Abstract
The recognition that AIDS originated as a zoonosis heightens public health concerns associated with human infection by simian retroviruses endemic in nonhuman primates (NHPs). These retroviruses include simian immunodeficiency virus (SIV), simian T-cell lymphotropic virus (STLV), simian type D retrovirus (SRV), and simian foamy virus (SFV). Although occasional infection with SIV, SRV, or SFV in persons occupationally exposed to NHPs has been reported, the characteristics and significance of these zoonotic infections are not fully defined. Surveillance for simian retroviruses at three research centers and two zoos identified no SIV, SRV, or STLV infection in 187 participants. However, 10 of 187 persons (5.3%) tested positive for SFV antibodies by Western blot (WB) analysis. Eight of the 10 were males, and 3 of the 10 worked at zoos. SFV integrase gene (int) and gag sequences were PCR amplified from the peripheral blood lymphocytes available from 9 of the 10 persons. Phylogenetic analysis showed SFV infection originating from chimpanzees (n = 8) and baboons (n = 1). SFV seropositivity for periods of 8 to 26 years (median, 22 years) was documented for six workers for whom archived serum samples were available, demonstrating long-standing SFV infection. All 10 persons reported general good health, and secondary transmission of SFV was not observed in three wives available for WB and PCR testing. Additional phylogenetic analysis of int and gag sequences provided the first direct evidence identifying the source chimpanzees of the SFV infection in two workers. This study documents more frequent infection with SFV than with other simian retroviruses in persons working with NHPs and provides important information on the natural history and species origin of these infections. Our data highlight the importance of studies to better define the public health implications of zoonotic SFV infections.
Retroviral zoonoses have received heightened public health attention because the origin of the human immunodeficiency virus types 1 and 2 (HIV-1 and -2) has been linked to cross-species transmission of simian immunodeficiency viruses (SIVs) from chimpanzees (Pan troglodytes) and sooty mangabeys (Cercocebus atys), respectively (10, 12, 32). SIV seroprevalence rates in naturally infected primates can reach 36%. Thus, human exposures to SIVs by hunting, butchering, or keeping infected primates as pets have been proposed as possible routes for human infections (12, 26).
Nonhuman primates (NHPs) are also natural hosts to several other exogenous retroviruses, including simian T-cell lymphotropic virus (STLV), simian type D retrovirus (SRV), and simian foamy virus (SFV) (8, 18, 20, 21). STLV type 1 (STLV-1) is found in at least 20 different Old World primate species, with seroprevalences ranging from 4 to 44% (8, 33). STLV-1 can be pathogenic and has been reported to be the cause of lymphomas in baboons (8, 37). Phylogenetic analysis suggests that multiple interspecies transmissions of STLV-1 to humans may have occurred. SRV infection can be highly prevalent (>90%) in Asian macaques and may result in an AIDS-like illness in this host (18).
While infections with SIV, STLV, or SRV can be restricted to particular geographic areas or to specific primate hosts, SFV infections are widespread among NHPs. Most primate species investigated thus far, including prosimians, New World monkeys, and Old World monkeys and apes, harbor SFV (15, 21). In captivity, more than 70% of adult NHPs are infected with SFV (15, 21), possibly reflecting the ease of transmissibility of this virus among NHPs in close contact. Phylogenetic analysis indicates species-specific viral lineages, suggesting a long-standing coexistence and coevolution between SFVs and their NHP host species (5, 13, 15, 29). However, evidence supporting the existence of a human-specific foamy virus (FV) is not available. A prototype FV isolated from a Kenyan patient in 1971, named the human FV (HFV), is phylogenetically a chimpanzee-like SFV (1, 14). The failure to identify HFV infection in several human populations has raised questions about the true origin of HFV and whether humans have a species-specific FV (2, 31).
NHPs are commonly used in biomedical research and are typical members of zoo collections around the world. Therefore, persons who work directly with these primates may be exposed to simian retroviruses. The identification of isolated infections with either SIV or SFV in occupationally exposed workers has suggested that occupational contact with NHPs may be associated with risks for transmission of simian retroviruses (16, 30, 34).
To assess the prevalence of zoonotic retrovirus infection of humans, we initiated a linked seroprevalence study of exposed workers in institutions in North America. Previous testing of 231 persons identified 4 (1.7%) infected with SFV of baboon (SFVBAB; three cases) or African green monkey (SFVAGM; one case) origin (13). In addition, evidence suggesting SRV infection was seen in two workers, with persistent seropositivity documented in one case (19). A subsequent anonymous serosurvey of zoo workers identified four (3%) SFV-seropositive persons among 133 workers whose jobs involved potential contact with NHPs (28). SFV screening of 46 exposed Canadian workers also identified two seropositive workers (4.3%), including one with a macaque-type SFV infection (4). While previous reports have helped identify a human population that is at risk for SFV and other zoonotic retroviral infections, many questions regarding the epidemiology and the natural history of these zoonotic infections remain unanswered. To date, published findings from different studies of 11 SFV-infected humans suggest asymptomatic infections; however, the limited number of cases, the short duration of follow-up, and the selection biases inherent in the enrollment of healthy workers to identify cases all limit the ability to identify potential disease associations (4, 13, 28, 30). Data are also not available to comparatively assess different SFV lineages for their relative infectivity, transmissibility, or pathogenic potential in humans. Therefore, additional studies are needed to better understand the natural history of SFV infections in humans and to assess the public health implications of these infections.
We report here new data from our ongoing surveillance of simian retroviruses among persons occupationally exposed to NHPs. We have identified 10 additional SFV-infected workers and characterized the primate species origin of these infections. In two cases, we provide the first evidence identifying the most likely source animal of the infection. This study highlights the relatively frequent cross-species transmission of simian retroviruses to humans.
MATERIALS AND METHODS
Study design.
Research institutions and zoological gardens in North America were invited to participate in our Institutional Review Board-approved, voluntary study of the seroprevalence of simian retrovirus infections among persons exposed to NHPs and their body fluids.
After consenting to join the study, participants completed a study questionnaire designed to determine their histories of work with and exposure to NHPs; participants also provided a blood specimen from which serum was obtained for serologic screening for SIV, STLV, SRV, and SFV. Persons with seropositive test results were interviewed regarding details of their exposure history and asked about their current general health status. Seropositive persons were also asked to provide fresh EDTA-treated blood specimens to obtain peripheral blood lymphocytes (PBLs) for PCR and virus isolation and to provide any available archived serum samples to determine the duration of seropositivity. Serum samples were stored at −20°C until used.
Animal specimens and determination of chimpanzee subspecies.
Fresh EDTA or sodium citrate-treated whole-blood specimens were obtained on an opportunistic basis from captive chimpanzees and a gorilla, in accordance with the animal care and use committees at each institution. PBLs were obtained by Ficoll-Hypaque centrifugation and DNA lysates were prepared as described previously (35). To avoid contamination, human and primate samples were processed separately and tested in laboratories in different buildings. To determine the subspecies of each chimpanzee, partial sequences of the hypervariable region 1 of the mitochondrial DNA (mtDNA) control region were amplified by using primers reported elsewhere (23; J. J. Ely, P. Gagneux, B. Dyke, W. H. Stone, W. M. Switzer, and W. H. Frels, unpublished data); the sequences were phylogenetically analyzed by using the neighbor-joining (NJ) method as described below and sequences available at GenBank.
Simian retrovirus serology.
Serum samples were screened for SFV antibodies by a Western blot (WB) assay that combines antigens from an African green monkey (SFVAGM) and a chimpanzee (SFVCPZ) into a single test, the combined-antigen WB assay (CA-WB), as previously described (15). This assay can detect antibodies to a wide range of SFV variants from monkey and ape species (15). Seroreactivity to both diagnostic Gag p68 and p72 monkey or p70 and p74 ape proteins were considered seropositive (15, 31). Serotyping to distinguish Old World monkey from ape-like SFV infections was performed on selected samples as previously described (28). Sera were also tested for antibodies to SIV, STLV, and SRV by using enzyme-linked immunosorbent assays and/or WB assays described in detail elsewhere (13, 19).
SFV PCR and sequence analysis.
Amplification of SFV proviral sequences was performed on PBL DNA using generic, nested primers from the viral integrase (int) region, as previously described (13, 31).
For selected samples, SFV gag sequences were also amplified by using primers specific for SFVCPZ to perform phylogenetic comparison using this more divergent region of the genome. The primers SPUGF1 [5′ GGC (A/G)C(G/A) GTT AT(A/T) CCT ATT CAG CAT 3′] and SPUGR1 [5′ TCG TCC TCG TCC TCC TCC GTA 3′] were used in a primary PCR product to amplify a 762-bp gag sequence. Five microliters of the primary PCR was used as a template for a nested PCR using the primers SPUGF2 [5′ TTG GCT (G/A)GG ACG AAT TGC TC 3′] and SPUGR2 [5′ GGT TGG TAA GTA CGG G(A/G)T CGA AGA 3′] to generate a 660-bp sequence. Standard PCR conditions were used for both rounds of amplification with the exception of an annealing temperature of 45°C and 40 cycles of amplification per round of PCR. Nested PCR products were electrophoresed in 1.8% agarose gels and visualized by ethidium bromide staining. PCR products were purified using the Qiaquick PCR purification kit (Qiagen Inc., Valencia, Calif.) and then sequenced in both directions by using a BigDye terminator cycle kit (Applied Biosystems, Foster City, Calif.) and an ABI 373 automated sequencer (Applied Biosystems).
Percent nucleotide divergence was determined with the GAP program in the Wisconsin sequence analysis package on a UNIX workstation (38). Sequences were aligned by using the ClustalW program (36), and the alignments were imported into either PAUP* (beta version 5.0) or MEGA (version 2.1) programs (17, 27). Distance-based trees were generated by using the Kimura two-parameter model in conjunction with the NJ and minimum-evolution (ME) methods in the MEGA program as previously described (15). Character-based tree-building methods were performed by using the maximum-likelihood (ML) procedures included in the PAUP* software as previously described (14, 27).
SFV isolation.
Viral isolation was attempted on selected SFV-seropositive persons by cocultivation of equal numbers of their PBLs and canine thymocyte (Cf2Th), BHK21, or Mus dunni cells as reported previously (13). Cultures were monitored every 3 to 4 days for syncytial cytopathic effect (CPE) typical of FV and for reverse transcriptase (RT) activity by using the Amp-RT assay, as performed elsewhere (13). When at least 50% of the cultures showed CPE, the cells were trypsinized, and DNA lysates were prepared and were screened for SFV int sequences by PCR as described above. SFV isolates from an orangutan (SFVPPY) and a baboon (SFV10BAB) were kindly provided by Paul Johnston and Richard Heberling, respectively, and an SFV isolated from a gorilla (SFVGGO) had been previously obtained by our lab. Primate-derived SFVs were propagated on Cf2Th cells and used as sequence controls in the phylogenetic analyses.
GenBank accession numbers.
The accession numbers for the new SFV integrase and gag sequences are AY195673 to AY195683, AY195685 to AY195698, and AY195699 to AY195719. The chimpanzee mtDNA sequences generated in the present study have the accession numbers AY195720 to AY195732.
RESULTS
Simian retrovirus seroprevalence in humans exposed to NHPs.
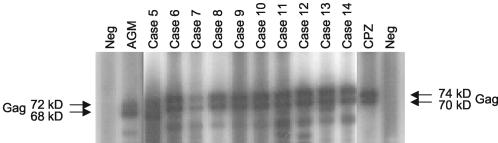
A total of 187 participants were tested in this study. One hundred eighty-six persons were from five institutions (three research institutions and two zoos), and one person had enrolled as an individual participant. Sera from all 187 participants were found to be negative for antibodies to SIV, SRV, and STLV. However, 9 of the 186 participants from institutions (4.84%) were found to be seropositive for SFV antibodies. These nine are referred to as subjects 5 and 7 to 14, according to the chronological order in which they were identified. The individual participant was also found to be SFV seropositive; this person is referred to as subject 6. Subjects 1 to 4, identified in the first phase of our surveillance study, have been described elsewhere (13). Figure 1 is a representative CA-WB result showing the seropositivity to the Gag doublet proteins of the 10 newly identified SFV-infected persons.
FIG. 1.
Detection of antibodies to SFV in infected workers by using the CA-WB assay. Neg, negative control sera. Seroreactivity to the diagnostic monkey or ape SFV Gag doublet proteins (p68/72 or p70/74, respectively) was observed in all SFV-infected specimens.
When these results are combined with previous findings (13), the overall SFV seroprevalence among persons enrolled in this study is 14 of 418 (3.35%). Three hundred seventy-five participants were employed by research institutions, and the overall seroprevalence rate among these persons was 11 of 375 (2.93%). However, SFV seroprevalence rates varied by institution and ranged from 0 to 11.11% (5 of 45 persons at one site). SFV seroprevalence at the two zoos was 1 of 17 (5.88%) and 2 of 26 (7.69%), for a combined seroprevalence of 3 of 43 (6.98%). These differences in infection rates between workers at zoos and research centers were not statistically significant (relative risk, 2.48; 95% confidence interval, 0.69 < relative risk < 8.19; Fisher's two-tailed P value = 0.165).
PCR and sequence analysis.
PBLs were available from 9 of the 10 persons with positive SFV CA-WB results. One person (subject 11) stopped participating in the study and did not provide an additional blood sample for PCR analysis. SFV int sequences were successfully amplified from the PBL DNA lysates from all 9 seropositive persons, and these sequences were then analyzed to determine the species origin of the SFV infection. Table 1 shows the percent nucleotide identities of the int sequences among all the new and previously identified subjects and the sequences from different SFV-infected NHPs. The SFV int sequences from all persons were unique except for sequences from subjects 8 and 10, who were from the same institution. SFV sequences from subject 5 had the highest sequence identity to those of SFV from baboons (87%), while those from subjects 6 to 10 and 12 to 14 were closest to sequences of SFV found in chimpanzees (93 to 98%).
TABLE 1.
Nucleotide identity of SFV integrase sequences (425 bp) among infected humans and NHPs
| Subject | % Nucleotide identity with SFV sequence froma:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Subject 1 | Subject 2 | Subject 3 | Subject 4 | Subject 5 | Subject 6 | AGM | BAB | MAC | CAT | CPZ | PPA | SPM | |
| 1 | 83 | 82 | 82 | 83 | 68 | 88 | 82 | 77 | 85 | 67 | 67 | 66 | |
| 2 | 96 | 85 | 86 | 67 | 82 | 96 | 76 | 82 | 66 | 69 | 62 | ||
| 3 | 85 | 86 | 68 | 82 | 97 | 77 | 82 | 67 | 69 | 62 | |||
| 4 | 96 | 67 | 81 | 86 | 77 | 81 | 67 | 68 | 64 | ||||
| 5 | 69 | 82 | 87 | 77 | 81 | 68 | 69 | 64 | |||||
| 6 | 68 | 68 | 68 | 70 | 93 | 82 | 64 | ||||||
| 7 | 69 | 68 | 68 | 69 | 70 | 93 | 68 | 68 | 70 | 70 | 96 | 84 | 67 |
| 8 | 68 | 68 | 68 | 69 | 69 | 92 | 68 | 68 | 70 | 69 | 97 | 83 | 67 |
| 9 | 68 | 69 | 69 | 70 | 69 | 93 | 69 | 69 | 71 | 71 | 98 | 84 | 67 |
| 10 | 68 | 68 | 68 | 69 | 69 | 92 | 68 | 68 | 70 | 69 | 97 | 83 | 67 |
| 12 | 68 | 69 | 69 | 69 | 69 | 92 | 69 | 69 | 70 | 70 | 98 | 84 | 67 |
| 13 | 68 | 69 | 69 | 70 | 69 | 92 | 68 | 68 | 70 | 71 | 98 | 83 | 67 |
| 14 | 68 | 69 | 69 | 70 | 69 | 92 | 68 | 68 | 70 | 71 | 98 | 83 | 67 |
AGM, African green monkey; BAB, baboon; MAC, macaque; CAT, C. atys (sooty mangabey); CPZ, chimpanzee; PPA, Pan paniscus (bonobo); SPM, spider monkey. Sequence data were not available for subject 11 because he stopped participation in the study. Subjects 1 to 4 were described previously (13). For each subject, the highest nucleotide identity with a sequence from an NHP is in bold.
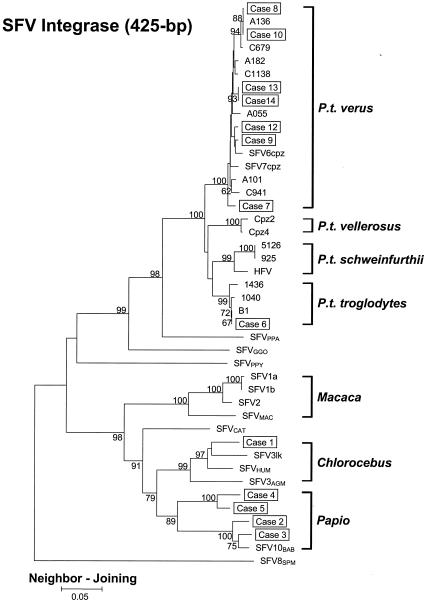
The int sequences from the nine subjects were then phylogenetically compared with those from subjects 1 to 4 and representative SFV-infected NHP species (Fig. 2). The int sequences all clustered in separate lineages by NHP species, suggesting a coevolution of host and SFV. Identical tree topologies were obtained with the ME and ML methods (data not shown). All int sequences from the nine new subjects were distinct from the sequences from subjects 1 to 4 (13). Sequences from subject 5 clustered with sequences from subject 4 in the baboon lineage, suggesting a baboon origin for their SFV infection. Sequences from eight subjects (subjects 6 to 10 and 12 to 14) clustered within the bootstrap-supported, monophyletic SFV chimpanzee clade and were distinct from the other monophyletic hominoid lineages containing SFV from Pan paniscus (bonobo or pygmy chimpanzee; SFVPPA), SFVGGO, or SFVPPY. These data indicate that chimpanzees were the source of SFV infections in these eight subjects.
FIG. 2.
Phylogenetic relationship of integrase sequences of SFV-infected workers and NHPs. The tree was derived by NJ analysis using 2 Kimura distances. The 13 cases of SFV infection are boxed. Samples were not available from subject 11. The subspecies origins for all 14 chimpanzee SFV sequences (B1, 1040, 1436, 1016, 1058, Cpz2, Cpz4, C941, A101, A055, C1138, A182, C679, and A136) are indicated. Virus origins: SFV6cpz and SFV7cpz, common chimpanzees (subspecies unknown); SFV1a, SFV1b, SFV2, and SFVMAC, macaques; SFV3AGM and SFV3lk, African green monkeys; SFVHUM, SFVAGM-infected human; SFVCAT, sooty mangabey (C. atys). Values on branch nodes represent the percentages of 1,000 bootstrap replicates, and only values greater than 60% are shown. The scale bar represents an evolutionary distance of 0.05 nucleotide per site. Trees were rooted by using the New World spider monkey (SFV8SPM) sequence.
Direct transmission of SFV from chimpanzees to workers and evidence of SFV coevolution with chimpanzee subspeciation.
We investigated whether SFVs in subjects 6, 8, and 10 and in chimpanzees which they reported having been injured by or having worked with were molecularly linked to each other by analyzing sequence identity and phylogenetic relatedness of their SFVs. In addition, SFV sequences from all four chimpanzee subspecies (Pan troglodytes troglodytes, P. t. verus, P. t. vellerosus, and P. t. schweinfurthii) were determined and included in the analysis to ensure appropriate interpretation of sequence relatedness between the subjects and chimpanzees and to evaluate whether SFV has coevolved at the subspecies level.
Subject 6 reported being severely bitten in 1977 by a chimpanzee (B1), with the injury requiring surgery. Serum samples archived from B1 since 1978, about 4 months after the reported injury, and sera archived from subject 6 since 1981 all tested positive for SFV antibodies. To assess whether chimpanzee B1 was the source of the infection in subject 6, we analyzed SFV int sequences from subject 6, B1, and 13 other chimpanzees. The 13 SFV sequences were obtained from three chimpanzees (C679, C941, and C1138) with which subject 6 had worked and from 10 chimpanzees (A055, A101, A136, A182, 1040, 1436, 925, 5126, Cpz2, and Cpz4) with which he had not worked. Four of the 13 chimpanzees (A055, A101, A136, and A182) were housed at a different institution, which employed subjects 7 to 11. Two chimpanzees were a mother-and-offspring pair (925 and 5126, respectively), representing a likely mother-to-infant SFV transmission. None of the study participants worked with chimpanzee 1040, 1436, 925, 5126, Cpz2, or Cpz4.
Phylogenetic analysis of mtDNA sequences from all 14 chimpanzees showed that they clustered into a monophyletic chimpanzee clade consisting of four distinct and bootstrap-supported (91 to 97 of 100) phylogroups (data not shown) represented by each of the four chimpanzee subspecies. The phylogenetic relatedness of the chimpanzee mtDNA sequences is similar to those reported by others (9-11). These results demonstrate that three chimpanzees were P. t. troglodytes (B1, 1040, and 1436), seven were P. t. verus (C679, C941, C1138, A055, A101, A136, and A182), two were P. t. vellerosus (Cpz2 and Cpz4), and two were P. t. schweinfurthii (925 and 5126) (data not shown).
Phylogenetic analysis of the int region showed that all 14 chimpanzee SFV sequences also clustered into a single monophyletic group comprising four major branches defined by each subspecies (Fig. 2). As observed with the mtDNA analysis, the four major SFV int clades consisted of sequences from the three P. t. troglodytes chimpanzees (B1, 1040, and 1436), the seven P. t. verus chimpanzees (C679, C941, C1138, A055, A101, A136, and A182), the two P. t. vellerosus chimpanzees (Cpz2 and Cpz4), and the two P. t. schweinfurthii chimpanzees (925 and 5126) (Fig. 2). These results suggest separate virus-host coevolution within each subspecies (Fig. 2). Identical tree topologies were obtained by using the ML and ME methods (data not shown).
The chimpanzee mother and offspring int sequences clustered tightly (bootstrap support of 100 of 100) in the P. t. schweinfurthii-specific lineage. The identical sequences found in this pair (Table 2) and the high phylogenetic relatedness indicate a mother-to-child SFV transmission. Interestingly, the int sequence from the prototype HFV also clustered in the P. t. schweinfurthii-specific clade, linking this isolate to this chimpanzee subspecies.
TABLE 2.
Nucleotide identities of SFV integrase (int) and gag sequences from selected SFVcpz-infected workers and SFV-infected chimpanzees (P. troglodytes)a
| Subject or chimpanzee | % Nucleotide identity
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
P. t. troglodytes
|
P. t. vellerosus, |
P. t. schweinfurthii
|
P. t. verus
|
|||||||||||||
| Subject 6 | B1 | 1436 | Cpz4 | HFV | 925b | Subject 7 | Subject 8 | Subject 9 | Subject 10 | A055 | A101 | A136 | A182 | C679 | C941 | |
| Subject 6 | 100 | 99 | 87 | 88 | 88 | 84 | 83 | 85 | 84 | 84 | 84 | 84 | 84 | 84 | 84 | |
| B1 | 100 | 99 | 87 | 88 | 88 | 84 | 83 | 85 | 84 | 84 | 84 | 84 | 84 | 84 | 84 | |
| 1436 | 98 | 98 | 87 | 88 | 87 | 84 | 83 | 85 | 84 | 82 | 84 | 84 | 85 | 84 | 84 | |
| Cpz4 | 92 | 92 | 92 | 84 | 84 | 80 | 81 | 81 | 82 | 82 | 83 | 83 | 83 | 81 | 81 | |
| HFV | 92 | 92 | 93 | 92 | 96 | 85 | 84 | 85 | 85 | 83 | 85 | 85 | 85 | 85 | 84 | |
| 1058 | 92 | 92 | 92 | 91 | 96 | 84 | 83 | 84 | 84 | 83 | 84 | 85 | 85 | 83 | 83 | |
| Subject 7 | 93 | 93 | 92 | 92 | 91 | 90 | 94 | 97 | 95 | 95 | 96 | 95 | 96 | 97 | 96 | |
| Subject 8 | 92 | 92 | 92 | 91 | 91 | 89 | 97 | 96 | 98 | 93 | 95 | 98 | 97 | 95 | 94 | |
| Subject 9 | 93 | 93 | 92 | 92 | 91 | 90 | 98 | 97 | 97 | 98 | 97 | 97 | 97 | 98 | 96 | |
| Subject 10 | 92 | 92 | 92 | 91 | 91 | 89 | 97 | 100 | 97 | 94 | 96 | 100 | 98 | 96 | 95 | |
| A055 | 92 | 92 | 92 | 92 | 90 | 90 | 97 | 97 | 98 | 97 | 98 | 96 | 97 | 96 | 98 | |
| A101 | 93 | 93 | 92 | 92 | 90 | 89 | 98 | 97 | 98 | 97 | 98 | 96 | 96 | 97 | 96 | |
| A136 | 92 | 92 | 92 | 91 | 91 | 89 | 97 | 100 | 97 | 100 | 97 | 97 | 98 | 96 | 95 | |
| A182 | 93 | 93 | 92 | 92 | 91 | 90 | 97 | 98 | 98 | 98 | 98 | 98 | 98 | 97 | 96 | |
| C679 | 92 | 92 | 92 | 91 | 91 | 90 | 97 | 99 | 97 | 99 | 97 | 97 | 99 | 98 | 97 | |
| C941 | 93 | 93 | 92 | 92 | 90 | 89 | 97 | 97 | 98 | 97 | 97 | 98 | 97 | 97 | 97 | |
Values below and above the diagonal are for int and gag, respectively. Results are grouped by chimpanzee subspecies as determined by phylogenetic analysis of their partial mitochondrial control region sequences. Boxes indicate the identities among SFVs originating from the same subspecies.
The int and gag sequences from the mother-offspring (925 and 5126) pair were identical, and thus, only the identities for the mother are shown.
The int sequences from subject 6 and chimpanzee B1 were identical (Table 2), and both clustered together with good bootstrap support in the P. t. troglodytes-specific group (Fig. 2). These int sequences were 1 to 2% divergent from other SFV sequences in this lineage (Table 2). The high genetic relatedness between the sequences of subject 6 and B1 suggests chimpanzee B1 as the source of the SFV infection of this worker.
We also found that subjects 8 and 10, who are coworkers, had int sequences identical to that of chimpanzee A136 but distant from all other SFV sequences (Table 2). Phylogenetic analysis of the int sequences of subjects 8 and 10 and chimpanzee A136 showed that all three sequences clustered together in the P. t. verus clade with a bootstrap support of 88 of 100, suggesting that A136 was the likely source of their SFV infections. Although both subjects 8 and 10 reported working with A136 and having contact with the body fluids of A136, neither person remembers a specific injury with this chimpanzee.
To investigate further the phylogenetic relationships obtained with the int gene, we obtained and analyzed sequences in the more variable and thus more discriminatory SFV gag region. Similar to the int sequences, all gag sequences clustered in separate lineages by NHP species, supporting the theory of the coevolution of host and SFV (Fig. 3). In addition, the SFV gag sequences from each chimpanzee subspecies again formed four distinct clusters within a monophyletic chimpanzee clade (Fig. 3), confirming the reliability of the phylogenetic relationships seen with the int sequences. A similar topology was obtained by using the ML method (data not shown).
FIG. 3.
Phylogenetic relationship of gag sequences of selected SFVCPZ-infected workers and NHPs. The tree was derived by NJ analysis using 2 Kimura distances. The eight cases of SFVCPZ infection are shown in boxes. The subspecies origin for 11 chimpanzee SFV sequences (B1, 1040, 1016, 1058, Cpz4, C941, A101, A055, A182, C679, and A136) is indicated. SFV1, macaque. Values on branch nodes represent the percentages of 1,000 bootstrap replicates, and only values greater than 60% are shown. The scale bar represents an evolutionary distance of 0.05 nucleotide per site.
As observed for the int region, gag sequences from subject 6 were indistinguishable from those of B1 (Table 2). The gag sequences from the mother-offspring pair were also identical. Phylogenetic analysis of the gag sequences from each of these pairs also showed that they clustered tightly together with high bootstrap support (Fig. 3).
Analysis of the gag sequences also clarified the sequence relatedness between chimpanzee A136 and subjects 8 and 10. The gag sequence from subject 10 clustered more tightly with A136 than with that of subject 8, suggesting that this chimpanzee is the likely source of infection for subject 10 and not for subject 8.
The HFV gag sequence, like the HFV int sequence, clustered in the P. t. schweinfurthii-specific clade (Fig. 3). These findings confirm a molecular link of this isolate to this chimpanzee subspecies.
SFV serotyping.
Since PBLs were not available from subject 11 for PCR and sequence analysis, we serotyped a serum sample from this person. Serum from subject 11 reacted more strongly against the SFVCPZ antigen than the SFVAGM antigen, suggesting a greater likelihood of infection with a chimpanzee-like SFV (data not shown).
SFV isolation and sequence analysis.
Virus isolation was attempted on PBLs from subject 5, infected with SFVBAB, and subject 6, infected with SFVCPZ, and from chimpanzee B1. CPE, RT, and proviral int sequences were observed for both subject 6 and B1 only in Cf2Th cultures at days 26 and 7, respectively (data not shown). Cell-free culture supernatant passages of isolates from both subject 6 and B1 to fresh Cf2Th cells again resulted in CPE, RT activity, and PCR detection of proviral int sequences (data not shown). Proviral int sequences from the passaged isolates were identical to those obtained from the PBLs of subject 6 and chimpanzee B1 (data not shown). None of the cultures from subject 5 showed any evidence of SFV isolation.
Histories, exposures, and duration of seropositivity.
Table 3 summarizes the case histories, reported exposures, and duration of seropositivity in the 10 subjects with newly identified cases of SFV infection. For completion, we also included a similar summary for previously reported subjects 1 to 4 (13). Eight of the 10 new subjects are men and two are women. The SFV-infected females (subjects 13 and 14) and one male (subject 12) all worked at zoos. The remaining seven SFV-infected men worked at research institutions. All workers reported 6 to 30 years of experience (mean, 20 years) working with a variety of Old and New World NHPs. Eight of the 10 newly identified SFV-infected subjects (subjects 6 to 11, 12, and 14) reported injuries involving specific NHPs. In contrast, subjects 5 and 13 could not remember any specific injuries. The injuries reported by subjects 6, 8, 9, and 14 were inflicted by chimpanzees, which is also the species from which their SFV infection originated. The SFV infection in subjects 7, 10, 12, and 13 also originated from chimpanzees. However, although each of these four subjects reported working with chimpanzees, none of them reported injuries involving chimpanzees.
TABLE 3.
Case histories, exposures, and duration of seropositivity in SFV-infected workersa
| Subject | Occupation | Sexb | Duration (yrs) and species of NHP exposure(s) | Reported injuriesc | Date of first seropositivityd | SFV origine |
|---|---|---|---|---|---|---|
| 1 | Animal caretaker | M | >20; AGM | Bitten twice by AGM | 1995 | AGM |
| 2 | Research scientist | M | >30; macaque, baboon, AGM, chimpanzee, gibbon, marmoset, bush baby | Cut with chimpanzee-contaminated glass tube (1970) | 1978 | Baboon |
| 3 | Animal care supervisor | M | >30; macaque, baboon, AGM, marmoset, owl monkey, tamarin | Severe baboon bite (before 1985) | 1988 | Baboon |
| 4 | Veterinarian | M | >20; macaque, baboon, AGM, chimpanzee, spider monkey, marmoset, capuchin, owl monkey | Severe baboon bite (1978) | 1994 | Baboon |
| 5 | Veterinarian | M | >29; macaque, baboon, AGM, Sykes's money, chimpanzee, gibbon, spider monkey, marmoset, capuchin, owl monkey, squirrel monkey, lemurs, wooly monkey, bush baby | None | 1979 | Baboon |
| 6 | Veterinarian | M | >25; macaque, baboon, AGM, chimpanzee, marmoset, squirrel monkey, bush baby | Severe chimpanzee bite requiring surgery | 1981 | Chimpanzee |
| 7 | Veterinarian | M | >20; macaque, chimpanzee, AGM, gorilla, orangutans, gibbons, spider monkey, capuchin, squirrel monkey | Bites, scratches, needlesticks, mucocutaneous (no dates or details) | 1990 | Chimpanzee |
| 8 | Animal caretaker | M | >14; macaque, baboon, AGM, chimpanzee, gorilla, orangutan, spider and squirrel monkeys | Rhesus scratch, bad chimpanzee bite to leg (1980-85) | 1985 | Chimpanzee |
| 9 | Animal caretaker | M | >6; macaque, baboon, AGM, chimpanzee, gorilla, orangutan, spider and squirrel monkeys, marmoset, capuchin | Chimpanzee scratches (1981) | 1980 | Chimpanzee |
| 10 | Animal care supervisor | M | >25; macaque, baboon, AGM, patas monkey, langur, chimpanzee, gorilla, orangutan, gibbon, spider, squirrel, owl and wooly monkeys, marmoset, capuchin | Needlestick with gorilla blood (1980); rhesus bite (1970), mucocutaneous (no details) | 1976 | Chimpanzee |
| 11 | Research technician | M | >10; macaque, baboon, AGM, chimpanzee, orangutan, spider and squirrel monkeys, marmoset, capuchin | Rhesus scratch (1994), AGM bite (1997) | NA | Chimpanzee-likef |
| 12 | Animal caretaker, zoo | M | >30; baboon, chimpanzee, gorilla, orangutan, gibbon, siamang, mandrill, drill, talapoin, spider monkey, marmoset, tamarin, woolly monkey | Bite, scratch, and needlestick with talapoin and DeBrazza's monkey blood | NA | Chimpanzee |
| 13 | Veterinary technician, zoo | F | >11; chimp, gorilla | None | NA | Chimpanzee |
| 14 | Animal caretaker, zoo | F | >10; baboon, chimpanzee, gorilla | Spit upon and scratched by chimpanzee | NA | Chimpanzee |
Subjects 1 to 4 have been described previously (13) and are included here for completeness. AGM, African green monkey.
M, male; F, female.
Self-reported occupational exposures.
First documented WB seropositivity from available archived serum or plasma samples. NA, specimen not available.
Source of SFV infection determined by phylogenetic analysis of proviral DNA sequences.
Based on SFV-type-specific WB.
Archived serum was available for six SFV-positive persons who worked at research institutions. Individuals were documented to have been seropositive for a period of 8 to 26 years (mean, 19.3 years; median, 22 years) (Table 3). The two earliest available sera from subjects 5 and 6 were collected in 1979 and 1981, respectively, and both were found to be WB positive (Table 3). For subjects 7 to 10, the last available WB-negative sera were collected in 1982, 1980, 1978, and 1975, respectively. The next sera available from subjects 7 to 10 were collected in 1990, 1985, 1980, and 1976, respectively, and all were found to be WB positive (Table 3). SFV infection may have occurred in subjects 7 to 10 during a period of about 8, 5, 2, and 1 year, respectively. Stored sera were not available from the zoo workers. Testing of archived sera from subjects 1 to 4 was reported previously (13).
Health status and secondary transmission.
With the exception of chronic diseases of aging for a few persons, all subjects considered themselves healthy at the time of identification of their SFV infection. To assess the risk of sexual transmission, fresh blood samples were obtained from the wives of three SFV-infected men. Two of the men were infected with SFVCPZ and one was infected with SFVBAB. Despite a documented exposure of 19 to 21 years (median and mean, 20 years), all three wives tested negative for SFV by both CA-WB and PCR. The spouses of three other men with baboon-type SFV infection previously tested negative also (13). Spouses of the remaining eight subjects were not available for testing.
Of all 14 seropositive workers identified in our surveillance, 11 (78.6%) reported donating blood more than once and included persons infected with SFVAGM, SFVBAB, or SFVCPZ genotypes. Testing of archived sera available from 7 persons showed that 6 (54.5%) of the 11 donors had donated blood after becoming seropositive. One seropositive person (9.1%) reported donating blood only before working with primates. Archived serum samples from the period of blood donation were not available for four persons, preventing clarification of their SFV serostatus at the time of donation.
DISCUSSION
The recognition that the HIV pandemic most likely originated from zoonotic SIV infections that adapted and was transmitted secondarily to become endemic in humans raises concerns about the introduction of a multitude of other simian retroviruses to humans. Our study demonstrates that SFV infection among persons occupationally exposed to NHPs is not a rare event. Serologic screening of 187 persons identified 10 new SFV-infected persons. Sequence analysis of PBLs available from nine persons indicated that the SFV originated from baboons in one person and chimpanzees in eight others. Although not definitive, serotyping also suggested a chimpanzee-like SFV in a tenth person. These data expand our previous findings and demonstrate that zoonotic SFV infections are more common than SIV, STLV, or SRV infections (13, 16, 19, 34).
The SFV-infected persons exhibited WB seropositivity documented to persist for up to 26 years. SFV sequences were amplified from the PBLs of all SFV-infected persons, including those found to have been seropositive for decades. These observations are consistent with long-standing persistent infections, and these results are similar to the viral persistence occurring in naturally SFV-infected NHPs (15, 31). The higher prevalence of SFV infection among exposed workers, compared with the prevalence of other simian retrovirus infections, may reflect an increased frequency of exposure to SFV, since SFV is the most prevalent retroviral infection among captive NHPs (15, 31). Alternatively, it may reflect a greater ease of transmission due to the presence of infectious SFV in the saliva and other body fluids of NHPs or a greater permissiveness of human cells for SFV than for other retroviruses (6, 21).
Consistent with our previous report, we confirm zoo workers as another occupational group at risk for SFV infection (28). Although the data are not statistically significant, our study suggests a higher rate of SFV infection among zoo keepers than among workers at research institutions. It is possible that zoo workers are at higher risk for SFV infection because personal protective equipment and biosafety training may not be as readily available or as carefully enforced at zoos as at research institutions. Alternatively, the higher SFV prevalence seen in zoo workers may be due to other factors, including greater exposure to SFV in body fluids like saliva or the performance of different types of procedures at zoos than at research institutions. Whether SFV infection is significantly higher in zoo workers will require confirmation with a larger sample size. Nonetheless, these preliminary findings argue for careful review of current biosafety practices standardly employed by zoo workers.
The SFV infection of these workers originated from primate species that are commonly used in research centers and zoos and that were handled by the SFV-infected workers. The majority of the workers identified in this study were infected with chimpanzee-type SFV but had reported exposure to and injuries from many other NHPs. Our methods preclude quantitative characterization of the relative frequency of contact between specific workers and specific NHPs. The predominance of SFVCPZ infection among these workers may reflect differences in the severity and frequency of exposure to SFVCPZ or sample selection biases in enrollment of workers. Alternatively, this observation may suggest an increased transmissibility of SFVCPZ to humans compared with other SFV variants due to the closer genetic relatedness between chimpanzees and humans. SFVCPZ may also be more transmissible because of higher viral loads in SFV-infected chimpanzees, since high viral loads are known to increase the transmission of other retroviruses (7, 22, 24). However, very little is known about the viral loads in naturally infected NHPs. Additional research is needed to better understand the reasons for the increased frequency of human infection with chimpanzee-type SFV among the studied population.
Our identification of SFVCPZ infection in four workers and SFVBAB infection in another person, all of whom did not report any specific injuries from either chimpanzees or baboons though they all worked directly with both of these NHPs, is significant. These results suggest that transmission of SFV to humans from exposure to NHP body fluids may occur more casually than previously thought. Thus, our findings reinforce the importance of adhering to appropriate biosafety precautions while working with NHPs, including using personal protective equipment (25).
Although SFV is nonpathogenic in naturally infected NHPs, the significance of SFV infection in humans is poorly defined. The introduction of SFV infections into humans is of concern because changes in the pathogenicity of simian retroviruses following cross-species infection are well documented, since both HIV-1 and HIV-2 emerged from benign SIV infections in the natural primate hosts (10, 26). We identify SFV-infected workers who report being in apparent good health after an average of 19 years of persistent infection, suggesting no abrupt change in viral pathogenicity. However, our observations to date are based on limited self-reported health information and cannot fully characterize these infections or the pathogenic potential of SFV in humans. Incidence of disease in SFV-infected persons may be low, may follow long latency periods, or may be associated with specific SFV clades that have not been identified here. In addition, the biases associated with recruitment from healthy worker populations further limit our ability to identify disease associations. Long-term follow-up of SFV-infected humans is needed and has been initiated by the Centers for Disease Control and Prevention in an attempt to better assess clinical outcomes of SFV infection.
We found that wives of one SFVBAB- and two SFVCPZ-infected workers remain uninfected despite years of intimate exposure. These data are consistent with our previous findings and raise the total to six uninfected wives (13). Collectively, these findings suggest that SFV is not transmitted easily among humans by intimate contact from male to female (13). Our study has identified the first reported SFV infections in women, thus suggesting that SFV may be spread through additional mechanisms: from mother to child or through sexual contact with an infected woman. However, spouses and children of these subjects were not available for testing to determine if SFV transmission occurs via these routes.
We also found that seven persons donated blood after the date they were retrospectively documented to be SFV seropositive, indicating the potential for secondary spread of SFV through blood donations. The presence of SFV-infected PBLs in the blood of all tested subjects also suggests a risk of secondary transmission by exposure to infected blood. However, a recent look-back study of recipients of blood components from an SFVCPZ-infected blood donor failed to identify evidence of SFV infection in two recipients of red cells, one recipient of filtered red cells, and one recipient of platelets (3). Nonetheless, more data are needed to better define the risks for SFV transmission through donated blood. We counsel all SFV-infected persons to refrain from donating blood or other biomaterials.
While the SFV phylogenetic relationships demonstrate coevolution with a diverse range of ape and monkey species, our analysis of mtDNA and SFV sequences from chimpanzees also indicates a coevolution of virus and host within each of the four chimpanzee subspecies. In addition, our finding that seven of the eight chimpanzee-like SFV sequences seen in the infected workers clustered with SFV from the P. t. verus subspecies, while one clustered with SFV from P. t. troglodytes, is consistent with the fact that more than 95% of all captive chimpanzees in the United States belong to the P. t. verus subspecies (Ely et al., unpublished).
To date, direct evidence of zoonotic transmission of simian retroviruses from known source animals to humans has not been documented. Instead, proof for such cross-species infections has been supported by indirect evidence, including phylogenetic relatedness between epidemiologically unlinked human and simian retroviruses (10, 12, 30, 33). We demonstrate in this study the high genetic relatedness between the SFV of subject 6 and that of chimpanzee B1, which had bitten him, thus establishing B1 as the likely source of the SFV infection in this worker. Likewise, our data linking the SFV of subject 10 to that of chimpanzee A136, with whom subject 10 had worked, suggests that this animal was the likely source of infection of subject 10. These observations provide the first direct evidence of zoonotic transfer of a simian retrovirus to humans.
While almost all NHP species investigated thus far harbor distinct and species-specific SFV lineages, evidence supporting the existence of a human-specific FV is not yet available. We show that the prototype FV, or HFV, first identified in a Kenyan patient in 1971 is an SFVCPZ variant from P. t. schweinfurthii, the chimpanzee subspecies in East Africa (1). This observation suggests that HFV most probably represents another cross-species infection from P. t. schweinfurthii and that human infection with SFV has been occurring for decades. In addition, serological testing of archived samples from the SFV-infected workers from the present study documents a WB-positive specimen from 1976, further supporting the finding that SFV transmission is not a recent occurrence. It is not understood why humans, despite being susceptible to FV infection and having a common evolution and long periods of cohabitation with NHPs, are not endemically infected with a distinct FV.
In conclusion, our study documents persistent SFV infections in persons occupationally exposed to NHPs and shows that simian retroviruses cross into humans more frequently than previously thought. This study also provides information on the natural history and species origin of these infections, supports appropriate attention to biosafety practices to prevent occupational infections, and highlights the importance of additional studies to better define the clinical outcome of these zoonotic infections.
Acknowledgments
We are grateful to all the study participants and the local project directors, who continue to help make this study a success. We thank Martine Peeters, Beatrice Hahn, and Feng Gao for providing chimpanzee DNA specimens; Althaf Hussain and Aprille Matthews for helping with the SFV WBs; Pascal Gagneux for providing the P. t. vellerosus mtDNA sequences and comments on an early draft; the veterinary and administrative staffs at many institutions for providing additional chimpanzee specimens; Harold McClure for providing a gorilla sample; and Paul Johnston and Richard Heberling for providing the SFVPPY and SFV10BAB isolates, respectively.
Use of trade names is for identification only and does not imply endorsement by the U.S. Department of Health and Human Services, the Public Health Service, or the Centers for Disease Control and Prevention.
REFERENCES
- 1.Achong, B. G., P. W. A. Mansell, M. A. Epstein, and P. J. Clifford. 1971. An unusual virus in cultures from a human nasopharyngeal carcinoma. J. Natl. Cancer Inst. 46:299-307. [PubMed] [Google Scholar]
- 2.Ali, M., G. P. Taylor, R. J. Pitman, D. Parker, A. Rethwilm, R. Cheingsong-Popov, J. N. Weber, P. D. Bieniasz, J. Bradley, and M. O. McClure. 1996. No evidence of antibody to human foamy virus in widespread human populations. AIDS Res. Hum. Retrovir. 12:1473-1483. [DOI] [PubMed] [Google Scholar]
- 3.Boneva, R. S., A. Grindon, S. Orton, W. M. Switzer, V. Shanmugam, A. Hussain, V. Bhullar, W. Heneine, M. Chamberland, T. M. Folks, and L. Chapman. 2002. Simian foamy virus infection in a blood donor. Transfusion 42:886-891. [DOI] [PubMed] [Google Scholar]
- 4.Brooks, J. I., E. W. Rud, R. G. Pilon, J. M. Smith, W. M. Switzer, and P. A. Sandstrom. 2002. Cross-species retroviral transmission from macaques to human beings. Lancet 360:387-388. [DOI] [PubMed] [Google Scholar]
- 5.Broussard, S. R., A. G. Comuzzie, K. L. Leighton, M. M. Leland, E. M. Whitehead, and J. S. Allan. 1997. Characterization of new simian foamy viruses from African nonhuman primates. Virology 237:349-359. [DOI] [PubMed] [Google Scholar]
- 6.Falcone, V., J. Leupold, J. Clotten, E. Urbanyi, O. Herchenroder, W. Spatz, B. Volk, N. Bohm, A. Toniolo, D. Neumann-Haefelin, and M. Schweizer. 1999. Sites of simian foamy virus persistence in naturally infected African green monkeys: latent provirus is ubiquitous, whereas viral replication is restricted to the oral mucosa. Virology 257:7-14. [DOI] [PubMed] [Google Scholar]
- 7.Fideli, U. S., S. A. Allen, R. Musonda, S. Trask, B. H. Hahn, H. Weiss, J. Mulenga, F. Kasolo, S. H. Vermund, and G. M. Aldrovandi. 2001. Virologic and immunologic determinants of heterosexual transmission of human immunodeficiency virus type 1 in Africa. AIDS Res. Hum. Retrovir. 17:901-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fultz, P. N. 1994. Simian T-lymphotropic virus type 1, p. 111-131. In J. A. Levy (ed.), The Retroviridae, vol. 3. Plenum Press, New York, N.Y. [Google Scholar]
- 9.Gagneux, P., M. K. Gonder, T. L. Goldberg, and P. A. Morin. 2001. Gene flow in wild chimpanzee populations: what genetic data tell us about chimpanzee movement over space and time. Philos. Trans. R. Soc. Lond. B 356:889-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao, F., E. Bailes, D. L. Robertson, Y. Chen, C. M. Rodenburg, S. F. Michael, L. B. Cummins, L. O. Arthur, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436-441. [DOI] [PubMed] [Google Scholar]
- 11.Gonder, M. K., J. F. Oates, T. R. Disotell, M. R. Forstner, J. C. Morales, and D. J. Melnick. 1997. A new west African chimpanzee subspecies? Nature 388:337. [DOI] [PubMed] [Google Scholar]
- 12.Hahn, B. H., G. M. Shaw, K. M. De Cock, and P. M. Sharp. 2000. AIDS as a zoonosis: scientific and public health implications. Science 287:607-614. [DOI] [PubMed] [Google Scholar]
- 13.Heneine, W., W. M. Switzer, P. Sandstrom, J. Brown, S. Vedapuri, C. A. Schable, A. S. Khan, N. W. Lerche, M. Schweizer, D. Neumann-Haefelin, L. Chapman, and T. M. Folks. 1998. Identification of a human population infected with simian foamy viruses. Nat. Med. 4:403-407. [DOI] [PubMed] [Google Scholar]
- 14.Herchenroder, O., R. Rolf, D. Loncar, E. K. Cobb, K. K. Murthy, J. Schneider, A. Mergia, and P. A. Luciw. 1994. Isolation, cloning and sequencing of simian foamy viruses from chimpanzees (SFV CPZ): high homology to human foamy virus (HFV). Virology 201:187-199. [DOI] [PubMed] [Google Scholar]
- 15.Hussain, A. I., V. Shanmugam, V. B. Bhullar, B. E. Beer, D. Vallet, A. Gautier-Hion, N. Wolfe, W. B. Karesh, A. M. Kilbourn, Z. Tooze, W. Heneine, and W. M. Switzer. 2003. Screening for simian foamy virus infection by using a combined antigen Western blot assay: evidence for a wide distribution among Old World primates and identification of four new divergent viruses. Virology 309:248-257. [DOI] [PubMed] [Google Scholar]
- 16.Khabbaz, R. F., W. Heneine, J. R. George, B. Parekh, T. Rowe, T. Woods, W. M. Switzer, H. M. McClure, M. Murphey-Corb, and T. M. Folks. 1994. Brief report: infection of a laboratory worker with simian immunodeficiency virus. N. Engl. J. Med. 330:172-177. [DOI] [PubMed] [Google Scholar]
- 17.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 18.Lerche, N. W. 1992. Epidemiology and control of simian type D retrovirus infection in captive macaques, p. 439-448. In S. Matano, R. H. Tuttle, H. Ishida, and M. Goodman (ed.), Topics in primatology, vol. 3. University of Tokyo Press, Tokyo, Japan. [Google Scholar]
- 19.Lerche, N. W., W. M. Switzer, J. L. Yee, V. Shanmugam, A. N. Rosenthal, L. E. Chapman, T. M. Folks, and W. Heneine. 2001. Evidence of infection with simian type D retrovirus in persons occupationally exposed to nonhuman primates. J. Virol. 75:1783-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lowenstine, L. J., and N. W. Lerche. 1988. Retrovirus infections of nonhuman primates: a review. J. Zoo Anim. Med. 19:168-187. [Google Scholar]
- 21.Meiering, C. D., and M. L. Linial. 2001. Historical perspective of foamy virus epidemiology and infection. Clin. Microbiol. Rev. 14:165-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mellors, J. W., C. R. Rinaldo, Jr., P. Gupta, R. M. White, J. A. Todd, and L. A. Kingsley. 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272:1167-1170. [DOI] [PubMed] [Google Scholar]
- 23.Morin, A. M., J. J. Moore, R. Chakraborty, L. Jin, J. Goodall, and D. S. Woodruff. 1994. Kin selection, social structure, gene flow, and the evolution of chimpanzees. Science 265:1193-1201. [DOI] [PubMed] [Google Scholar]
- 24.Nagai, M., K. Usuku, W. Matsumoto, D. Kodama, N. Takenouchi, T. Moritoyo, S. Hashiguchi, M. Ichinose, C. R. M. Bangham, S. Izumo, and M. Osama. 1998. Analysis of HTLV-1 proviral load in 202 HAM/TSP patients and 243 asymptomatic HTLV-1 carriers: high proviral load strongly predisposes to HAM/TSP. J. Neurovirol. 4:586-593. [DOI] [PubMed] [Google Scholar]
- 25.National Research Council. 2003. Occupational health and safety in the care and use of nonhuman primates. [Online.] http://search.nap.edu/books/030908914X/html/. [PubMed]
- 26.Peeters, M., V. Courgnaud, B. Abela, X. Pourrut, F. Bibollet-Ruche, S. Loul, F. Liegeosis, C. Butel, D. Koulagna, E. Mpoudi-Ngole, G. M. Shaw, B. H. Hahn, and E. Delaporte. 2002. Risk to human health from a plethora of simian immunodeficiency viruses in primate bushmeat. Emerg. Infect. Dis. 8:451-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rogers, J. S., and D. L. Swofford. 1999. Multiple local maxima for likelihoods of phylogenetic trees: a simulation study. Mol. Biol. Evol. 16:1079-1085. [DOI] [PubMed] [Google Scholar]
- 28.Sandstrom, P. A., K. O. Phan, W. M. Switzer, T. Fredeking, L. Chapman, W. Heneine, and T. M. Folks. 2000. Simian foamy virus infection among zoo keepers. Lancet 355:551-552. [DOI] [PubMed] [Google Scholar]
- 29.Schweizer, M., and D. Neumann-Haefelin. 1995. Phylogenetic analysis of primate foamy viruses by comparison of pol sequences. Virology 207:577-582. [DOI] [PubMed] [Google Scholar]
- 30.Schweizer, M., V. Falcone, J. Gange, R. Turek, and D. Neumann-Haefelin. 1997. Simian foamy virus isolated from an accidentally infected human individual. J. Virol. 71:4821-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schweizer, M., R. Turek, H. Hahn, A. Schliephake, K. O. Netzer, G. Eder, T. M. Reinhard, A. Rethwilm, and D. Neuman-Haefelin. 1995. Markers of foamy virus infections in monkeys, apes, and accidentally infected humans: appropriate testing fails to confirm suspected foamy virus prevalence in humans. AIDS Res. Hum. Retrovir. 11:161-170. [DOI] [PubMed] [Google Scholar]
- 32.Sharp, P. M., D. L. Robertson, F. Gao, and B. H. Hahn. 1994. Origins and diversity of human immunodeficiency viruses. AIDS 8:S27-S42. [Google Scholar]
- 33.Slattery, J. P., G. Franchini, and A. Gessain. 1999. Genomic evolution, patterns of global dissemination, and interspecies transmission of human and simian T-cell leukemia/lymphotropic viruses. Genome Res. 9:525-540. [PubMed] [Google Scholar]
- 34.Sotir, M., W. Switzer, C. Schable, J. Schmitt, C. Vitek, and R. F. Khabbaz. 1997. Risk of occupational exposure to potentially infectious nonhuman primate materials and to simian immunodeficiency virus. J. Med. Primatol. 26:233-240. [DOI] [PubMed] [Google Scholar]
- 35.Switzer, W. M., D. Pieniazek, P. Swanson, H. H. Samdal, V. Soriano, R. M. Khabbaz, J. E. Kaplan, R. B. Lal, and W. Heneine. 1995. Phylogenetic relationship and geographic distribution of multiple human T-cell lymphotropic virus type II subtypes. J. Virol. 69:621-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voevodin, A., E. Samilchuk, H. Schatzl, E. Boeri, and G. Franchini. 1996. Interspecies transmission of macaque simian T-cell leukemia/lymphoma virus type 1 in baboons resulted in an outbreak of malignant lymphoma. J. Virol. 70:1633-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Womble, D. D. 2000. GCG: the Wisconsin package of sequence analysis programs. Methods Mol. Biol. 132:3-22. [DOI] [PubMed] [Google Scholar]