Abstract
Proteins regulate gene expression by controlling mRNA biogenesis, localization, translation and decay. Identifying the composition, diversity and function of mRNPs (mRNA protein complexes) is essential to understanding these processes. In a global survey of S. cerevisiae mRNA binding proteins we identified 120 proteins that cross-link to mRNA, including 66 new mRNA binding proteins. These include kinases, RNA modification enzymes, metabolic enzymes, and tRNA and rRNA metabolism factors. These proteins show dynamic subcellular localization during stress, including assembly into stress granules and P-bodies (Processing-bodies). CLIP (cross-linking and immunoprecipitation) analyses of the P-body components Pat1, Lsm1, Dhh1 and Sbp1 identified sites of interaction on specific mRNAs revealing positional binding preferences and co-assembly preferences. Taken together, this work defines the major yeast mRNP proteins, reveals widespread changes in their subcellular location during stress, and begins to define assembly rules for P-body mRNPs.
Introduction
The control of cytoplasmic mRNA function is dictated by the interactions of mRNA with the core translation, localization, and mRNA degradation machinery, as well as sequence specific regulatory proteins. Regulation at the level of mRNA is important for cells to respond rapidly to environmental changes1. Issues in understanding post-transcriptional regulation include determining the spectrum of mRNA binding proteins (RBPs) and how they interact with specific mRNAs, as well as understanding how such proteins, individually or in combination, affect mRNA function. We set out to determine the major mRNA binding proteins in Saccharomyces cerevisiae and to identify some of the basic principles of mRNA-protein interaction.
Yeast cells represent an ideal system for determining the principles of mRNP formation and function. A substantial number of yeast mRNA binding proteins have been identified from studies of the mechanisms of mRNA biogenesis, localization, translation and degradation. Studies of a more global nature in yeast have met with modest success. Purification of mRNPs under native conditions was unable to significantly enrich mRNA binding proteins over the general cellular population of proteins2. Genome wide protein-RNA interactions studies in vitro suggested additional RNA binding proteins, some of which have been verified in vivo2.
Recent and historical experiments have utilized cross-linking of proteins to mRNAs in vivo and purification of the mRNA under denaturing conditions to identify mRNA binding proteins3–5. We have now applied such methods to yeast to identify the major mRNA binding proteins under conditions of stress. We performed these experiments under glucose deprivation because conditions used to cross-link proteins to RNAs in vivo can trigger a stress response6 and we wanted the cells to be in a defined state. Moreover, post-transcriptional control is important during stress and involves changes in translation, mRNA degradation, and the localization of mRNPs into stress granules and P-bodies7, which are related to a large family of RNA granules, including maternal mRNP granules, neuronal mRNP granules, and some RNP granules associated with neurodegenerative diseases8. Thus, an analysis of mRNPs under stress should yield additional information about post-transcriptional control.
Here we undertake a characterization of yeast mRNPs by identifying the major mRNA binding proteins of yeast. We also identify widespread relocalization of mRNA binding proteins during stress and characterize the mRNA binding sites of P-body proteins, defining principles by which these proteins assemble into mRNPs.
Results
Identification of Yeast mRNA Binding Proteins
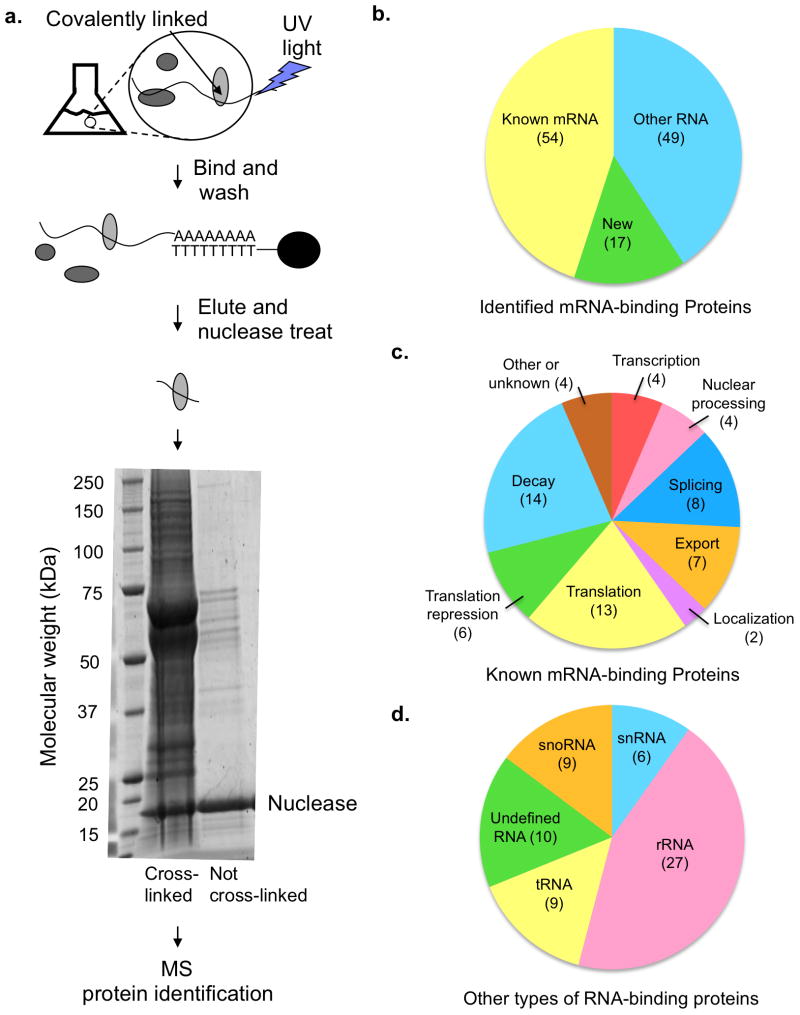
To identify proteins directly interacting with mRNAs, we developed a method similar to those recently used in HeLa cells and human embryonic kidney cells4,5. In this method, previously referred to as “in vivo capture of RBPs”, “interactome capture” and “identification of mRNA interacting proteins”, proteins are directly cross-linked to mRNAs in vivo using UV light, after which mRNA is purified under denaturing conditions via its poly(A) tail (Fig. 1a). After elution from an oligo(dT) column, the RNA-protein complexes are RNase treated, separated by SDS-PAGE, and the protein composition analyzed by LC MS-MS. We examined proteins cross-linking to mRNAs under conditions of glucose deprivation stress for reasons described above.
Figure 1.
Description of the in vivo RBP capture procedure and identified mRNA binding proteins. (a) A schematic of in vivo capture of RBPs to identify mRNA binding proteins including an SDS PAGE gel stained with SYPRO Ruby dye. (b–d) Proteins identified by mass spectrometry (MS). Numbers in parentheses in parts (b–d) indicate the number of proteins in each category. Some proteins are found in multiple categories. (b) The distribution of 120 identified poly(A) RNA binding proteins between known mRNA binding proteins, new RNA binding proteins and new mRNA binding proteins previously known to interact with other types of RNA. (c) The functional categories of previously known mRNA binding proteins identified by the RBP capture assay. (Total of 54 proteins) (d) The types of RNA, other than mRNA, bound by the newly identified proteins (total of 49 proteins). For proteins closely associated with RNA biology, but for which a specific RNA binding partner has not been identified, or which are predicted to bind RNA based on structural homology to RNA binding proteins, the category “undefined RNA” has been used.
We found that cross-linking enhanced the amount of protein purifying with the mRNA in comparison to the control sample that had not been cross-linked (Fig. 1a). This indicates that the observed proteins were predominantly those that had cross-linked directly to the poly(A)+ RNA. Following two biological replicates (five technical replicates) we identified 120 proteins reproducibly and statistically enriched in the UV cross-linked sample, thus defining the dominant proteins in yeast mRNPs (Supplementary Table 1 and methods).
Distribution of Major Yeast mRNA Binding Proteins
The 120 proteins that co-purified with mRNA include 54 proteins previously shown to bind mRNA or to be intimately involved in mRNA biology (Fig. 1b), demonstrating that this method is successful at identifying mRNA binding proteins (Supplementary Table 1, examples shown in Table 1). These proteins come from all stages of mRNA metabolism, including transcription, splicing, export, localization, translation and decay (Fig. 1c). We identified both nuclear and cytoplasmic proteins, indicating that the assay is capable of identifying proteins from various regions of the cell in a variety of mRNPs.
Table 1.
Examples of mRNA binding proteins identified by mass spectrometry
| Category | Gene | Systematic name | Function |
|---|---|---|---|
|
| |||
| Known mRNA binding protein | Tif4631 | YGR162W | Translation initiation |
| Khd1 | YBL032W | Localization | |
| Xrn1 | YGL173C | Decay | |
| Puf3 | YLL013C | Translation repression | |
|
| |||
| tRNA binding protein | Gln4 | YOR168W | tRNA synthetase |
| Dus3 | YLR401C | tRNA modification | |
|
| |||
| Ribosome biogenesis | Nop56 | YLR197W | Ribosome biosynthesis |
| Nsr1 | YGR159C | Ribosome biosynthesis | |
| Cbf5 | YLR175W | Ribosome biosynthesis | |
|
| |||
| Other functions | Ksp1 | YHR082C | Kinase |
| Ste20 | YHL007C | Kinase | |
| Cys4 | YGR155W | Cysteine biosynthesis | |
| Imd3 | YLR432W | GTP biosynthesis | |
| Pol2 | YNL262W | DNA polymerase | |
We identified a number of proteins that are known to bind RNA but not known to interact with mRNA (Fig. 1b, 1d, Table 1), including five tRNA synthetases and two tRNA modification enzymes (Pus1 and Dus3). Instances of tRNA synthetases modifying the stability or translation of mRNAs have been described9–11. Multiple tRNA synthetases interacting with mRNAs suggests that this is a more common phenomenon for yeast than previously understood. The presence of tRNA modification proteins in the mRNA binding pool suggests that these proteins might specifically modify individual mRNAs to modulate mRNA fate.
A major category of mRNA binding proteins identified was ribosome-processing proteins. Twenty-one of these proteins were enriched in our assay, suggesting that mRNA binding is a common secondary role for proteins involved in ribosome processing. One possibility is that these proteins were identified due to interactions with contaminating ribosomes. Three lines of evidence argue against this scenario. First, two of these proteins, Nop56 and Nsr1, are verified mRNA binding proteins12. Second, the highly abundant ribosome structure proteins are not identified in the RBP capture assay (with the exception of Rps20A). Finally, Ash1 mRNA has been observed to pass through the nucleolus during its maturation13. Potential mRNA binding activity of ribosome biogenesis factors suggests that there may be considerable cross talk between mRNA regulation and ribosome processing.
We identified proteins with no previously known RNA binding activity (Table 1). These include Vma1, a subunit of the vacuolar ATPase. Two proteins involved in DNA metabolism were identified (Pol2 and Rfa1). Various metabolic proteins were observed (Imd2, Imd3, Imd4, Cys4, Cpr1). Proteins that have QN rich domains (for example Psp1) and might play a role in stress granules or P-body formation were also found14. Finally, we identified the Ste20 and Ksp1 kinases.
Recent surveys in mammalian cell lines have used similar methods to identify mRNA binding proteins4,5. Ninety-two of the 120 proteins identified in this study have human orthologs or similar human proteins, and 72 (78%) of these human orthologs have been shown to interact with mRNA by similar assays (Supplementary Table 2). This defines a conserved core of mRNA binding proteins from yeast to humans. As expected this list contains a number of canonical mRNA binding factors. Strikingly, the list of conserved mRNA binding factors also includes all 21 of the ribosome biogenesis factors identified here, indicating that the interaction between rRNA processing proteins and mRNA is conserved. Several other proteins that have not been associated with mRNA biology through biochemical work are also conserved mRNA binding proteins. These include the tRNA modification enzymes Dus3 and Pus1 and two tRNA synthetases (Tys1 and Hts1) as well as the peptidyl-prolyl cis-trans isomerase Cpr1. The conserved interaction between these proteins and mRNA suggests that their mRNA binding activity has an important biological function that has not yet been characterized.
Intracellular Distribution of mRNA Binding Proteins
We identified the mRNA binding proteins described above under stress conditions, thus they may function in post-transcriptional stress response pathways. A conserved aspect of the eukaryotic stress response is the aggregation of non-translating mRNPs into stress granules and P-bodies15. We monitored the localization of these proteins in both the presence and absence of stress to reveal if regulation of mRNA under stress is spatially restricted in the cell or if it occurs in diverse compartments. Additionally, this data could determine the possibility of proteins being in the same mRNP, as components of the same mRNP would have similar intracellular localization patterns.
We looked at the subcellular location of the enriched proteins using appropriate strains from the library of C terminally GFP tagged yeast proteins. Due to either the unavailability of the GFP strain or inadequate signal, 13 of the 120 enriched proteins could not be observed. These localization experiments revealed the following key points.
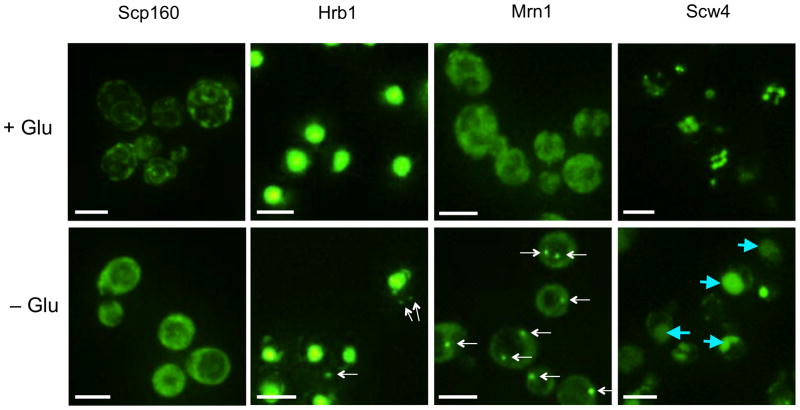
First, we observed that the mRNP proteins are found in various cellular regions under log phase growth and stress conditions including two proteins (Scp160 and Bfr1) preferentially associated with the ER (Fig. 2 and Table 2). Despite the diversity in the localization of these proteins, we mainly observed localization in one of four compartments under stress conditions: the nucleus, stress granules, P-bodies, or diffuse in the cytosol. As proteins located in different compartments are less likely to be parts of the same mRNP, we conclude that there are at least four discrete types of mRNPs under these conditions. As the protein pool is different in these four compartments, it is likely that mRNA within separate compartments would be subject to distinct functional consequences.
Figure 2.
Fluorescence microscopy images to show localization of mRNA binding proteins fused to GFP under log phase growth (+Glu) and glucose deprivation conditions (−Glu). All white scale bars indicate 5 μm. Foci are indicated by white arrows. Vacuoles are indicated by blue arrows.
Table 2.
Localization of mRNA binding proteins during log phase growth and under glucose deprivation
| +Glu | −Glu | Proteins |
|---|---|---|
| Nucleus | Nucleus | Cbf5, Dus3, Hrp1, Mak21, Mlp1, Mtr4, Mud2, Nab2, Nan1, Noc2, Nop4, Nop56, Nop58, Nop7, Nop9, Nrd1, Nsr1, Nug1, Pap2, Pol2, Prp8, Puf6, Pus1, Pwp2, Rat1, Rpb2, Rrp5, Sof1, Spt5, Sto1, Tho2, Urb1, Utp20 |
| Nucleus | Nucleus and P-body or stress granule | Gbp2, Hrb1, Mex67, Npl3 |
| Nucleus | Nucleus and other foci | Prp43 |
| Nucleus and other foci | Nucleus and other foci | Rfa1 |
| Nucleusand cytoplasm | Nucleus and cytoplasm | Cpr1, Pbp2, Nam8, Sen1 |
| Cytoplasm | Cytoplasm | Bem2, Dbp1*, Gln4, Gus1, Hts1, Imd2, Imd3, Imd4, Lsg1, Nab3*, New1, Nip1, Rrp12, Ski2, Ste20*, Sup35, Tif3, Tif32, Tys1, Ubp3, Yll032c |
| Cytoplasm | Cytoplasm and P-body or stress granule | Bre5, Dhh1, Eap1, Ecm32, Gis2, Hek2, Ksp1, Mrn1, Nab6, Ngr1, Pab1, Pat1, Pbp1, Psp1, Psp2, Pub1, Puf2, Puf3, Puf4, Puf5, Sbp1, Slf1, Slh1, Sro9, Tae2, Tif4631, Tif4632, Upf1, Upf3, Xrn1, Ybr238c, Ygr250c |
| Cytoplasm | Cytoplasm and other foci | Pin4 |
| Cytoplasm | Cell Wall | Puf1 |
| Cytoplasm and other foci | Cytoplasm and other foci | Cys4 |
| Cytoplasmic vesicle or golgi | Vacuole | Scw4 |
| Endoplasmic reticulum | Cytoplasm | Bfr1, Scp160 |
| Mitochondria | Mitochondria | Msc6, Mss116, Rmd9 |
| Vacuole and other foci | Vacuole and other foci | Vma1 |
| Unidentified organelle | Unidentified organelle | Tif31 |
Secondly, we observed that glucose starvation induced intracellular relocalization in 41 of the 107 (38%) mRNA binding proteins tested (Table 2). Consistent with prior work, the majority of mRNP protein re-localization was to P-bodies or stress granules (see below). However, we observed novel relocalizations of mRNP proteins, including disassociation from the ER (Bfr1 and Scp160) and movement into the vacuole (Scw4) (Fig. 2). Interestingly, nuclear proteins involved in rRNA processing remained in the nucleolus under stress conditions. One possibility is that under stress the binding of these proteins to specific mRNAs retains the mRNA in the nucleolus. Consistent with this idea, evidence suggests that at least some yeast mRNAs pass through the nucleolus during biogenesis13.
Thirdly, we identified 14 new components of yeast stress granules and P-bodies by examining the co-localization of each of the GFP fusion proteins that accumulated in cytoplasmic foci with known markers of stress granules (Pub1-mCherry) or P-bodies (Edc3-mCherry) following glucose deprivation. The exact composition of stress granules and P-bodies can depend upon the specific stress applied to the cells16–18. Thus, we may miss some proteins that assemble into stress granules or P-bodies under different stresses.
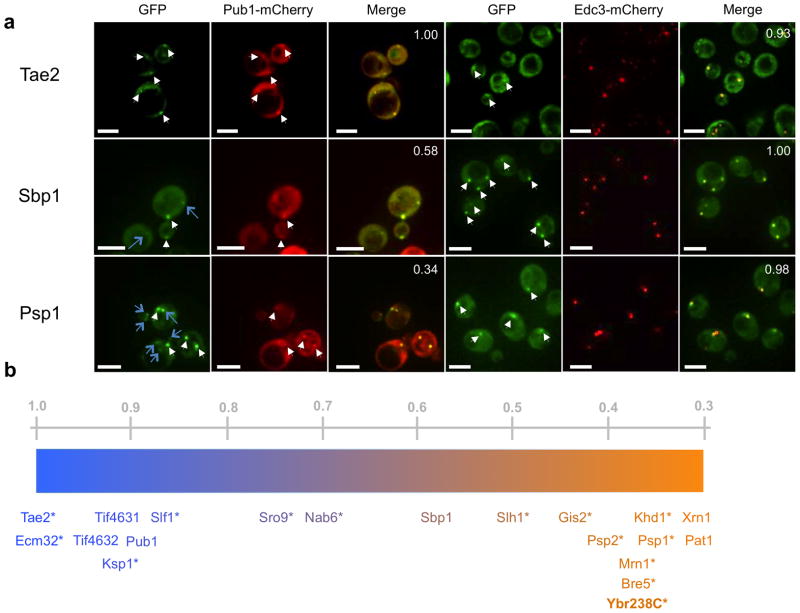
Since yeast stress granules and P-bodies can spatially overlap and are likely to represent a continuum of mRNP states, we used a quantitative assay to assess if an individual protein was more prevalent in stress granules or P-bodies. In this assay, for each new mRNP protein accumulating in cytoplasmic foci, we determined the fraction of GFP foci that co-localized with Pub1-mCherry or with Edc3-mCherry in separate experiments (Fig. 3a). While P-body and stress granule components show high degrees of co-localization with Edc3; stress granule factors are likely to overlap with Pub1 more often than P-body components (Supplementary Fig. 1). Thus, components having greater than 67% overlap with Edc3 were considered to be either P-body or stress granule factors. Certain proteins like Pin4 and Rfa1 did not clear this criterion, and are categorized as components of “Other Foci” (Table 2, Supplementary Fig. 1). To quantify the P-body or stress granule like feature of the remaining proteins, they are represented on a gradient in Figure 3b based on their overlap with Pub1-mCherry. This analysis identified new P-body factors including: Khd1, known for its role in Ash1 localization; translational regulators Gis2 and Mrn1; suppressors of DNA polymerase mutations Psp1 and Psp2; ubiquitin protease cofactor Bre5, and mitochondrial membrane protein YBR238C (Fig. 3b). New stress granule factors include the polysome associated proteins Tae2, Ecm32 and Slf1; and kinase Ksp1 (Fig. 3b). Ste20 and Dbp1 were sporadically found to associate with granules and may be weak granule components.
Figure 3.
Categorization of new granule components. (a) Fluorescence microscopy of granule components of interest (Tae2, Sbp1 and Psp2) tagged with GFP. P-body marker Edc3 and stress granule marker Pub1 are tagged with mCherry. All white scale bars indicate 5 μm. White arrows indicate co-localizing GFP and mCherry foci. Blue arrows indicate GFP foci that do not co-localize with mCherry foci. Numbers in the upper right hand corner indicate the fraction of GFP foci that co-localize with mCherry foci averaged over three images. (b) Proteins are characterized as P-body or stress granule-like along a gradient based on fluorescence microscopy data, some of which are shown in (a). Orange indicates P-body-like characteristics and blue indicates stress granule-like features. Newly identified granule factors are marked with an asterisk. Numbers above the gradient represent fractional co-localization of granules with Pub1-mCherry granules averaged over three images.
Analysis of mRNAs Bound to Granule Proteins
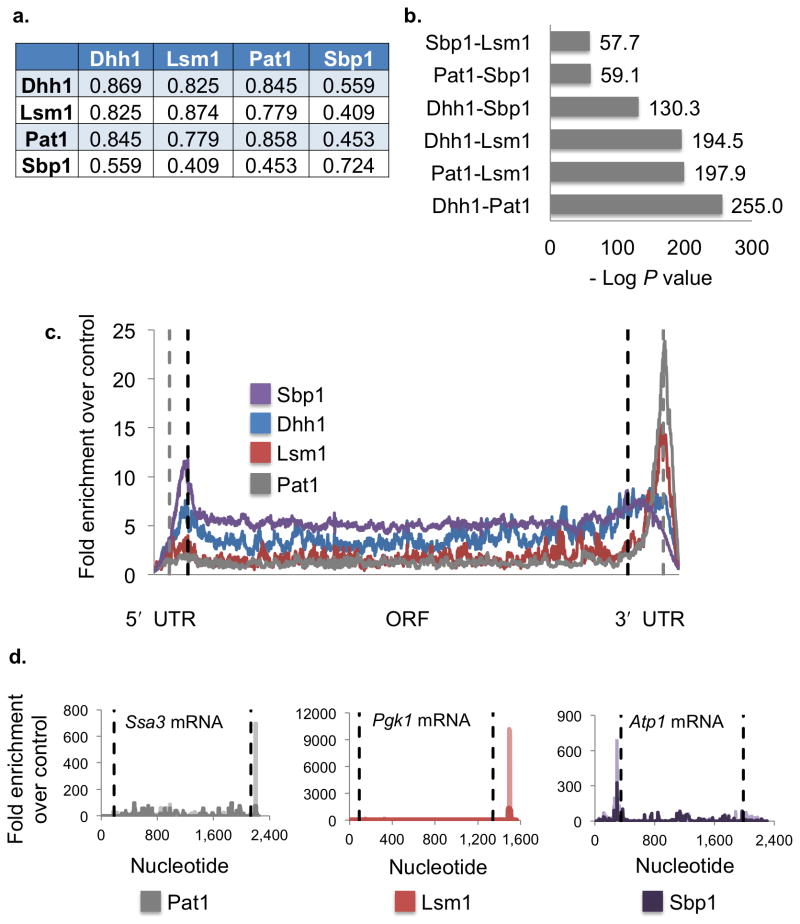
Using the above analysis we identified the major mRNP components and demonstrated that they are localized in different subcellular regions before and during stress responses, suggesting there are multiple types of mRNPs. To investigate how specific mRNPs are organized and on which mRNAs, we have begun an analysis of the interaction of mRNA with proteins in P-bodies. Specifically, we utilized CLIP (see methods) to map the interaction sites of the P-body components Pat1, Lsm1, Dhh1, and Sbp1 on mRNAs19. Sbp1 can be contrasted to the others since it is found in both P-bodies and stress granules (Fig. 3a and 3b). CLIP experiments were performed in duplicate and both mRNA targets and peak locations on transcripts showed significant levels of reproducibility (Fig. 4a, Supplementary Fig. 2a). Sites of binding were identified in two classes. We defined a rigorous set of peaks (referred to as Tier 1, Supplementary Table 3) using a False Discovery Rate (FDR) of < 1%, and requiring at least 20-fold enrichment of peak height over overlapping or proximal peaks from profiles generated using a fragmented mRNA library (see methods). We are highly confident that Tier 1 peaks represent bona fide sites of mRNA-protein interaction. However, since this standard is likely to exclude some real sites of binding, we also identified binding sites using an FDR of <2% and a twofold enrichment over proximal control peaks, (Tier 2 sites, Supplementary Table 3). Analyses were then carried out with both sets of peaks. Since the results were similar for both sets, we present the analyses for Tier 1 peaks, revealing the following key observations.
Figure 4.
P-body proteins bind to a shared group of mRNAs with positional specificity. (a) Pearson correlation coefficients for CLIP sequence counts of co-localized peaks between two data sets. Comparison with self indicates Pearson correlation coefficient for degree of similarity between the two replicates. (b) Bar graph showing degrees of similarity between the mRNA targets of Dhh1, Lsm1, Pat1 and Sbp1 identified by CLIP as −log P-values. (c) The enrichment of CLIP sequence tags over the control averaged across all bound mRNAs for each protein is shown. Sbp1 targets are shown in purple, Dhh1 in blue, Lsm1 in red, and Pat1 in gray. Limits of the open reading frame (ORF) are indicated by dashed black lines. Ends of the UTRs (untranslated regions) and beginnings of the extension regions are indicated by dashed gray lines. 50 nucleotide extensions regions were added to both ends of the transcript to account for annotation errors. Lengths are scaled to the average 5′ UTR, ORF and 3′ UTR lengths over the entire genome. (d) Plots showing enrichment of CLIP sequence tags over the control sequence data for individual mRNAs. Pat1 data is shown in gray for Ssa3 mRNA, Lsm1 in red for Pgk1 mRNA and Sbp1 is shown in purple for Atp1 mRNA. Dashed black lines indicate the limits of the open reading frame. Replicate data sets are shown in darker shades for each trace. Additional CLIP data sets for other proteins on the same mRNA are shown in Supplementary Fig. 2b.
First, we observed that all four proteins showed substantial overlap in the set of bound mRNAs with high scores for statistical significance (Fig. 4b). Replicate CLIP data sets demonstrate the highest levels of similarity, indicating that the data is repeatable and that similarity does not appear due to non-specific mRNA background (Supplementary Fig. 2a). This is consistent with these proteins tending to co-assemble on mRNAs that accumulate in P-bodies and with previous biochemical and genetic data since Pat1 directly interacts with both the Lsm1–7 complex and Dhh1 (ref. 20); and Sbp1 promotes mRNA decapping in a Dhh1-dependent manner21. An additional trend is that Pat1, Lsm1 and Dhh1 share a higher degree of similarity to one another than to Sbp1 (Fig. 4b). This is consistent with the fact that Pat1, Lsm1 and Dhh1 associate more with P-bodies, while Sbp1 also has substantial presence in stress granules (Fig. 3b)19.
Second, based on the analysis of the genome wide distribution of binding sites we observed positional bias in the sites of mRNA interaction of these proteins (Fig. 4c). Pat1 and Lsm1 both preferentially bind the 3′ end of the mRNA (Fig. 4c, 4d and Supplementary Fig. 2b, 2c), which is consistent with biochemical experiments showing that this complex prefers to bind oligoadenylated 3′ ends22 and protects the 3′ end from trimming in vivo23. However, it should be noted that Pat1 and Lsm1 also interact with mRNAs in the ORF and 5′ UTR (Supplementary Fig. 3). These positional biases are significant, as 63% of all Pat1 binding sites and 51% of all Lsm1 binding sites are in the 3′ UTR whereas based on average 3′ UTR to total mRNA length ratio, this number would be ~10% by chance (p<0.0001 for both Pat1 and Lsm1). Analysis of sites of interactions by DREME24 (Discriminative DNA Motif Discovery v. 4.8.1) fails to identify any strong consensus sequence suggesting that the binding of Pat1 and Lsm1 to mRNAs may be more strongly dictated by the 3′ end and oligo(A) tail. Similar analysis of Dhh1 binding sites also failed to yield a consensus sequence.
In contrast to Lsm1 and Pat1, Sbp1 shows bias towards the 5′ UTR (Fig. 4c, 4d and Supplementary Fig. 2b, 2c), which may be explained by its direct binding to eIF4G25. This interaction might explain why Sbp1 can also be observed in stress granules (Fig. 3a and 3b). Analysis of Sbp1 peaks demonstrates enrichment in TCTTC/G (p=5.9×10−10). However this consensus is only present in 18.1% of the Sbp1 Tier 1 peaks and therefore makes a relatively minor contribution to the overall occupancy of this protein.
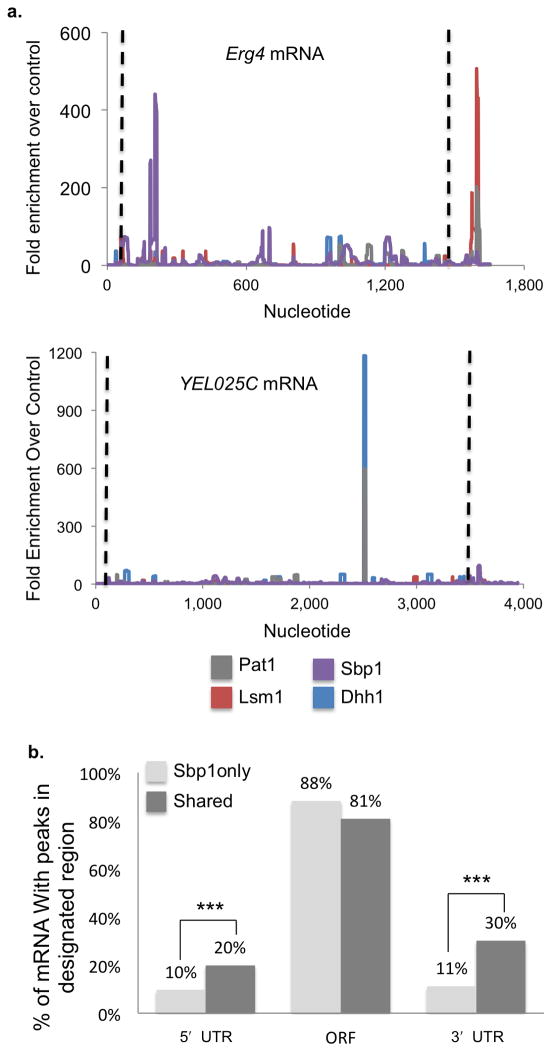
A third interesting observation is the occurrence of co-assembly of some proteins on the mRNA. This was assessed by identifying co-localized peaks and comparing the number of sequence reads26 (see methods). As expected, we observed that replicate experiments for individual proteins generally showed the highest degree of similarity (Fig. 4a, Supplementary Fig. 4a, 4b). The most significant overlap between different proteins was observed amongst Pat1, Lsm1 and Dhh1 (Fig. 4a), which is consistent with biochemical experiments showing strong physical interactions between these proteins20,21,27. We interpret this observation to suggest that the direct physical interaction of Pat1 with Dhh1 and (or) the Lsm1–7 complex frequently leads to local co-assembly on the mRNA. We also observed a certain degree of overlap between Dhh1 and Sbp1 peaks (Fig. 4a), consistent with Sbp1 having functional interactions with Dhh1 (ref. 21). In contrast, the correlation between Sbp1 and Pat1 or Lsm1 peaks, where there is no evidence for a direct physical or functional interaction, is lower (Fig. 4a). Examples of the overlap of individual protein peaks on specific mRNAs are shown in Figure 5a and Supplementary Figure 4c.
Figure 5.
Interactions between P-body proteins influence mRNA binding. (a) Plots showing enrichment of CLIP sequence tags over the control sequence data for individual mRNAs Erg4 and YEL025C. Dhh1 is shown in blue, Lsm1 in red, Pat1 in gray and Sbp1 in purple. Dashed black lines indicate the limits of the open reading frame. (b) Graph illustrating the percent of mRNA with CLIP sequence tag peaks in the 5′ UTR, open reading frame and 3′ UTR for Sbp1 target mRNA bound only by Sbp1 (light gray) or by other factors as well (“shared,” dark gray). Percentages do not add up to 100% as individual mRNA may have peaks in multiple regions. *** indicates P-value <0.0001 for unpaired Student’s t-test, n=755 mRNA for Sbp1 only, 294 mRNA for Shared.
The co-assembly of individual proteins on the mRNA implies that these proteins may influence each others binding to the mRNA. In this light, we observed that the location of Sbp1 binding sites in the mRNA is influenced by Dhh1, Lsm1 and Pat1, such that Sbp1 is more likely to bind the or 3′ UTRs in the presence of binding sites for any of these factors (Fig. 5b, Supplementary Fig. 5a, 5b, 5c). Specifically, the percentage of Sbp1 targets with a binding site in the 5′ UTR doubles from 10% to 20% for those mRNA that are shared targets. The effect is greater in the 3′ UTR, where there is a three-fold increase in Sbp1 binding (30% vs. 11%). These observations demonstrate that the preferred site of Sbp1 binding is influenced by the presence of other components of the mRNP.
A final trend is the variation in peak number per mRNA for the four proteins. Both Pat1 and Lsm1, which have the strongest positional bias, have a high percentage of Tier 1 mRNA targets with only a single peak (87.7% and 84.3% respectively). This suggests that each P-body mRNA typically contains only one Pat1 or Lsm1–7 complex. In contrast, only 53.2% of Sbp1 mRNA targets contain a single peak. Dhh1 shows a moderate level of specificity, with 65.2% of mRNAs having a single peak. Thus Dhh1 and Sbp1 either bind in a less specific manner, such that the position of those proteins on an mRNA would be variable, or multiple copies of these proteins are bound to an mRNA. The latter possibility is consistent with the fact that Dhh1 and Sbp1 are more abundant (42,900 and 12,800 copies per cell respectively) than Pat1 and Lsm1 in the cell (626 and 3,490 copies per cell respectively)28.
Discussion
In this work we applied three methods to understand mRNP structure and composition in yeast: zero distance cross-linking and mass spectrometry to identify mRNA binding proteins, fluorescence microscopy to identify the location of these proteins, and CLIP to characterize the nature of mRNA binding of several proteins. Via these methods we have revealed some basic principles of protein-mRNA interactions as discussed below.
The RBP capture assay identified 120 proteins that compose the major yeast mRNA binding proteins under glucose deprivation conditions. Several important findings come from this list. First, nearly half are known mRNA binding proteins, indicating that this method robustly identifies mRNA binding proteins. Second, many of the proteins interact with other areas of RNA metabolism, suggesting considerable cross talk between various areas of RNA biology, particularly with ribosome biosynthesis. Third, a large percentage of these mRNA binding proteins are conserved between yeast and mammals. Fourth, proteins unrelated to RNA biology were identified. In this category, interactions between DNA biology and metabolic enzymes have been identified, consistent with similar mammalian surveys4,5. One protein (Vma1) with a role in vacuole biology was also identified. This protein may target specific mRNAs to vacuoles for degradation. During ribophagy, 60S ribosomal subunits are degraded in the vacuole29, and recent work from our lab has linked vacuole biology to granule formation (J.R. Buchan and R. Parker unpublished). We also identified two kinases (Ste20 and Ksp1) as mRNA binding proteins. One interesting possibility is that these kinases could specifically regulate proteins associated with the mRNAs that they bind. This type of cis-regulation within an mRNP would be an effective mechanism for altering the fate of an mRNA in response to environmental stimuli. This model is supported by a role for Ste20 in controlling mRNA degradation and stress granule formation during oxidative stress30. Alternatively, RNA binding could modulate the activity of these kinases.
Some mRNA binding proteins will be missed by our analysis. Proteins will be missed if they are in poor geometry to cross-link to mRNA. For example, while we observe eIF4G1, eIF4G2 and Sto1, the large subunits of the cytoplasmic and nuclear cap binding complexes, we did not detect Cbc2 or eIF4E, perhaps because being bound to the cap presents a small region of RNA for cross-linking. We anticipate that proteins expressed at low levels or only binding a few mRNAs are missed in our analyses. For example, we did not observe Sgn1 and She2, which bind few mRNAs (10 and 22 respectively), and are estimated to be expressed at relatively low levels12. Such proteins could be identified by deeper mass spectroscopy analysis. Our data is also missing components of the decay machinery that preferentially bind mRNA after deadenylation, including Dcp1–Dcp2, Edc3, the Lsm1–7 complex, and the exosome31. Ccr4–Pop2 may be absent because stress inhibits deadenylation32, or because cross-linking of Ccr4–Pop2 to the poly(A) tail interferes with binding to oligo(dT). It is notable that we did observe many decay factors (e.g. Pat1, Dhh1, Sbp1, Upf1, Upf3) that would be expected to interact with poly(A)+ mRNAs33.
By examining the subcellular location of mRNP proteins during glucose deprivation, we observed a large-scale rearrangement of mRNA binding proteins, with 38% changing localization pattern in response to stress. This change in localization is likely to reflect some change in mRNP function. These changes revealed several facts. First, the major change is aggregation into P-bodies or stress granules. This suggests that these aggregates may be major sites of mRNA control under stress. However, as mRNA binding proteins are not limited to these granules, there are likely other important sites of mRNP regulation under these conditions. Second, we identified 14 new members of these granules. Some of these new members are post-translational modification proteins (a kinase, Ksp1, and a ubiquitin protease cofactor, Bre5) that could potentially modify the ability of some mRNPs to enter or exit these granules. An additional protein that was occasionally found to associate with granules, Ste20, has been implicated in stress granule formation, suggesting that it might be involved in targeting specific mRNAs to granules, which is consistent with its role in stress granule formation30. Third, nearly all translation related factors tested entered granules, suggesting that many proteins involved in translation enter granules under stress conditions. Fourth, we identified novel changes in protein localization associated with stress response. Such changes include movement into the vacuole, exit from the nucleus and aggregation into novel foci. In sum, subcellular rearrangement of mRNPs is a major and global response to stress.
In the final part of this work we identified the mRNAs bound by P-body associated proteins Pat1, Lsm1, Dhh1 and Sbp1 using CLIP. These proteins bind a highly overlapping list of mRNAs, reflecting their co-localization to granules in vivo. The extent of similarity of their mRNA targets is consistent with known physical and genetic interactions between these proteins. Lsm1, Pat1 and Dhh1, which share an intricate network of physical interactions20, also have the most significant level of overlapping targets (Fig. 4b). Moreover, the mRNAs cross-linked to Sbp1 are most related to those interacting with Dhh1 (Fig. 4b), consistent with Sbp1 enhancing the ability of Dhh1 to stimulate decapping21, and the fact that Sbp1 and Dhh1 both co-localize in stress granules to some extent34 (Fig. 3).
A surprising observation to come from the identification of binding sites for these four granule associated proteins is that none of them have strong sequence specificity. Rather, all four proteins have positional specificity relative to mRNA landmarks. Pat1 and Lsm1 co-localize at the very 3′ end of mRNAs, while Sbp1 (and to a lesser extent Dhh1) demonstrates a preference for the 5′ UTR. This mode of binding allows for a broad set of targets, a potentially desirable effect for proteins affecting the metabolism of many mRNAs. Such positional preference can have clear functional advantages (particularly for roles in suppressing translation initiation, decapping, deadenylation, etc.). One envisions that positional preference for mRNA binding proteins is either dictated by end specific features (such as a cis-diol or oligo(A) tail at the 3′ end), or by other position specific protein interactions. One candidate for such a linker protein is eIF4G, a canonical translation initiation factor and part of the cap-binding complex. Recent work has demonstrated that eIF4G is able to bind Sbp1 (ref. 25). This interaction could tether Sbp1 to the 5′ region of an mRNA, its preferred binding site.
We observed evidence of co-assembly of proteins on mRNA. Most strikingly, we observed that the peaks of Pat1 overlapped strongly with the peaks of Dhh1 and Lsm1 (Fig. 4a). This is consistent with strong physical interactions between Pat1 and these proteins20,22,27. The simplest interpretation of these observations is that the direct interactions between these proteins can lead to local co-assembly on the mRNA. A corollary of this interpretation is that individual proteins can affect either the recruitment of other proteins, or their binding site. Consistent with that view, it is known that Pat1 is required for the recruitment of the Lsm1–7 complex to P-bodies, and presumably to mRNAs35. Moreover, we observe that the presence of Dhh1, Lsm1, or Pat1 on the mRNA can alter the preferred location of the Sbp1 protein (Fig. 5b). An important area for future research will be to determine how the binding patterns of each mRNP component influences the localization and function of others.
The co-assembly and influence of mRNP components on the binding of one another highlights two principles. First, the interaction between proteins and mRNA is highly complex and not solely determined by sequence specificity. Thus it is important to take the entire mRNP structure into account when predicting binding sites, rather than relying solely on sequence and (or) mRNA structure. Second, it is likely that function of mRNA binding proteins may vary as a function of other protein factors within the mRNP. For instance, Pat1 may have different activities when co-assembled with Dhh1 than with the Lsm1–7 complex. This combinatorial ability could lead to a wide variety of functional consequences for mRNAs bound to a smaller number of proteins.
Here we have begun to gather global information about mRNP structure and function. We have established the major components of yeast mRNPs, determined that re-localization is one mechanism by which post-transcriptional control of mRNA fate may occur and found that the mRNA targets of granule associated proteins are an overlapping set and that positional specificity and mRNP environment are important determinants of binding. In the future it will be of great interest to synthesize the increasing body of protein-mRNA interaction data being produced by a variety of investigators to create a global model for mRNP structure. Such a model could both elucidate the principles by which mRNPs form as well as predict the structure of mRNPs, and potentially the fates of mRNAs, under various conditions.
Online Methods
In vivo Capture of RBPs Procedure
Cells were pelleted and resuspended in 1X PBS for 30 minutes. Cells were exposed to 1,200 mJ/cm2 of 254 nm UV in Stratalinker 1,800 (Stratagene) with two 2-minute breaks on ice and gentle mixing. Control cells were incubated in PBS but not UV treated. Cells were then resuspended in Lysis Buffer (10 mM Tris pH 7.4, 600 mM NaCl, 10 mM EDTA, 0.2% SDS, 10 mM vanadyl complex (New England Bioscience), 2 mM DTT, complete EDTA-free cocktail tablet (Roche)) frozen in pellets and lysed in a ball mill grinder (Retsch PM200). Lysed cells was resuspended in additional lysis buffer and thawed on ice. Lysate was clarified at 2,300×g for 5 minutes. This soft pellet was rinsed in lysis buffer and re-spun. Supernatants from the two spins were combined. One gram of oligo(dT) cellulose (Sigma-Alrdrich) was rinsed in water, 20 mL of 0.1 M NaOH then equilibrated in lysis buffer. Oligo(dT) cellulose and lysate were mixed and rocked at room temperature for 1 hour. The cellulose was spun down at 1,000 rpm for 1 minute and the supernatant was removed. The cellulose was resuspended in Lysis Buffer and gently poured into a column then washed with 20 mL of Lysis Buffer, 30 mL of Wash Buffer A (10 mM Tris pH 7.4, 150 mM NaCl, 1% SDS, 10 mM EDTA, 2 mM DTT) and 30 mL Wash Buffer B (10 mM Tris pH 7.4, 150 mM NaCl, 10 mM EDTA, 2 mM DTT). The poly(A) RNA was eluted in 1 mL fractions of TE pH 7.5 heated to 65 °C. Fractions containing RNA were pooled and digested with micrococcal nuclease (NEB) at 37 °C for 15 minutes. Reactions were quenched with EGTA. 0.2% SDS and 300 mM NaCl were added to prevent protein aggregation and samples were concentrated to ~20 μL in 0.5 mL 10 kD MWCO concentrators (Millipore). Samples were run on a 4–12% NuPAGE Novex acrylamide gel (Life Sciences) at 150 V for ~1.5 hours and stained with Sypro ruby dye (Biorad) then imaged on a phosphorimager (Typhoon 9410, Molecular Dynamics). Lanes were cut into 5 pieces with approximately equal amounts of protein for MS analysis. Two biological replicates of this procedure were performed.
Tandem mass spectrometry coupled to liquid chromatography (LC-MS-MS)
Excised Sypro Ruby-stained protein gel bands (or regions of bands) following 1D SDS-PAGE were digested with trypsin (10 μg/mL) at 37°C overnight. LC-MS-MS analysis of in-gel trypsin digested-proteins1 was carried out using a LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) equipped with an Advion nanomate ESI source (Advion), following ZipTip (Millipore) C18 sample clean-up according to the manufacturer’s instructions. Peptides were eluted from a C18 pre-column (100-μm ID × 2 cm, Thermo Fisher Scientific) onto an analytical column (75-μm ID × 10 cm, C18, Thermo Fisher Scientific) using a 5–20% gradient of solvent B (acetonitrile, 0.1% formic acid) over 65 minutes, followed by a 20–35% gradient of solvent A over 25 minutes, all at a flow rate of 400 nl/min. Solvent A consisted of water and 0.1% formic acid. Data dependent scanning was performed by the Xcalibur v 2.1.0 software2 using a survey mass scan at 60,000 resolution in the Orbitrap analyzer scanning m/z 350–1,600, followed by collision-induced dissociation (CID) tandem mass spectrometry (MS-MS) of the fourteen most intense ions in the linear ion trap analyzer. Precursor ions were selected by the monoisotopic precursor selection (MIPS) setting with selection or rejection of ions held to a ± 10 ppm window. Dynamic exclusion was set to place any selected m/z on an exclusion list for 45 seconds after a single MS-MS. All MS-MS spectra were searched against a Saccharomyces cerevisiae protein database downloaded July 29, 2011 from UniProtKB (http://www.uniprot.org/uniprot/?query=taxonomy:4932) using Thermo Proteome Discoverer 1.2 (Thermo Fisher Scientific). At the time of the search, the Saccharomyces cerevisiae protein database contained 34,577 entries. Proteins were identified at 95% confidence with XCorr scores3 as determined by a reversed database search.
Only proteins identified by two or more unique peptides, each with a 99% or higher level of confidence, were included in the analysis. From this pool those that were enriched over the non-cross-linked control by two-fold or greater either in the number of peptides identifying them or in the total signal area associated with that protein in both replicates were considered to be positive hits. The list of enriched proteins does not correlate with protein abundance.
Microscopy and Image Analysis
Available strains carrying GFP tagged proteins were obtained from the Life TechnologiesTM Yeast GFP Fusion Collection. These strains were grown to 0.4 – 0.6 OD600 in complete minimal medium. The culture was then split into two equal halves. One half was used to observe localization under log phase conditions. Other half was spun down, rinsed with complete minimal medium without glucose, and then re-suspended in medium without glucose for 30 min before microscopy. Imaging and image processing was done as described (Buchan et al. 2008)4. All GFP fusion proteins that aggregate into foci with or without glucose starvation stress, were transformed with Pub1-mCherry (pRP 1661) and Edc3-mCherry (pRP 2148) separately to check for co-localization with stress granules and P-bodies respectively. Glucose starvation and co-localization experiments were done as in Buchan et al. 2008 with the following differences: First, glucose starvation was performed for 15 min4. Second, 10 Z stacks were taken for each image.
Image quantification was done by manually counting foci for three independent images for each protein.
CLIP: UV-crosslinking, immunoprecipitation and library construction
Untagged and TAP-tagged Pat1, Lsm1, Dhh1 and Sbp1 strains were obtained (Open Biosystems). Strains were grown in YEPD at 30° C to mid-log and re-suspended in PBS for 10 minutes to induce P-bodies. Stressed cells were UV-crosslinked at 0.8–1.2J/cm2. Cell lysates were partially clarified at 4,000 rpm and digested with RNAse A (Sigma). TAP-tagged proteins were pulled-down with Rabbit IgG conjugated Dynabeads (Invitrogen) and washed in lysis buffer with 1M urea to reduce non-specific binding. Purified mRNPs were radio-labeled with 32P ATP and resolved on a 4–12% NuPAGE gel (Life Technologies) and transferred onto Protran nitrocellulose (Whatman) membranes. Desired bands were excised and treated with proteinase K (Roche). RNA fragments were isolated, decapped and cloned into RNA libraries following standard protocols5,6. CLIP assays were performed in duplicate and mRNA targets determined correlated well (see Supplementary Fig. 2a). Control data was obtained by similarly creating a small RNA library from purified poly(A) mRNA.
CLIP: Data collection and analysis
Small RNA libraries were sequenced with an Illumina cassava 1.8 pipeline and raw sequences were processed using FASTX-toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) before mapping to the S288C yeast genome (SGD) using Novoalign (http://www.novocraft.com/main/page.php?s=novoalign) or Bowtie 2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml). Reads from duplicated experiments were combined for subsequent data analysis. Total sequence reads were: Dhh1=15,425,458 (replicate 1=11,205,516; replicate 2=4,219,942); Lsm1=2,478,368 (replicate 1=1,376,056; replicate 2=1,102,312); Pat1=11,807,063 (replicate 1= 9,092,555; replicate 2=2,714,508); Sbp1=17,062,164 (replicate 1=14,003,980; replicate 2=3,058183). Unique sequence reads for each protein were: Dhh1= 3,470,728 (replicate 1=2,671,981; replicate 2=805,402); Lsm1=426,279 (replicate 1=295,841; replicate 2=129,472); Pat1=1,865,516 (replicate 1=1,523,345, replicate 2=345,672); Sbp1=13,786,228 (replicate 1=12,016,153; replicate 2 =1,763,965). Identical reads are treated as independent for all analysis. For mRNA analysis, annotated transcription start and stop sites were obtained from a published database7. Profiles of the transcriptome were generated with 50 nt extensions at both 5′ and 3′ ends of each mRNA. These extensions were trimmed in cases where they overlapped with annotated transcripts. When overlap occurred between an ORF and tRNA, snRNA or rRNA the mRNA was discarded. Sequence counts in the resulting transcripts were summed. Counts were normalized to reflect the depth of sequencing for each protein by multiplying the counts across all transcripts by (total counts in control)/(total counts for sample). Perl scripts were used to identify significant Tier 1 and Tier 2 peaks (Perl 5.12.13). Ratio of protein signal to control was taken as (signal +1)/(control+1) to account for cases with 0 signal in control. Average plots were made by normalizing the highest peak in an mRNA to 100. 5′ UTRs, ORFs and 3′ UTRs were individually scaled to the average size of these regions calculated for the yeast genome prior to averaging, for visualization. Consensus sequence was identified by submitting peak sequences to DREME8 (MEME suite) online at meme.sdsc.edu/meme/cgi-bin/dreme.cgi.
Statistical Analysis
For significance of overlap between two lists in Figure 4b and Supplementary Figure 2a, Z-scores were calculated by taking the ratio of the difference between the actual extent of overlap and overlap by random chance to the standard deviation obtained from the null hypothesis distribution. Hypergeometric distribution was used as null hypothesis. −log P values were calculated from Z-scores.
To assess the statistical significance of the positional specificities of P-body proteins, a Chi-square test was performed to calculate P values.
To assess the significance of differences in Figure 5c and Supplementary Figure 5b P values were calculated using unpaired Student’s t-test.
Yeast Growth Conditions and Additional Statistical Analyses are described in Supplementary Note.
Supplementary Material
Acknowledgments
Mass spectrometry and proteomics data were acquired by the Arizona Proteomics Consortium supported by National Institute of Environmental Health Science grant ES06694 (the S.W.E.H.S.C.), National Institutes of Health, National Cancer Institute grant CA023074 (the A.Z.C.C.) and by the BIO5 Institute of the University of Arizona. The Thermo Fisher LTQ Orbitrap Velos mass spectrometer was provided by National Institutes of Health, National Center for Research Resources grant 1S10 RR028868-01 (G.T.). J.R. Buchan for assistance with microscopy. C. Decker and other members of the Parker lab for helpful discussions. Funding from National Institutes of Health grant 7R37 GM045443 (R.P.), Howard Hughes Medical Institute grant (R.P.) and Leukemia and Lymphoma Society fellowship 5687-13 (S.F.M.).
Footnotes
Author Contributions
R.P., S.F.M., S.J. and M.S. designed the project. S.F.M. performed the in vivo RBP capture experiments and CLIP analysis, S.J. did the microscopy and CLIP analysis and M.S. performed the CLIP experiments and analysis. R.P., S.F.M. and S.J. wrote the paper.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 2.Tsvetanova NG, Klass DM, Salzman J, Brown PO. Proteome-wide search reveals unexpected RNA-binding proteins in Saccharomyces cerevisiae. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0012671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matunis MJ, Matunis EL, Dreyfuss G. PUB1: a major yeast poly(A)+ RNA-binding protein. Molecular and cellular biology. 1993;13:6114–6123. doi: 10.1128/mcb.13.10.6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castello A, et al. Insights into RNA Biology from an Atlas of Mammalian mRNA-Binding Proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 5.Baltz AG, et al. The mRNA-Bound Proteome and Its Global Occupancy Profile on Protein-Coding Transcripts. Molecular Cell. 2012;46:674–690. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 6.Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 8.Dewey CM, et al. TDP-43 aggregation in neurodegeneration: Are stress granules the key? Brain Res. 2012 doi: 10.1016/j.brainres.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nogueira T, de Smit M, Graffe M, Springer M. The relationship between translational control and mRNA degradation for the Escherichia coli threonyl-tRNA synthetase gene. Journal of Molecular Biology. 2001;310:709–722. doi: 10.1006/jmbi.2001.4796. [DOI] [PubMed] [Google Scholar]
- 10.Frugier M, Giegé R. Yeast aspartyl-tRNA synthetase binds specifically its own mRNA. Journal of Molecular Biology. 2003;331:375–383. doi: 10.1016/s0022-2836(03)00767-8. [DOI] [PubMed] [Google Scholar]
- 11.Sampath P, et al. Noncanonical function of glutamyl-prolyl-tRNA synthetase: gene-specific silencing of translation. Cell. 2004;119:195–208. doi: 10.1016/j.cell.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 12.Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 2008;6:e255. doi: 10.1371/journal.pbio.0060255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du TG, et al. Nuclear transit of the RNA-binding protein She2 is required for translational control of localized ASH1 mRNA. EMBO reports. 2008;9:781–787. doi: 10.1038/embor.2008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Decker CJ, Teixeira D, Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buchan JR, Parker R. Eukaryotic Stress Granules: The Ins and Outs of Translation. Molecular Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buchan JR, Yoon JH, Parker R. Stress-specific composition, assembly and kinetics of stress granules in Saccharomyces cerevisiae. Journal of Cell Science. 2011;124:228–239. doi: 10.1242/jcs.078444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoyle NP, Castelli LM, Campbell SG, Holmes LEA, Ashe MP. Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J Cell Biol. 2007;179:65–74. doi: 10.1083/jcb.200707010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grousl T, et al. Robust heat shock induces eIF2alpha-phosphorylation-independent assembly of stress granules containing eIF3 and 40S ribosomal subunits in budding yeast, Saccharomyces cerevisiae. Journal of Cell Science. 2009;122:2078–2088. doi: 10.1242/jcs.045104. [DOI] [PubMed] [Google Scholar]
- 19.Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nissan T, Rajyaguru P, She M, Song H, Parker R. Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Molecular Cell. 2010;39:773–783. doi: 10.1016/j.molcel.2010.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Segal SP, Dunckley T, Parker R. Sbp1p affects translational repression and decapping in Saccharomyces cerevisiae. Molecular and cellular biology. 2006;26:5120–5130. doi: 10.1128/MCB.01913-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chowdhury A, Mukhopadhyay J, Tharun S. The decapping activator Lsm1p-7p-Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA. 2007;13:998–1016. doi: 10.1261/rna.502507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He W, Parker R. The yeast cytoplasmic LsmI/Pat1p complex protects mRNA 3′ termini from partial degradation. Genetics. 2001;158:1445–1455. doi: 10.1093/genetics/158.4.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bailey TL. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics. 2011;27:1653–1659. doi: 10.1093/bioinformatics/btr261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rajyaguru P, She M, Parker R. Scd6 targets eIF4G to repress translation: RGG motif proteins as a class of eIF4G-binding proteins. Molecular Cell. 2012;45:244–254. doi: 10.1016/j.molcel.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoell JI, et al. RNA targets of wild-type and mutant FET family proteins. Nat Struct Mol Biol. 2011 doi: 10.1038/nsmb.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coller JM, Tucker M, Sheth U, Valencia-Sanchez MA, Parker R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA. 2001;7:1717–1727. doi: 10.1017/s135583820101994x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 29.Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10:602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- 30.Yoon JH, Choi EJ, Parker R. Dcp2 phosphorylation by Ste20 modulates stress granule assembly and mRNA decay in Saccharomyces cerevisiae. J Cell Biol. 2010;189:813–827. doi: 10.1083/jcb.200912019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tharun S, Parker R. Targeting an mRNA for decapping: displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs. Molecular Cell. 2001;8:1075–1083. doi: 10.1016/s1097-2765(01)00395-1. [DOI] [PubMed] [Google Scholar]
- 32.Hilgers V, Teixeira D, Parker R. Translation-independent inhibition of mRNA deadenylation during stress in Saccharomyces cerevisiae. RNA. 2006;12:1835–1845. doi: 10.1261/rna.241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coller J, Parker R. Eukaryotic mRNA decapping. Annu Rev Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- 34.Swisher KD, Parker R. Localization to, and Effects of Pbp1, Pbp4, Lsm12, Dhh1, and Pab1 on Stress Granules in Saccharomyces cerevisiae. PLoS ONE. 2010;5:e10006. doi: 10.1371/journal.pone.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teixeira D, Parker R. Analysis of P-body assembly in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:2274–2287. doi: 10.1091/mbc.E07-03-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 1.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 2.Andon NL, et al. Proteomic characterization of wheat amyloplasts using identification of proteins by tandem mass spectrometry. Proteomics. 2002;2:1156–1168. doi: 10.1002/1615-9861(200209)2:9<1156::AID-PROT1156>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 3.Qian WJ, et al. Probability-based evaluation of peptide and protein identifications from tandem mass spectrometry and SEQUEST analysis: the human proteome. J Proteome Res. 2005;4:53–62. doi: 10.1021/pr0498638. [DOI] [PubMed] [Google Scholar]
- 4.Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol. 2008;183:441–455. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 6.Wang Z, Tollervey J, Briese M, Turner D, Ule J. CLIP: construction of cDNA libraries for high-throughput sequencing from RNAs cross-linked to proteins in vivo. Methods. 2009;48:287–293. doi: 10.1016/j.ymeth.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 7.Nagalakshmi U, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bailey TL. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics. 2011;27:1653–1659. doi: 10.1093/bioinformatics/btr261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.