SUMMARY
Cell growth and differentiation are controlled by growth factor receptors coupled to the GTPase Ras. Oncogenic mutations disrupt GTPase activity leading to persistent Ras signaling and cancer progression. Recent evidence indicates that monoubiquitination of Ras leads to Ras activation. Mutation of the primary site of monoubiquitination impairs the ability of activated K–Ras to promote tumor growth. To determine the mechanism of human Ras activation we chemically ubiquitinated the protein and analyzed its function by NMR, computational modeling, and biochemical activity measurements. We established that monoubiquitination has little effect on Ras GTP binding, GTP hydrolysis, or exchange factor activation, but severely abrogates the response to GTPase activating proteins in a site–specific manner. These findings reveal a new mechanism by which Ras can trigger persistent signaling in the absence of receptor activation or an oncogenic mutation.
INTRODUCTION
Ras plays a central role in cell growth, differentiation, and apoptosis and is a member of a large superfamily of guanine nucleotide binding proteins whose activity is regulated by cycling between inactive GDP–bound and active GTP–bound states1. Conformational changes associated with the GDP– and GTP–bound states are localized primarily to two regions, Switch I (residues 30–37) and Switch II (60–76), and these conformational changes direct specific interactions with regulators and effectors2,3. Ras effectors recognize the GTP–bound state of Ras with higher affinity than the GDP–bound state, and these effectors serve to initiate downstream signaling events. Ras has weak intrinsic GTPase activity, but it does not act alone4. The guanine nucleotide state of Ras is regulated by two distinct types of protein modulators, which act in opposition to one another. Guanine nucleotide exchange factors (GEFs) facilitate exchange of GDP with GTP to promote Ras activation5 whereas GTPase–activating proteins (GAPs) stimulate the hydrolysis of GTP and Ras deactivation6. Ras is the most prevalent oncogene found in human cancer; about 30% of human tumors contain an activating Ras mutation7,8. Most commonly, transforming Ras mutations decrease the sensitivity of the protein to GAP–mediated regulation9.
While the roles of GEFs and GAPs have been extensively characterized, it is less clear how some post–translational modifications, like monoubiquitination, contribute to Ras function and signaling. Monoubiquitination is a dynamic and reversible modification that can orchestrate cellular events including DNA repair, gene expression, endocytosis, and nuclear export10. Emerging evidence suggests that monoubiquitination regulates large and small GTPases, including Ras11–14. Monoubiquitination of K–Ras at position 147 has been shown to promote tumorigenesis15; mutation of oncogenic K–Ras to prevent monoubiquitination (RasK147L) impaired its ability to promote tumor growth when ectopically expressed in NIH 3T3 mouse fibroblasts. These findings suggest that Ras activity and signaling are modulated by monoubiquitination, in the manner of an oncogenic mutation or receptor stimulus. Left unresolved is the mechanism by which monoubiquitination leads to activation of Ras.
Here, we set out to identify the molecular mechanism through which Ras activity is regulated by monoubiquitination. We show that monoubiquitination at position 147 does not alter the intrinsic biochemical properties of Ras, but severely disrupts regulation of Ras by GAPs. This effect is specific to monoubiquitination at position 147 and is not observed when Ras is monoubiquitinated at other adjacent lysines. The loss of GAP–mediated hydrolysis accounts for the accumulation of Ras–GTP in vivo. Thus, as with oncogenic mutations of Ras, monoubiquitination renders the protein resistant to GAP–mediated regulation.
RESULTS
Monoubiquitination of Ras
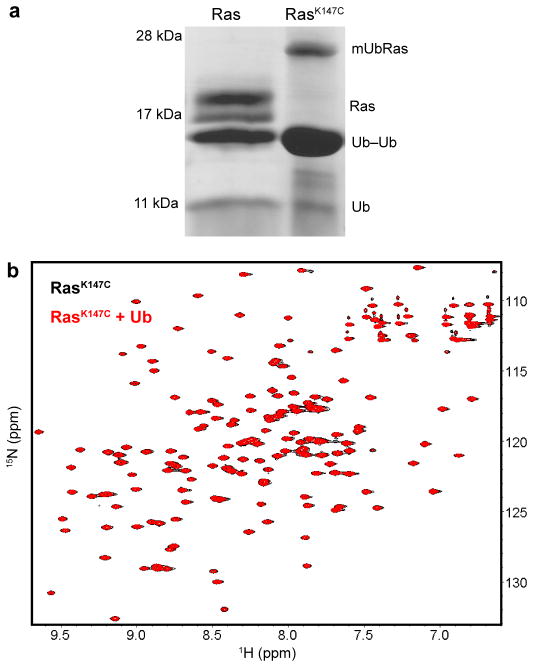
We conducted a series of in vitro studies to elucidate the mechanism of Ras regulation by monoubiquitination. These studies required fully ubiquitinated protein that was exclusively modified at Lys147 and in quantities sufficient for detailed biochemical and biophysical analysis. Recent investigations of monoubiquitinated substrates and ubiquitinating enzymes employed multiple methods of direct chemical ligation to generate the protein–Ubiquitin linkage16–21. In our approach, we replaced the native Ubiquitin linkage with a disulfide bond between a substituted cysteine at position 147 of Ras (RasK147C) and a cysteine at the carboxyl–terminus (c–terminus) of Ubiquitin (UbiquitinG76C). A surface accessible cysteine (Cys118) in Ras was replaced with serine to avoid unwanted modification (RasC118S, hereafter “Ras”). We previously showed that the C118S mutation did not alter Ras structure or biochemical properties22. The chemical ligation method does not require complicated intermediate chemical or enzymatic steps but instead provides a simple, specific approach to ubiquitination. The disulfide ligation strategy, using a more complicated cysteamine intermediate, was validated in previous studies of Proliferating Cell Nuclear Antigen (PCNA), where it was shown that chemically and enzymatically monoubiquitinated PCNA exhibit identical catalytic properties17. As seen in Figure 1a, we drove Ubiquitin modification of Ras at position 147 to completion by the addition of a ten–fold excess of UbiquitinG76C at pH 8.0. We conducted our experiments using H–Ras (1–166), for which the biochemical and structural (NMR and X–Ray) properties are best established, but corroborated the results using K–Ras (1–166) as indicated. All three mammalian isoforms H–, K–, and N–Ras show similar biochemical properties in the absence of the hypervariable c–terminus23,24. Furthermore, we used immunoprecipitation assays to show that, in the absence of c–terminal modification, monoubiquitination still leads to an increase in the GTP–bound population of H–Ras or K–Ras in HEK293T cells (Supplementary Figure 1).
Figure 1.
Monoubiquitination of Ras. (a) Reaction of UbiquitinG76C with Ras or a RasK147C mutant, under non–reducing conditions. The product of the reaction contains mUbRas, Ras, Ubiquitin–Ubiquitin dimer (Ub–Ub), and free Ubiquitin (Ub). (b) HSQC spectra of 15N–RasK147C bound to Mg–GDP in the absence (black) and presence (red) of ten–fold excess free Ubiquitin.
Downstream effectors of Ras, like Raf, have a Ubiquitin–like fold25. Thus, we considered whether Ubiquitin could bind to Ras in the manner of an effector. To this end, we used NMR in the presence of free unlabeled Ubiquitin to determine whether Ubiquitin altered spectral features associated with Ras backbone amides. A 1H–15N 2D HSQC overlay of 15N–enriched H–RasK147C in the absence and presence of Ubiquitin is shown in Figure 1b. The assignments for H Ras (1–166) were previously determined26 and we verified the shifted backbone amide resonances of H–RasK147 using 3D HNCACB data (Supplementary Fig. 2). Comparison of the position and intensity of the backbone amide resonances indicates that Ras is not altered by the presence of free Ubiquitin. In support of these observations, as shown in Figure 2a–b and Supplementary Figure 3, we found that the intrinsic rate of GDP dissociation and GTP hydrolysis were unaffected by the presence of Ubiquitin or Ubiquitin dimers in solution. These results indicated that Ras did not specifically interact with Ubiquitin. Therefore, for subsequent analyses, we did not separate monoubiquitinated Ras (mUbRas) from free Ubiquitin.
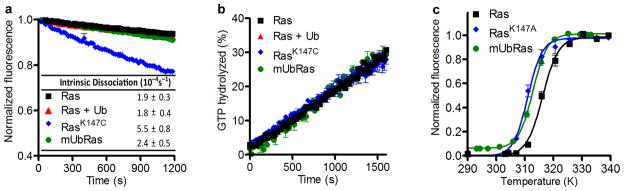
Figure 2.
Monoubiquitinated Ras retains intrinsic stability and activity. (a) Intrinsic nucleotide dissociation rates for Ras, RasK147C, and mUbRas loaded with MANT–GDP. Dissociation was monitored following the addition of unlabeled GDP by the decrease in fluorescence emission over time. Data were fit to an exponential dissociation curve, and the results are the mean ± s.d. (n=4). (b) Intrinsic single–turnover GTP hydrolysis for Ras, RasK147C, and mUbRas. Hydrolysis was initiated by the addition of Mg2+ and monitored by the change in fluorescence of Flippi when bound to free phosphate. Data were converted to a phosphate concentration using a standard curve. The concentration of phosphate equal to 100% GTP hydrolyzed was determined in the presence of GAP. Results are the mean ± s.d. (n=6). (c) Thermal stability of Ras, RasK147A, and mUbRas measured by 4–Fluoro–7–aminosulfonylbenzofurazan (ABD–F) incorporation as a function of temperature. The data were normalized using the maximum fluorescence intensity. Results are the mean ± s.d. (n=4).
Monoubiquitinated Ras Retains Intrinsic GTPase Activity
Previous computational studies predicted that the stability of a ubiquitinated substrate depends on the site of ubiquitination and type of Ubiquitin–Ubiquitin linkage27. To determine if monoubiquitination alters Ras, we compared the thermal stability of unmodified Ras and mUbRas. To this end, we employed the Quantitative Cysteine Reactivity (fQCR) assay28, which uses a cysteine reactive dye to measure rates of protein unfolding as a function of temperature. Because Ubiquitin does not have any native cysteines, it is invisible by this method. As shown in Figure 2c, we found that monoubiquitination decreases the thermal stability of Ras by 3.5 degrees (43.1± 0.2 °C, 39.2 ± 0.3 °C, and 39.6 ± 0.2 °C for Ras, RasK147A, and mUbRas, respectively), a change that is not likely to have a substantial effect on this otherwise highly stable protein in vivo. These data suggest that, despite the size of Ubiquitin, monoubiquitination at position 147 does not lead to thermal destabilization of Ras.
While monoubiquitination of Ras does not substantially affect thermal stability, it could alter intrinsic activity. For example, it is possible that ubiquitination impairs guanine nucleotide binding, similar to mutations at the adjacent residue, Ala146 (refs. 29,30). To measure rates of nucleotide dissociation, we equilibrated Ras and mUbRas with the fluorescent analog N–methylanthraniloyl (MANT)–GDP and measured fluorescence over time in the presence of excess unlabeled GDP. We observed a slight increase (2–3 fold) in the intrinsic rate of nucleotide dissociation for RasK147C as compared to native Ras, while the rate for mUbRas was unaltered (Fig. 2a). This result suggests that ubiquitination of Ras does not have the same impact on nucleotide binding as a point mutation at the same residue.
We next sought to establish whether monoubiquitination alters the intrinsic rate of GTP hydrolysis. To this end we measured single turnover GTP hydrolysis using Flippi, a fluorescent sensor that detects free phosphate31. As shown in Figure 2b and Supplementary Figure 4, neither mutation of Lys147 nor monoubiquitination of Ras affected the intrinsic rate of GTP hydrolysis (calculated as 0.012 ± 0.002 min−1 for all variants). Taken together, these results indicate that monoubiquitination does not alter the activity of Ras, and that another mechanism must account for the accumulation of mUbRas in the GTP–bound state in vivo.
Chemical Ubiquitination Mimics Native Ubiquitination
We found that monoubiquitination does not alter the intrinsic biochemical activity of Ras, even though mUbRas accumulates in the GTP–bound state in vivo. Previous studies have shown that chemically ubiquitinated PCNA functions similarly to the enzymatically ubiquitinated protein17. To further establish that chemical ubiquitination of Ras is a good mimic of native ubiquitination, we built computational models of Ubiquitin ligated to Ras.
To create the model, we used a recently developed module of the Rosetta protein modeling software suite32,33 that samples the conformational space available to ligated proteins. We modified Rosetta to consider disulfide and native isopeptide ubiquitination linkages and generated model structures of mUbRas using both linkages. We generated these models without the use of experimentally–derived constraints.
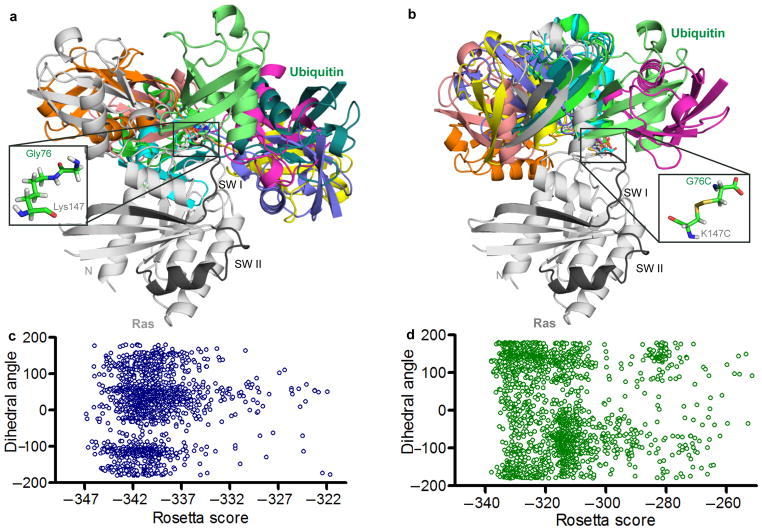
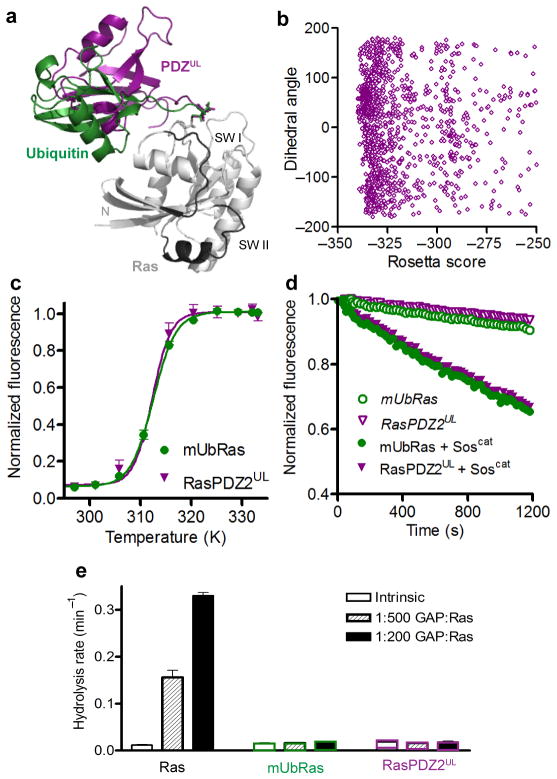
Shown in Figure 3a and 3b are the ten lowest scoring structures of each type of linkage, sorted by Rosetta total score from populations of approximately 2000 models. Comparison of these models indicates that the two systems behave similarly; Ubiquitin samples a wide range of conformations when ligated to Ras and all Ubiquitin positions are allowed at low energy scores. (Figure 3c and 3d). This modeling result suggests that chemical ubiquitination is a good surrogate for native ubiquitination of Ras. The data also suggest that Ubiquitin does not bind with high affinity to any single site on Ras, which is consistent with our findings that Ubiquitin does not specifically interact with Ras when the two proteins are free in solution.
Figure 3.
Rosetta model of native and chemical ubiquitination of Ras. (a) The ten lowest scoring Rosetta models of the native linkage of Ras monoubiquitination at position 147 lacking constraints to bias the model. Ras (5P21) is shown in grey with switch regions (SWI and SWII) highlighted in black. Ubiquitin (1UBQ) conformers shown in colors. Inset: native linkage between Ras Lys147 and Ubiquitin G76. (b) The ten lowest scoring Rosetta models of the chemical linkage of Ras monoubiquitination at position 147 lacking any constraints to bias the model. Ras and Ubiquitin colored as in panel a. Inset: chemical linkage between Ras K147C and Ubiquitin G76C. (c) The distribution of Ubiquitin orientations relative to Ras plotted against Rosetta energy scores for the native linkage. The Y axis shows the dihedral angle, in degrees, of the torsional angle between the center of mass of Ubiquitin, the linking Ras residue (147), the center of mass of Ras and an arbitrary Ras reference atom. (d) The distribution of Ubiquitin orientations relative to Ras plotted against Rosetta energy scores for the chemical linkage. Axes are the same as described in panel c.
Monoubiquitination Affects the Switch Regions of Ras
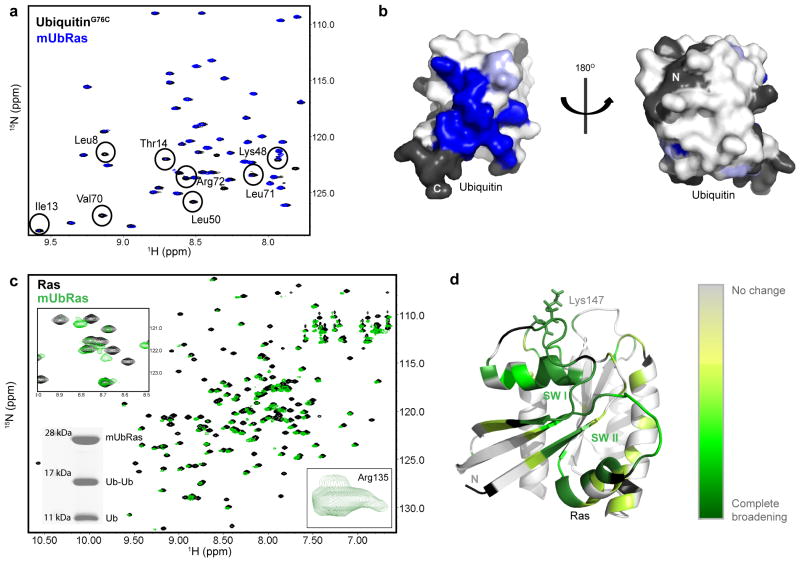
Results obtained from computational modeling suggest that there is no single preferred interaction between Ubiquitin and Ras. To test this prediction experimentally, we used NMR to examine spectral differences between Ubiquitin and Ras upon monoubiquitination. First, we 15N–enriched UbiquitinG76C and examined the 1H–15N 2D HSQC spectrum of Ubiquitin when ligated to RasK147C. By this method we observed partial to complete resonance broadening of eleven backbone amides, but no substantial chemical shifts within Ubiquitin (Fig. 4a). By mapping these spectral changes onto the structure of Ubiquitin in Figure 4b, it is evident that one face of Ubiquitin is primarily altered upon ligation with Ras. A possible explanation for the inability to detect a subset of Ubiquitin amide resonances is that Ras ligation restricts conformational sampling, leading to exchange broadening.
Figure 4.
Surfaces of Ras and Ubiquitin affected by monoubiquitination. (a) HSQC spectra of 15N–UbiquitinG76C free (black) or ligated to RasK147C (blue). Residues that broaden are labeled based on previous assignments40. (b) Space filling model of the structure of Ubiquitin (1UBQ) with residues that show decreased intensity when ligated to Ras (blue). Residues with no information are colored black. (c) HSQC spectra of 15N–RasK147C bound to Mg–GDP alone (black) and when monoubiquitinated (green). Inset (Top): enhancement of one expanded region showing residues that broaden and disappear. Inset (Bottom Left): SDS–PAGE gel showing integrity of mUbRas sample after HSQC analysis. Inset (Bottom Right): close up of Arg135, which exhibits multiple populations. (d) Mapping of Ras backbone amides that disappear upon monoubiquitination onto the structure of Ras. Darker green indicates more appreciable broadening (primarily in the SW I and SW II). Residues with no information are colored black.
We next reversed our labeling scheme and 15N–enriched Ras prior to ligation with Ubiquitin and collected a 1H–15N 2D HSQC spectrum. As shown in Figure 4c, a number of NH peaks broaden in mUbRas relative to the unmodified protein. Eighty–four of the 137 detectable backbone amide resonances dispersed across mUbRas exhibited multiple populations rather than a single, Lorentzian shaped peak (Supplementary Figure 5). The multiple populations indicate that Ubiquitin adopts more than one position relative to Ras on a timescale detectable by NMR. We also observed a substantial number of residues that broadened and, in some cases, could no longer be detected in mUbRas (Fig. 4c). The broadened peaks primarily localize to the switch regions (Fig. 4d). In the NMR spectra of Ras–GTP, backbone amides associated with residues in Switch I and Switch II are not detectable because they are in intermediate exchange on the NMR timescale34. The observed broadening in the GDP–bound state of mUbRas suggests a change in backbone dynamics, possibly due to conformational exchange or dynamic sampling of the switch regions. Since Ras regulators and effectors interact through the switch regions, monoubiquitination could alter the population of active Ras by changing how mUbRas interacts with regulators.
Monoubiquitination of Ras Inhibits GAP–Mediated Hydrolysis
In cells, the nucleotide–bound state of Ras is regulated both by GEFs, which increase the rate of GDP dissociation, and GAPs, which enhance the rate of GTP hydrolysis. Our NMR data suggest that monoubiquitination affects the switch regions of Ras, which in turn could alter interactions with GEFs and GAPs. Thus the increased GTP–bound population of mUbRas in vivo could be caused by either an increased sensitivity to GEFs or decreased sensitivity to GAPs.
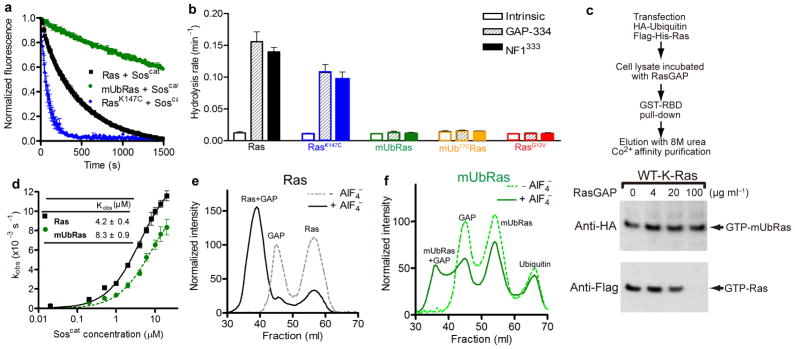
We first determined if the rate of GEF–mediated GDP dissociation is altered when Ras is monoubiquitinated. For these experiments, we equilibrated RasK147C, mUbRas, and Ras with MANT–GDP, and measured the rate of GDP dissociation in the presence of a catalytic fragment from the Ras GEF, Sos (Soscat)35. While the rate of GEF–mediated GDP dissociation was faster for RasK147C than Ras, the percent increase compared to the intrinsic rate of dissociation was the same, indicating that mutation at position 147 does not change the overall sensitivity of Ras to GEF–mediated regulation. However, we observed a decrease in the rate of GEF–mediated nucleotide dissociation for mUbRas compared to unmodified Ras (Fig. 5a).
Figure 5.
Monoubiquitination decreases the sensitivity of Ras to downregulation by GAPs. (a) Nucleotide dissociation reaction for Ras, RasK147C, and mUbRas loaded with MANT–GDP in the presence of a 1:1 molar ratio of Ras to Soscat. Data were fit to an exponential dissociation curve, and the results are the mean ± s.d. (n=4). (b) Single–turnover GTP hydrolysis for Ras, RasK147C, mUbRas, mUbRas ubiquitinated with Ub77C (mUb77CRas), and RasG12V in the presence of NF1333 or GAP–334 at a molar ratio of 1:500 GAP:Ras. Results are the mean ± s.d. (n=6). (c) Immunoblotting of GTP-bound Ras and GTP-bound mUbRas in cell extract in the presence of increasing concentrations of RasGAP. Anti–Flag and anti–HA antibodies reveal the relative fraction of total Ras and mUbRas, respectively. (d) Titration of Ras with Soscat. Experiments were performed as described panel a, except the concentration of Soscat was varied while Ras was held constant at 0.2 μM. Data plotted as a function of the Soscat concentration. Results are the mean ± s.d. (n=3). (e) Gel filtration of Ras and NF1333 in the absence (dotted line) and presence (solid line) of AlF4−. (f) Gel filtration of mUbRas and NF1333 in the absence (dotted line) and presence (solid line) of AlF4−.
We next considered the effect of Ras monoubiquitination on GAP–mediated hydrolysis. To this end we compared the rate of GTP hydrolysis for Ras and mUbRas in the presence of the catalytic domains of two GAPs, NF1 (NF1333) and p120GAP(GAP–334)4,36. At a GAP–to–Ras ratio of 1:500, we observed an order of magnitude increase in the rate of GTP hydrolysis for unmodified Ras relative to the intrinsic rate of GTP hydrolysis. No increase in the rate of GTP hydrolysis was observed for mUbRas in the presence of the same GAP–to Ras ratio (Fig. 5b). Therefore, mUbRas is insensitive to GAP–mediated regulation, similar to an oncogenic RasG12V mutation9. We obtained similar results using K–Ras (Supplementary Figure 6), indicating that the effects of monoubiquitination on Ras are not isoform–specific.
To verify that the differences between the enzymatic and chemical ubiquitination linkers (seven bonds and five bonds, respectively) do not alter GAP–responsiveness, we placed an additional cysteine at the c–terminus of Ubiquitin (UbiquitinC77), thereby creating a linker one bond longer than the native linker. We measured the rate of GAP–mediated GTP hydrolysis and observed that the response of Ras ligated to UbiquitinC77 was identical to Ras ligated to UbiquitinG76C (Fig. 5b). These results indicate that variations in the linker length on this scale (1–2 bonds) do not influence the sensitivity of mUbRas to GAP downregulation.
To validate the use of an in vitro system to dissect the mechanism of Ras regulation, we measured the sensitivity of mUbRas to GAP–mediated hydrolysis in a cellular reconstitution system. We immunoprecipitated Ras from HEK293T cells and compared the sensitivity of the monoubiquitinated and unmodified fractions of Ras to regulation by GAP. As seen in Figure 5c, monoubiquitinated K–Ras is less sensitive than the unmodified protein to GAP–mediated GTP hydrolysis. These data support our in vitro findings that monoubiquitination increases the population of active, GTP–bound Ras through a defect in sensitivity to GAP–mediated regulation.
To determine if the reduced response to regulators is due to a change in binding affinity for mUbRas, we first measured the extent to which monoubiquitination disrupts the interaction between Ras and Soscat (Fig. 5d). Results from these analyses indicated that the binding affinity between mUbRas and Soscat is 8.3 ± 0.9 μM, which is half the observed binding affinity between Ras and Soscat (4.2 ± 0.4 μM), consistent with the small reduction in the rate of GDP dissociation observed. However, a decrease in the rate of GDP dissociation would favor the GDP–bound state of Ras. Thus, the minor differences in GEF binding do not account for the accumulation of Ras–GTP in vivo.
To determine whether ubiquitination also leads to a reduction in GAP binding affinity, we compared the ability of Ras and mUbRas to bind to NF1333 in the presence of AlF4−. As seen in Figure 5e, in the presence of AlF4− almost 100% of NF1333 bound to Ras, which was present in slight excess. In contrast, about 50% of the NF1333 bound to mUbRas under the same conditions (Fig. 5f), which suggests that the binding affinity between GAP and mUbRas is reduced relative to unmodified Ras. While monoubiquitination affects both GEF–and GAP–mediated activity, the GAP defect has a greater influence on the enzyme kinetics and as such is predicted to have a dominant effect on the contribution to GTP–bound Ras. These data are supported by kinetic modeling of the respective GEF and GAP defects shown in Supplementary Figure 7. Taken together, our data reveal a substantial reduction in GAP activity as a consequence of Ras monoubiquitination, which accounts for the accumulation of activated Ras in vivo.
Modifying Ras with PDZ2 Impairs GAP–Mediated Hydrolysis
Our computational and NMR data suggest that Ubiquitin does not form a specific, high–affinity interaction with Ras. If this observation is correct, then modification of Ras with any protein similar to Ubiquitin should also impair GAP–mediated hydrolysis. As a test of this model, we chemically ligated Ras to PDZ2 (RasPDZ2), a 9 kDa protein with a Ubiquitin–like fold but no obvious sequence similarity to Ubiquitin37. We replaced the unstructured c–terminal extension of PDZ2, defined as the region after the folded domain ends in the crystal structure, with that of Ubiquitin (PDZ2UL). Therefore, all differences between PDZ2UL and Ubiquitin are contained in the folded regions of the two proteins.
Modeling of PDZUL on Ras shows that PDZ adopts a similar spread of possible conformations as Ubiquitin (Fig. 6a and 6b), suggesting that it could have an impact on Ras activity that is comparable to that of Ubiquitin ligation. As seen in Figure 6c, mUbRas and RasPDZ2UL have identical melting temperatures, indicating that neither ubiquitination nor PDZ2UL ligation substantially alters the thermal stability of Ras. Similar to ubiquitination, PDZ2UL ligation does not alter intrinsic Ras nucleotide dissociation rates, and GEF–mediated dissociation is reduced to the same extent as for mUbRas (Fig. 6d). Finally, RasPDZ2UL retains intrinsic GTP hydrolysis activity, but is insensitive to GAP–mediated GTP hydrolysis (Fig. 6e). These data indicate that non–specific interactions between Ras and Ubiquitin are responsible for the insensitivity of mUbRas to GAPs.
Figure 6.
Modification of Ras with PDZ2 resembles modification with Ubiquitin. (a) Rosetta model of Ras (5P21) in grey modified at position 147 with Ubiquitin (1UBQ) in green and PDZUL (3LNX) in purple. (b) The distribution of PDZUL orientations relative to Ras plotted against Rosetta energy scores for the chemical linkage. This plot follows the scheme of Figure 3B. (c) Thermal stability of Ras and RasPDZ2 with the Ubiquitin linker (RasPDZ2UL) measured by ABD–F incorporation as a function of temperature. Results are the mean ± s.d. (n=4). (d) Nucleotide dissociation reaction for RasPDZ2UL and mUbRas loaded with MANT–GDP in the absence and presence of a 1:1 molar ratio of Ras to Soscat. Results are the mean ± s.d. (n=4). (e) Single–turnover GTP hydrolysis for Ras, RasPDZ2UL, and mUbRas in the presence of GAP–334 at a molar ratio of 1:500 and 1:200 GAP:Ras. Results are the mean ± s.d. (n=6).
The Effect of Ubiquitination is Site–Specific
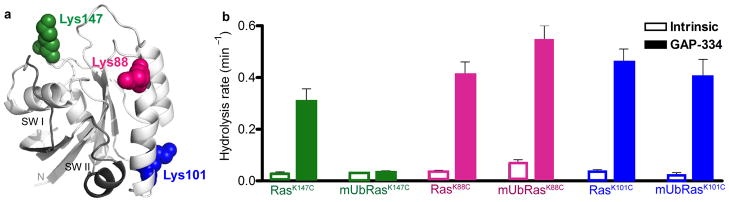
Finally, while the GAP insensitivity of modified Ras is not specific to Ubiquitin, it could be specific to modification at position 147. To address this possibility, we chose two other lysines on Ras that were not identified as sites of monoubiquitination in the mass spectrometry screen of monoubiquitinated K–Ras15. We chose position 88 because it is near the switch regions of Ras, similar to position 147, and could have a similar effect as monoubiquitination at position 147. We chose position 101 because the side chain is oriented toward the opposite face of Ras and would likely be less disruptive to the Ras active site and switch regions if monoubiquitinated (Figure 7a).
Figure 7.
The impaired GAP–sensitivity of mUbRas is site–specific. (a) Ribbon diagram of Ras–GDP (1CRR) with the switch regions highlighted in black and the side chains of Lys147, Lys88, and Lys101 represented as spheres in green, fuchsia, and blue, respectively. (b) Single–turnover GTP hydrolysis for Ras mutated and ubiquitinated at position 147, 88, or 101 in the absence and presence of GAP–334 at a molar ratio of 1:200 GAP:Ras. Results are the mean ± s.d. (n=4).
Using our Rosetta models, we found that monoubiquitination of Ras at position 88 or 101 does not cause steric clashes with GAP–334, similar to monoubiquitination at position 147 (Supplementary Figure 8). To determine whether monoubiquitination at position 88 or 101 could affect GAP–mediated regulation, we mutated each of these residues to cysteine, modified them with UbiquitinG76C, and measured the effect of Ubiquitination on intrinsic and GAP–mediated GTP hydrolysis. As seen in Figure 7b, ubiquitination at position 88 or 101 of Ras does not affect the intrinsic or GAP–mediated rate of GTP hydrolysis. Only monoubiquitination at position 147 impairs GAP–mediated hydrolysis, indicating that the outcome of monoubiquitination is site–specific.
In summary, we used a combination of biochemical, structural, and computational approaches to uncover the mechanism of Ras regulation by monoubiquitination. Our data indicate that ubiquitination activates Ras by impairing the catalytic efficiency of Ras GAPs. Furthermore, the most commonly ubiquitinated position in vivo, position 147, is the only lysine tested that impairs GAP–mediated hydrolysis. More broadly, our findings reveal how monoubiquitination promotes sustained signaling and cell transformation.
DISCUSSION
It was established recently that monoubiquitination increases the proportion of Ras that is in the activated (GTP–bound) state, that monoubiquitination enhances association with the downstream effectors Raf and PI3–Kinase, and that mutation of the primary site of monoubiquitination impairs oncogenic Ras–mediated tumorigenesis. Here we show that monoubiquitination decreases the sensitivity of Ras to GAP–mediated hydrolysis. A major advance was our ability to easily generate mUbRas, modified at a single site, in a form suitable for detailed biophysical studies. This chemical ligation strategy will likely be useful for the study of other monoubiquitinated proteins. Surprisingly, monoubiquitination did not alter the intrinsic activity of Ras, despite the size of the modification. Our modeling and NMR analyses indicated that Ubiquitin dynamically samples a broad surface area of Ras that alters switch region dynamics. These results led us to examine the effect of monoubiquitination on the interaction of Ras with its cognate GEF and GAPs, which also target the switch domains. The analysis revealed that monoubiquitination abrogates GAP–mediated GTP hydrolysis. All other activities, including the ability to bind regulators, were largely preserved and our kinetic modeling suggests that the GAP defect will dominate. Furthermore, this outcome was specific to monoubiquitination at position 147. Thus our work establishes an entirely new mode of Ras activation in which signaling is sustained even in the absence of hormone stimulus or oncogene mutation.
It will be interesting to determine how monoubiquitination affects other signaling proteins including other Ras–family GTPases. Known targets of monoubiquitination include K–Ras15, H–Ras, and N–Ras11. Monoubiquitination disrupts interactions of Rap2A with effector proteins and inhibits the ability of Rap2A to promote dendrite development12. Monoubiquitination has also been observed in Rac1, although the biological consequence of this modification is not yet known13,14. Our chemical ligation strategy and multi–dimensional approach will be useful for the study of these targets, particularly in cases where the relevant ubiquitin ligase has not been identified.
Another question concerns the role of the preferred site of monoubiquitination, Lys147 (ref. 15). Whereas ubiquitination of this site has severe consequences for GAP function, targeted ubiquitination of two other candidate sites left GAP–mediated hydrolysis unperturbed. Lys147 is part of the SAX motif, and a lysine at the third position in this motif is conserved in Ras proteins across species, as well as in other Ras–family GTPases including RhoA, RhoB, RhoC, Rap, Ral, Rab, Rheb and Ran38. It has been shown previously that mutation of the highly conserved adjoining residue, Ala146, leads to enhanced GDP exchange, GTP loading and cellular transformation29,30,39. In contrast, we have shown that a mutation of the ubiquitination site itself, Lys 147, has little effect on nucleotide binding or regulator–mediated activity. We speculate that the lysine has been conserved to allow regulation through monoubiquitination. It will be interesting to determine whether other members of the Ras subfamily are also ubiquitinated at this position and whether ubiquitination in such cases leads to sustained activation.
Conclusions
We have demonstrated that monoubiquitination of Ras impedes the function of GTPase activating proteins. Key to our analysis was the ability to generate monoubiquitinated protein, modified at a single residue, suitable for biophysical analysis. Through multi–disciplinary computational, structural and biochemical approaches we identified a novel mechanism of Ras activation, one that is independent of any oncogenic mutation or a sustained receptor stimulus. Given the established importance of Ras in the control of cell growth and differentiation, our findings may reveal opportunities for new pharmaceuticals that target the ubiquitination machinery.
ONLINE METHODS
Protein Purification
The Ras domains (1–166) of H–Ras and K–Ras were expressed in the pQlinkH vector (Addgene) with a histidine purification tag in Escherichia coli BL21 (DE3) RIPL cells (Stratagene: La Jolla, CA). Proteins were purified following standard Qiagen nickel affinity purification procedures. The His tag was cleaved overnight with Tobacco Etch Virus. Ras proteins were further purified by removal of uncleaved protein using Ni–NTA agarose beads (Qiagen). The final product was > 95% pure by SDS–PAGE. Proteins were stored in 20 mM HEPES, pH 8.0, 50 mM NaCl, 500 μM TCEP, 50 μM GDP and 5 mM MgCl2.
Soscat (John Kuriyan; University of California, Berkeley) was purified as previously described35. The catalytic domains of p120GAP (GAP–334) 4 and NF1 (NF1333)36, were expressed in pQlinkH and purified as described for Ras. Purified proteins were stored in 20 mM HEPES, pH 8.0, 50 mM NaCl and 500 μM TCEP. Full length UbiquitinG76C and hPTPe–PDZ2 ULG97C (c–terminal residues KGQSPC replaced with the Ubiquitin residues VLRLRGC)37 were expressed in the pQlinkH vector system and purified as described for Ras. Proteins were stored in 20 mM HEPES, pH 8.0, 50 mM NaCl and 500 μM TCEP.
Ligation of UbiquitinG76C and PDZ2 ULG97C to RasK147C
The chemical ligation strategy used to link Ras to UbiquitinG76C or PDZ2ULG97C was adapted from Merkley et al.16. Briefly, a ten–fold excess of UbiquitinG76C or PDZ2 ULG97C was added to RasK147C and dialyzed into 20 mM Tris, pH 8.0, 50 mM NaCl, 5 mM MgCl2, and 50 μM GDP at 4 °C overnight. The amount of disulfide complex formation was determined by non–reducing SDS–PAGE and considered complete by the absence of modified Ras.
Thermal Stability of Ras
The fast quantitative cysteine reactivity (fQCR) method28 was employed to measure changes in Ras thermal stability. Briefly, 2 μM Ras was incubated with 1 mM 4–fluoro–7–aminosulfonylbenzofurazan (ABD–F, Anaspec) at pH 7.0 in the presence of 20 μM GDP and 2 mM MgCl2 at the desired temperature for five minutes. Fluorescent intensity was measured on a PHERAstar plate reader (BMG Labtech). The data were normalized and fit to determine the temperature at which half the protein was unfolded, representing the Tm. Results are the mean ± s.d. (n=3).
Ras Nucleotide Dissociation and Hydrolysis Assays
The rate of nucleotide dissociation was measured using MANT–GDP (BioLog: San Diego, CA) as previously reported41,42. Briefly, MANT–GDP–bound Ras (2 μM) was added to 1 mL assay buffer (50 mM Tris, pH 7.4, 50 mM NaCl and 5 mM MgCl2) and exchange initiated by addition of 2 mM GDP. MANT–GDP dissociation was measured as a change in fluorescence intensity over time (excitation: 360 nm, emission: 440 nm) (LS50B Perkin–Elmer Luminescence Spectrometer). Fluorescence data were fit in GraphPad Prism (GraphPad Software; San Diego, CA) to a one–phase exponential decay curve. For GEF–mediated dissociation and binding, 200 nM Ras and 0.2 μM to 20 μM Soscat were used. The nucleotide dissociation rate was plotted as a function of Soscat concentration and fit to one site binding to determine the binding affinity between Ras or mUbRas and Soscat. Results are the mean ± s.d. (n=4).
Single–turnover GTP hydrolysis assays were performed as previously described43, except that the phosphate binding protein Flippi 5U (Addgene) was used to detect inorganic phosphate released upon GTP hydrolysis31. Flippi 5U was purified as previously described31. All buffers were made phosphate free by dialysis with 1 U nucleoside phosphorylase (Sigma, USA) and 2 mM inosine (Sigma, USA). For GAP–mediated hydrolysis, 50 μM Ras and 0.1 μM (1:500) and 0.25 μM (1:200) NF1333 and GAP–334 were used. The ratio of fluorescence emission was measured at 480 nm and 530 nm with an excitation of 435 nm on a SpectraMax M5 (Molecular Devices). Fluorescence ratios were converted to phosphate concentrations using a standard curve. Hydrolysis curves were fit in GraphPad Prism (GraphPad Software; San Diego, CA) to a one–phase exponential association curve. Results are the mean ± s.d. (n=6).
GAP binding was monitored as previously described44. Briefly, 50 μM Ras and 40 μM NF1333 were mixed in the presence or absence of AlF4− (10 mM NaF, 450 μM AlCl3) on ice. The sample was run on an S75 column in 30 mM Tris, pH 7.5 and 5 MM MgCl2. Data was normalized to the amount of free NF1333 in the absence of AlF4−.
GST–RBD Immunoprecipitation Assay
Ras activation was measured as described previously15. Flag–His–K–Ras or the c–terminal mutants Flag–His–K–RasC185S and Flag–His–H–RasC186S were co–expressed with HA–Ubiquitin in HEK293T cells. The cells were rinsed with cold PBS and lysed with Buffer A (0.5% NP–40, 40 mM HEPES [pH 7.4], 150 mM NaCl, 10% glycerol, 1 mM DTT, 1 μg ml−1 leupeptin, 2 μg ml−1 aprotinin, 1 μg ml−1 pepstatin A, 100 μM AEBSF, Halt phosphatase inhibitor cocktail (Thermo Scientific), 10 mM iodoacetamide (IAA) and 5 mM N–ethylmaleimide (NEM)). The soluble fraction from the cell lysates were isolated by centrifugation at 13,000 rpm for 10 min, split, and subjected to anti–Flag agarose immunoprecipitation or incubated with 10 μg of GSH–Sepharose bound GST–Raf–RBD in the presence of 1 mg ml−1 BSA for 30 min as described previously15. The immunoprecipitated proteins were washed three times with Buffer A and eluted by the addition of 8 M urea. To ensure detection of mUbRas, a secondary purification on Co2+ Talon metal affinity chromatography beads (Clontech) was performed. Flag–His–Ras was eluted with sample buffer containing 50 mM EDTA. For the GAP sensitivity assay, bacterially produced GAP–334 was incubated with the cell lysate for 20 min at room temperature and subjected to analysis using GST–Raf–RBD.
NMR Experiments
For NMR studies, 15N–and 15N,13C–enriched samples of Ras and Ubiquitin were produced using standard protocols in M–9 minimal media26. 1H–15N 2D HSQC experiments were conducted on a Varian 700 MHz with a cryoprobe in 20 mM MOPs, pH 6.8, 100 mM NaCl, 5 mM MgCl2, 0.01% sodium azide, 10% D2O, 1 mM DPTA, and 2 mM GDP at 25° C and with 500 μM protein.
Rosetta Modeling
The modeling strategy used was adapted from Saha et al.32. The previous protocol modeled a thioester linkage between Ubiquitin Gly76 and cysteine on a Ubiquitin E2 enzyme. Modifications include altering the linkage type to disulfide or isopeptide linkages and replacing the pre–existing system–specific constraints with optional command–line defined constraints. Further modifications include the reporting of specific Ubiquitin–Ras residue pair distances and Ubiquitin–Ras positional metrics used to quantify mUbRas conformational ensembles. Also added was the ability to include arbitrary nonmoving atoms in the simulation, used to include the guanine nucleotide, magnesium ion, and in some cases, GAP during simulations. A chemically conjugated model of Ras and Ubiquitin was created and the torsion angles within the linker region were modeled while sampling side chain conformations throughout the interface.
For the isopeptide linker, protocol UBQ_Gp_LYX–cterm was used. Torsions allowed to vary included: the chi angles of Lys147 of Ras (sampled from Rosetta’s implementation of Dunbrack’s 2002 rotamer library45,46), the isopeptide bond, and both phi and psi for the Gly76, Gly75, and Arg74 of Ubiquitin. For the disulfide linker, protocol UBQ_Gp_CYD–CYD was used. Torsions sampled include the chi angles for K147C on Ras and G76C on Ubiquitin (from the Dunbrack library and explicit sampling of chi 2), the disulfide bond, phi of Ubiquitin G76C, and both phi and psi for Gly75 and Arg74 of Ubiquitin.
Sampling was performed with a standard Rosetta Metropolis–Monte Carlo search protocol45. For each combination of ligand, attachment chemistry, and Ras attachment location, the protocol was run for 2400 hours on a 2.66 MHz chip. This produces about 1500–3000 structures using 20,000 Monte Carlo cycles per trajectory.
Supplementary Material
Acknowledgments
We thank A. Lee (University of North Carolina at Chapel Hill (UNC–CH), Chapel Hill, North Carolina, USA) for supplying the PDZ2 domain. We would also like to thank D. Isom (UNC–CH, Chapel Hill, North Carolina, USA) for his fQCR expertise and input on data analysis. This work was supported by NIH grants R01CA089614 (S.L.C.), R01GM073180–06S1 (H.G.D.), R01GM073960 and R01GM073151 (B.K.), and R01GM41890 and P01CA117969 (L.C.C.). S.M.L. was supported by the Program in Molecular and Cellular Biophysics and NIH grant T32GM008570. A.T.S. was supported, in part, by the Japanese Society for the Promotion of Science Research Fellowship for Research Abroad, Kanae Foundation for Research Abroad, and a Genentech Fellowship.
Footnotes
AUTHOR CONTRIBUTIONS
R.B. performed biochemical and NMR studies. S.M.L. and B.K. conducted Rosetta modeling. R.B. and E.M.W. performed site–specific ubiquitination assays. A.T.S., J.C.W., and L.C.C. performed cell lysate experiments and kinetic modeling. R.B., H.G.D., and S.L.C. designed experiments, analyzed data, and wrote the manuscript.
References
- 1.Cox AD, Der CJ. The dark side of Ras: regulation of apoptosis. Oncogene. 2003;22:8999–9006. doi: 10.1038/sj.onc.1207111. [DOI] [PubMed] [Google Scholar]
- 2.Kjeldgaard M, Nyborg J, Clark BF. The GTP binding motif: variations on a theme. FASEB J. 1996;10:1347–68. [PubMed] [Google Scholar]
- 3.Herrmann C. Ras–effector interactions: after one decade. Curr Opin Struct Biol. 2003;13:122–129. doi: 10.1016/s0959-440x(02)00007-6. [DOI] [PubMed] [Google Scholar]
- 4.Scheffzek K, et al. The Ras–RasGAP Complex: Structural Basis for GTPase Activation and Its Loss in Oncogenic Ras Mutants. Science. 1997;277:333–339. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- 5.Sprang S. GEFs: master regulators of G–protein activation. Trends Biochem Sci. 2001;26:266–267. doi: 10.1016/s0968-0004(01)01818-7. [DOI] [PubMed] [Google Scholar]
- 6.Sprang SR. G proteins, effectors and GAPs: structure and mechanism. Curr Opin Struct Biol. 1997;7:849–856. doi: 10.1016/s0959-440x(97)80157-1. [DOI] [PubMed] [Google Scholar]
- 7.Bos JL. Ras Oncogenes in Human Cancer: A Review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 8.Adjei AA. Blocking Oncogenic Ras Signaling for Cancer Therapy. J Natl Cancer Inst. 2001;93:1062–1074. doi: 10.1093/jnci/93.14.1062. [DOI] [PubMed] [Google Scholar]
- 9.Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9:517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hicke L. Protein regulation by monoubiquitin. Nat Rev Mol Cell Biol. 2001;2:195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- 11.Jura N, Scotto–Lavino E, Sobczyk A, Bar–Sagi D. Differential Modification of Ras Proteins by Ubiquitination. Mol Cell. 2006;21:679–687. doi: 10.1016/j.molcel.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 12.Kawabe H, et al. Regulation of Rap2A by the Ubiquitin Ligase Nedd4–1 Controls Neurite Development. Neuron. 2010;65:358–372. doi: 10.1016/j.neuron.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nethe M, Hordijk PL. The role of ubiquitylation and degradation in RhoGTPase signalling. J Cell Sci. 2010;123:4011–4018. doi: 10.1242/jcs.078360. [DOI] [PubMed] [Google Scholar]
- 14.Visvikis O, et al. Activated Rac1, but not the tumorigenic variant Rac1b, is ubiquitinated on Lys 147 through a JNK–regulated process. FEBS J. 2008;275:386–396. doi: 10.1111/j.1742-4658.2007.06209.x. [DOI] [PubMed] [Google Scholar]
- 15.Sasaki AT, et al. Ubiquitination of K–Ras Enhances Activation and Facilitates Binding to Select Downstream Effectors. Sci Signal. 2011;4:ra13. doi: 10.1126/scisignal.2001518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merkley N, Barber KR, Shaw GS. Ubiquitin Manipulation by an E2 Conjugating Enzyme Using a Novel Covalent Intermediate. J Biol Chem. 2005;280:31732–31738. doi: 10.1074/jbc.M505205200. [DOI] [PubMed] [Google Scholar]
- 17.Chen J, Ai Y, Wang J, Haracska L, Zhuang Z. Chemically ubiquitylated PCNA as a probe for eukaryotic translesion DNA synthesis. Nat Chem Biol. 2010;6:270–272. doi: 10.1038/nchembio.316. [DOI] [PubMed] [Google Scholar]
- 18.Virdee S, et al. Traceless and Site–Specific Ubiquitination of Recombinant Proteins. J Am Chem Soc. 2011;133:10708–10711. doi: 10.1021/ja202799r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar KS, Spasser L, Ohayon S, Erlich LA, Brik A. Expeditious Chemical Synthesis of Ubiquitinated Peptides Employing Orthogonal Protection and Native Chemical Ligation. Bioconjugate Chem. 2011;22:137–143. doi: 10.1021/bc1004735. [DOI] [PubMed] [Google Scholar]
- 20.Eger S, et al. Generation of a Mono–ubiquitinated PCNA Mimic by Click Chemistry. ChemBioChem. 2011;12:2807–2812. doi: 10.1002/cbic.201100444. [DOI] [PubMed] [Google Scholar]
- 21.Purbeck C, Eletr ZM, Kuhlman B. Kinetics of the Transfer of Ubiquitin from UbcH7 to E6AP. Biochemistry. 2009;49:1361–1363. doi: 10.1021/bi9014693. [DOI] [PubMed] [Google Scholar]
- 22.Mott HR, Carpenter JW, Campbell SL. Structural and Functional Analysis of a Mutant Ras Protein That Is Insensitive to Nitric Oxide Activation. Biochemistry. 1997;36:3640–3644. doi: 10.1021/bi962790o. [DOI] [PubMed] [Google Scholar]
- 23.Reuther GW, Der CJ. The Ras branch of small GTPases: Ras family members don’t fall far from the tree. Curr Opin Cell Biol. 2000;12:157–65. doi: 10.1016/s0955-0674(99)00071-x. [DOI] [PubMed] [Google Scholar]
- 24.Takai Y, Sasaki T, Matozaki T. Small GTP–binding proteins. Physiol Rev. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- 25.Wohlgemuth S, et al. Recognizing and Defining True Ras Binding Domains I: Biochemical Analysis. J Mol Biol. 2005;348:741–758. doi: 10.1016/j.jmb.2005.02.048. [DOI] [PubMed] [Google Scholar]
- 26.Campbell–Burk S, Domaillea P, Starovasnik M, Boucher W, Laue E. Sequential assignment of the backbone nuclei of c–H–ras(1–166) GDP using a novel 4D NMR strategy. J Biomol NMR. 1992;2:639–646. doi: 10.1007/BF02192852. [DOI] [PubMed] [Google Scholar]
- 27.Hagai T, Levy Y. Ubiquitin not only serves as a tag but also assists degradation by inducing protein unfolding. PNAS. 2010;107:2001–2006. doi: 10.1073/pnas.0912335107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Isom DG, Marguet PR, Oas TG, Hellinga HW. A miniaturized technique for assessing protein thermodynamics and function using fast determination of quantitative cysteine reactivity. Proteins: Struct, Funct and Bioinf. 2011;79:1034–1047. doi: 10.1002/prot.22932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janakiraman M, et al. Genomic and Biological Characterization of Exon 4 KRAS Mutations in Human Cancer. Cancer Res. 2010;70:5901–5911. doi: 10.1158/0008-5472.CAN-10-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feig LA, Cooper GM. Relationship among guanine nucleotide exchange, GTP hydrolysis, and transforming potential of mutated ras proteins. Mol Cell Biol. 1988;8:2472–2478. doi: 10.1128/mcb.8.6.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gu H, et al. A novel analytical method for in vivo phosphate tracking. FEBS Lett. 2006;580:5885–5893. doi: 10.1016/j.febslet.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saha A, Lewis S, Kleiger G, Kuhlman B, Deshaies RJ. Essential Role for Ubiquitin–Ubiquitin–Conjugating Enzyme Interaction in Ubiquitin Discharge from Cdc34 to Substrate. Mol Cell. 2011;42:75–83. doi: 10.1016/j.molcel.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leaver–Fay A, et al. Chapter nineteen–Rosetta3: An Object–Oriented Software Suite for the Simulation and Design of Macromolecules. In: Michael LJ, Ludwig B, editors. Methods Enzymol. Vol. 487. Academic Press; 2011. pp. 545–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ito Y, et al. Regional Polysterism in the GTP–Bound Form of the Human c–Ha–Ras Protein. Biochemistry. 1997;36:9109–9119. doi: 10.1021/bi970296u. [DOI] [PubMed] [Google Scholar]
- 35.Sondermann H, et al. Structural Analysis of Autoinhibition in the Ras Activator Son of Sevenless. Cell. 2004;119:393–405. doi: 10.1016/j.cell.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 36.Scheffzek K, et al. Structural analysis of the GAP–related domain from neurofibromin and its implications. EMBO J. 1998;17:4313–4327. doi: 10.1093/emboj/17.15.4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kozlov G, Gehring K, Ekiel I. Solution Structure of the PDZ2 Domain from Human Phosphatase hPTP1E and Its Interactions with C–Terminal Peptides from the Fas Receptor. Biochemistry. 2000;39:2572–2580. doi: 10.1021/bi991913c. [DOI] [PubMed] [Google Scholar]
- 38.Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J Cell Sci. 2005;118:843–846. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- 39.Edkins S, et al. Recurrent KRAS Codon 146 mutations in human colorectal cancer. Cancer Biology & Therapy. 2006;5:928–932. doi: 10.4161/cbt.5.8.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weber PL, Brown SC, Mueller L. Sequential proton NMR assignments and secondary structure identification of human ubiquitin. Biochemistry. 1987;26:7282–7290. doi: 10.1021/bi00397a013. [DOI] [PubMed] [Google Scholar]
- 41.Lenzen C, Cool RH, Wittinghofer A. Analysis of intrinsic and CDC25–stimulated guanine nucleotide exchange of p21ras–nucleotide complexes by fluorescence measurements. Methods Enzymol. 1995;255:95–109. doi: 10.1016/s0076-6879(95)55012-7. [DOI] [PubMed] [Google Scholar]
- 42.Lenzen C, Cool RH, Prinz H, Kuhlmann J, Wittinghofer A. Kinetic analysis by fluorescence of the interaction between Ras and the catalytic domain of the guanine nucleotide exchange factor Cdc25Mm. Biochemistry. 1998;37:7420–7430. doi: 10.1021/bi972621j. [DOI] [PubMed] [Google Scholar]
- 43.Shutes A, Der CJ. Real–time in vitro measurement of intrinsic and Ras GAP–mediated GTP hydrolysis. Methods Enzymol. 2006;407:9–22. doi: 10.1016/S0076-6879(05)07002-3. [DOI] [PubMed] [Google Scholar]
- 44.Ahmadian MR, Mittal R, Hall A, Wittinghofer A. Aluminium fluoride associates with the small guanine nucleotide binding proteins. FEBS Lett. 1997;408:315–318. doi: 10.1016/s0014-5793(97)00422-5. [DOI] [PubMed] [Google Scholar]
- 45.Rohl CA, Strauss CEM, Misura KMS, Baker D. Protein Structure Prediction Using Rosetta. In: Ludwig B, Michael LJ, editors. Methods in Enzymology. Vol. 383. Academic Press; 2004. pp. 66–93. [DOI] [PubMed] [Google Scholar]
- 46.Dunbrack RL, Jr, Karplus M. Backbone–dependent Rotamer Library for Proteins Application to Side–chain Prediction. J Mol Biol. 1993;230:543–574. doi: 10.1006/jmbi.1993.1170. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.