Abstract
Macromolecular crowding inside cells affects the thermodynamic and kinetic properties of proteins. The scaled particle theory (SPT) has played an important role toward establishing a qualitative picture for the effects of crowding. However, SPT-based modeling lacks molecular details. Molecular dynamics simulations overcome this limitation, but at great computational cost. Here we present a theoretical method for modeling crowding at the atomic level. The method makes it possible to achieve exhaustive conformational sampling in modeling crowding effects and to tackle challenges posed by large protein oligomers and by complex mixtures of crowders.
I. INTRODUCTION
The total protein and RNA concentrations inside cells reach 300–400 g/l [1] and the macromolecules together are estimated to occupy over 30% of cellular volume. The crowded environments are expected to have profound effects on the thermodynamics and kinetics of protein folding, binding, aggregation, and other more complex biological events [2]. Numerous in vitro experiments now support this expectation [3–20]. Historically the scaled particle theory (SPT) [21] has played an important role toward establishing a qualitative picture for the effects of crowding [2, 22]. However, SPT-based modeling of crowding effects lacks molecular details.
This limitation has been overcome by recent molecular dynamics (MD) simulations of crowding, via two approaches [12, 23–30]. In the direct simulation approach, a protein molecule is placed inside a box of crowders, and the motions of the protein and the crowders are followed simultaneously. The computational demands presented by such large simulation systems have necessitated the use of coarse-grained models [23–25, 27, 29].
We have developed a “postprocessing” approach for simulations of crowding [12, 26, 28, 30], whereby the protein is simulated in the absence of crowders and the conformations thus sampled are reweighted according to crowding-induced changes in chemical potential. The postprocessing approach makes it possible to represent the protein at the atomic level, but the calculation of crowding-induced changes in chemical potential, by devising a procedure akin to Widom’s insertion method [31], still requires significant computer times. Here we present a theoretical method for calculating crowding-induced changes in chemical potential for atomistic proteins. This method yields accurate results but at substantially reduced computational cost, opening the possibility of more exhaustive conformational sampling in modeling crowding effects. Moreover, it enables computational studies of challenging problems presented by large protein oligomers, on which effects exerted by crowding are especially dramatic [3, 4, 15–20], and by complex mixtures of many species of crowders, which are required for realistically mimicking the intracellular milieu.
II. THEORY AND IMPLEMENTATION
Our method is based on the fundamental measure theory (FMT) [32–34], which is a type of density functional theory for fluids but specialized to mixtures of convex hard particles. Unlike the SPT, the FMT does not rely on any assumption about the shapes of the convex particles, and derives the SPT for the special case of spherical particles [33]. When a test protein is randomly placed into a distribution of convex crowders, the FMT predicts the increase in chemical potential of the test protein as [33]
| (1) |
where kB is Boltzmann’s constant and T is the absolute temperature; vp, sp, and lp are the volume, surface area, and linear size of the test protein; Πc is the osmotic pressure of the crowders, and γc and κc are the corresponding quantities for surface tension and bending rigidity; and φ is the total volume fraction of the crowders. The latter quantities are expressed in terms of the weighted number densities of the different species of crowders:
| (2) |
where cα is the number density of species α crowders, and lα, sα, and vαare their linear size, surface area, and volume, respectively. The results are
| (3) |
| (4) |
| (5) |
The FMT has been applied to convex particles with relatively simple geometric shapes [33].
We generalize the FMT to test proteins represented at the atomic level and refer to our method the generalized fundamental measure theory (GFMT). A hint of the GFMT appeared when we found that Δμ obtained in our previous work [12, 26] by the insertion procedure could be fitted to Eq. (1). However, the fitting did not produce a predictive method, because it was not clear how vp, sp, and lp could be calculated. An atomistic protein is not a convex particle. But, we realized that, regardless of whether the test particle is convex, what matters to Δμis the spatial region excluded to the crowders (the magnitude of Δμdepends on both the size and the shape of this region). For a convex test particle, this region is the same as the physical space occupied by the particle. However, for a non-convex test particle, the particular region excluded to a crowder depends on the latter’s size and shape.
In this work, we model the test protein at the atomic level but the crowders as spheres (Fig. 1a), like in previous studies using MD simulations [12, 23–27]. For spherical crowders at a given radius, the test protein’s region of exclusion is enclosed by what we refer to as the crowder-exclusion surface. This surface is the same as the solvent-exclusion surface, except here the solvent probe is a crowder. (The solvent-exclusion surface when calculated with a 1.4-Å solvent probe is better known as the molecular surface [35]; it consists of contact parts, which are convex, and reentrant parts, which are concave.) It is obvious to identify vp as the volume enclosed by the crowder-exclusion surface and sp as the area of this surface. This identification is similar in spirit to the morphometric approach of Roth et al. [36]. For the linear size lp, we looked for a quantity that would measure how far the different parts of the crowder-exclusion surface are dispersed in space. Such a quantity seemed to be provided by the radius of gyration, rg, of points uniformly distributed on the surface. We therefore assumed that lp ∝ rg. To determine the proportionality constant, we noted that, for a spherical test protein, lp is known to be its radius, and rg is also calculated to be the radius; therefore we set the proportionality constant to be 1, i.e., lp = rg. (Further details on the calculation of rg, including a comparison of rg and the FMT result for lp can be found in the supplementary information [37].) In contrast to the fact that the morphometric approach has no predictive power, we show below that Eq. (1), with vp, sp, and lp calculated according to the crowder-exclusion surface, gives accurate predictions of Δμ.
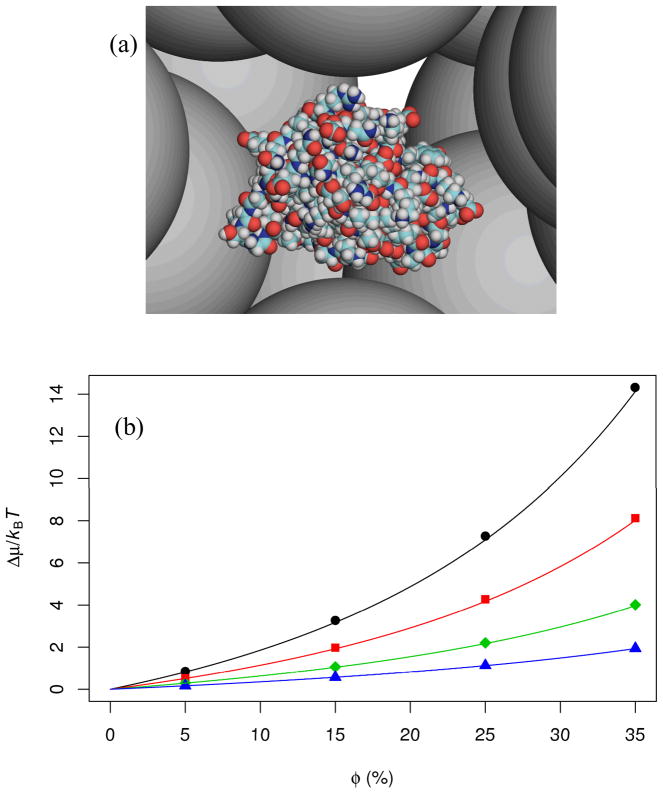
FIG. 1.
(a) All-atom representation of the barnase-barstar complex surrounded by crowders. (b) Results ofΔμ for the protein complex. Symbols are obtained in our previous work [26] by the insertion procedure, with results for crowder radii of 15, 20, 30, and 50 Å shown as circles, squares, diamonds, and triangles, respectively, while curves are predictions of the GFMT. In all GFMT calculations, 10,000 directions were used to define the crowder-exclusion surface; the cubic grid representing the excluded volume had a spacing of 0.5 Å. At each crowder size, the calculation of vp, sp, and lp per protein conformation took ~10 s of CPU time on an AMD Opteron 2382 processor; subsequently using vp, sp, and lp in Eq. (1) to calculate Δμ at different crowder volume fractions required negligible amounts of time. In comparison, the aggregate CPU times for four crowder volume fractions at each crowder size were 80–160 s per snapshot of crowder configurations by the insertion procedure; to reduce statistical errors, the calculations for each protein conformation were repeated on 10–100 snapshots of crowder configurations, bringing the total CPU times per protein conformation at a given crowder size to 800–16000 s. Overall the GFMT gained a speed up of ~102 to 103-fold over the insertion procedure.
Existing codes for generating the molecular surface are either too slow or do not work at all for the range of probe radius of interest here. We therefore devised a simple and fast procedure to generate the crowder-exclusion surface. The basic idea is to place a crowder probe at many positions (totaling N) around the test protein, where the crowder comes into contact with the test protein. These positions are along rays with uniformly separated directions emanating from the geometric center of the test protein. Along each ray the crowder is positioned at the farthest point where it is tangent to at least one protein atom. From this position, the ray is traced backward for a length equal to the crowder radius, arriving at the crowder surface. That final location is taken to be a sample point on the crowder-exclusion surface. The solid angle around each of the N directions is 4π/N. If the radial distance of sample point i is ri, then the surface area, Δsi, accorded to the sample point is . Because the ri and hence Δsi values are not uniform, the sample points are not uniformly distributed on the crowder-exclusion surface. When averaging over the surface, Δsi must be introduced as a weight.
To calculate vp, the volume inside the sphere with radius equal to the largest value of ri is discretized into a cubic grid. A voxel in the grid is eliminated when either it is farther away from the protein center than any part of the crowder probe or it is inside the crowder probe; this test is carried out when the crowder probe is placed at each of the N positions. The remaining voxels make up vp. The calculation of sp follows an algorithm proposed by Windreich et al. [38]. Basically, each voxel comprising vp is assigned a weight according to how its six faces are buried by its neighbors. (Our vp and sp results are virtually identical to those obtained from an existing method [39] but require substantially less CPU times; see supplementary information [37].) Finally lp is calculated as the radius of gyration of sample points on the crowder-exclusion surface:
| (6) |
where rij are the distances between sample points and the weight factors Δsi account for the non-uniform distribution of the sample points.
In our postprocessing approach to crowding simulations [12, 26], the motions of the test protein and those of the crowders are followed in two separate simulations. For each representative conformation of the protein in a given state, Δμ is then calculated by the insertion procedure over different snapshots of crowder configurations. Finally the results for exp(−Δμ/kBT) are averaged over protein conformations and over crowder configurations. With the GFMT, the crowder simulation is no longer needed. Instead, for each protein conformation, we calculate vp, sp, and lp for the given crowder size and then use Eq. (1) to calculate Δμ.
III. APPLICATIONS
We illustrate the predictive power of the GMFT for crowding effects on two central biophysical problems: the folding equilibria of individual proteins and the binding equilibria of proteins pairs. The test systems, cytochrome b562 in the folded and unfolded states and barnase and barstar in the unbound and bound states, are the same as those in our previous work presenting the postprocessing approach [26]; the same protein conformations generated in the absence of crowders are used here (additional information is found in supplementary information [37]). In Table 1 we list the averaged values of vp, sp, and lp (over conformations) for the test systems, calculated for three crowder sizes ranging from 15 to 50 Å. The averaged vp, sp, and lp increase with increasing crowder size, as expected. Note that we do not use these averaged values in Eq. (1) to calculate the final Δμ; rather, we use the vp, sp, and lp values from the different protein conformations to obtain individual results forΔμand then average the results for exp(−Δμ/kBT). The agreement between the GFMT predictions on Δμand those obtained in our previous work [26] by the insertion procedure is illustrated in Fig. 1b.
Table 1.
Average values of vp, sp, and lp for the test systems
| Proteins | vp(Å3)a | sp(Å2)a | lp(Å)a |
|---|---|---|---|
| Folded cytochrome b562 | 21022.4; 22861.1; 24013.8 | 4462.2; 4525.9;4586.0 | 21.1; 21.4; 21.8 |
| Unfolded cytochrome b562 | 25674.3; 29931.6; 32897.8 | 5711.0; 5875.4; 6026.7 | 24.5; 25.4; 25.9 |
| Barnase | 21393.7; 23301.2; 24377.7 | 4326.7; 4409.4; 4473.2 | 19.7; 20.2; 20.5 |
| Barstar | 17132.1; 18203.7; 18809.9 | 3569.2; 3617.4; 3654.4 | 17.6; 17.9; 18.1 |
| Barnase-barstar complex | 39011.1; 42311.1; 44266.2 | 6577.8; 6673.7; 6752.6 | 24.5; 25.2; 25.6 |
Averaged values for vp, sp, and lp are listed for three crowder sizes: 15 Å; 30 Å; and 50 Å.
In vitro experiments [5–9] and MD simulations [23, 25, 26, 29] have found modest enhancements of protein folding stability by crowding; similar effects have been found on the binding stability of small proteins [12]. Figure 2 shows that, when the difference in Δμis taken between the folded and unfolded states of cytochrome b562 or between the bound and unbound states of barnase and barstar, the modest increases in folding stability and binding stability found by the postprocessing approach in our previous work [26] are still predicted well by the GFMT.
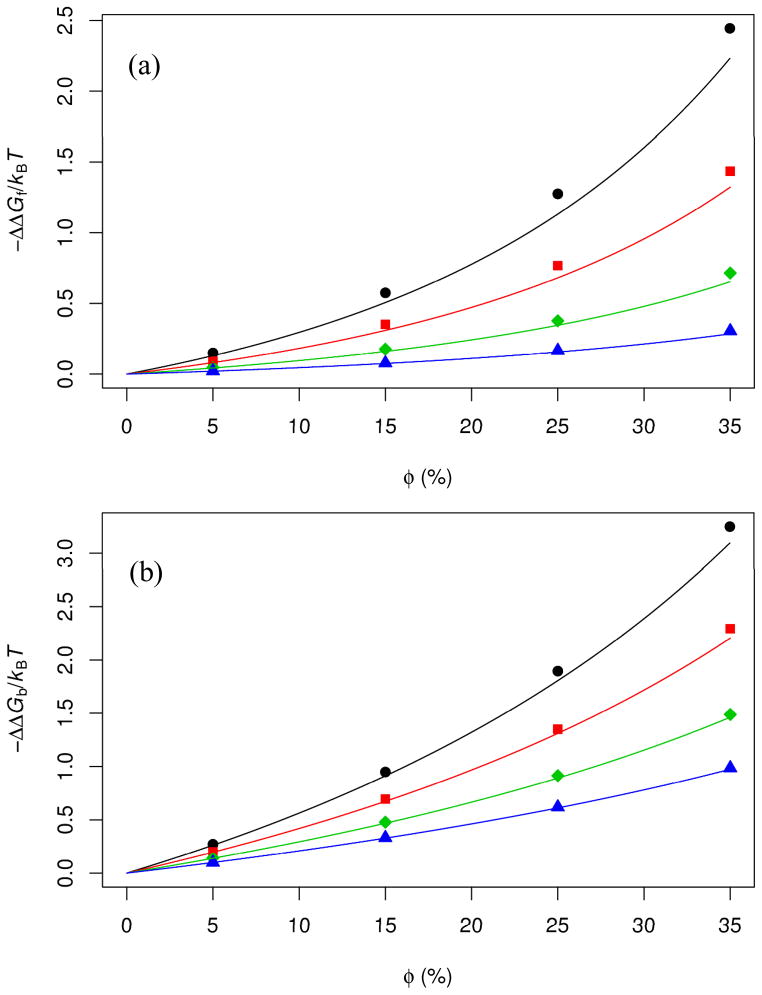
FIG. 2.
Effects of crowding on the (a) folding free energy of cytochrome b562 and (b) binding free energy of barnase and barstar. Symbols are obtained in our previous work [26] by the insertion procedure, while curves are predictions of the GFMT. The different curves represent the different crowder radii listed in Fig. 1b.
The concentrated intracellular milieu is comprised of many species of macromolecules, each at a low concentration. As a step toward realistically mimicking in vivo environments, we have studied in in vitro experiments the effect of a mixture of two crowding agents on folding stability [9]. By using the postprocessing approach for simulations of crowding, we also studied the effect of mixing crowders of two different sizes [26]. As shown by Table 1, the vp, sp, and lp parameters for atomistic proteins depend on crowder size. To signify this dependence, we denote by vpα the volume of the test protein excluded to a species α crowder, and similarly by spα and lpα the crowder-species specific area and linear size. When a mixture of crowders is present, we propose the following mixing rule:
| (7) |
where q = v, s, or l. Figure 3 shows that the resulting predictions for the effects of a mixture of 15-Å and 50- Å crowders on folding stability are in good agreement with results obtained by the postprocessing approach in our previous work [26]. As we emphasized previously [26], the stabilization effect of the mixture is greater than the sum of the two constituents. This non-additive effect of mixed crowding is captured by the GFMT.
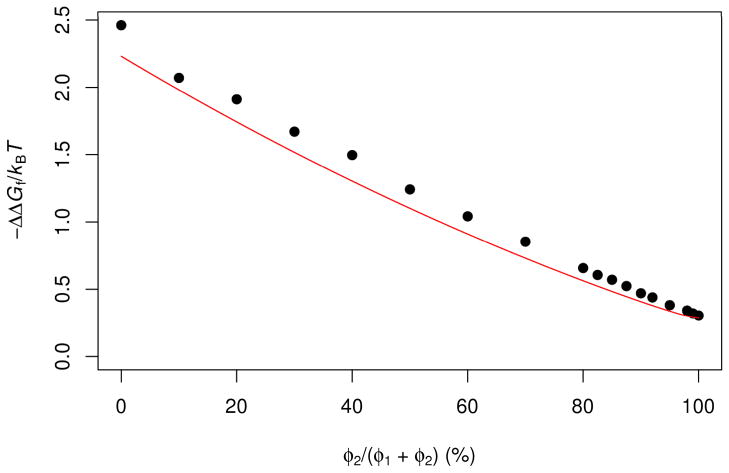
FIG. 3.
Effects of mixed crowding on the folding free energy of cytochrome b562. Two species of crowders, with 15-Å and 50- Å radii, are mixed at different ratios. Symbols are obtained in our previous work [26] by the insertion procedure, while curves are predictions of the GFMT.
We have demonstrated that the GFMT gains significant computational speed over the insertion procedure without loss of accuracy. The speed up may be particularly useful for exhaustive sampling of states (such as the unfolded state of a protein; Qin et al, to be published) comprised of vastly different conformations. More importantly, the GFMT applies equally well to situations where the insertion procedure faces tremendous challenges. Examples of such situations are large protein oligomers and mixtures of many crowder species. A particularly large oligomeric complex is the ribosome, which carries out the essential function of protein synthesis. There is experimental evidence that macromolecular crowding may significantly shift the association/dissociation equilibrium of the large and small subunits of the ribosome [4]; our preliminary calculation with the GFMT indicates that macromolecular crowding may stabilize the ribosome against dissociation by as much as 40 kBT. The GFMT can similarly be used to model at the atomic level protein aggregation and polymerization under crowding; there the effects of crowding have been found to be especially dramatic [3, 15–20]. We will also be able to model mixtures of many crowder species, as illustrated in Fig. 4. Future work will be directed at further development of the GFMT when crowders are also represented at the atomic level.
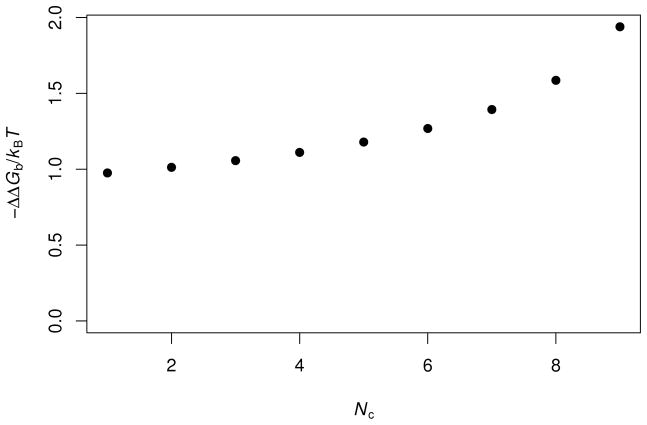
FIG. 4.
GMFT predictions for the effects on the binding free energy of barnase and barstar by increasingly complex mixtures of crowders. The total volume fraction of crowders is fixed at 35%. Nc denotes the number of crowder species. At Nc = 1, all crowders have a 50-Å radius. At Nc = 2, crowders with 50- Å and 45- Å radii are mixed. Each successive increase in Nc introduces a new species that is 5 Å smaller than the previous smallest crowders. At Nc = 9, the crowder radii range from 10 to 50 Å. At each Nc, all the species have the same volume fraction.
Supplementary Material
Acknowledgments
This work was supported in part by Grant GM058187 from the National Institutes of Health.
References
- 1.Zimmerman SB, Trach SO. J Mol Biol. 1991;222:599. doi: 10.1016/0022-2836(91)90499-v. [DOI] [PubMed] [Google Scholar]
- 2.Zhou H-X, Rivas G, Minton AP. Annu Rev Biophys. 2008;37:375. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuller RS, Kaguni JM, Kornberg A. Proc Natl Acad Sci U S A. 1981;78:7370. doi: 10.1073/pnas.78.12.7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zimmerman SB, Trach SO. Nucleic Acids Research. 1988;16:6309. doi: 10.1093/nar/16.14.6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qu Y, Bolen DW. Biophys Chem. 2002;101–102:155. doi: 10.1016/s0301-4622(02)00148-5. [DOI] [PubMed] [Google Scholar]
- 6.Spencer DS, et al. J Mol Biol. 2005;351:219. doi: 10.1016/j.jmb.2005.05.029. [DOI] [PubMed] [Google Scholar]
- 7.Ai X, et al. J Am Chem Soc. 2006;128:3916. doi: 10.1021/ja057832n. [DOI] [PubMed] [Google Scholar]
- 8.Roberts A, Jackson SE. Biophys Chem. 2007;128:140. doi: 10.1016/j.bpc.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 9.Batra J, Xu K, Zhou HX. Proteins. 2009;77:133. doi: 10.1002/prot.22425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan J-M, et al. Protein Sci. 2008;17:2156. doi: 10.1110/ps.037325.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Homouz D, et al. Proc Natl Acad Sci U S A. 2008;105:11754. doi: 10.1073/pnas.0803672105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Batra J, et al. Biophys J. 2009;97:906. doi: 10.1016/j.bpj.2009.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drenckhahn D, Pollard TD. J Biol Chem. 1986;261:12754. [PubMed] [Google Scholar]
- 14.Kuttner YY, et al. J Am Chem Soc. 2005;127:15138. doi: 10.1021/ja053681c. [DOI] [PubMed] [Google Scholar]
- 15.Shtilerman MD, Ding TT, Lansbury PT., Jr Biochemistry. 2002;41:3855. doi: 10.1021/bi0120906. [DOI] [PubMed] [Google Scholar]
- 16.Uversky VN, et al. FEBS Lett. 2002;515:99. doi: 10.1016/s0014-5793(02)02446-8. [DOI] [PubMed] [Google Scholar]
- 17.Hatters DM, Minton AP, Howlett GJ. Journal of Biological Chemistry. 2002;277:7824. doi: 10.1074/jbc.M110429200. [DOI] [PubMed] [Google Scholar]
- 18.del Alamo M, Rivas G, Mateu MG. J Virol. 2005;79:14271. doi: 10.1128/JVI.79.22.14271-14281.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rotter M, et al. J Mol Biol. 2005;347:1015. doi: 10.1016/j.jmb.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 20.Snoussi K, Halle B. Biophysical Journal. 2005;88:2855. doi: 10.1529/biophysj.104.055871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lebowitz JL, Helfand E, Praestgaard E. J Chem Phys. 1965;43:774. [Google Scholar]
- 22.Minton AP. Methods Enzymol. 1998;295:127. doi: 10.1016/s0076-6879(98)95038-8. [DOI] [PubMed] [Google Scholar]
- 23.Cheung MS, Klimov D, Thirumalai D. Proc Natl Acad Sci U S A. 2005;102:4753. doi: 10.1073/pnas.0409630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minh DD, et al. J Am Chem Soc. 2006;128:6006. doi: 10.1021/ja060483s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stagg L, et al. Proc Natl Acad Sci U S A. 2007;104:18976. doi: 10.1073/pnas.0705127104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin S, Zhou H-X. Biophys J. 2009;97:12. doi: 10.1016/j.bpj.2009.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pincus DL, Thirumalai D. J Phys Chem B. 2009;113:359. doi: 10.1021/jp807755b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin S, et al. J Phys Chem Lett. 2010;1:107. doi: 10.1021/jz900023w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mittal J, Best RB. Biophys J. 2010;98:315. doi: 10.1016/j.bpj.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tjong H, Zhou H-X. Biophys J. 2010 in press. [Google Scholar]
- 31.Widom B. J Chem Phys. 1963;39:2802. [Google Scholar]
- 32.Rosenfeld Y. Phys Rev Lett. 1989;63:980. doi: 10.1103/PhysRevLett.63.980. [DOI] [PubMed] [Google Scholar]
- 33.Oversteegen SM, Roth R. J Chem Phys. 2005;122:214502. doi: 10.1063/1.1908765. [DOI] [PubMed] [Google Scholar]
- 34.Wu J, Li Z. Annu Rev Phys Chem. 2007;58:85. doi: 10.1146/annurev.physchem.58.032806.104650. [DOI] [PubMed] [Google Scholar]
- 35.Richards FM. Annu Rev Biophys Bioeng. 1977;6:151. doi: 10.1146/annurev.bb.06.060177.001055. [DOI] [PubMed] [Google Scholar]
- 36.Roth R, Harano Y, Kinoshita M. Phys Rev Lett. 2006;97:078101. doi: 10.1103/PhysRevLett.97.078101. [DOI] [PubMed] [Google Scholar]
- 37.See EPAPS Document No.xxx for further details on the calculation of the geometric parameters vp, sp, and rg and on the proteins studied here. For more information on EPAPS, see http://www.aip.org/pubservs/epaps.html
- 38.Windreich G, Kiryati N, Lohmann G. Pattern Recog. 2003;36:2531. [Google Scholar]
- 39.Voss NR, et al. J Mol Biol. 2006;360:893. doi: 10.1016/j.jmb.2006.05.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.