Abstract
The field of phylogeography has received a lot of attention for its application to molecular evolution and geographic migration of species. More recent work has included infectious diseases especially zoonotic RNA viruses like influenza and rabies. Phylogeography of viruses has the potential to advance surveillance at agencies such as public health departments, agriculture departments, and wildlife agencies. However, little is known about how these agencies could use phylogeography for applied surveillance and the integration of animal and human sequence data. Here, we highlight its potential to support ‘translational public health’ that could bring sequence data to the forefront of surveillance. We focus on swine influenza H3N2 because of the recent link to a variant form in humans. We discuss the implications to applied surveillance and the need for an integrated biomedical informatics approach for adoption at agencies of animal and public health.
Keywords: Molecular Sequence Data, Phylogeography, Epidemiology, Zoonoses, Influenza A Virus, H3N2 Subtype
1. Introduction
There is a tremendous amount of sequence data being generated by bench science and curated in electronic databases (Sarkar, 2007), however little effort has been done to create informatics systems to integrate this data into public health or animal agencies (Agriculture, Wildlife, etc.) for surveillance. This is likely due to a combination of factors including a lack of informatics skills at these agencies, and a lack of perceived benefit for combining this data for practical purposes. The use of sequence data can support ‘translational public health’ ((Mirhaji, 2009), p. 159), in which data generated from bench science research can help inform public health decision making (Mirhaji, 2009).
Phylogeography is a field that can address translational public health. This work focuses on the geographical lineages of species such as vertebrates or viruses (Avise, 2000) and uses sequence data along with geographical information as the foundations of this science. There has been a growing interest in phylogeography of zoonotic RNA viruses (Holmes, 2004; Lemey et al., 2009; Wallace and Fitch, 2008) because of their often shorter genomes and rapid mutations (Holmes, 2004). However, this domain has rarely been integrated as a trusted resource at health agencies to support zoonotic surveillance. For example, in considering infectious zoonotic diseases, questions related to disease migration over geographic areas, virus population growth, and risk of genetic shift to human-adapted strains, cannot adequately be addressed using only summarized state reportable disease data. For infectious diseases, answering these questions can enhance surveillance because it goes beyond simple numerical summarization and examines virus migration patterns and the relationship between geography, population, and health (infection).
Prior phylogeography research of zoonotic viruses demonstrates its potential for population health surveillance. For example, Wallace and Fitch (Wallace and Fitch, 2008) studied influenza H5N1 in various animal hosts. The authors examined migration over Europe, Asia, and Africa using genomic sequence data and phylogeography. They found that many of the H5N1 strains originated in southern China and spread to Indonesia, Japan, Thailand, and Vietnam, likely as the result of commercial trade (Wallace and Fitch, 2008). The analysis also demonstrated certain areas were successful at filtering out new strains of H5N1, while still enduring outbreaks of older strains of the disease (Wallace and Fitch, 2008). This type of analysis of can enhance surveillance of infectious agents and population health control measures (Wallace and Fitch, 2008). The information gained from their work likely would not have occurred if the authors used only reportable disease data.
In another study, Biek et al. (Biek et al., 2007) examined the phylogeography of rabies virus in raccoons in the Eastern United States and linked an initial outbreak in the 1970’s to an expansion resulting in the current epizootic condition in the mid-Atlantic geographic area. As part of their analysis, the authors layered their phylogeographic tree onto a geographic map in order to visualize the dispersion of the virus across these states (Biek et al., 2007).
While the literature highlights the potential for phylogeography to support surveillance of zoonotic disease, its use at the state agency level is rare. Its absence is related to both the lack of understanding of the potential to support surveillance and the need for public health informatics systems to integrate sequence data with traditional reportable disease data. To our knowledge, there is currently no translational public health informatics system at a health department or agriculture department. One application with potential is the SUPRAmap project by Janies et al. (Janies et al., 2010) which is a web application that allows the user to combine genomic, evolutionary, geospatial and temporal data for biogeography. The application can take raw sequence data, aligned sequences, complete trees, geographical data, and data files that describe the variables (leaf nodes) (Janies et al., 2010). It can be used as a webservice, or downloaded as a desktop application. Also by Janies et al. is Routemap (Janies, 2012), developed at Ohio State University. The system produces phylogeographical models by allowing users to submit sequences through the website and receive a geographically annotated (Keyhole Markup Language) file for viewing in Google Earth. Finally, Driscoll et al. developed Disease View, a database of host-pathogen interactions that also includes HealthMap (Brownstein and Freifeld, 2007) for geospatial mapping of cases from news sources (Driscoll et al., 2011).
In this study, we will highlight the potential for phylogeography as a translational science for state public health decision-making. Here, our focus is on state-level surveillance rather than global or national. We use swine influenza A H3N2 as a case study because of its zoonotic potential and the recent emergence of pandemic 2009 H1N1 (2009pH1N1) into specific segments of its genome (2012b). From 2010 to 2012, the United States Department of Agriculture through their Swine Influenza Virus Surveillance Program has reported that 211 positive cases of swine H3N2 have been identified from October 2010 to July 12 with 41% of these recent cases identified as having lineage from 2009pH1N1 (USDA, 2012). The discovery of human cases of influenza variant H3N2v in Maine, Indiana, and Pennsylvania (Canfield, 2011) demonstrate the potential for 2009pH1N1 to mix with other subtypes of the influenza A virus. Currently, 224 human cases have been reported since July 2012 alone with direct contact to swine as the most likely source (often from fairs) (CDC, 2012). In a July 2012 outbreak at a fair in Indiana, four individuals were infected with H3N2v and subsequent testing of 12 swine revealed all were positive for H3N2 (Blanton, 2012). Thus, understanding the genomic history of this subtype and the relationship between geographic models of dispersion and new cases of disease could be an important asset to helping population level surveillance of among animals and humans.
2. Material and methods
In phylogeography, the two influenza surface proteins, hemagglutinin (HA) and neuraminidase (NA) are often studied because of their rapid transformations over a short time period. Thus, for this analysis, we focused on those two genes. We searched the National Institute of Allergy and Infectious Disease’s (NIAID) influenza research database (IRD) (NIAID, 2011) a public sequence database of influenza viruses A, B, and C. We used the terms: influenza type = A, strain = H3N2, gene segment = HA, host = swine, time period = 2009–2012, country = USA, and No Duplicates. This resulted in 181 HA sequences. In order to get detailed geographic information, we wrote a Java script that used the NCBI E-utilities web service (Sayers, 2008) to extract geographic information from the corresponding GenBank record of each of the sequences. None of the records had information beyond a state name such as a county or town. Thus, we used the geographical information in the strain name (such as A/swine/NY/A01104005/2011(H3N2)) to identify the location.
For the neuraminidase (NA) data set, the same search strategy was used except the gene was changed to NA. This resulted in 156 sequences.
Both sets of sequences were preprocessed then submitted as FASTA files to ZooPhy (Scotch et al., 2010), a bioinformatics framework for zoonotic phylogeography developed at Arizona State University (ASU). ZooPhy integrates separate bioinformatics software into a single framework including: ClustalW (Higgins and Sharp, 1988; Thompson et al., 1994) for sequence alignment, jModeltest (Posada, 2009; Posada, 2011) for analysis of substitution models, and BEAST (2011; Drummond and Rambaut, 2007) for molecular evolution and phylogeography using a Bayesian approach.
The result of a ZooPhy run is a single maximum clade credibility (MCC) tree and its corresponding parameters that can be used to infer spatial dispersion of a genetic lineage (in this case, the lineage of a virus). The MCC can be thought of as the ‘best’ representation of genetic characterization. For ZooPhy, the default length of the Bayesian run is 10,000,000 steps. We analyzed the log statistics after completion and decided to increase the length to 50,000,000 in order to increase quality of the model parameters. The final MCC tree for both HA and NA data sets generated 45,000 trees. This was done outside of ZooPhy using the Saguaro high-performance computer (ASU, 2012) at ASU as well as TreeAnnotator (2012a) (part of the BEAST package) by specifying a 10% burn-in and 0.65 posterior probability threshold.
2.1 Statistical Phylogeography
For our approach, we modeled the work of Lemey et al. (Lemey et al., 2009) who studied the phylogeography of avian influenza H5N1 on a pandemic scale. In addition to generating sets of trees, we also provided statistics to explain the model and the diffusion process of the virus. For example, we estimated the number of non-zero rates of diffusion between locations (states) in the model. We determined the routes (e.g. A→B) that had the strongest support by employing the Bayes Factor test (Lemey et al., 2009). The Bayes Factor was calculated for both HA and NA using a separate software application SPREAD (Bielejec et al., 2011) which also generates a keyhole markup language (KML) file for viewing the geographic migration in Google Earth.
We also used two additional statistical phylogeography approaches including the Association Index (AI) and the Kullback-Leibler (KL). The AI tests the hypothesis that taxons (i.e. tips in a tree) with a given trait (e.g. location A) are no more likely to share that trait with adjoining taxa than with other taxa in the set (Parker; Parker et al., 2008). We used the program Bayesian Tip-Significance testing (BaTS) (Parker, 2008) to calculate the AI. The other statistic, the KL, measures divergence between the root state prior and posterior probability for each MCC tree. For phylogeography, the root is equivalent to the origin in the diffusion process. Thus the KL will measure the statistical power that location A is the origin of the H3N2 spread and thus is an estimation of how well the model uses the data to explain the likely root location (Lemey et al., 2009). For the Kullback-Leibler, we used the program Matlab version 2011a (MathWorks, 2012) and a program written by Razavi (Razavi, 2012).
3. Results
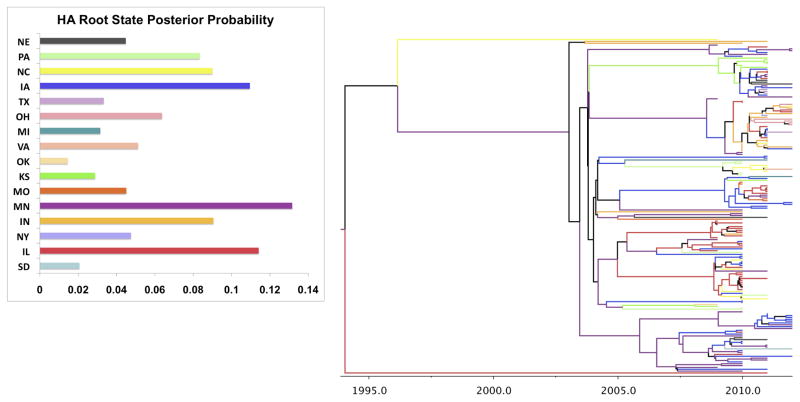
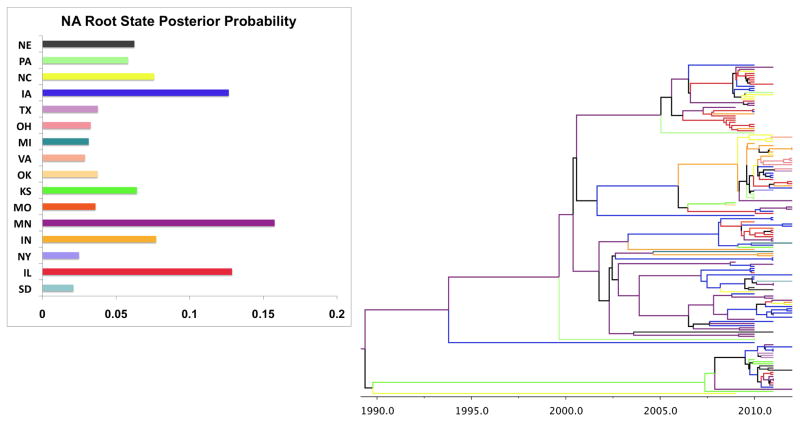
The HA root state posterior probability and the phylogeographic MCC tree is shown in Figure 1, while the NA results are provided in Figure 2. Both genes are in agreement that Minnesota is the origin of the swine influenza H3N2 migration. The roots are close in age with the NA root emerging slightly later (deeper) in time than the HA (1989 vs. 1994). Thus while the earliest observed cases in our dataset were from 2009, the model estimates that the age of the initial divergence of swine H3N2 strains occurs much earlier. In addition, both models suggest that Minnesota dominates the branches indicating it as key site for diffusion to the other states. Thus while the models suggest it as the likely origin, it also continues to serve as a site for dispersion well after the initial divergence (in 1989 or 1994). Both trees also highlight Iowa as having a large role in H3N2 dispersion. This state contains several branches (blue) along the interior of both trees, potentially highlighting it as a ‘secondary epicenter’ in the virus’s spread.
Figure 1. HA Root state posterior probability and maximum clade credibility (MCC) phylogeographic tree.
Minnesota has the highest probability (0.1316) of being the origin (root) of the swine H3N2 evolution.
Figure 2. NA Root state posterior probability and maximum clade credibility (MCC) phylogeographic tree.
Like HA, Minnesota has the highest probability (0.1576) of being the origin (root) of the swine H3N2 evolution.
Table 1 shows additional statistical phylogeography metrics including the Association Index (AI) and the Kullback-Leibler (KL). Here, the observed AI values and the corresponding 95% Highest Probability Density (HPD) are also extremely high (> 1). This suggests that there is a phylogeographic relationship in the lineage of the two H3N2 genes and that geography played a role in the transmission of the virus. These produced statistically significant p-values (both trees < 0.01). The KL on the other hand estimates divergence of prior and posterior probabilities of the root state and were fairly low for both trees (0.18 vs. 0.19). Here, we used a fixed prior for each tree (1/K, where K is the number of unique geographic states) and the posterior estimates reported in Figures 1 and 2. The small numbers indicate the phylogeographic models are able to generate root state posteriors that are not very different from the underlying priors and thus achieve a moderately low statistical power (Lemey et al., 2009). Thus while both trees indicate that Minnesota is the origin, the statistical power of this certainty is low and should be carefully considered.
Table 1.
Kullback-Leibler and Association Index statistics for the two phylogeography models.
Statistically significant (p-value <0.05).
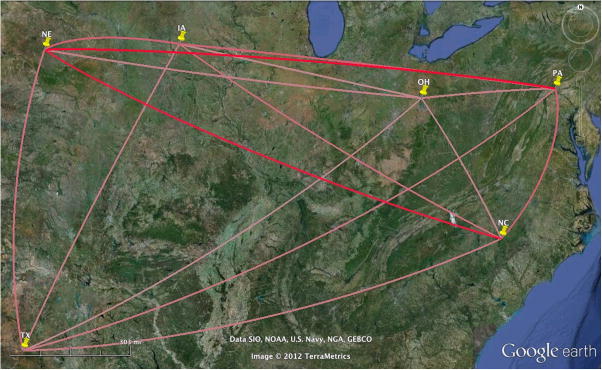
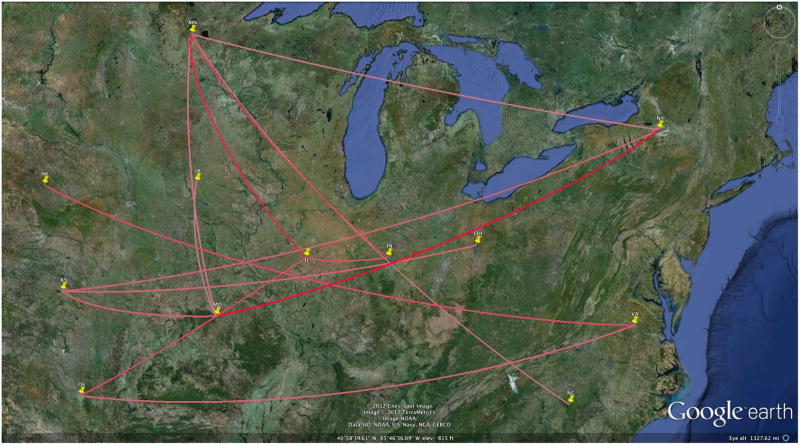
The most significant rates are shown in Figure 3 and 4 through the Bayes factor (BF) test using a cutoff of 3. The darker color indicates stronger support. For the HA map, PA → NE (East to West) pathway has the highest support with an extremely large Bayes Factor of 23,001. The Bayes Factor analysis for the NA sequences identified New York → Missouri as the highest. Thus the maps indicate similarity in relation to paths moving from East to West especially amongst the midwestern states. While a BF of 3 represents a cutoff, there is a disparity between the set of remaining significant diffusion rates. As explained in Lemey (Lemey et al., 2009), the Bayes Factor (BF) values represent the difference between the posterior and prior probabilities that the rates between two locations (A → B) are non-zero. Thus routes with extremely high BF likely have a large difference between the prior and posterior odds that a migration exists from point A to point B.
Figure 3. Significant dispersion routes for the HA tree using the Bayes factor (BF) test for significant non-zero rates.
A cutoff of BF = 3 shows fifteen routes with the darker the color indicating the higher BF (pink to red). The Pennsylvania → Nebraska route has the strongest support (BF=23,001).
Figure 4. Significant dispersion routes for the NA tree.
A cutoff of BF = 3 shows fourteen routes with the darker the color indicating the higher BF (pink to red). Fourteen routes are shown and like the HA tree, the New York -→ Missouri route has the strongest support (BF= 118).
4. Discussion
The phylogeographic models presented here highlight the potential of this science to support surveillance at agriculture, wildlife, and public health agencies. Here, we focused on swine influenza H3N2, however many different infectious diseases, especially RNA viruses, are suitable for molecular evolution analysis over a relatively short time period (although bacterial agents such as Borrelia burgdorferi have also been studied (Hoen et al., 2009). For example, recent efforts have been published that analyzed the geographic migration of H5N1 among avian and human populations over only a few years of data (Fusaro et al., 2010; Haase et al., 2010; Lam et al., 2008). In addition, phylogeography has been applied to other viruses beyond influenza such as rabies (Biek et al., 2007), West Nile Virus (Zehender et al., 2011), and Hantavirus (Lam et al., 2008).
4.1 Application to Population Health Agencies
The application of phylogeography as a translational tool for public health decision-making is not well understood. Like translational medicine that uses data from the bench to bedside for clinical care, the same can be true for population health. Here, genetic sequence data of viruses can be utilized for surveillance of infectious diseases using disciplines such as phylogeography. In addition, the combination of traditional public health data (i.e. counts of observed cases) with evolutionary models offers the potential to enhance surveillance even further. By determining the origin of the outbreak (e.g. Minnesota) and the temporal and spatial migration, epidemiologists can be more informed about public health interventions. For example, interventions to block the virus at the source can limit the exposure to other geographical areas (2007). In addition, if virus migration routes are predicted, there is a better chance at isolating the strain that can then be used to make a vaccine (2007). In our example, the prediction of migration routes (East to West) as well as secondary epicenters can enable animal health agencies to monitor feral swine as well as transportation of domestic swine across state boundaries.
4.2 Biomedical Informatics for Translational Public Health
Work needs to be done to understand the current use and need of bioinformatics resources such as GenBank and molecular evolution software at state-level health agencies. We hypothesize that utilization is low largely because of the difficulties in designing a usable system that is translatable to an audience that is: 1) not experienced in bioinformatics and dealing with large sequence databases, and 2: must use the knowledge about the past to make inferences about current and future population risk. Thus, since phylogeography makes inferences about the past, additional knowledge must be embedded into a system that promotes an understanding of the implications seen in the maps of dispersion routes and how they relate to current and future population needs. Thus, systematic usability studies are an essential component to the successful development of phylogeography systems. In addition, dispersion models must be combined with traditional public health data (i.e. counts and rates) to relate these estimates to observed cases. Without these steps, it is unlikely that translational public health systems will be adopted by agencies of human and animal health.
The authors recognize several limitations with this work including the use of state-level geography to infer geographic dispersion. We utilized the centroid latitude and longitude for each state and this likely does not reflect the true location of each swine host that was represented in the sequences. Our previous work highlights the lack of sufficient geographical metadata in GenBank and the need for biomedical informatics approaches to enhance the quality of this key element of phylogeography (Scotch et al., 2011). This is especially important if a state (likely a large area such as Texas, Alaska, or California) needs to focus on internal spread from one part of the state to another. This would be nearly impossible to do with only state-level information about the location of the host. For our example, we feel that the model is still a reasonable estimation of interstate dispersion of the virus.
Another limitation is that we did not compare other approaches of molecular evolution including maximum likelihood and maximum parsimony. Our purely Bayesian discrete model has limitations including inferring the migration paths by only considering the observed locations (of the swine). For example, if a state had only one strain in a data set, removing that one strain would eliminate it completely from the model. Thus it would not be considered in the dispersion history. A different approach based on a continuous model attempts to impute states in the migration that are not observed (Lemey et al., 2010). This work could be valuable for zoonotic disease surveillance in the absence of known geospatial metadata.
5. Conclusion
The purpose of this study was to highlight the potential for phylogeography as a science for enhancing zoonotic disease surveillance. This work can support translational public health by brining sequence data from databases to the forefront of public health decision-making. At Arizona State University, we are developing ZooPhy, a platform to streamline phylogeographic analysis of zoonotic diseases with the intention of bringing it to health agencies. In this paper, we utilize ZooPhy along with additional software to demonstrate how the molecular evolution and phylogeography of swine influenza H3N2 has spread within the United States.
More work is needed to explore the needs and current use of bioinformatics resources at state agencies of health and the careful design of this framework in order to support decision making about current and future animal and human population risk.
Supplementary Material
Highlights
Phylogeography can be used for studying the spread of zoonotic viruses.
We focus on swine influenza H3N2 because of the recent variant form in humans.
We used Bayesian phylogeographic models and statistical phylogeography.
Minnesota was the origin and the Iowa was a secondary center for spread.
Sequence data generated from bench research can support zoonotic surveillance.
Acknowledgments
This research was supported by National Institutes of Health/National Library of Medicine (NLM) grant R00LM009825 (to MS).
Abbreviations
- ASU
Arizona State University
- IRD
Influenza Research Database
- KML
keyhole markup language
- HPD
highest probability density
- AI
Association Index
- KL
Kullback-Leibler
- MCC
maximum clade credibility
- NIAID
National Institute of Allergy and Infectious Diseases
- HA
Hemagglutinin
- NA
Neuraminidase
- BF
Bayes Factor
- IA
Iowa
- IL
Illinois
- IN
Indiana
- KS
Kansas
- MI
Michigan
- MN
Minnesota
- MO
Missouri
- NE
Nebraska
- NY
New York
- NC
North Carolina
- OH
Ohio
- OK
Oklahoma
- PA
Pennsylvania
- SD
South Dakota
- TX
Texas
- VA
Virginia
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Phylogeography Tracks Bird Flu’s Long March. Scitizen.com (Ed.); [Google Scholar]
- 2.BEAST [Google Scholar]
- 3.TreeAnnotator. 2012a. [Google Scholar]
- 4.Update: Influenza A (H3N2)v Transmission and Guidelines — Five States, 2011. Morbidity and Mortality Weekly Report (MMWR) 2012b;60:1741–1744. [PubMed] [Google Scholar]
- 5.ASU. Saguaro. 2012. [Google Scholar]
- 6.Avise JC. Phylogeography: the history and formation of species. Harvard University Press; Cambridge, Mass: 2000. [Google Scholar]
- 7.Biek R, Henderson JC, Waller LA, Rupprecht CE, Real LA. A high-resolution genetic signature of demographic and spatial expansion in epizootic rabies virus. Proc Natl Acad Sci U S A. 2007;104:7993–7998. doi: 10.1073/pnas.0700741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bielejec F, Rambaut A, Suchard MA, Lemey P. SPREAD: spatial phylogenetic reconstruction of evolutionary dynamics. Bioinformatics. 2011;27:2910–2912. doi: 10.1093/bioinformatics/btr481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanton L. Notes from the Field: Outbreak of Influenza A (H3N2) Virus Among Persons and Swine at a County Fair — Indiana, July 2012. MMWR. 2012;61:561. [PubMed] [Google Scholar]
- 10.Brownstein JS, Freifeld CC. HealthMap: the development of automated real-time internet surveillance for epidemic intelligence. Euro Surveill. 2007;12 doi: 10.2807/esw.12.48.03322-en. E071129 071125. [DOI] [PubMed] [Google Scholar]
- 11.Canfield C. New swine flu virus sickens 5 children in 3 states. Associated Press; 2011. [Google Scholar]
- 12.CDC. Still Linked to Pig Exposure. 2012. More H3N2v Cases Reported. [Google Scholar]
- 13.Driscoll T, Gabbard JL, Mao C, Dalay O, Shukla M, Freifeld CC, Hoen AG, Brownstein JS, Sobral BW. Integration and visualization of host-pathogen data related to infectious diseases. Bioinformatics. 2011;27:2279–2287. doi: 10.1093/bioinformatics/btr391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fusaro A, Nelson MI, Joannis T, Bertolotti L, Monne I, Salviato A, Olaleye O, Shittu I, Sulaiman L, Lombin LH, Capua I, Holmes EC, Cattoli G. Evolutionary dynamics of multiple sublineages of H5N1 influenza viruses in Nigeria from 2006 to 2008. J Virol. 2010;84:3239–3247. doi: 10.1128/JVI.02385-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haase M, Starick E, Fereidouni S, Strebelow G, Grund C, Seeland A, Scheuner C, Cieslik D, Smietanka K, Minta Z, Zorman-Rojs O, Mojzis M, Goletic T, Jestin V, Schulenburg B, Pybus O, Mettenleiter T, Beer M, Harder T. Possible sources and spreading routes of highly pathogenic avian influenza virus subtype H5N1 infections in poultry and wild birds in Central Europe in 2007 inferred through likelihood analyses. Infect Genet Evol. 2010;10:1075–1084. doi: 10.1016/j.meegid.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 17.Higgins DG, Sharp PM. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 18.Hoen AG, Margos G, Bent SJ, Diuk-Wasser MA, Barbour A, Kurtenbach K, Fish D. Phylogeography of Borrelia burgdorferi in the eastern United States reflects multiple independent Lyme disease emergence events. Proc Natl Acad Sci U S A. 2009;106:15013–15018. doi: 10.1073/pnas.0903810106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holmes EC. The phylogeography of human viruses. Mol Ecol. 2004;13:745–756. doi: 10.1046/j.1365-294x.2003.02051.x. [DOI] [PubMed] [Google Scholar]
- 20.Janies DA. Routemap. 2012. [Google Scholar]
- 21.Janies DA, Voronkin IO, Das M, Hardman J, Treseder TW, Studer J. Genome informatics of influenza A: from data sharing to shared analytical capabilities. Anim Health Res Rev. 2010;11:73–79. doi: 10.1017/S1466252310000083. [DOI] [PubMed] [Google Scholar]
- 22.Lam TT, Hon CC, Pybus OG, Kosakovsky Pond SL, Wong RT, Yip CW, Zeng F, Leung FC. Evolutionary and transmission dynamics of reassortant H5N1 influenza virus in Indonesia. PLoS Pathog. 2008;4:e1000130. doi: 10.1371/journal.ppat.1000130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemey P, Rambaut A, Drummond AJ, Suchard MA. Bayesian phylogeography finds its roots. PLoS Comput Biol. 2009;5:e1000520. doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemey P, Rambaut A, Welch JJ, Suchard MA. Phylogeography takes a relaxed random walk in continuous space and time. Mol Biol Evol. 2010;27:1877–1885. doi: 10.1093/molbev/msq067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MathWorks. Matlab v. 2011a. 2012. [Google Scholar]
- 26.Mirhaji P. Public Health Surveillance Meets Translational Informatics: A Desiderata. Journal of the Association for Laboratory Automation. 2009;14:157–170. [Google Scholar]
- 27.NIAID. Influenza Research Database. 2011. [Google Scholar]
- 28.Parker J. BaTS Manual. [Google Scholar]
- 29.Parker J. Bayesian Tip-Significance testing (BaTS) 2008. [Google Scholar]
- 30.Parker J, Rambaut A, Pybus OG. Correlating viral phenotypes with phylogeny: accounting for phylogenetic uncertainty. Infect Genet Evol. 2008;8:239–246. doi: 10.1016/j.meegid.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 31.Posada D. Selection of models of DNA evolution with jModelTest. Methods Mol Biol. 2009;537:93–112. doi: 10.1007/978-1-59745-251-9_5. [DOI] [PubMed] [Google Scholar]
- 32.Posada D. jModeltest: phylogenetic model averaging. 2011. [DOI] [PubMed] [Google Scholar]
- 33.Razavi N. Kullback-Leibler Divergence. 2012. [Google Scholar]
- 34.Sarkar IN. Biodiversity informatics: organizing and linking information across the spectrum of life. Brief Bioinform. 2007;8:347–357. doi: 10.1093/bib/bbm037. [DOI] [PubMed] [Google Scholar]
- 35.Sayers E. E-utilities Quick Start, Entrez Programming Utilities Help [Internet] National Center for Biotechnology Information; Bethesda, MD: 2008. [Google Scholar]
- 36.Scotch M, Mei C, Brandt C, Sarkar IN, Cheung K. At the intersection of public-health informatics and bioinformatics: using advanced Web technologies for phylogeography. Epidemiology. 2010;21:764–768. doi: 10.1097/EDE.0b013e3181f534dd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scotch M, Sarkar IN, Mei C, Leaman R, Cheung KH, Ortiz P, Singraur A, Gonzalez G. Enhancing phylogeography by improving geographical information from GenBank. J Biomed Inform. 2011;44(Suppl 1):S44–47. doi: 10.1016/j.jbi.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.USDA. Swine Influenza Surveillance: USDA Swine Influenza Surveillance Update. 2012. [Google Scholar]
- 40.Wallace RG, Fitch WM. Influenza A H5N1 immigration is filtered out at some international borders. PLoS One. 2008;3:e1697. doi: 10.1371/journal.pone.0001697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zehender G, Ebranati E, Bernini F, Lo Presti A, Rezza G, Delogu M, Galli M, Ciccozzi M. Phylogeography and epidemiological history of West Nile virus genotype 1a in Europe and the Mediterranean basin. Infect Genet Evol. 2011;11:646–653. doi: 10.1016/j.meegid.2011.02.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.