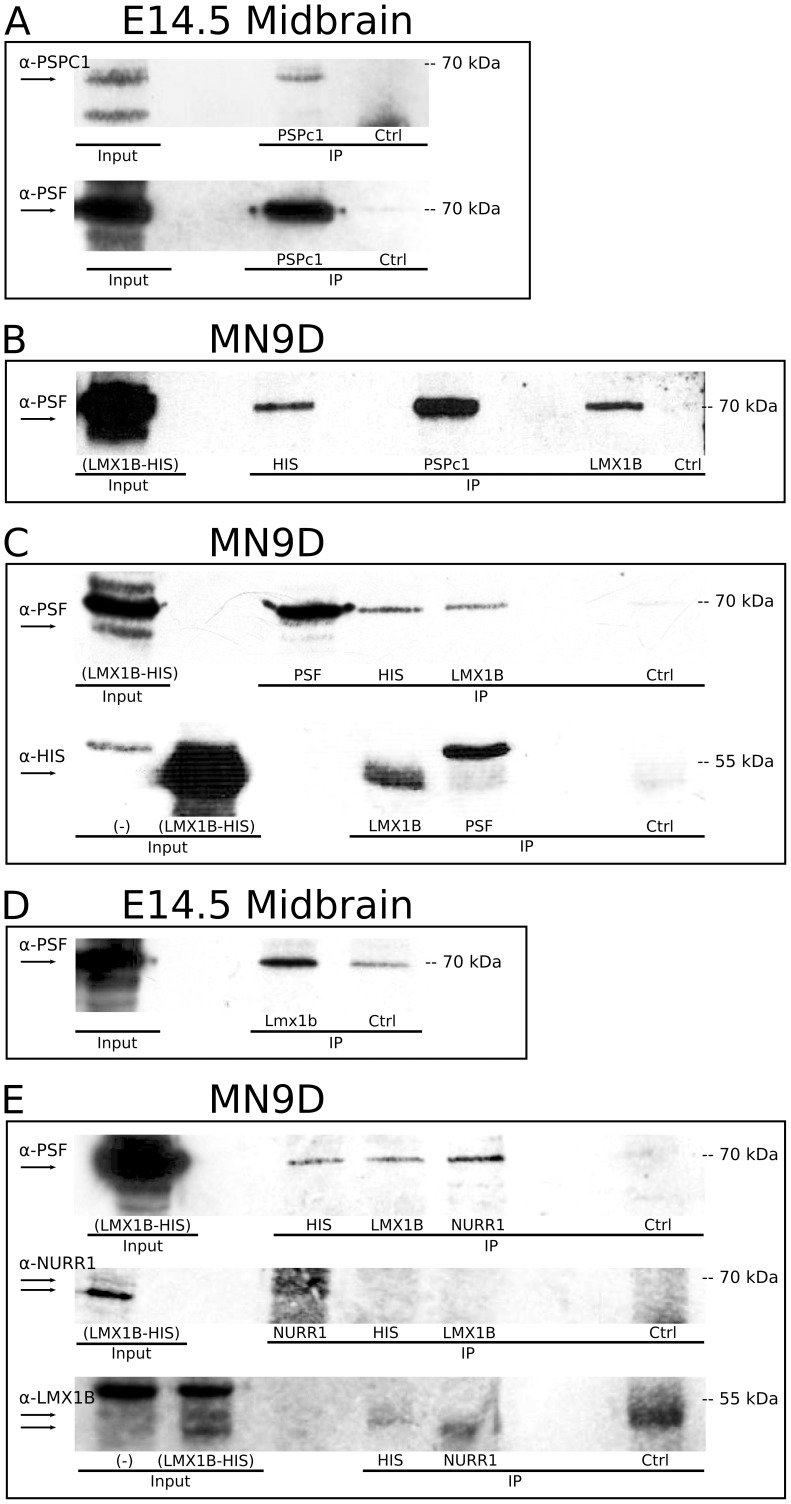
We first confirmed the described interaction between PSPC1 and PSF, in vitro and in vivo. IP experiments with anti-PSPC1 were performed on total lysate from MN9D cells, and E14.5 dissected midbrain material. Western blot analysis revealed successful pull down of endogenous PSPC1, and more importantly, we validated that PSF indeed co-immunoprecipitated with PSPC1, suggesting a clear physical interaction under these conditions (FIG. 4A). Similar IP experiments on MN9D cell lysates confirmed the PSF/PSPC1 interaction (FIG. 4B).
Figure 4. Possible interaction between PSPC1 and PSF, and between PSF and LMX1B.
(A) PSPC1 is detected in E14.5 midbrain neurons, and has an interaction with PSF in these cells. (B) An interaction with PSF is shown in HIS IP, PSPC1 IP and LMX1B IP, in MN9D cells overexpressing LMX1B-HIS. (C ) Again, PSF interacts with LMX1B, as shown in a HIS and LMX1B IP. LMX1B IP reveals clear pull-down of LMX1B-HIS, which also observed in the PSF IP, and in lower amount in the control IP. (D) PSF interacts with LMX1B in developing midbrain neurons. (E) Confirmation of the interaction of PSF with LMX1B(-HIS), and with NURR1. NURR1 IP reveals pull down of the protein, however this is not shown in HIS or LMX1B IP. Moreover, in a NURR1 IP, no LMX1B could be observed. Ctrl, control IP with normal host serum; IP, immunoprecipitation; (-), MN9D cells transfected with control(empty) vector; (LMX1B-HIS) MN9D cells transfected with LMX1B-HIS overexpression vector.
As we have demonstrated that PSF interacts with PSPC1, we additionally aimed to test the possible interaction of LMX1B with PSF. Importantly, HIS and LMX1B IP experiments on MN9D cell lysates showed a clear interaction with PSF (FIG. 4B,C). In addition, when performing an IP against PSF, LMX1B-HIS protein could be detected (FIG. 4C). To determine whether the interaction of LMX1B with PSF also exists in vivo, IP was performed on E14.5 dissected midbrain material (FIG. 4D). In agreement with the observed interaction in vitro, we could clearly detect PSF protein after pull-down of LMX1B (FIG. 4D), thereby confirming that this interaction also exists in the developing midbrain.
As mentioned above, in a previous study, a Nurr1 transcriptional complex was identified in which PITX3 and NURR1 showed an interaction with PSF [20]. Since we found interactions between LMX1B and PSF, in cells and in E14.5 midbrain tissue, we hypothesized that LMX1B might be involved in this transcriptional complex as well. We therefore analyzed a possible direct interaction between LMX1B and NURR1. IP was performed on LMX1B-HIS OE MN9D cell lysate, with pull-down of NURR1, HIS and LMX1B (FIG. 4E). We were able to confirm the binding of PSF to all three immunoprecipitated proteins. However, the direct interaction with NURR1 was not detected in HIS or LMX1B IP experiments. After pull-down of HIS-tagged protein, two LMX1B-HIS products were observed, however in the NURR1 IP, these bands were absent (FIG. 4E). Altogether, despite the identified interactions of LMX1B with PSF, and PSF with NURR1, we were not able to confirm direct binding of LMX1B with NURR1. It is still possible that LMX1B is involved in the Nurr1 transcriptional complex, but likely via indirect interaction through PSF.
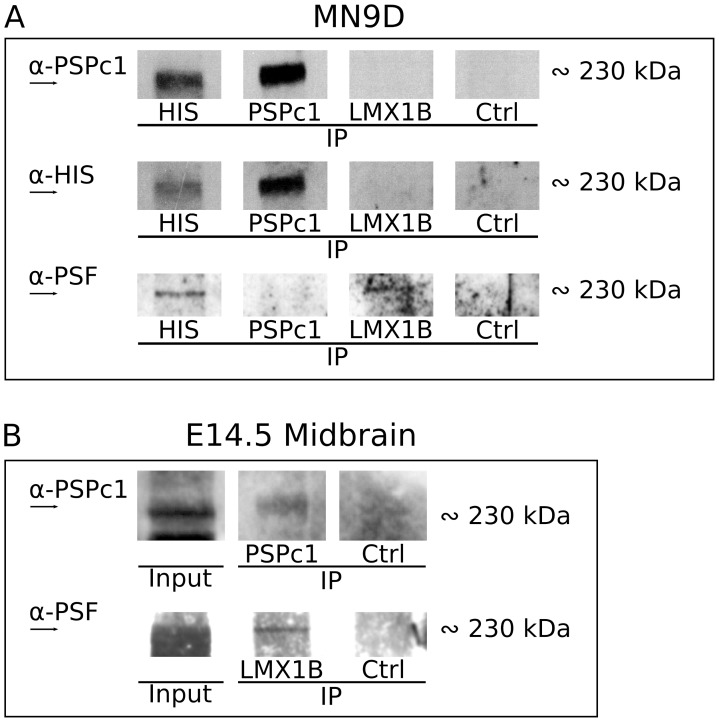
Identification of a 230 kDa Protein Complex Containing PSPC1, PSF and LMX1B
In contrast to the absence of PSPC1 protein in LMX1B-(HIS) IP experiments, at the expected size, a clear protein band was observed at approximately 230 kDa (FIG. 5A). At this position, a protein band was observed after Western analysis with a PSPC1 antibody and an LMX1B antibody in HIS IP experiments and PSPC1 IP experiments, respectively (FIG. 5A). This suggests that the hypothesized interaction between PSPC1 and LMX1B-(HIS) might exist in a larger complex, that is detected at 230 kDa. Surprisingly, at the 230 kDa position, also PSF was detected in a HIS IP, and in an LMX1B IP. However, we were not able to detect this PSF protein at 230 kDa in a PSPC1 IP (FIG. 5A). The presence of these proteins at 230 kDa, make it tempting to hypothesize that one or more protein complexes exists, containing PSPC1 (70–65 kDa), PSF (100, 95 and 75 kDA) and LMX1B-(HIS) (55–50 kDa), and that this complex is strong enough to remain intact in Western analysis. Since this result might suggest an alternative complex of LMX1B, PSPC1 and PSF we aimed to validate this complex in vivo. E14.5 dissected midbrains were used for IP of PSPC1 and LMX1B. Western analysis demonstrated a specific PSPC1 positive signal at 230 kDa, indicating that we could pull-down this PSPC1-containing protein complex, in vivo (FIG. 5B). Interestingly, also the presence of PSF at 230 kDa was confirmed, in LMX1B IP experiments (FIG. 5B).
Figure 5. Identification of a 230 kDa complex containing LMX1B(-HIS), PSPC1 and PSF.
(A) PSPC1 protein is identified in a protein band of approximately 230 kDa, in a PSPC1 IP and in a HIS IP, but not in an LMX1B IP. When immunoblotting for HIS, a protein of the same size is detected in these IP experiments, and again not in an LMX1B IP. After immunoblotting for PSF, of the same blot, PSF is detected at 230 kDa in the HIS IP, faintly in the PSPC1 IP, and in the LMX1B IP. (B) Analysis of 230 kDa proteins in vivo, in E14.5 dissected midbrain tissue. PSPC1 pull-down reveals PSPC1 protein at 230 kDa, suggesting that the 230 kDa protein complex exist in vivo. At the same height, PSF is detected when immunoprecipitating for LMX1B. Ctrl, control IP with normal host serum; IP, immunoprecipitation.
To conclude, PSF and PSPC1, together with LMX1B(-HIS) were identified through IP experiments in a protein complex of 230 kDa, in vitro and in vivo.
LMX1B Interacts with TMPO and HSP70
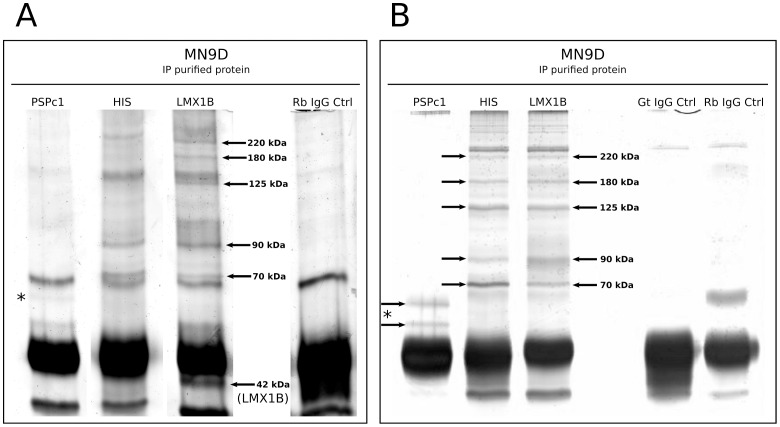
In order to further examine other possible interactions of unknown proteins with LMX1B we used a different purification set-up based on IP experiments with antibodies against PSPC1, HIS and LMX1B and analyzed this on silver-stained gels (FIG. 6). Compared to the initially used Ni-NTA purification method, the IP experiments yielded very clean results in terms of protein background. PSPC1 IP resulted in two clear protein bands that were absent in the control IP (FIG. 6, asterisks). Since they range between 60 and 65 kDa, they may represent two PSPC1 protein isoforms, that were detected before on Western blot. Most interestingly, HIS and LMX1B IP samples display protein separation patterns that were almost identical between the different IP samples. Several protein bands were clearly and specifically observed in all IP experiments and were absent in the control IP. To further analyze some of these proteins, all promising bands were excised and subjected to mass spectrometry analysis (The Proteome Factory, Berlin) (FIG. 6A,B, arrows). The Mascot results of this analysis are shown in table 2, 3 and 4.
Figure 6. LMX1B co-immunoprecipitated protein identification.
(A) IP against HIS, PSPC1 and LMX1B on LMX1B-HIS OE MN9D cell lysate, followed by separation on silver-stained SDS gel. The PSPC1 IP shows two protein bands that might represent both PSPC1 isoforms. Five protein bands of the LMX1B IP were excised and used for mass spectrometry analysis (arrows). In addition, the 42 kDa protein was taken along, as a positive control, and was later confirmed as LMX1B protein. (B) Second IP experiment against HIS, PSPC1 and LMX1B on LMX1B-HIS OE MN9D cell lysate, followed by separation on silver-stained (FireSilver kit, Proteome Factory, Berlin) SDS gel, confirming and improving the pattern as described in the first IP experiment (A). The PSPC1 IP shows two protein bands that likely represent both PSPC1 isoforms, and they were used for mass spectrometry analysis. Five differential protein bands of the LMX1B and HIS IP were excised and used mass spectrometry analysis (arrows). Gt IgG Ctrl, goat IgG control; Rb IgG Ctrl, rabbit IgG control.
Table 2. Mascot search results of proteins from LMX1B IP purified excised protein bands.
| Excised fragment | Protein name | Uniprot name | Mass | Score | Peptide match |
| 220 kDa | Keratin 15 | KRT15 | 49086 | 70 | 3 |
| Dystrophia myotonica-containing WD repeat motif | DMWD | 65572 | 23 | 1 | |
| 180 kDa | Keratin 15 | KRT15 | 49086 | 59 | 1 |
| Keratin 75 | KRT75 | 42357 | 46 | 2 | |
| 125 kDa | Myosin 1c | MYO1C | 118082 | 113 | 6 |
| 95 kDa | Lamina-associated polypeptide 2, isoforms alpha/zeta | LAP2/TMPO | 75285 | 169 | 5 |
| Proto-oncogene protein C-ros | ROS1 | 10510 | 40 | 1 | |
| FERM and PDZ domain-containing protein 1 | FRMPD1 | 169102 | 40 | 1 | |
| Cyclic AMP specific phosphodiesterase | PDE4D5A | 23890 | 33 | 1 | |
| 70 kDa | Keratin 8 | KRT8/Card2 | 54514 | 104 | 1 |
| Heat shock protein 70 | HSP70 | 70793 | 51 | 3 | |
| X-ray repair complementing defective repair in Chinese hamster cells 6 | KU70/Xrcc6 | 69442 | 26 | 3 | |
| 42 kDa | LIM homeobox transcription factor 1 beta | LMX1B | 41541 | 81 | 5 |
| Actin related protein 3 | ACTR3 | 47327 | 67 | 3 | |
| Histocompatibility 2, class II antigen E beta 2 | H2-EB2 | 29003 | 19 | 1 | |
| Integrin beta-4 | ITGB4 | 101086 | 19 | 1 |
Table 3. Mascot search results of proteins from HIS IP purified excised protein bands.
| Excised fragment | Protein name | Uniprot name | Mass | Score | Peptide match |
| 220 kDa | Analysis failed | – | – | – | – |
| 180 kDa | Titin isoform N2-A | TTN | 3713700 | 60 | 26 |
| Glucocorticoid receptor DNA-binding factor 1 | GRLF1 | 170285 | 52 | 5 | |
| mCG142711 | mCG142711 | 76465 | 30 | 7 | |
| 125 kDa | Titin isoform N2-A | TTN | 3713700 | 41 | 18 |
| Msx2-interactin protein | SPEN | 398292 | 32 | 4 | |
| 90 kDa | Titin isoform N2-A | TTN | 3713700 | 35 | 17 |
| Cardiomyopathy-associated protein 5 | CMYA5 | 412787 | 32 | 7 | |
| 70 kDa | Axonemal dynein heavy chain | DNAHC | 474513 | 31 | 7 |
| 65 kDa | Titin | TTN | 3764005 | 38 | 18 |
| Coiled-coil domain-containing 15 | CCDC15 | 94848 | 31 | 2 | |
| 60 kDa | Leucine-rich repeat-containing protein 16b | LRRC16B | 150322 | 37 | 2 |
| Family with sequence similarity 186, member A | FAM186A | 194430 | 31 | 3 |
Table 4. Mascot search results of proteins from LMX1B-2 IP purified excised protein bands.
| Excised fragment | Protein name | Uniprot name | Mass | Score | Peptide match |
| 220 kDa | Analysis failed | – | – | – | – |
| 180 kDa | DEAH (Asp-Glu-Ala-His) box polypeptide 9 | DHX9/DDX9 | 131636 | 70 | 5 |
| 125 kDa | Keratin 8 | KRT8 | 54514 | 38 | 1 |
| 90 kDa | Lamina-associated polypeptide 2 isoform alpha | LAP2/TMPO | 75122 | 334 | 13 |
| 70 kDa | Heat shock protein 70 | HSP70 | 70793 | 175 | 4 |
As mentioned above, we hypothesized that the 60 and 65 kDa bands might represent PSPC1 protein. Yet, mass spectrometry analysis failed to confirm this. The proteins identified in these bands yielded very low scores, each with a value of approximately 30 and only two peptide queries matched per sample (data not shown). Therefore the identified proteins likely were background proteins or false hits.
Of the HIS IP experiment, five protein bands were analyzed (FIG. 6B,arrows and Table 3). Unfortunately, the 220 kDa protein analyzes failed, and furthermore, several other protein bands resulted in the identification of Titin protein (Table 3). This might be a contamination of the sample. Nonetheless, the unique peptide numbers matched, were extremely high, and from literature, it is known that Titin, which is the largest protein currently known, has at least one polyhistidine stretch in one of its domains [21], [22]. Likely, the IP with anti-HIS antibody resulted in the pull-down of (part of) this Titin protein, due to high affinity for the poly-histidine stretch. Therefore, in this study, we only focused on the other proteins identified. One protein displayed both a relatively high protein score (52), and high number of peptide queries matched (5): Glucocorticoid receptor DNA-binding factor 1 (GRLF1) (Table 3; 180 kDa protein band). Based on the number of unique peptide matches, mCG142711, Msx2-interacting protein (SPEN), cardiomyopathy-associated protein 5 (CMYA5) and axonemal dynein heavy chain (DNAHC), are interesting as well. Notably, these two identified proteins do not match in theoretical size compared to the position in the silver stained gel. However, the Mascot-data indicated that these proteins are significant hits.
Importantly, of the two separate LMX1B IP experiments, five protein bands were analyzed of each IP. Additionally, as a control, we also sent a 42 kDa protein band, suspected to be LMX1B, for analysis, and this was indeed confirmed with 5 peptide queries matched and a relatively high score of 81 (FIG. 6A and Table 2, 42 kDa band). The two separate LMX1B IP experiments (FIG. 6A,B, arrows) resulted in the mass spectrometry identification of several highly interesting proteins. Based on the combination of 6 unique peptide-queries matched and a relatively high score of 113, Myosin 1c (MYO1C) can be considered as a promising potential binding partner of LMX1B (Table 2; 125 kDa sample). The second LMX1B IP resulted in the identification of DEAH (Asp-Glu-Ala-His) box polypeptide 9 (DHX9/DDX9). The Mascot analysis revealed a number of 5 peptide-queries matched and a high score. However, the true size of this protein is 132 kDa whilst it was identified in the 180 kDa sample (Table 4). Intriguingly, when comparing the two LMX1B IP experiments, two protein hits were found in both (Table 2 and 4). Heat shock protein 70 (HSP70) was identified in the two separate 70 kDa protein bands, with peptide-queries matched of respectively 3 and 4, which is lower than the threshold of 5 that we were applying for the selection of the other proteins. However, the individual scores are high and the fact that this protein is identified in two different samples makes it very likely that this protein is a potential interactor of LMX1B. Finally, the most interesting protein that was identified, in both 95 kDa samples, is Lamina-associated polypeptide 2 (LAP2/TMPO). In both IP experiments, it displayed the highest scores among all significant proteins identified, and moreover, in the second LMX1B IP, 13 unique peptide-queries were matching with TMPO.
Taken together, several proteins were identified with a Mascot score above a cut-off of 50, and with a peptide-queries match of preferably 5 or higher: GRLF1, DHX9 and MYO1C. The two novel proteins HSP70 and TMPO were identified in two separate experiments and may therefore represent the most reliable results.
Discussion
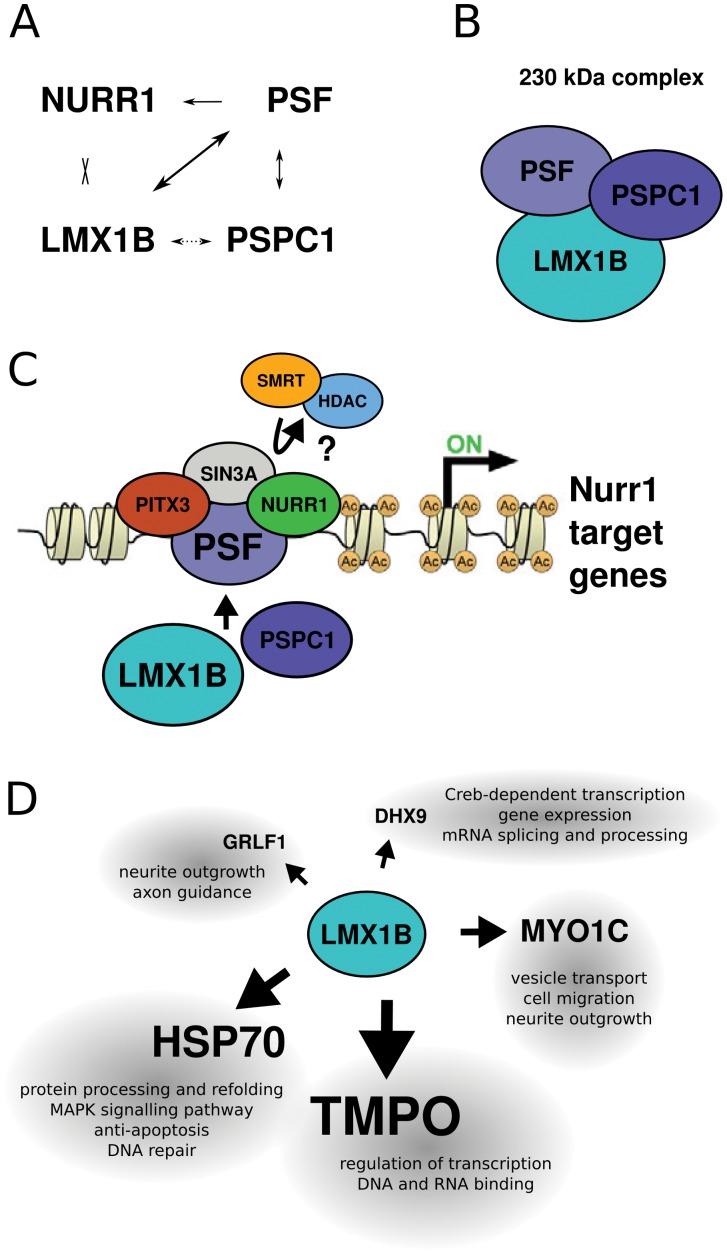
To acquire a better understanding of the functional role of Lmx1b in mdDA development, we aimed to identify proteins that have a physical interaction with LMX1B, and we initiated this with an open screen for direct interactors. HIS-tagged affinity purification of LMX1B protein, followed by mass spectrometry resulted in the identification of Paraspeckle protein PSPC1. However, by means of IP, we initially could not confirm an interaction between LMX1B and PSPC1. Importantly, a 230 kDa protein complex was identified in several IP experiments, that contained PSPC1, PSF and LMX1B (FIG. 7B). So, despite the lack of a confirmed single interaction between LMX1B and PSPC1, we were able to confirm an interaction between these proteins in a larger complex, also containing PSF, a known interactor of PSPC1 and confirmed here. PSPC1 belongs to the family of DBHS (Drosophila behavior and human splicing) proteins, that have several conserved domains in common, of which one is the NONO/Paraspeckle (NOPS) domain [23]. Within this highly conserved DBHS domain, the three DBHS proteins PSPC1, NONO and PSF share 70% sequence identity, and the domain is essential for homodimerization and heterodimerization of the three proteins with each other [17], [19]. Recently, PSF was identified as a crucial co-factor in a Nurr1 transcriptional complex, during mdDA development [20]. The finding of PSPC1 in a 230 kDa complex with LMX1B and PSF made us hypothesize that, to perform transcriptional functions in mdDA development, LMX1B might be part of this Nurr1 transcriptional complex, via PSPC1 and PSF (FIG. 7C). Importantly, in this study we validated that LMX1B and PSF have a physical interaction, in the identified larger protein complex, but also at the single protein level. To further address a possible interaction of LMX1B in this Nurr1 transcriptional complex, we investigated a potential, direct interaction between LMX1B and NURR1, but were unable to confirm this (FIG. 7A). However, it is still possible that LMX1B is involved in the transcriptional complex, but not through direct interaction with NURR1.
Figure 7. Schematic representation of suggested protein interactions with LMX1B.
(A) Interactions were identified by means of immunoprecipitation experiments. Direct interactions between NURR1 and PSF, between PSF and LMX1B and between PSF and PSPC1 were found. (LMX1B)-HIS interaction with PSPC1 was identified in a 230 kDa complex only. (B) Western analysis revealed interactions between PSF, PSPC1 and LMX1B at 230 kDa, suggesting a complex of this size containing all three proteins. (C) The Nurr1 transcriptional complex (adapted from [20]. We hypothesize that LMX1B might have a function in this complex as well, by physically binding to PSF, maybe via PSPC1. (D) Novel direct interactors of LMX1B identified in a screen in which IP experiments were analyzed on silver-stained protein gels and by mass-spectrometry analysis.
Novel Potential LMX1B Interacting Proteins
In addition to the screen via affinity purification, a second set-up was used to find novel interacting proteins of LMX1B (FIG. 7D). For this, IP purification of LMX1B, HIS and PSPC1 protein was performed, followed by SDS-PAGE, silver-staining and mass spectrometry analysis. The clear confirmation of LMX1B, which was taken along as a positive control, indicates that the set-up is valid for purifying proteins and identifying them by means of mass spectrometry.
A 125 kDa protein band resulted in the identification of Myosin 1c protein (MYO1C), that has a known size 118 kDa. Mass spectrometry analysis showed that 6 different peptide-queries all matched with MYO1C, and also the Mascot score was relatively high, strongly indicating that this identified protein is a true hit. The Myosin protein 1 family consist of actin-based molecular motors, which have various functions in vesicle transport, and docking to the plasma membrane [24], [25]. The protein is expressed ubiquitously [26], and expression in the brain is mainly found in neurons [25]. Based on online expression databases, Myo1c mRNA is found throughout the brain with enriched expressions in the dorsal midbrain, in the developing midbrain oculomotor complex region. A more general function of Myo1c is cargo delivery and membrane trafficking, and the protein has been linked to cell migration and neurite outgrowth [25], [26], making it a promising protein for further investigation of its role in binding to LMX1B, and a possible function in mdDA development.
In addition, we identified Glucocorticoid receptor DNA-binding factor 1 (GRLF1; 170 kDa), and DEAH (Asp-Glu-Ala-His) box polypeptide 9 (DHX9/DDX9/RHA; 132 kDA) which is smaller than the expected 180 kDa based on the observed separation on gel (silver stain). DHX9, also known as RNA helicase A (RHA), has a role in Creb-dependent transcription, but has additionally been implicated in nuclear export of unspliced viral RNA’s [27]. DEAH helicase domains, like homeobox domains, are involved in ATP-DEPENDENT chromatin remodeling or bind DNA and post-translationally modified nucleosomes, in either way influencing gene expression [28]. GRLF1 (p190RhoGAP) functions in neurite outgrowth [29] and mice lacking GRLF1 show defects in axon guidance and fasciculation [30]. Furthermore, GRLF1 interacts with plexins, and is essential in semaphorin signaling to the actin cytoskeleton [31]. Both genes show high mRNA levels in the developing brain and are clearly expressed in the developing midbrain, and in the adult mdDA system (online expression databases: Genepaint.org and the Allen Brain Atlas).
Finally, two proteins were identified in two different IP experiments simultaneously. Heat-shock protein 70 (HSP70/HSPA1B), was identified from 70 kDa protein bands. The fact that this protein is identified twice in separate experiments, enhances the possibility that this represents a true Lmx1b interacting protein. It is a chaperone protein that is highly expressed in response to cell stress [32]. The protein has been implicated in brain development and prevents protein misfolding and aggregation associated with neurodegenerative diseases like Alzheimer’s disease and Parkinson’s disease [32]. That study also showed that administration of this protein to mdDA neurons can rescue the cells from apoptosis.
The other potential interactor, Lamina-associated polypeptide 2 (LAP2/TMPO), was identified in the 90 kDa protein bands of the two separate LMX1B IP experiments. In both mass spectrometry analyzes, a high peptide-queries match and score was found. Lap2/Tmpo encodes seven mouse thymopoietin (TMPO) mRNA transcripts, that encode ubiquitously expressed nuclear proteins [33]. Not much is known about the exact function of the protein and no literature about any role in neuronal development was found. The gene is expressed in the developing brain, but in low levels. However, it is expressed in higher levels in the ventricular zone, partially overlapping with the most caudal dorsal mdDA neuronal field (online expression databases).
The double-independent identification of the above described binding partners of LMX1B, under the described conditions, suggest that these proteins are able to interact with LMX1B and modify its functional aspects. The exact role of these proteins in Lmx1b function awaits analysis through in vivo manipulation, determining which part of the Lmx1b phenotype may be depending on a specific LMX1B protein-protein interaction event.
Funding Statement
This work was supported by a VICI-grant (no. 865.09.002 to M.P. Smidt) and by a TI Pharma grant (no. T5-207, The Netherlands) The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Smidt MP, Asbreuk CH, Cox JJ, Chen H, Johnson RL, et al. (2000) A second independent pathway for development of mesencephalic dopaminergic neurons requires Lmx1b. Nat Neurosci 3: 337–341 doi:10.1038/73902 [DOI] [PubMed] [Google Scholar]
- 2. Burbach JPH, Smits S, Smidt MP (2003) Transcription factors in the development of midbrain dopamine neurons. Ann N Y Acad Sci 991: 61–68. [DOI] [PubMed] [Google Scholar]
- 3. Smidt MP, Smits SM, Burbach JPH (2003) Molecular mechanisms underlying midbrain dopamine neuron development and function. Eur J Pharmacol 480: 75–88. [DOI] [PubMed] [Google Scholar]
- 4. Guo C, Qiu H-Y, Huang Y, Chen H, Yang R-Q, et al. (2007) Lmx1b is essential for Fgf8 and Wnt1 expression in the isthmic organizer during tectum and cerebellum development in mice. Development 134: 317–325 doi:10.1242/dev.02745 [DOI] [PubMed] [Google Scholar]
- 5. Adams KA, Maida JM, Golden JA, Riddle RD (2000) The transcription factor Lmx1b maintains Wnt1 expression within the isthmic organizer. Development 127: 1857–1867. [DOI] [PubMed] [Google Scholar]
- 6. Yan CH, Levesque M, Claxton S, Johnson RL, Ang S-L (2011) Lmx1a and lmx1b function cooperatively to regulate proliferation, specification, and differentiation of midbrain dopaminergic progenitors. J Neurosci 31: 12413–12425 doi:10.1523/JNEUROSCI.1077-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lin W, Metzakopian E, Mavromatakis YE, Gao N, Balaskas N, et al. (2009) Foxa1 and Foxa2 function both upstream of and cooperatively with Lmx1a and Lmx1b in a feedforward loop promoting mesodiencephalic dopaminergic neuron development. Dev Biol 333: 386–396 doi:10.1016/j.ydbio.2009.07.006 [DOI] [PubMed] [Google Scholar]
- 8. Nakatani T, Kumai M, Mizuhara E, Minaki Y, Ono Y (2010) Lmx1a and Lmx1b cooperate with Foxa2 to coordinate the specification of dopaminergic neurons and control of floor plate cell differentiation in the developing mesencephalon. Dev Biol 339: 101–113 doi:10.1016/j.ydbio.2009.12.017 [DOI] [PubMed] [Google Scholar]
- 9. Metzakopian E, Lin W, Salmon-Divon M, Dvinge H, Andersson E, et al. (2012) Genome-wide characterization of Foxa2 targets reveals upregulation of floor plate genes and repression of ventrolateral genes in midbrain dopaminergic progenitors. Development 139: 2625–2634 doi:10.1242/dev.081034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Marini M, Bongers EMHF, Cusano R, Di Duca M, Seri M, et al. (2003) Confirmation of CLIM2/LMX1B interaction by yeast two-hybrid screening and analysis of its involvement in nail-patella syndrome. Int J Mol Med 12: 79–82. [PubMed] [Google Scholar]
- 11. Marini M, Giacopelli F, Seri M, Ravazzolo R (2005) Interaction of the LMX1B and PAX2 gene products suggests possible molecular basis of differential phenotypes in Nail-Patella syndrome. Eur J Hum Genet 13: 789–792 doi:10.1038/sj.ejhg.5201405 [DOI] [PubMed] [Google Scholar]
- 12. Tzchori I, Day TF, Carolan PJ, Zhao Y, Wassif CA, et al. (2009) LIM homeobox transcription factors integrate signaling events that control three-dimensional limb patterning and growth. Development 136: 1375–1385 doi:10.1242/dev.026476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhao Y, Kwan K-M, Mailloux CM, Lee W-K, Grinberg A, et al. (2007) LIM-homeodomain proteins Lhx1 and Lhx5, and their cofactor Ldb1, control Purkinje cell differentiation in the developing cerebellum. Proc Natl Acad Sci USA 104: 13182–13186 doi:10.1073/pnas.0705464104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Agulnick AD, Taira M, Breen JJ, Tanaka T, Dawid IB, et al. (1996) Interactions of the LIM-domain-binding factor Ldb1 with LIM homeodomain proteins. Nature 384: 270–272 doi:10.1038/384270a0 [DOI] [PubMed] [Google Scholar]
- 15. Choi HK, Won LA, Kontur PJ, Hammond DN, Fox AP, et al. (1991) Immortalization of embryonic mesencephalic dopaminergic neurons by somatic cell fusion. Brain Res 552: 67–76. [DOI] [PubMed] [Google Scholar]
- 16. Hermanson E, Joseph B, Castro D, Lindqvist E, Aarnisalo P, et al. (2003) Nurr1 regulates dopamine synthesis and storage in MN9D dopamine cells. Exp Cell Res 288: 324–334. [DOI] [PubMed] [Google Scholar]
- 17. Fox AH, Bond CS, Lamond AI (2005) P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol Biol Cell 16: 5304–5315 doi:10.1091/mbc.E05-06-0587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kuwahara S, Ikei A, Taguchi Y, Tabuchi Y, Fujimoto N, et al. (2006) PSPC1, NONO, and SFPQ are expressed in mouse Sertoli cells and may function as coregulators of androgen receptor-mediated transcription. Biol Reprod 75: 352–359 doi:10.1095/biolreprod.106.051136 [DOI] [PubMed] [Google Scholar]
- 19. Myojin R, Kuwahara S, Yasaki T, Matsunaga T, Sakurai T, et al. (2004) Expression and functional significance of mouse paraspeckle protein 1 on spermatogenesis. Biol Reprod 71: 926–932 doi:10.1095/biolreprod.104.028159 [DOI] [PubMed] [Google Scholar]
- 20. Jacobs FMJ, van Erp S, van der Linden AJA, von Oerthel L, Burbach JPH, et al. (2009) Pitx3 potentiates Nurr1 in dopamine neuron terminal differentiation through release of SMRT-mediated repression. Development 136: 531–540 doi:10.1242/dev.029769 [DOI] [PubMed] [Google Scholar]
- 21. Zou P, Gautel M, Geerlof A, Wilmanns M, Koch MHJ, et al. (2003) Solution scattering suggests cross-linking function of telethonin in the complex with titin. J Biol Chem 278: 2636–2644 doi:10.1074/jbc.M210217200 [DOI] [PubMed] [Google Scholar]
- 22. Labeit S, Kolmerer B (1995) Titins: Giant Proteins in Charge of Muscle Ultrastructure and Elasticity. Science 270: 293–296 doi:10.1126/science.270.5234.293 [DOI] [PubMed] [Google Scholar]
- 23. Bond CS, Fox AH (2009) Paraspeckles: nuclear bodies built on long noncoding RNA. J Cell Biol 186: 637–644 doi:10.1083/jcb.200906113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bose A, Robida S, Furcinitti PS, Chawla A, Fogarty K, et al. (2004) Unconventional myosin Myo1c promotes membrane fusion in a regulated exocytic pathway. Mol Cell Biol 24: 5447–5458 doi:10.1128/MCB.24.12.5447-5458.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sherr EH, Joyce MP, Greene LA (1993) Mammalian myosin I alpha, I beta, and I gamma: new widely expressed genes of the myosin I family. J Cell Biol 120: 1405–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brandstaetter H, Kendrick-Jones J, Buss F (2012) Myo1c regulates lipid raft recycling to control cell spreading, migration and Salmonella invasion. Journal of Cell Science. Available:http://www.ncbi.nlm.nih.gov/pubmed/22328521. Accessed 2012 Mar 7. [DOI] [PMC free article] [PubMed]
- 27. Li J, Tang H, Mullen T-M, Westberg C, Reddy TR, et al. (1999) A role for RNA helicase A in post-transcriptional regulation of HIV type 1. PNAS 96: 709–714 doi:10.1073/pnas.96.2.709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pardo M, Lang B, Yu L, Prosser H, Bradley A, et al. (2010) An expanded Oct4 interaction network: implications for stem cell biology, development, and disease. Cell Stem Cell 6: 382–395 doi:10.1016/j.stem.2010.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jeon C-Y, Kim H-J, Morii H, Mori N, Settleman J, et al. (2010) Neurite outgrowth from PC12 cells by basic fibroblast growth factor (bFGF) is mediated by RhoA inactivation through p190RhoGAP and ARAP3. J Cell Physiol 224: 786–794 doi:10.1002/jcp.22184 [DOI] [PubMed] [Google Scholar]
- 30. Brouns MR, Matheson SF, Settleman J (2001) p190 RhoGAP is the principal Src substrate in brain and regulates axon outgrowth, guidance and fasciculation. Nat Cell Biol 3: 361–367 doi:10.1038/35070042 [DOI] [PubMed] [Google Scholar]
- 31. Barberis D, Casazza A, Sordella R, Corso S, Artigiani S, et al. (2005) p190 Rho-GTPase activating protein associates with plexins and it is required for semaphorin signalling. J Cell Sci 118: 4689–4700 doi:10.1242/jcs.02590 [DOI] [PubMed] [Google Scholar]
- 32. Nagel F, Falkenburger BH, Tönges L, Kowsky S, Pöppelmeyer C, et al. (2008) Tat-Hsp70 protects dopaminergic neurons in midbrain cultures and in the substantia nigra in models of Parkinson’s disease. J Neurochem 105: 853–864 doi:10.1111/j.1471-4159.2007.05204.x [DOI] [PubMed] [Google Scholar]
- 33. Berger R, Theodor L, Shoham J, Gokkel E, Brok-Simoni F, et al. (1996) The characterization and localization of the mouse thymopoietin/lamina-associated polypeptide 2 gene and its alternatively spliced products. Genome Res 6: 361–370. [DOI] [PubMed] [Google Scholar]