Abstract
Over a decade has passed since the elucidation of the first X-ray crystal structure of any complex II homolog. In the intervening time, the structures of five additional integral-membrane complex II enzymes and three homologs of the soluble domain have been determined. These structures have provided a framework for the analysis of enzymological studies of complex II superfamily enzymes, and have contributed to detailed proposals for reaction mechanisms at each of the two enzyme active sites, which catalyze dicarboxylate and quinone oxidoreduction, respectively. This review focuses on how structural data have augmented our understanding of catalysis by the superfamily.
Keywords: Complex II, succinate dehydrogenase, succinate:quinone oxidoreductase, fumarate reductase, quinol:fumarate reductase, x-ray crystallography
1. Introduction
During respiration, living cells use oxidation-reduction reactions to harvest energy from their environment and convert this into a convenient chemical form, most frequently ATP. This commonly requires the formation of a transmembrane electrochemical gradient as an intermediate step. Mitochondrial aerobic respiration catalyzes oxidative phosphorylation, which is just one example of an energy-harvesting bioenergetic pathway. In oxidative phosphorylation, electrons from environmental chemicals (food) are transferred into the electron transport chain and eventually to molecular oxygen. Mitochondrial aerobic respiration uses five membrane-spanning complexes for this process, and electrons enter the electron transport chain through either complex I or complex II. Complex II bridges the processes of oxidative phosphorylation with the Krebs cycle, thus linking these two energy harvesting processes in higher organisms. In addition, membrane-spanning homologs of complex II participate in both aerobic respiration and bacterial anaerobic respiration with fumarate as the terminal electron acceptor [1].
2. Structures of complex II enzymes
2.1 Global architecture of the complex II superfamily
Complex II enzymes couple two distinct chemical reactions: the reversible oxidoreduction of succinate and fumarate, catalyzed in a soluble domain, and the reversible oxidoreduction of quinol and quinone, which is catalyzed in a membrane-spanning domain. Members of the superfamily have increased reaction rates and improved catalytic efficiency when catalyzing the reaction in one direction, and have been given distinct names to reflect this [2]. Succinate:quinone oxidoreductases (SQRs, succinate dehydrogenases, SdhABCD) are kinetically advantaged to oxidize succinate and reduce quinone, a reaction important for aerobic respiration. Conversely, quinol:fumarate reductases (QFRs, fumarate reductases, FrdABCD) are kinetically advantaged to catalyze the reverse reaction, i.e. to reduce fumarate and oxidize quinol (Table 1).
Table 1.
Kinetic parameters of succinate oxidation and fumarate reduction by SQR and QFR enzymes. Comparison of kcat, Km, and catalytic efficient reveals a distinct kinetic advantage for each enzyme in one direction. Numbers are taken from Maklashina (2006) J. Biol. Chem.
| Succinate oxidation |
Fumarate reduction |
|||||
|---|---|---|---|---|---|---|
| kcat (s−1) | Kmsucc (μM) | kcat/Km (μM−1s−1) | kcat (s−1) | Kmfum (μM) | kcat/Km (μM-1s−1) | |
| E. coli SQR | 110.0±4.0 | 110±10 | 1000 | 2.0±0.2 | 100±12 | 20 |
| E. coli QFR | 30.0±0.5 | 550±38 | 54 | 250±10 | 30±0.5 | 232 |
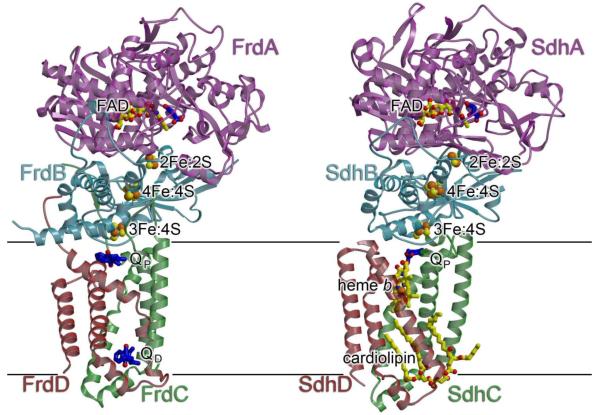
No matter the preferred direction of the reaction, complex II superfamily enzymes share a global architecture with a large soluble domain and a smaller integral-membrane domain (Fig. 1). The soluble domain contains two polypeptide chains; a large flavoprotein (Fp; SdhA or FrdA; ~600 amino acids) that covalently attaches flavin adenine nucleotide (FAD) and is organized around a Rossmann fold, and an iron-sulfur protein (Ip; SdhB or FrdB; ~250 amino acids) coordinates a 2Fe:2S, a 3Fe:4S, and a 4Fe:4S cluster and has domains related to bacterial and plant-type ferredoxins. The complex II enzymes are grouped into a superfamily based upon sequence similarity between the SdhA/FrdA (flavoprotein) and SdhB/FrdB (iron-sulfur protein) subunits of the soluble domain, which generally contain >40% sequence identity. By contrast, the membrane-spanning subunits (SdhC/FrdC and SdhD/FrdD) differ substantially across the superfamily (Fig. 2). Indeed, the membrane-spanning domain of complex II enzymes may contain 0, 1 or 2 membrane-spanning subunits with 5 or 6 transmembrane helices and 0, 1, or 2 b-type hemes. As a result, it has been hypothesized that the membrane-spanning regions have distinct evolutionary origins. Thus, the complex II superfamily has been further divided into four subfamilies (types A-E) that are classified according to the number of transmembrane polypeptides and b-type hemes within the membrane spanning region [3, 4]. More recent phylogenetic analysis based upon the sequence of the iron-sulfur protein (SdhB or FrdB) is consistent with the evolutionary grouping of these subtypes but suggests further evolutionary subgroups in subfamilies A and C [4].
Fig. 1. Overview of structures of complex II.
Shown are (A) the E. coli QFR in complex with fumarate (PDB ID: 3P4P [34]) and (B) the E. coli SQR in complex with oxaloacetate (PDB ID: 1NEK [7]) from the same perspective. The E. coli SQR has been demonstrated to be a trimer, but for clarity, only the monomer is shown. In each case, the flavoprotein (FrdA/SdhA) is colored purple, the iron sulfur protein (FrdB/SdhB) is colored light blue, and the membrane-spanning subunits (FrdC&FrdD/SdhC&SdhD) are colored red and green. Cofactors are shown with yellow carbons and bonds. Substrates or inhibitors are shown with dark blue carbons and bonds and include the substrate, fumarate (bound to the E. coli QFR), and inhibitor, oxaloacetate (bound to the E. coli SQR), within the dicarboxylate interconversion active site of the flavoprotein, and substrate menaquinol (bound to the E. coli QFR) and ubiquinone (bound to the E. coli SQR) within each Q-site. Inspection of these two structures shows that the soluble domains are similar in structure and cofactor composition while the membrane domains differ in both. In panel (A), the positions of the menaquinol molecules in the figure were modeled from their experimentally identified positions in the structure of the E. coli QFR in complex with citrate (PDB ID: 1L0V [5]).
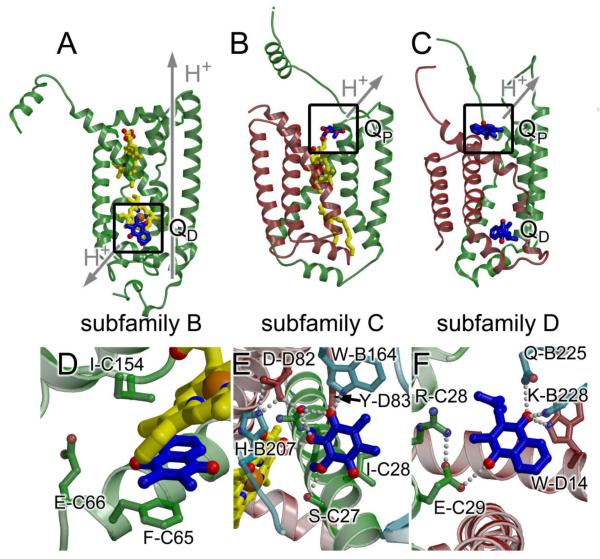
Fig. 2. Membrane spanning subunits from complex II subfamilies.
(A) W. succinogenes QFR (PDB ID: 2BS4 [6]; subfamily B) (B) Avian SQR (PDB ID: 1YQ3 [9]; subfamily C) (C) E. coli QFR (PDB ID 1L0V: [5]; subfamily D). Figure panels (A) - (C) were made following alignment of the flavoprotein of each complex, thus observed differences in helix packing do not reflect differences in the view. The functional Q-site is highlighted with a box. Coloring is the same as is in Fig. 1. (D) - (F) A close-up view of the functional Q-site for (D) W. succinogenes QFR; (E) Avian SQR (for consistency with the text, the numbering is for the E. coli SQR enzyme, and can be cross-referenced to the avian numbering in Table 3); (F) E. coli QFR. In each case, the view has been rotated such that interacting residues are clearly observed. In (D), the rotation is 70° around the x-axis and 30° around the y-axis. In (E) the rotation is 180° around the x-axis and 60° around the y-axis. In (F), the rotation is 30° around the x-axis, 180° around the y-axis, and 30° around the z-axis. Residues within 5 Å are highlighted and hydrogen-bonding interactions are shown with grey dots. There are no hydrogen-bonding interactions between the W. succinogenes QFR and the 2,3-dimethyl-1,4-napthoquinone molecule. Instead, π-stacking with the side chain Phe-C65 and with the heme porphyrin may contribute to binding.
To date, the X-ray crystal structures of six complex II superfamily enzymes have been determined [5-10]. These structures represent three subfamilies: subfamily B (one membrane-spanning polypeptide with five helices and two b-type hemes, exemplified by the Wolinella succinogenes QFR [6]), type C (two membrane spanning polypeptides that each have three membrane-spanning helices, exemplified by the E. coli [7], porcine [8], and avian [9] SQRs, and the Ascaris suum QFR [10]), and subfamily D (two membrane spanning polypeptides each containing three helices and no b-type heme, exemplified by the E. coli QFR [5]). Despite the sequence and cofactor differences of the membrane-spanning domain, at a global level the members of these subfamilies appear structurally similar (Fig. 1), and the membrane-spanning regions between subfamily B and subfamily C have surprisingly high structural similarity (for example, an RMS deviation of 1.7 Å between W. succinogenes FrdC and avian SdhC/D over 130 Cα atoms) despite a lack of detectable sequence identity when using automated sequence alignments [11].
An important impact of this global architecture is the formation and separation of the two distinct active sites (Fig. 1), one within the soluble domain that catalyzes the 2H+/2e- interconversion of succinate and fumarate (dicarboxyate interconversion site; Fig. 3), and one within the membrane spanning regions where quinone and quinol (collectively termed “Q” here if an oxidation state is not implied) are interconverted (Q-site; Fig. 2D-F). These two active sites are separated by ~40 Å, but chemical turnover between them is coupled since electrons that are the products of one reaction become the substrates of the second reaction. Electrons are transferred between these active sites via the three Fe:S clusters coordinated to the SdhB or FrdB subunit (Fig. 1).
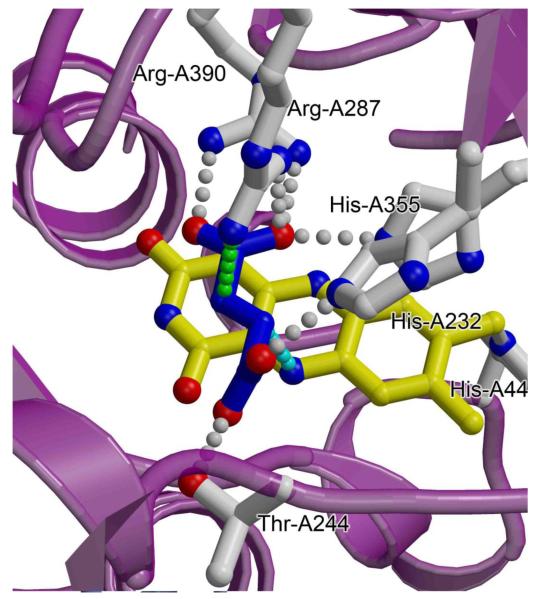
Fig. 3. The dicarboxylate-interconverting active site of complex II enzymes.
A close-up view of the FAD-containing active site of the E. coli SQR (PDB ID: 2WDQ; [36]) is shown with the position of the substrate modeled by removing the O2 from the malate-like intermediate. Backbone atoms of the flavoprotein are colored purple, side chains discussed in-text are highlighted with grey carbons and bonds, the fumarate is shown with blue carbons and bonds, and the FAD is shown with yellow carbons and bonds. Hydrogen-bonding interactions between side chains and the substrate are shown with grey dotted lines. The proposed pathway of hydride transfer between the N5 of the FAD and the C2 of substrate is shown with a cyan dotted line, and the proposed pathway of proton transfer is shown with a green dotted line. Side chain numbering corresponds to the E. coli QFR.
Intriguingly, X-ray crystal structures have demonstrated that the oligomerization state of complex II proteins differs both across the superfamily and across subfamilies with “monomers” [5, 8-10] (where a monomer is defined as the SdhA/B/C/D or FrdA/B/C/(D)), dimers [6], and trimers [7] all observed to date. The impact of the differences in oligomerization is not yet understood, however it is clear that the chemical reaction proceeds through the monomer. Thus, oligomerization may have evolved independently in different subfamily members to promote protein stability, or perhaps as a method to improve the packing of these enzymes into the biological membrane.
2.2 Structures of quinol:fumarate reductases
X-ray crystal structures of QFRs from E. coli [5] (Fig. 1) and W. succinogenes [6] provided the first high-resolution information about the complex II superfamily and represent subfamilies D and B, respectively. The crystal structure of the QFR from the eukaryotic parasite A. suum has only recently been determined [10] and belongs to subfamily C. As anticipated, these three enzymes each have similar folds in the soluble domains. Interestingly, the membrane spanning domains, where menaquinol oxidation takes place, are all arranged around a four-helix bundle [12, 13]. However, there are striking differences in the structures of the membrane spanning domains (Fig. 2A-C) that reflect these being classified into different subfamilies. In specific, these differ in the number of membrane-spanning polypeptides (two for the E. coli and A. suum enzymes, one for the W. succinogenes enzyme), number of membrane-spanning helices (six for the E. coli and A. suum enzymes, five for the W. succinogenes enzyme), the presence of b-type heme (absent for the E. coli enzyme, one b-type heme in the A. suum enzyme, two b-type hemes in the W. succinogenes enzyme), the location of the Q-site (proximal for the E. coli and A. suum enzymes, distal for the W. succinogenes enzyme), and the identity of the quinol used as the electron donor (menaquinol in the E. coli and W. succinogenes enzymes and rhodoquinol in the A. suum enzyme).
2.3 Structures of succinate:quinone oxidoreductases and mitochondrial complex II
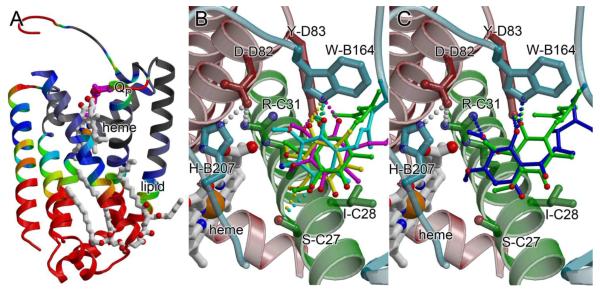
The structure of the E. coli SQR was the first subfamily C SQR to be determined (Fig. 1) [7]. This was followed several years later with the determination of the structures of two mitochondrial complex II enzymes, those of the porcine [8] and avian [9] SQR complexes, both purified from native tissue. Each of these enzymes contains four polypeptide chains. As anticipated by the classification within the complex II superfamily, the soluble domains of bacterial SQR and mitochondrial complex IIs are similar in fold. However, the membrane-spanning subunits of bacterial and mitochondrial SQRs also display structural similarity to each other (and to the A. suum QFR, despite the kinetically favored direction of the reaction), reflecting the grouping of these enzymes into a subfamily. For example, the SdhC and SdhD subunits of porcine and avian SQR are structurally nearly identical, with an rms deviation of Cα atoms 0.8 Å for the SdhC subunit and 0.5 Å for the SdhD subunit. The membrane-spanning regions of the vertebrate mitochondrial enzymes each display somewhat greater structural deviations from the A. suum, QFR and the E. coli SQR, the latter of which contains a large insert between TM1 and TM2 of the SdhC subunit as compared to the other three subfamily C members. However, these structural differences in the A. suum QFR and the E. coli SQR are located away from the functional regions of the protein (Fig. 4A). Indeed, the architecture immediately surrounding the b-type heme and Q-site (Fig. 4B,C, Table 3) are very strongly conserved across the structurally characterized enzymes in subfamily C [11].
Fig. 4. The membrane-spanning domain of subfamily C enzymes.
(A) Maximum deviations (in Å) of Cα atoms from the E. coli, porcine, and avian SQR and the A. suum QFR mapped onto the membrane-spanning subunits of the E. coli SQR (PDB ID: 2WDQ; [36]). Regions where the deviation of Cα atoms is 0.5-1.5 Å are colored black; 1.5-2.5 Å are colored grey, 2.5-3.5 Å are colored blue, 3.5-4.5 Å are colored green, 4.5-5.5 Å are colored yellow, and > 5.5 Å are colored red. The regions of greatest structural difference (red) are located on the distal side of the membrane, and regions near the heme and Q-site are well conserved, with rms deviations around 1Å. (B) The location of ubiquinone from porcine SQR (magenta), avian SQR (yellow), E. coli SQR (green), and rhodoquinol from A. suum QFR (cyan) are superimposed within the binding pocket of the avian SQR (PDB ID:1YQ3 [9]). Chains are colored using the same scheme as in Fig. 3, and side chain numbering is for the E. coli SQR, but can be cross-referenced to other enzymes in Table 3. (C) The location of ubiquinone (green) and the inhibitor atpenin-A5 (blue) modeled within the binding pocket of the avian SQR (PDB ID:1YQ3 [9]) highlights the difference in location between the Q1 and Q2 binding sites.
Table 3.
Selected residues lining the Q-site in subfamily C enzymes
| E. coli SQR | Porcine SQR | Avian SQR | A. suum QFR | Notes |
|---|---|---|---|---|
| B-subunit | ||||
| Pro-B160 | Pro-B169 | Pro-B169 | Pro-B193 | |
| Trp-B164 | Trp-B173 | Trp-B173 | Trp-B197 | Nε H-bonds to O1 of Q |
| Arg-B205 | Arg-B214 | Arg-B214 | Ly-B238 | |
| His-B207 | His-B216 | His-B216 | His-B240 | May H-bond to deeply-bound Q |
| C-subunit | ||||
| Ser-C27 | Ser-C42 | Ser-C39 | Ser-C72 | Oγ H-bonds to Q in some positions |
| Ile-C28 | Ile-C43 | Ile-C40 | Gly-C73 | |
| Arg-C31 | Arg-C46 | Arg-C43 | Arg-C76 | Nη1 may H-bond to deeply-bound Q |
| D-subunit | ||||
| Asp-D82 | Asp-D90 | Asp-D57 | Asp-D106 | |
| Tyr-D83 | Tyr-D91 | Tyr-D58 | Tyr-D107 | OH H-bonds to O1 of Q |
2.3 Soluble homologs of SdhA/FrdA (the flavoprotein)
Soluble homologs of the complex II flavoprotein catalyze α,β-dehydrogenase reactions. Of the known flavoprotein homologs, the soluble fumarate reductase from Shewanella firgidimarina is the best characterized both structurally and biochemically [14-23], with significant structural work on both the soluble fumarate reductase from Shewanella putrefaciens [24] and the l-aspartate oxidase from E. coli [25, 26] also reported. Neither the Fcc3 fumarate reductases nor l-asparate oxidase contain sequence regions similar to the iron protein or the membrane subunits of the complex II superfamily. Instead, both of the Shewanella soluble fumarate reductases contain a single polypeptide chain with sequence analysis predicting a domain similar to the flavoprotein (FrdA/SdhA) subunit of the integral-membrane complex II enzymes fused to a tetra-heme cytochrome. As a result, these soluble fumarate reductases are also known as Fcc3 cytochromes. By contrast, the E. coli l-aspartate oxidase, which catalyzes the oxidation of l-aspartate as the first step in NAD+ biosynthesis [27], does not contain a peripheral domain housing redox cofactors. Despite these architectural differences, investigations of the soluble flavoprotein homologs have been key for elucidating mechanisms of dicarboxylate oxidoreduction in complex II. These investigations have been aided by high expression levels, purification protocols that do not require detergents, reasonably straightforward biophysical properties, and the ability of crystals of these flavoprotein homologs to diffract to high-resolution.
3. Structure-based mechanism of dicarboxylate interconversion
3.1 Overall chemical reaction
Prior to the availability of any structure of a complex II family member, the basic chemical steps of fumarate reduction by QFRs had been delineated. Briefly, fumarate reduction proceeds via a two-step mechanism. A hydride is first transferred from the N5 atom of reduced FAD to the C2 atom of the fumarate double bond (Fig. 3). In the subsequent step, a proton is transferred from a side chain to the C3 of the fumarate carbanion, completing the reaction. The catalytically active form of the enzyme is regenerated when FAD undergoes a 2e-/1H+ reduction. In the integral-membrane complex II family members, the electrons for the FAD reduction are derived from menaquinol oxidation and are transferred from the Q-site to the dicarboxylate site via the Fe:S clusters in the iron protein subunit. Re-protonation of the FAD likely uses a protein-based proton shuttle.
The succinate oxidation reaction is less well characterized, but should proceed by approximately the same reaction steps in the reverse order, with the exception that an anionic flavin semiquinone is detectable during enzyme turnover [28]. An important aspect of succinate oxidation by complex II family members is the requirement for the FAD to be covalently attached to the Nε atom of a conserved histidine residue via an 8α-N(3)-histidyl-linkage [29, 30], which raises the reduction potential of the cofactor ~150 mV (from −219 mV to −55 mV in E. coli) [31]. This covalent linkage is only observed in true complex II family members and not in the soluble flavoprotein homologs. The mechanism of covalent attachment of the FAD to the protein hasn’t been delineated, but FAD insertion may be facilitated by a complex II-specific assembly factor termed Sdh5 in yeast [32], and SdhE in bacteria [33].
The structures of complex II family members have allowed the dicarboxylate interconversion reactions to be placed into the context of the active site architecture and have identified specific side chain residues required for substrate binding, transition state formation, and proton transfer. Structurally, the mechanism of fumarate reduction has been better investigated than that of succinate oxidation. The delineation of the geometric details of fumarate reduction in the complex II superfamily draws heavily upon structure-based enzymology of the easily expressed and biochemically manipulated E. coli QFR and the S. frigidimarina Fcc3 soluble fumarate reductase.
3.2 Substrate binding and alignment
Crystallographic studies have helped develop hypotheses both for the approximate geometry of the reaction intermediate and for the formation of the transition state. SQRs, QFRs, and soluble flavoprotein homologs have been crystallized in the presence of fumarate [6, 34] or other dicarboxylates [8, 9, 14-17, 21, 24, 25, 34-36]. These structures revealed that the side chains of two histidines, two arginines, and one threonine and three backbone amide nitrogens are all within hydrogen-bonding distance of dicarboxylate (Table 2). Thus, any of these could contribute to substrate binding or transition state formation. For simplicity, the E. coli QFR numbering will be referred to in this text regardless of the protein where the original experiment was performed, but each amino acid identity can be cross-referenced to the appropriate numbering for selected enzymes with available structures in Table 2. All of the side chains within the active site (His-A232, Thr-A244, Arg-A287, His-A355, and Arg-A390,) have been investigated by site-directed mutagenesis. Site-specific substitution of residues equivalent to His-A232 His-A355 or Arg-A390 alters both kcat and the Km for fumarate [14, 15, 37], suggesting that they could be involved in turnover. However, crystallographic studies on the S. frigidimarina Fcc3 soluble fumarate reductase indicated that the influence the histidine residues on kcat was likely the result of active site reorganization [14]. Thus, one may speculate that these residues of the FAD-binding domain instead contribute to the binding of substrate. By comparison, Thr-A244 and Arg-A287 appeared to be of primary importance for the catalytic mechanism as monitored by assessing the influence of substitution on kcat/Km [15, 37, 38].
Table 2.
Selected residues lining the dicarboxylate active site in complex II flavoprotein homologs with available structures
| Residues located on the FAD-binding domain (lock-and-key, substrate binding residues) | ||||||||
|---|---|---|---|---|---|---|---|---|
| E. coli | E. coli | Porcine | Avian | A. suum | W. succinogenes | S. frigidimarina | L-Asp Oxidase | Proposed role |
| QFR | SQR | SQR | SQR | QFR | QFR | Fcc3 | ||
| Side chains | ||||||||
| His-A232 | His-A242 | His-A254 | His-A253 | His-A276 | His-A257 | His-365 | His-244 | Substrate binding |
| His-A355 | His-A354 | His-A365 | His-A364 | His-A387 | Arg-A369 | His-504 | His-351 | Substrate binding |
| Arg-A390 | Arg-A399 | Arg-A409 | Arg-A408 | Arg-A432 | Arg-A404 | Arg-544 | Arg-386 | Substrate binding, polarization |
| Backbone | ||||||||
| Gly-A50 | Gly-A51 | Gly-A63 | Gly-A62 | Gly-A85 | Gly-A49 | Gly-170 | Gly-51 | Substrate binding |
| Ser-A393 | Gly-A402 | Ala-A412 | Ala-A411 | Ala-A435 | Gly-A407 | Gly-547 | Ser-389 | Substrate binding |
| Residues located on the capping domain (induced fit, catalytic residues) | ||||||||
|---|---|---|---|---|---|---|---|---|
| E. coli | E. coli | Porcine | Avian | A. suum | W. succinogenes | S. frigidimarina | L-Asp Oxidase | Proposed role |
| QFR | SQR | SQR | SQR | QFR | QFR | Fcc3 | ||
| Side chains | ||||||||
| Thr-A244 | Thr-A254 | Thr-A266 | Thr-A265 | Thr-A288 | Thr-A269 | Thr-377 | Thr-259 | Transition state stabilization |
| Arg-A287 | Arg-A286 | Arg-A298 | Arg-A297 | Arg-A320 | Arg-A301 | Arg-402 | Arg-290 | Proton shuttle, substrate polarization |
| Backbone | ||||||||
| Glu-A245 | Glu-A255 | Glu-A267 | Glu-A266 | Glu-A289 | Glu-A270 | Glu-378 | Glu-260 | Transition state stabilization |
Co-structures of complex II homologs with fumarate [6, 34] have revealed that the substrate binds with the C2-C3 double bond aligned along the C(4a)-N5 bond of the FAD cofactor [34] (Fig. 1). This parallel alignment of bonds has been predicted to maximize the HOMO-LUMO overlap via an orbital steering mechanism [34], and is one of several mechanisms proposed to facilitate hydride transfer from the N5 of FAD. Interestingly, molecules that are commonly classified as competitive inhibitors but can act as alternative substrates show a similar alignment of the transformed bond along the C(4a)-N5 of the FAD; in comparison, competitive inhibitors that are not acted upon by the enzyme do not display this bond alignment [34].
3.3 Formation of a twisted reaction intermediate
The central double bond of fumarate normally constrains this dicarboxylate to a planar conformation. However, co-structures of fumarate bound to the E. coli, W. succinogenes, or A. suum QFR showed a distinct twisting of the carboxylate groups with respect to each other [6, 10, 34] (Fig. 1A). A similarly twisted conformation is observed in the many co-structures of complex II superfamily enzymes [8, 9, 34-36] or soluble flavoprotein homologs [14-17, 21, 24, 25] with dicarboxylate competitive inhibitors (Fig. 1B). A twisted conformation of fumarate is hypothesized to strain the double bond to make it more amenable to reduction [6, 34, 38] and is a second mechanism proposed to promote hydride transfer.
One controversial hypothesis for how the enzyme twists fumarate involves interdomain movement. Like many enzyme active sites, the dicarboxylate interconverting active site of complex II and specifically the location of both Thr-A244 and Arg-A287 is at the interface between two domains. In the complex II flavoprotein, these domains have been termed the FAD-binding domain and the capping domain [5]. The presence of FAD and dicarboxylate appears to influence the orientation of these two domains with respect to each other in the various crystal structures of complex II and flavoprotein homologs [9], with the structures lacking bound substrate or cofactor generally having the more open interdomain orientations [6, 23, 26], while those with inhibitors or substrate bound generally having intermediate or closed interdomain orientations [5, 9, 35, 39].
The most open conformation is observed in the structure of l-aspartate oxidase crystallized without the FAD cofactor [26] and in this conformation a tunnel is opened between domains. This tunnel should allow easy access of the substrate to the dicarboxylate active site. In the open conformation, the binding pocket lined by residues orthologous to His-A232 His-A355 and Arg-A390 is pre-formed, but residues Thr-A244 and Arg-A287 are not aligned for catalysis.
Both the likelihood of interdomain motion and the influence of Thr-A244 and Arg-A287 on the catalytic mechanism have been investigated. The earliest evidence for the importance of interdomain alignment during catalysis was performed prior to the determination of the structure of any complex II family member. This early study identified a reactive thiol equivalent to Cys-A247 in the E. coli QFR. The modification of the residue equivalent to Cys-A247 with sulfhydryl reagents showed significant inhibition of enzyme activity, and this residue could be protected from modification by the inclusion of substrates [40-45]. This motivated the prediction that Cys-A247 was an active site residue. Mutagenesis studies later demonstrated that Cys-A247 was not, in fact, required for enzymatic function [46, 47], however, substitution of the adjacent Arg-A248 resulted in a loss of catalytic activity [47]. The structure of the E. coli QFR [5] later demonstrated that these amino acids are not within the active site, but instead located at the interface between the FAD-binding and capping domains.
Further evidence for interdomain alignment being important for catalysis comes from the examination of disease-associated mutations in the human SQR. The mutation of the amino acid equivalent in position to Gly-A464, which is located between the FAD-binding and capping-domains and is predicted to influence interdomain dynamics [11], is associated with a debilitating neurodegenerative disease known as Leigh’s syndrome in humans [48]. The patient with this substitution had reduced succinate oxidase activity in fibroblasts suggesting that altering the interdomain association influences enzyme turnover.
These studies are consistent with site-directed mutagenesis of the E. coli QFR that alters Thr-A234 of the interdomain hinge (Fig. 5B). A structure of the FrdA T234A variant demonstrated that this mutation stabilized of the open conformation of the enzyme, which is anticipated to be catalytically incompetent since the active site residues Thr-A244 and Arg-A287 would not be appropriately aligned [9]. Indeed this mutation reduced catalytic efficiency 400-fold [38].
Finally, the introduction of disulfide bonds between the FAD-binding and capping domains of the S. frigidimarina Fcc3 fumarate reductase to stabilize the closed conformation showed a 4-fold reduction in kcat and an 11-fold reduction of kcat/Km as compared to the wild-type enzyme, consistent with reduced access of substrate to the closed state [19], but retention of the alignment of the active site residues. Each of these lines of evidence is consistent with interdomain rearrangements accompanying catalysis, and specifically with the open state of the enzyme lacking proper alignment of the active site residues and therefore being catalytically compromised. However, catalytic activity was not fully abrogated in any of these studies, which may reflect limitations of the experimental design or that the open orientation of the two domains is instead important for FAD-insertion.
Regardless of putative enzymatic motions over the course of the reaction coordinate, the amino acids of the capping domain are only aligned for catalysis when the FAD-binding and capping domains are closed around the substrate. Studies on the E. coli QFR demonstrate that mutation of Thr-A244 reduced kcat/Km 34,000-fold, consistent with a role for this side chain in the catalytic mechanism [38]. Combined with interdomain closure, it was proposed that Thr-A244 torques the substrate into the twisted conformation. Residual activity of the FrdA T244A enzyme variant is observed and may reflect the contribution of the backbone amide nitrogen of Glu-A245 to the stabilization of the twisted reaction intermediate.
3.4 Polarization of the bound substrate
After binding within the active site, negative charge is directed from the C2 carbon toward the C3-C4 end of the dicarboxylate by the positively charged protein side chains [15, 21]. Both Arg-A287 and Arg-A390 may contribute to this polarization. The guanido group of Arg-A390 in particular is conserved in position and interacts with the C4 end of the molecule. By comparison, in various crystal structures, the position of Arg-A287 is somewhat variable [12], but the side chain guanido Nη1 and Nη2 nitrogens are commonly positioned near the C3 and carboxylate of C4, respectively. Site-directed substitution of the residue equivalent to Arg-A287 on the S. frigidimarina Fcc3 identified that, subsequent to hydride transfer, the side chain acts as a Lewis acid to stabilize the negative charge of the carbanion at the C3 position [15].
3.5 Proton transfer from an active site arginine to the carbanion reaction intermediate
A side-chain exhibiting a neutral pKa value (7.3 in the E. coli QFR, 7.5 in the E. coli SQR [28]) is proposed to mediate proton transfer to the carbanion C3. Using the E. coli l-aspartate oxidase [37] and the S. frigidimarina soluble Fcc3 fumarate reductase [14-16] as model systems, the influence of site-specific variation of each active site residue on both structure and activity was assessed. This demonstrated that substitution of the residue equivalent to Arg-A287 to a non-ionizable residue reduced kcat to undetectable levels, while substitution with an ionizable residue partially restored activity [15, 37]. This suggests that in addition to its role in stabilizing the carbanion, Arg-A287 likely acts as the proton shuttle.
Like Thr-A244, Arg-A287 is located on the capping domain, and an appropriate interdomain orientation is likely important for the correct alignment of this side chain. Indeed, previous analysis of the structures of complex II flavoprotein homologs shows that the position of Arg-A287 with respect to bound substrate or inhibitor in the active site is dependent upon the interdomain orientation, and may reflect this side chains being mobile during catalytic turnover [12]. Side chain movement is commonly observed in proton shuttling residues, and is consistent with the role of Arg-A287 as the proton shuttle in this system. Furthermore, if interdomain movement does occur during catalysis, Arg-A287 would likely be solvent exposed at the end of each catalytic cycle, facilitating reprotonation.
3.5 Summary of dicarboxylate interconversion in complex II enzymes
In summary, substrate dicarboxylate enters the active site between domains and binds to a pre-organized site comprising three histidine side chains, one arginine side chain, and two amide nitrogens. Binding into this site aligns the C2-C3 bond of substrate along the C(4a)-N5 bond of the FAD cofactor. Interdomain closure aligns the Arg-A287 and Thr-A244 side chains and the Glu-A245 amide nitrogen. The latter two of which twist the substrate into a strained reaction intermediate while the positive charges of Arg-A287 and Arg-A390 polarize the substrate. All of these geometric factors may facilitate hydride transfer from FAD, forming the carbanion intermediate. Arg-A287 then acts as a proton shuttle, completing the reaction cycle. Interestingly, this reaction mechanism combines the lock-and-key binding of substrate to pre-organized residues within the FAD-binding domain with induced-fit by the residues of the capping domain, specifically the side chain of Thr-A244, the backbone amide of Glu-A245, and the side chain of Arg-A287, as the domains change orientation.
4. Mechanisms of quinone and quinol interconversion
4.1 Overview of quinone/quinol binding and oxidoreduction
Various Q species can act as electron carriers between bioenergetic proteins, hence the mechanism of 2H+/2e− oxidoreduction of quinone and quinol has been characterized in several distinct systems. Q molecules have two chemical positions for the oxidoreduction, the O1 and the O4 position, each of which can donate or accept 1H+ and 1e-. In complex II enzymes, the presence of EPR-detectable semiquinone intermediates during turnover is consistent with the step-wise removal of protons and electrons [49, 50]. As a result, there are at least two possibilities for the protons to be transferred to bulk solvent, i.e. either the Q substrate could bind to a site that contains two proton shuttles, one on each side of the binding pocket, that transfer protons at different rates, or the Q substrate could bind to a site that contains a single proton shuttle, and following the removal of one proton, the substrate could move to align the second position with the same proton shuttle.
With either mechanism, Q movement is likely to accompany the reaction cycle. This makes a structural characterization of the reaction challenging since mobile species are difficult to observe by X-ray crystallography. As a result, structural characterization of complex II enzymes bound to competitive inhibitors, site directed mutagenesis, and computational modeling have all been used to identify or verify the location of the Q-sites in complex II superfamily enzymes. This has identified spatially distinct Q-binding sites in the different subfamilies that have been termed QP (for proximal) and QD (for distal). Even given the verification by complementary techniques, the exact location of Q-binding is not perfectly defined in any of these enzymes, and the identification of the proton shuttles has either required substantial complementary experimentation, or has (sometimes) not been proposed.
4.2 Menaquinol oxidation and ubiquinone reduction by the E. coli QFR
The E. coli QFR contains two integral-membrane polypeptide chains (FrdC and FrdD), each with three membrane-spanning helices. Site-directed mutagenesis, fluorescence quench titrations, and EPR spectroscopy are consistent with the presence of a single functional Q-site coupled to fumarate reduction [51]. X-ray crystallographic studies identified two regions of electron density consistent with Q binding within the membrane [5] (Fig. 2C) and termed QP and QD, however, only one of these sites, QP, has a location consistent with direct electron transfer to the soluble domain [52] (Fig. 2F). This site is lined with residues from the FrdB, FrdC, and FrdD subunits and is located within 8 Å of the electron-transferring 3Fe:4S cofactor [5]. QP was confirmed as a functional site with further X-ray crystallographic studies [53] of the E. coli QFR in complex with the quinol-blocking inhibitors heptyl quinolone N-oxide and 1-(p-chlorophenyl)ethyl] 4,6-dinitrophenol [54, 55]. Oxidation of menaquinol at QP would release protons to the side of the membrane where fumarate reduction occurs, which results in an electro-neutral process [56].
The E. coli QFR can bind either mena-Q or ubi-Q at the QP site [54], however, menaquinol oxidation is believed to be physiologically coupled to the fumarate reduction reaction in the enzyme. Investigation of the mechanism of menaquinol oxidation has combined structural and spectroscopic measurements. Both FTIR spectroscopic measurements and the crystal structure indicate that the O1 of the bound mena-Q forms hydrogen-bonding contacts with Gln-B225Nε2, Lys-B228 Nζ and Trp-D14 Nε1, while the O4 forms a single hydrogen-bonding contact to Glu-C29 Oε1.
Available evidence is most consistent with the presence of two separate proton shuttles for menaquinol oxidation at QP in the E. coli QFR. Mutation of Glu-C29 to a non-ionizable residue results in the formation of an EPR-detectable menasemiquinone radical [49, 51]. This suggests that one proton shuttle removes a proton from the O1 of the menaquinol, but that enzyme turnover cannot be completed. The most parsimonious explanation is that Glu-C29 normally acts as the proton shuttle from the O4 position. The only ionizable residue interacting with the O1 position is Lys-B228. While this suggests that Lys-B228 is the proton shuttle from the O1 position, this hasn’t been rigorously demonstrated, and an alternative is that the Q moves during turnover, such that the proton is shuttled from the O1 position via another, as yet unidentified side chain prior to the interaction between the menaquinol and Lys-B228.
When the E. coli QFR performs succinate oxidation, it is believed to couple this reaction to ubiquinone reduction. Here, it has been suggested that the O1 position of ubiquinone maintains the same hydrogen-bonding contacts as menaquinol (i.e. to Gln-B225 Nε2, Lys-B228 Nζ and Trp-D14 Nε1). However, the quinone ring rotates slightly such that the O4 position hydrogen-bonds to the side chain of Arg-D28 (rather than Glu-C29) within the membrane [57]. Consistent with this, the mutation of Glu-C29 to a non-ionizable residue results in protein that is fully functional for ubiquinone reduction but deficient in menaquinol oxidation [57]. This minimally indicates that the proton shuttle for menaquinol oxidation differs from that used for ubiquinone reduction, and strongly suggests Arg-D28 acts as the proton shuttle for ubiquinone reduction. The limited resolution of the structures of the E. coli QFR has prevented the accurate placement of water molecules within the Q-site, such that the pathway of proton transfer from either menaquinol oxidation or ubiquinone reduction to bulk solvent has not yet been further mapped.
4.3 Menaquinol oxidation by the W. succinogenes QFR
Structural mapping of the Q-site and proton shuttle pathway in the W. succinogenes QFR originates with a crystal structure of this enzyme in complex with 2,3-dimethyl-1,4-napthoquinone [58]. This redox-active molecule is structurally similar to the menaquinone head group, and the corresponding quinol can support QFR activity in in vitro assays. The 2,3-dimethyl-1,4-napthoquinone binds within the FrdC subunit in a location that is distal to the soluble domain (QD) (Fig. 2A). The location of the menaquinol binding site within the W. succinogenes QFR was verified using a combination of computational modeling and site-directed mutagenesis of Glu-C66 [59, 60], a residue located adjacent to the binding site for 2,3-dimethyl-1,4-napthoquinone and between the QD-site and the positive side of the membrane (Fig. 2D), and the only obvious candidate for a proton shuttle from this site. The loss of activity in the FrdC E66Q variant led to the proposal that the identified QD site was functionally relevant and that the side chain of Glu-C66 acted as a proton shuttle. The location of this proton shuttle would be anticipated to release protons to the positive side of the membrane, which would contribute to the formation of the transmembrane electrochemical gradient. However, the energy of transferrin electrons from menaquinol to fumarate is theoretically insufficient to support proton pumping.
This apparent conundrum was reconciled with investigation of a second integral-membrane glutamate residue, Glu-C180, which is located in the middle of the membrane-spanning region and also demonstrated to be critical for enzyme activity [58, 61, 62]. Multiple studies are consistent with Glu-C180 and the heme propionates participating in a second, distinct proton shuttling pathway that brings protons back along the gradient to the negative side of the membrane [58, 60, 61, 63-65] (Fig. 2A). When acting in conjunction with proton shuttling from Glu-C66, this second proton shuttling pathway renders the W. succinogenes QFR reaction cycle energetically neutral. The presence of two distinct proton shuttles with opposing directions and mediated by glutamates (E) is termed the “E-pathway hypothesis” and may be important for function in enzymes within the B subfamily of complex II enzymes [59].
4.4 Rhodoquinol oxidation by A. suum QFR
The A. suum QFR couples fumarate reduction to rhodoquinol oxidation. In the structure, a rhodo-Q was identified at a site classified as QP (Fig. 4B), and co-crystallization of the A. suum QFR with the fungicide and competitive inhibitor flutolanil confirms this as a functional Q-site [10]. The location of this site strongly suggests that it is the immediate electron donor to the 3Fe:4S cluster.
The structural understanding of the A. suum enzyme lags behind that of both the E. coli and W. succinogenes QFRs by 13 years, and further in-depth studies of the structural basis for rhodoquinol oxidation by the A. suum have not yet been performed. However, the classification of this enzyme into subfamily C (Fig. 2B,E, Fig. 4) and the similarity between the QP-sites in the A. suum QFR and all other subfamily C members suggests that rhodoquinol oxidation might proceed by the reverse mechanism as ubiquinone reduction in subfamily C SQRs (described below in section 4.5).
4.5 Ubiquinone reduction by subfamily C SQRs
Nearly all of the detailed structure-function studies aimed at identification of the mechanism of Q oxidoreduction in subfamily C enzymes have been performed on the E. coli SQR. However, the strong sequence and structural conservation surrounding both the heme and the QP site in subfamily C enzymes (Fig. 4A, Table 3) suggests that results from the biochemical investigations of the easily-manipulated E. coli enzyme are applicable to the remaining members of the subfamily. For simplicity, the E. coli SQR side chain numbering will be used in-text, but can be cross-referenced to other subfamily C complex II enzymes in Table 3.
In subfamily C enzymes, the Q-site is classified as Qp (Fig. 2B) and physiologically couples the reduction of ubiquinone to the oxidation of succinate, thus contributing to the formation of the reduced quinone pool. The binding site for the ubiquinone head group is located at the interface between the membrane and soluble regions of SQR and is lined with residues from the SdhB, SdhC, and SdhD subunits. Only the first two isoprene groups of the ubiquinone is observed in crystal structures, suggesting that the majority of the hydrophobic tail is not specifically bound to the protein. The location of the Q-site in subfamily C SQRs was identified with co-crystallization of ubiquinone with E. coli [7], avian [9], and porcine [8] SQRs, and verified via co-crystallization studies of subfamily C enzymes with competitive inhibitors of Q-binding, including 2-thenoyltrifluoroacetone [8], carboxin [9, 36], pentacholorophenol [36], atpenin-A5 [66], or 2-(1-methyl-hexyl)-4,6-dinitrophenol [7]. Examination of the placement of ubi-Q within the structures of subfamily C complex II enzymes (Fig. 4B) reveals differences in positioning of the head group in each structure despite a strongly conserved architecture of the surrounding binding pocket [7-10]. It is not clear if this difference in placement reflects real differences in binding or if this instead reflects challenges in substrate placement within ambiguous crystallographic electron density. Interestingly, the O1 of ubi-Q makes conserved hydrogen-bonding contacts to Trp-B173 Nε and Tyr-D83 OH in all structures available to date. Corroborating this placement are studies of E. coli SQR where alteration of Tyr-D83 by site-directed mutagenesis results in a loss of an EPR-detectable semiquinone signal [50]. However, only the placement of ubi-Q within the avian SQR model (and rhodo-Q within the A. suum QFR) is consistent with any hydrogen-bonding partner for the O4 of position, which is to the side chain analogous to Ser-C27. Site-directed substitution of Ser-C27 results in the loss of the EPR-detectable semiquinone signal [50], and suggests that Ser-C27 is important for stabilization of this radical.
Of note is the structure between the E. coli SQR and atpenin-A5 [66] (Fig. 4C). This competitive inhibitor has been demonstrated to bind more deeply within the QP-site than ubiquinone, prompting computational modeling studies of the native substrate within the Q-site. These calculations were consistent with Q being able to bind in two distinct locations, with Q1 observed in the crystal structures available to-date, and Q2 closely superimposing with the atpenin-A5 binding location. In the Q2 position, the O3 methoxy position of ubiquinone is proposed to approach and hydrogen-bond to the side chain of His-B207, while the O4 would form a hydrogen-bond with Ser-C27. Attempts to validate the Q2 position with site-directed mutagenesis of His-B207 show that substitution of this side chain has limited effects on enzyme turnover [67]. Furthermore, this position is commonly mutated in fungi resistant to carboxin [68]. Taken together, this suggests that the Q2 position may be of limited importance for catalytic activity, but that this part of the Q-site is required for the binding of some competitive inhibitors of ubiquinone catalysis.
It has remained challenging to propose a clear side-chain mediated proton shuttle from either the Q1 or the Q2-position, motivating the suggestion that ordered water molecules were major components of the proton-shuttling pathway. However, site-directed mutagenesis of side chains proposed to order water molecules near the Q-site have had measurable but limited effects on Q turnover [36, 69]. An alternative proposal is that the Tyr-D83 and Trp-B173 are sufficiently close to the surface of the phospholipid bilayer that they may be exposed to solvent and not require an additional proton shuttling pathway. Clearly, further investigation of Q-turnover in subfamily C enzymes is warranted.
Subfamily C enzymes contain a single b-type heme within the membrane spanning subunits. The location of the b-type heme is off-pathway from the electron transfer cofactors and this cofactor is not involved in direct electron transfer between the dicarboxylate active site and the Q-site [7]. Indeed, the converse is true, electrons appear to require transfer through the to Q-site if they are to reduce heme [50]. Investigation of a variant E. coli SQR lacking the b-type heme shows reduced stability of the complex when extracted into detergent, but retains all other physiological properties of the enzyme, including redox cycling [70]. Further, the lowering of the reduction potential or complete absence of the b-type heme does not increase the formation of reactive radical species [70, 71], which suggests that the heme does not act as a sink for excess electrons. Accordingly, the functional significance of this cofactor remains somewhat mysterious.
5. Summary and Conclusions
The wealth of information from 14 years of structural investigations on complex II enzymes has complemented 100 years of biochemical investigation on complex II enzymes and has contributed to the development of detailed mechanistic proposals for catalysis at the multiple active sites. The conserved oxidoreduction of succinate and fumarate is common to all complex II superfamily members, and as anticipated, the investigation of dicarboxylate oxidoreduction in any homolog has yielded insights into the mechanisms of complex II superfamily. By comparison, the mechanism for Q oxidoreduction appears to be somewhat unique in enzymes belonging to each subfamily, reflecting how independent evolution has resulted in a diverse set of solutions for a single chemical problem.
Acknowledgements
Work from the author’s laboratory has been funded by the National Institutes of Health grant GM079419, the Ellison Medical Foundation AG-NS-0325-06, and pilot funds from the Vanderbilt Center for Molecular Toxicology NIH P30 ES000267.
Abbreviations
- SQR
succinate:quinone oxidoreductase
- QFR
quinol:fumarate reductase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Kroger A. Fumarate as terminal acceptor of phosphorylative electron transport. Biochim Biophys Acta. 1978;505:129–145. doi: 10.1016/0304-4173(78)90010-1. [DOI] [PubMed] [Google Scholar]
- [2].Hirsch CA, Rasminsky M, Davis BD, Lin EC. A Fumarate Reductase in Escherichia Coli Distinct from Succinate Dehydrogenase. J Biol Chem. 1963;238:3770–3774. [PubMed] [Google Scholar]
- [3].Hagerhall C, Hederstedt L. A structural model for the membrane-integral domain of succinate: quinone oxidoreductases. FEBS Lett. 1996;389:25–31. doi: 10.1016/0014-5793(96)00529-7. [DOI] [PubMed] [Google Scholar]
- [4].Lemos RS, Fernandes AS, Pereira MM, Gomes CM, Teixeira M. Quinol:fumarate oxidoreductases and succinate:quinone oxidoreductases: phylogenetic relationships, metal centres and membrane attachment. Biochim Biophys Acta. 2002;1553:158–170. doi: 10.1016/s0005-2728(01)00239-0. [DOI] [PubMed] [Google Scholar]
- [5].Iverson TM, Luna-Chavez C, Cecchini G, Rees DC. Structure of the Escherichia coli fumarate reductase respiratory complex. Science. 1999;284:1961–1966. doi: 10.1126/science.284.5422.1961. [DOI] [PubMed] [Google Scholar]
- [6].Lancaster CR, Kroger A, Auer M, Michel H. Structure of fumarate reductase from Wolinella succinogenes at 2.2 A resolution. Nature. 1999;402:377–385. doi: 10.1038/46483. [DOI] [PubMed] [Google Scholar]
- [7].Yankovskaya V, Horsefield R, Tornroth S, Luna-Chavez C, Miyoshi H, Leger C, Byrne B, Cecchini G, Iwata S. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science. 2003;299:700–704. doi: 10.1126/science.1079605. [DOI] [PubMed] [Google Scholar]
- [8].Sun F, Huo X, Zhai Y, Wang A, Xu J, Su D, Bartlam M, Rao Z. Crystal structure of mitochondrial respiratory membrane protein complex II. Cell. 2005;121:1043–1057. doi: 10.1016/j.cell.2005.05.025. [DOI] [PubMed] [Google Scholar]
- [9].Huang LS, Sun G, Cobessi D, Wang AC, Shen JT, Tung EY, Anderson VE, Berry EA. 3-nitropropionic acid is a suicide inhibitor of mitochondrial respiration that, upon oxidation by complex II, forms a covalent adduct with a catalytic base arginine in the active site of the enzyme. J Biol Chem. 2006;281:5965–5972. doi: 10.1074/jbc.M511270200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Shimizu H, Osanai A, Sakamoto K, Inaoka DK, Shiba T, Harada S, Kita K. Crystal structure of mitochondrial quinol-fumarate reductase from the parasitic nematode Ascaris suum. J Biochem. 2012 doi: 10.1093/jb/mvs051. [DOI] [PubMed] [Google Scholar]
- [11].Iverson TM, Maklashina E, Cecchini G. Structural Basis for Malfunction in Complex II. J Biol. Chem. 2012 Aug 17; doi: 10.1074/jbc.R112.408419. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Iverson TM, Luna-Chavez C, Schroder I, Cecchini G, Rees DC. Analyzing your complexes: structure of the quinol-fumarate reductase respiratory complex. Curr Opin Struct Biol. 2000;10:448–455. doi: 10.1016/s0959-440x(00)00113-5. [DOI] [PubMed] [Google Scholar]
- [13].Lancaster CR, Kroger A. Succinate: quinone oxidoreductases: new insights from X-ray crystal structures. Biochim Biophys Acta. 2000;1459:422–431. doi: 10.1016/s0005-2728(00)00180-8. [DOI] [PubMed] [Google Scholar]
- [14].Doherty MK, Pealing SL, Miles CS, Moysey R, Taylor P, Walkinshaw MD, Reid GA, Chapman SK. Identification of the active site acid/base catalyst in a bacterial fumarate reductase: a kinetic and crystallographic study. Biochemistry. 2000;39:10695–10701. doi: 10.1021/bi000871l. [DOI] [PubMed] [Google Scholar]
- [15].Mowat CG, Moysey R, Miles CS, Leys D, Doherty MK, Taylor P, Walkinshaw MD, Reid GA, Chapman SK. Kinetic and crystallographic analysis of the key active site acid/base arginine in a soluble fumarate reductase. Biochemistry. 2001;40:12292–12298. doi: 10.1021/bi011360h. [DOI] [PubMed] [Google Scholar]
- [16].Mowat CG, Pankhurst KL, Miles CS, Leys D, Walkinshaw MD, Reid GA, Chapman SK. Engineering water to act as an active site acid catalyst in a soluble fumarate reductase. Biochemistry. 2002;41:11990–11996. doi: 10.1021/bi0203177. [DOI] [PubMed] [Google Scholar]
- [17].Pankhurst KL, Mowat CG, Miles CS, Leys D, Walkinshaw MD, Reid GA, Chapman SK. Role of His505 in the soluble fumarate reductase from Shewanella frigidimarina. Biochemistry. 2002;41:8551–8556. doi: 10.1021/bi020155e. [DOI] [PubMed] [Google Scholar]
- [18].Pankhurst KL, Mowat CG, Rothery EL, Hudson JM, Jones AK, Miles CS, Walkinshaw MD, Armstrong FA, Reid GA, Chapman SK. A proton delivery pathway in the soluble fumarate reductase from Shewanella frigidimarina. J Biol Chem. 2006;281:20589–20597. doi: 10.1074/jbc.M603077200. [DOI] [PubMed] [Google Scholar]
- [19].Rothery EL, Mowat CG, Miles CS, Mott S, Walkinshaw MD, Reid GA, Chapman SK. Probing domain mobility in a flavocytochrome. Biochemistry. 2004;43:4983–4989. doi: 10.1021/bi030261w. [DOI] [PubMed] [Google Scholar]
- [20].Rothery EL, Mowat CG, Miles CS, Walkinshaw MD, Reid GA, Chapman SK. Histidine 61: an important heme ligand in the soluble fumarate reductase from Shewanella frigidimarina. Biochemistry. 2003;42:13160–13169. doi: 10.1021/bi030159z. [DOI] [PubMed] [Google Scholar]
- [21].Taylor P, Pealing SL, Reid GA, Chapman SK, Walkinshaw MD. Structural and mechanistic mapping of a unique fumarate reductase. Nat Struct Biol. 1999;6:1108–1112. doi: 10.1038/70045. [DOI] [PubMed] [Google Scholar]
- [22].Wardrope C, Mowat CG, Walkinshaw MD, Reid GA, Chapman SK. Fumarate reductase: structural and mechanistic insights from the catalytic reduction of 2-methylfumarate. FEBS Lett. 2006;580:1677–1680. doi: 10.1016/j.febslet.2006.02.016. [DOI] [PubMed] [Google Scholar]
- [23].Bamford V, Dobbin PS, Richardson DJ, Hemmings AM. Open conformation of a flavocytochrome c3 fumarate reductase. Nat Struct Biol. 1999;6:1104–1107. doi: 10.1038/70039. [DOI] [PubMed] [Google Scholar]
- [24].Leys D, Tsapin AS, Nealson KH, Meyer TE, Cusanovich MA, Van Beeumen JJ. Structure and mechanism of the flavocytochrome c fumarate reductase of Shewanella putrefaciens MR-1. Nat Struct Biol. 1999;6:1113–1117. doi: 10.1038/70051. [DOI] [PubMed] [Google Scholar]
- [25].Bossi RT, Negri A, Tedeschi G, Mattevi A. Structure of FAD-bound L-aspartate oxidase: insight into substrate specificity and catalysis. Biochemistry. 2002;41:3018–3024. doi: 10.1021/bi015939r. [DOI] [PubMed] [Google Scholar]
- [26].Mattevi A, Tedeschi G, Bacchella L, Coda A, Negri A, Ronchi S. Structure of L-aspartate oxidase: implications for the succinate dehydrogenase/fumarate reductase oxidoreductase family. Structure. 1999;7:745–756. doi: 10.1016/s0969-2126(99)80099-9. [DOI] [PubMed] [Google Scholar]
- [27].Nasu S, Wicks FD, Gholson RK. L-Aspartate oxidase, a newly discovered enzyme of Escherichia coli, is the B protein of quinolinate synthetase. J Biol Chem. 1982;257:626–632. [PubMed] [Google Scholar]
- [28].Maklashina E, Iverson TM, Sher Y, Kotlyar V, Andrell J, Mirza O, Hudson JM, Armstrong FA, Rothery RA, Weiner JH, Cecchini G. Fumarate reductase and succinate oxidase activity of Escherichia coli complex II homologs are perturbed differently by mutation of the flavin binding domain. J Biol Chem. 2006;281:11357–11365. doi: 10.1074/jbc.M512544200. [DOI] [PubMed] [Google Scholar]
- [29].Weiner JH, Dickie P. Fumarate reductase of Escherichia coli. Elucidation of the covalent-flavin component. J Biol Chem. 1979;254:8590–8593. [PubMed] [Google Scholar]
- [30].Walker WH, Singer TP. Identification of the covalently bound flavin of succinate dehydrogenase as 8-alpha-(histidyl) flavin adenine dinucleotide. J Biol Chem. 1970;245:4224–4225. [PubMed] [Google Scholar]
- [31].Blaut M, Whittaker K, Valdovinos A, Ackrell BA, Gunsalus RP, Cecchini G. Fumarate reductase mutants of Escherichia coli that lack covalently bound flavin. J Biol Chem. 1989;264:13599–13604. [PubMed] [Google Scholar]
- [32].Hao HX, Khalimonchuk O, Schraders M, Dephoure N, Bayley JP, Kunst H, Devilee P, Cremers CW, Schiffman JD, Bentz BG, Gygi SP, Winge DR, Kremer H, Rutter J. SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma. Science. 2009;325:1139–1142. doi: 10.1126/science.1175689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].McNeil MB, Clulow JS, Wilf NM, Salmond GP, Fineran PC. SdhE is a conserved protein required for flavinylation of succinate dehydrogenase in bacteria. J Biol Chem. 2012;287:18418–18428. doi: 10.1074/jbc.M111.293803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Tomasiak TM, Archuleta TL, Andrell J, Luna-Chavez C, Davis TA, Sarwar M, Ham AJ, McDonald WH, Yankovskaya V, Stern HA, Johnston JN, Maklashina E, Cecchini G, Iverson TM. Geometric restraint drives on- and off-pathway catalysis by the Escherichia coli menaquinol:fumarate reductase. J Biol Chem. 2011;286:3047–3056. doi: 10.1074/jbc.M110.192849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Huang LS, Shen JT, Wang AC, Berry EA. Crystallographic studies of the binding of ligands to the dicarboxylate site of Complex II, and the identity of the ligand in the “oxaloacetate-inhibited” state. Biochim Biophys Acta. 2006;1757:1073–1083. doi: 10.1016/j.bbabio.2006.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ruprecht J, Yankovskaya V, Maklashina E, Iwata S, Cecchini G. Structure of Escherichia coli succinate:quinone oxidoreductase with an occupied and empty quinone-binding site. J Biol Chem. 2009;284:29836–29846. doi: 10.1074/jbc.M109.010058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Tedeschi G, Ronchi S, Simonic T, Treu C, Mattevi A, Negri A. Probing the active site of L-aspartate oxidase by site-directed mutagenesis: role of basic residues in fumarate reduction. Biochemistry. 2001;40:4738–4744. doi: 10.1021/bi002406u. [DOI] [PubMed] [Google Scholar]
- [38].Tomasiak TM, Maklashina E, Cecchini G, Iverson TM. A threonine on the active site loop controls transition state formation in Escherichia coli respiratory complex II. J Biol Chem. 2008;283:15460–15468. doi: 10.1074/jbc.M801372200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lancaster CR, Gross R, Simon J. A third crystal form of Wolinella succinogenes quinol:fumarate reductase reveals domain closure at the site of fumarate reduction. Eur J Biochem. 2001;268:1820–1827. [PubMed] [Google Scholar]
- [40].Ackrell BA, Cochran B, Cecchini G. Interactions of oxaloacetate with Escherichia coli fumarate reductase. Arch Biochem Biophys. 1989;268:26–34. doi: 10.1016/0003-9861(89)90561-4. [DOI] [PubMed] [Google Scholar]
- [41].Unden G, Kroger A. An essential sulfhydryl group at the substrate site of the fumarate reductase of Vibrio succinogenes. FEBS Lett. 1980;117:323–326. doi: 10.1016/0014-5793(80)80972-0. [DOI] [PubMed] [Google Scholar]
- [42].Robinson JJ, Weiner JH. Molecular properties of fumarate reductase isolated from the cytoplasmic membrane of Escherichia coli, Can. J Biochem. 1982;60:811–816. doi: 10.1139/o82-101. [DOI] [PubMed] [Google Scholar]
- [43].Kenney WC. The reaction of N-ethylmaleimide at the active site of succinate dehydrogenase. J Biol Chem. 1975;250:3089–3094. [PubMed] [Google Scholar]
- [44].Vinogradov AD, Gavrikova EV, Zuevsky VV. Reactivity of the sulfhydryl groups of soluble succinate dehydrogenase. Eur J Biochem. 1976;63:365–371. doi: 10.1111/j.1432-1033.1976.tb10238.x. [DOI] [PubMed] [Google Scholar]
- [45].Vinogradov AD, Winter D, King TE. The binding site for oxaloacetate on succinate dehydrogenase. Biochem Biophys Res Commun. 1972;49:441–444. doi: 10.1016/0006-291x(72)90430-5. [DOI] [PubMed] [Google Scholar]
- [46].Hederstedt L, Heden LO. New properties of Bacillus subtilis succinate dehydrogenase altered at the active site. The apparent active site thiol of succinate oxidoreductases is dispensable for succinate oxidation. Biochem J. 1989;260:491–497. doi: 10.1042/bj2600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Schroder I, Gunsalus RP, Ackrell BA, Cochran B, Cecchini G. Identification of active site residues of Escherichia coli fumarate reductase by site-directed mutagenesis. J Biol Chem. 1991;266:13572–13579. [PubMed] [Google Scholar]
- [48].Parfait B, Chretien D, Rotig A, Marsac C, Munnich A, Rustin P. Compound heterozygous mutations in the flavoprotein gene of the respiratory chain complex II in a patient with Leigh syndrome. Hum Genet. 2000;106:236–243. doi: 10.1007/s004390051033. [DOI] [PubMed] [Google Scholar]
- [49].Hagerhall C, Magnitsky S, Sled VD, Schroder I, Gunsalus RP, Cecchini G, Ohnishi T. An Escherichia coli mutant quinol:fumarate reductase contains an EPR-detectable semiquinone stabilized at the proximal quinone-binding site. J Biol Chem. 1999;274:26157–26164. doi: 10.1074/jbc.274.37.26157. [DOI] [PubMed] [Google Scholar]
- [50].Tran QM, Rothery RA, Maklashina E, Cecchini G, Weiner JH. The quinone binding site in Escherichia coli succinate dehydrogenase is required for electron transfer to the heme b. J Biol Chem. 2006;281:32310–32317. doi: 10.1074/jbc.M607476200. [DOI] [PubMed] [Google Scholar]
- [51].Rothery RA, Seime AM, Spiers AM, Maklashina E, Schroder I, Gunsalus RP, Cecchini G, Weiner JH. Defining the Q-site of Escherichia coli fumarate reductase by site-directed mutagenesis, fluorescence quench titrations and EPR spectroscopy. Febs J. 2005;272:313–326. doi: 10.1111/j.1742-4658.2004.04469.x. [DOI] [PubMed] [Google Scholar]
- [52].Page CC, Moser CC, Chen X, Dutton PL. Natural engineering principles of electron tunnelling in biological oxidation-reduction. Nature. 1999;402:47–52. doi: 10.1038/46972. [DOI] [PubMed] [Google Scholar]
- [53].Iverson TM, Luna-Chavez C, Croal LR, Cecchini G, Rees DC. Crystallographic studies of the Escherichia coli quinol-fumarate reductase with inhibitors bound to the quinol-binding site. J Biol Chem. 2002;277:16124–16130. doi: 10.1074/jbc.M200815200. [DOI] [PubMed] [Google Scholar]
- [54].Maklashina E, Cecchini G. Comparison of catalytic activity and inhibitors of quinone reactions of succinate dehydrogenase (Succinate-ubiquinone oxidoreductase) and fumarate reductase (Menaquinol-fumarate oxidoreductase) from Escherichia coli. Arch Biochem Biophys. 1999;369:223–232. doi: 10.1006/abbi.1999.1359. [DOI] [PubMed] [Google Scholar]
- [55].Yankovskaya V, Sablin SO, Ramsay RR, Singer TP, Ackrell BA, Cecchini G, Miyoshi H. Inhibitor probes of the quinone binding sites of mammalian complex II and Escherichia coli fumarate reductase. J Biol Chem. 1996;271:21020–21024. doi: 10.1074/jbc.271.35.21020. [DOI] [PubMed] [Google Scholar]
- [56].Ohnishi T, Moser CC, Page CC, Dutton PL, Yano T. Simple redox-linked proton-transfer design: new insights from structures of quinol-fumarate reductase. Structure. 2000;8:R23–32. doi: 10.1016/s0969-2126(00)00098-8. [DOI] [PubMed] [Google Scholar]
- [57].Maklashina E, Hellwig P, Rothery RA, Kotlyar V, Sher Y, Weiner JH, Cecchini G. Differences in protonation of ubiquinone and menaquinone in fumarate reductase from Escherichia coli. J Biol Chem. 2006;281:26655–26664. doi: 10.1074/jbc.M602938200. [DOI] [PubMed] [Google Scholar]
- [58].Lancaster CR, Sauer US, Gross R, Haas AH, Graf J, Schwalbe H, Mantele W, Simon J, Madej MG. Experimental support for the “E pathway hypothesis” of coupled transmembrane e− and H+ transfer in dihemic quinol:fumarate reductase. Proc Natl Acad Sci U S A. 2005;102:18860–18865. doi: 10.1073/pnas.0509711102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Haas AH, Lancaster CR. Calculated coupling of transmembrane electron and proton transfer in dihemic quinol:fumarate reductase. Biophys J. 2004;87:4298–4315. doi: 10.1529/biophysj.104.042945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Lancaster CR, Gorss R, Haas A, Ritter M, Mantele W, Simon J, Kroger A. Essential role of Glu-C66 for menaquinol oxidation indicates transmembrane electrochemical potential generation by Wolinella succinogenes fumarate reductase. Proc Natl Acad Sci U S A. 2000;97:13051–13056. doi: 10.1073/pnas.220425797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Haas AH, Sauer US, Gross R, Simon J, Mantele W, Lancaster CR. FTIR difference spectra of Wolinella succinogenes quinol:fumarate reductase support a key role of Glu C180 within the “E-pathway hypothesis” of coupled transmembrane electron and proton transfer. Biochemistry. 2005;44:13949–13961. doi: 10.1021/bi051011d. [DOI] [PubMed] [Google Scholar]
- [62].Madej MG, Nasiri HR, Hilgendorff NS, Schwalbe H, Lancaster CR. Evidence for transmembrane proton transfer in a dihaem-containing membrane protein complex. Embo J. 2006;25:4963–4970. doi: 10.1038/sj.emboj.7601361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Lancaster CR. Wolinella succinogenes quinol:fumarate reductase-2.2-A resolution crystal structure and the E-pathway hypothesis of coupled transmembrane proton and electron transfer. Biochim Biophys Acta. 2002;1565:215–231. doi: 10.1016/s0005-2736(02)00571-0. [DOI] [PubMed] [Google Scholar]
- [64].Mileni M, Haas AH, Mantele W, Simon J, Lancaster CR. Probing heme propionate involvement in transmembrane proton transfer coupled to electron transfer in dihemic quinol:fumarate reductase by 13C- labeling and FTIR difference spectroscopy. Biochemistry. 2005;44:16718–16728. doi: 10.1021/bi051034s. [DOI] [PubMed] [Google Scholar]
- [65].Lancaster CR, Haas AH, Madej MG, Mileni M. Recent progress on obtaining theoretical and experimental support for the “E-pathway hypothesis” of coupled transmembrane electron and proton transfer in dihaem-containing quinol:fumarate reductase. Biochim Biophys Acta. 2006;1757:988–995. doi: 10.1016/j.bbabio.2006.05.012. [DOI] [PubMed] [Google Scholar]
- [66].Horsefield R, Yankovskaya V, Sexton G, Whittingham W, Shiomi K, Omura S, Byrne B, Cecchini G, Iwata S. Structural and computational analysis of the quinone-binding site of complex II (succinate-ubiquinone oxidoreductase): a mechanism of electron transfer and proton conduction during ubiquinone reduction. J Biol Chem. 2006;281:7309–7316. doi: 10.1074/jbc.M508173200. [DOI] [PubMed] [Google Scholar]
- [67].Ruprecht J, Iwata S, Rothery RA, Weiner JH, Maklashina E, Cecchini G. Perturbation of the quinone- binding site of complex II alters the electronic properties of the proximal [3Fe-4S] iron-sulfur cluster. J Biol Chem. 2011;286:12756–12765. doi: 10.1074/jbc.M110.209874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Scalliet G, Bowler J, Luksch T, Kirchhofer-Allan L, Steinhauer D, Ward K, Niklaus M, Verras A, Csukai M, Daina A, Fonne-Pfister R. Mutagenesis and functional studies with succinate dehydrogenase inhibitors in the wheat pathogen Mycosphaerella graminicola. PLoS One. 2012;7:e35429. doi: 10.1371/journal.pone.0035429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Cheng VW, Johnson A, Rothery RA, Weiner JH. Alternative sites for proton entry from the cytoplasm to the quinone binding site in Escherichia coli succinate dehydrogenase. Biochemistry. 2008;47:9107–9116. doi: 10.1021/bi801008e. [DOI] [PubMed] [Google Scholar]
- [70].Tran QM, Rothery RA, Maklashina E, Cecchini G, Weiner JH. Escherichia coli succinate dehydrogenase variant lacking the heme b. Proc Natl Acad Sci U S A. 2007;104:18007–18012. doi: 10.1073/pnas.0707732104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Tran QM, Fong C, Rothery RA, Maklashina E, Cecchini G, Weiner JH. Out of plane distortions of the heme b of Escherichia coli succinate dehydrogenase. PLoS One. 2012;7:e32641. doi: 10.1371/journal.pone.0032641. [DOI] [PMC free article] [PubMed] [Google Scholar]