Abstract
Cryo-electron microscopy (cryo-EM) is increasingly becoming a mainstream technology for studying the architecture of cells, viruses and protein assemblies at molecular resolution. Recent developments in microscope design and imaging hardware, paired with enhanced image processing and automation capabilities, appear poised to further advance the effectiveness of cryo-EM methods. These developments promise to increase the speed and extent of automation and to improve the resolutions that can be achieved, rendering this technology capable of determining a wide variety of biological structures. Additionally, established modalities for structure determination, such as X-ray crystallography and nuclear magnetic resonance spectroscopy, are being routinely integrated with cryo-EM density maps to achieve atomic-resolution models of complex, dynamic molecular assemblies. In this review, which is directed towards readers who are not experts in cryo-EM methodology, we provide an overview of emerging themes in the application of this technology to the investigation of diverse questions in biology and medicine. We discuss the ways in which these methods are being used to study structures of macromolecular assemblies that range in size from whole cells to small proteins. Finally, we include a description of how the structural information obtained by cryo-EM is deposited and archived in a publicly accessible database.
What is cryo-EM?
Over the past decade, the phrase “cryo-electron microscopy”, often abbreviated as “cryo-EM”, has evolved to encompass a broad range of experimental methods. At the core, each of these is based upon the principle of imaging radiation-sensitive specimens in a transmission electron microscope under cryogenic conditions. In biology, applications of cryo-EM now span a wide spectrum, ranging from imaging intact tissue sections and plunge-frozen cells to individual bacteria, viruses and protein molecules. Cryo-electron tomography, single-particle cryo-electron microscopy, and electron crystallography are all sub-disciplines of cryo-EM that have been used successfully to analyze biological structures in different contexts. These methods have been used singly as well as in hybrid approaches, where the information from electron microscopy is combined with complementary information obtained using X-ray crystallographic and NMR spectroscopic approaches.
A large number of reviews are published annually highlighting the rapid strides being made in the cryo-EM field [1–14]. These reviews feature detailed methodological advances in specific sub-disciplines such as tomography or single-particle imaging, and report new insights into biological mechanisms revealed through the application of cryo-EM technology. The present review has a different focus and is directed towards students, non-specialists, and researchers in other disciplines who seek a primer on the overall landscape of cryo-EM and an appreciation of the range of biological systems that can be investigated using this technology. Our purpose, therefore, is to use selected examples to provide an introduction to cryo-EM that demystifies the technology and conveys a sense of excitement about this growing area of structural biology.
We begin the review with a brief introduction to imaging using an electron microscope, including the principles underlying data collection and the distinction between cryo-EM and other electron microscope applications that involve imaging under non-cryogenic conditions. In subsequent sections, we highlight different cryo-EM applications that are used to investigate structures ranging in size from cells and viruses to small molecular complexes. We detail the use of cryo-electron tomography to analyze one-of-a-kind entities, such as whole cells, viruses and even molecular complexes, employed with and without the use of sub-volume averaging. Then we introduce single-particle averaging, the process of combining large numbers of 2D projection images to determine 3D structures. And finally, we discuss how averaging methods can be used to determine structures of proteins, at near-atomic resolution, when they are present in ordered assemblies such as two-dimensional crystals. We end with a brief description of the Electron Microscopy Data Bank (EMDB; emdatabank.org), which, together with the Protein Data Bank (PDB; pdb.org), serves as a publicly available repository for EM data at all resolutions.
Imaging with an electron microscope
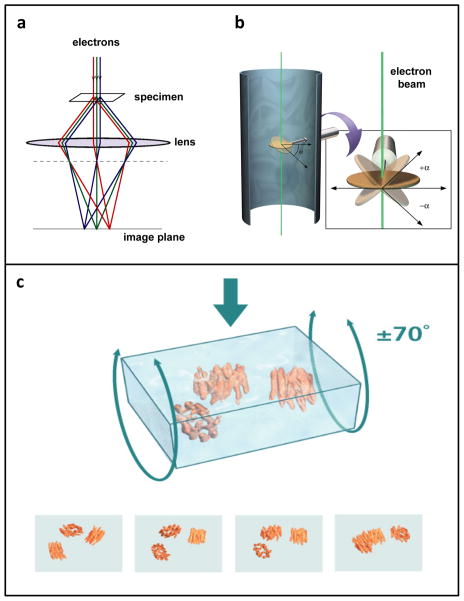
Imaging biological objects in an electron microscope is, in principle, analogous in some respects to light-microscopic imaging of cell and tissue specimens mounted on glass slides. In light microscopy, visible photons serve as the source of radiation; once they pass through the specimen, they are refracted through glass optical lenses to form an image. In electron microscopy, the radiation is electrons, emitted by a source that is housed under a high vacuum, and then accelerated down the microscope column at accelerating voltages of typically 80–300 kV. After passing through the specimen, scattered electrons are focused by the electromagnetic lenses of the microscope (Figure 1a). The most significant difference between optical microscopy and electron microscopy, however, lies in the resolving power of the two methods. Resolution is directly influenced by the wavelength of the imaging radiation source: the shorter the wavelength, the higher the attainable resolution. The resolution possible with visible light (wavelengths ~ 4000–7000 Å; 1 Å = 10−10 m), is significantly less than that achieved with electron sources in a typical transmission electron microscope (wavelength of ~ 0.02 Å for operation at 300 kV). Thus, the wavelength of electron radiation itself does not impose a limit on the resolution that could be obtained when imaging biological macromolecules. The possibility of imaging biological structures with electrons was demonstrated convincingly in 1975 with the determination of the structure of bacteriorhodopsin at ~ 7 Å resolution [15]. These pioneering studies led to the eventual determination of near atomic resolution maps for bacteriorhodopsin [16, 17], and other membrane proteins such as aquaporin [18]. These studies further laid the foundation for recent progress in generation of atomic resolution models for numerous icosahedral viruses [19]. Even higher resolutions have been attained for non-organic samples, which can withstand considerably higher electron doses than biological materials during imaging without loss of structural integrity. For example, individual gold atoms can be directly visualized in high-end electron microscopes from most manufacturers, and the 3D structures of gold nanoparticles have been obtained at atomic resolution using electron tomography [20].
Figure 1.
Image formation in the electron microscope. (a) Schematic illustrating image formation in an electron microscope, highlighting the similarities between electron and optical microscopy. (b) Schematic illustrating the principle of data collection for electron tomography. As the specimen is tilted relative to the electron beam, a series of images is taken of the same field of view. (c) Rendering of selected projection views generated during cryo-electron tomography as a vitrified film (formed by rapidly freezing a thin aqueous suspension) is tilted relative to the electron beam. To reconstruct the three-dimensional volume, a set of projection images is “smeared” out along the viewing directions to form back-projection profiles. The images are combined computationally to recover the density distribution of the object. Figure adapted from [110].
So why is it not possible to routinely image individual proteins, viruses and cells in their native state directly in an electron microscope at atomic resolution? The primary reason this is challenging is because of the extensive damage that results from the interaction of electrons with organic matter. Electron irradiation leads to the breaking of chemical bonds and creation of free radicals, which in turn cause further secondary damage [21–23]. One way to mitigate electron-induced sample damage is to use the method of negative staining, in which accessible molecular surfaces are coated with reagents containing heavy atoms, such as uranyl acetate, that are much less radiation-sensitive than organic matter. Because these stains do not penetrate into biological samples, they essentially make a cast of the specimen surface, a high contrast “relief” of the surface, albeit at the expense of internal structural information and with the potential for artifacts such as sample flattening. Nevertheless, this has been a common mode of specimen preparation for many decades in conventional transmission electron microscopy, and has been routinely used for visualization of cells, viruses and proteins, yielding, in the latter case, structures at resolutions of ~ 40 Å to ~ 20 Å [24, 25]. Images of unstained samples or samples that are embedded in sugars such as trehalose, which acts to stabilize proteins in aqueous solution and to increase the image contrast [26], can be obtained at sufficiently low electron doses to yield internal structural information. However, the lower electron doses result in images with a poor signal-to-noise ratio, as unstained biological samples are composed primarily or entirely of low atomic number elements that scatter electrons poorly. Therefore, the difficulty in high-resolution imaging of unstained specimens is that electron doses that are low enough to minimize radiation damage and preserve the specimen generate noisy images, while electron doses that are high enough to get a good signal-to-noise ratio lead to unacceptable levels of specimen damage.
Two approaches are used in modern “high-resolution” electron microscopy to solve this problem. The first approach involves “cryo-electron microscopy”, in which imaging is performed using frozen specimens maintained at either liquid nitrogen or liquid helium temperatures. Almost four decades ago, measurements on the decay of diffraction intensities at room temperature versus cryogenic temperatures established that cryo-electron microscopy could reduce the effects of radiation damage [22]. The development of methods to rapidly freeze (vitrify) biological specimens in a layer of glass-like ice [27, 28], which could then be imaged at liquid nitrogen and/or helium temperatures, paved the way for this method to become commonly used [10, 29]. Plunge-freezing aqueous solutions into a cryogen, such as liquid ethane cooled by liquid nitrogen, is a method used to prepare specimens for cryo-EM applications in tomography, single-particle imaging and for helical and two-dimensional crystals. Imaging at liquid nitrogen temperatures reduces the extent of radiation damage by as much as 6-fold compared to ambient temperatures [28]. This means that, for the same amount of radiation damage, an image can be recorded at cryogenic temperatures with a much higher electron dose [30, 31]. Both liquid nitrogen and liquid helium have been used successfully to obtain 3D reconstructions at near-atomic resolution; a careful comparison of the two cryogens indicate that they show similar protective effects in resolutions between ~ 4–20 Å [32], although the choice of cryogen has long been under debate [28, 33]. Another way to increase the signal-to-noise ratio involves averaging images of a large number of identical units, in an approach that has an intellectual connection to the way in which scattering of X-rays by billions of molecules is averaged to obtain structural information in X-ray crystallography. This technique was first used in the context of helical assemblies [30] and two-dimensional protein crystals imaged at room temperature [31], as well as under cryogenic conditions [27]. Single-particle imaging and electron tomography of individual molecular complexes also invoke the principle of averaging multiple images collected from frozen-hydrated specimens of molecular complexes. These two ideas, namely the concept of imaging at cryogenic temperatures and the concept of averaging multiple low-dose images, form the basis of modern high-resolution biological electron microscopy. The application of these principles to biological imaging is discussed in the following sections, although we will note instances where they are not always used in concert.
Cryo-electron tomography: cellular and sub-cellular structures
One approach to obtaining 3D structures of macromolecular assemblies using electron microscopy is tomography [1, 34, 35], in which a series of images is collected, with each image taken at a different tilt relative to the direction of the incident electron beam (Figure 1b). For imaging thicker specimens such as large viruses or whole bacterial or eukaryotic cells, images are generally collected with the aid of an imaging filter that improves contrast by eliminating inelastically scattered electrons. These images can then be combined computationally, using a strategy similar to that used in computerized axial tomography (CAT), to generate “tomograms”, essentially 3D images of the specimen. Two general types of strategies exist for the further analysis of tomographic data. When specific molecular entities can be clearly identfied within cells or on the surface of viruses, or when molecular complexes are imaged in vitro, averaging methods can be employed to obtain more detailed structural information than is present in the original tomograms. However, many cellular sub-structures (or organelles in the case of mammalian cells) are morphologically heterogeneous, necessitating comparison across multiple tomograms to identify patterns in structural variation.
Electron tomographic imaging requires that specimens be thin enough for the incident electron beam to be transmitted, typically not much more than ~ 0.5 microns even for microscopes capable of operation at 300 kV. Accelerating voltages of 200–300 kV enable greater sample penetrance of the electron beam [34] than microscopes operating at lower voltages. Samples that are sufficiently thin can be negatively stained for room temperature tomography or plunge-frozen under native conditions for cryo-electron microscopy. Thicker samples, such as tissues or cell pellets, require more pretreatment [13]. Classically, samples destined for room temperature tomography are chemically fixed, dehydrated with sequential graded steps of solvent exchange, plastic-embedded and cured to form a block. The block is cut into sections using a diamond knife, and the sections are treated with stains such as uranyl acetate or lead citrate to generate contrast. Thicker samples can also be high pressure-frozen or slam-frozen to promote formation of vitreous, unstructured ice, rather than crystalline ice, which disrupts native cellular architecture. Frozen samples then undergo freeze-substitution for dehydration and fixation, followed by low-temperature plastic embedding, thin sectioning and staining, as above. When necessary, samples of bacterial cells [36] or mammalian cells [37, 38] can also be thinned under cryogenic conditions using focused ion beam milling. Fortunately, many bacteria have thicknesses around 0.5 μm, thus reducing the need for significant sample manipulation. This has made bacterial tomography a particularly productive area for the application of cryo-electron tomography (reviewed in [3]).
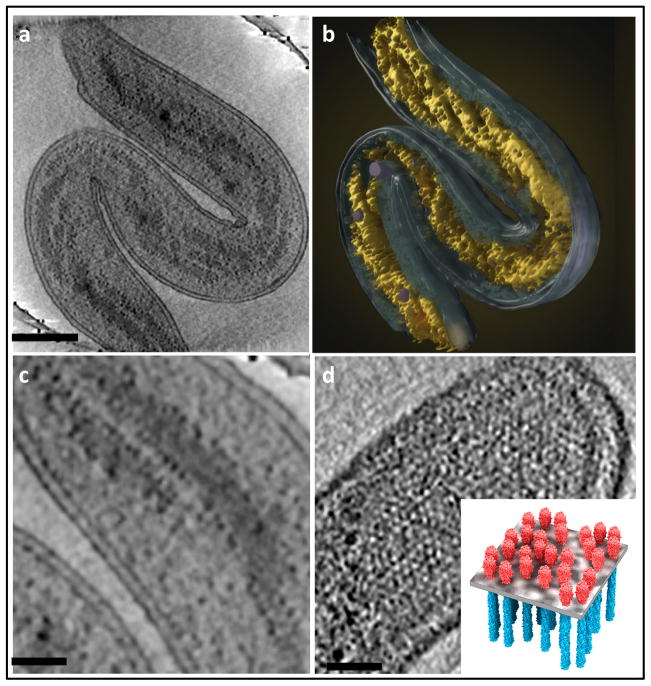
Numerous insights have emerged from these tomographic studies, such as the organization of the bacterial cytoskeleton [13, 39], flagellar motor [40], lateral arrangement of membrane protein assemblies [41], visualization of DNA ejection in Bacillus anthracis spore-binding phage [42] and of cell-cell fusion [43], to name just a few examples. Another illustration of the power of tomographic imaging in unraveling internal bacterial structures is the recent discovery of the spiral organization of the bacterial nucleoid [44] in Bdellovibrio bacteriovorus cells (shown in Figure 2). In addition to revealing the twisted nucleoid structure, the tomograms also reveal the localization of ribosomes at the interface between the nucleoid and the cytosol, as well as the ordered arrangement of the chemoreceptor assemblies in the inner membrane.
Figure 2.
Use of cryo-electron tomography to image the interior architecture of intact bacterial cells. (a,b) Illustration of spiral architecture of the nucleoid in Bdellovibrio bacteriovorus showing (a) a 210 Å thick tomographic slice through the 3D volume of a cell and (b) a 3D surface rendering of the same cell, with the spiral nucleoid highlighted (yellow). (c) Higher magnification view of a tomographic slice through the cell, showing well-separated nucleoid spirals and ribosomes (dark dots) distributed at the edge of the nucleoid. (d) Expanded views of 210 Å thick tomographic slices, showing top-views of polar chemoreceptor arrays. A schematic model (inset) illustrates the spatial arrangement of the chemoreceptor arrays in the plane of the membrane. Scale bars: 2000 Å in (a) and 500 Å in (c,d). Figure adapted from [44].
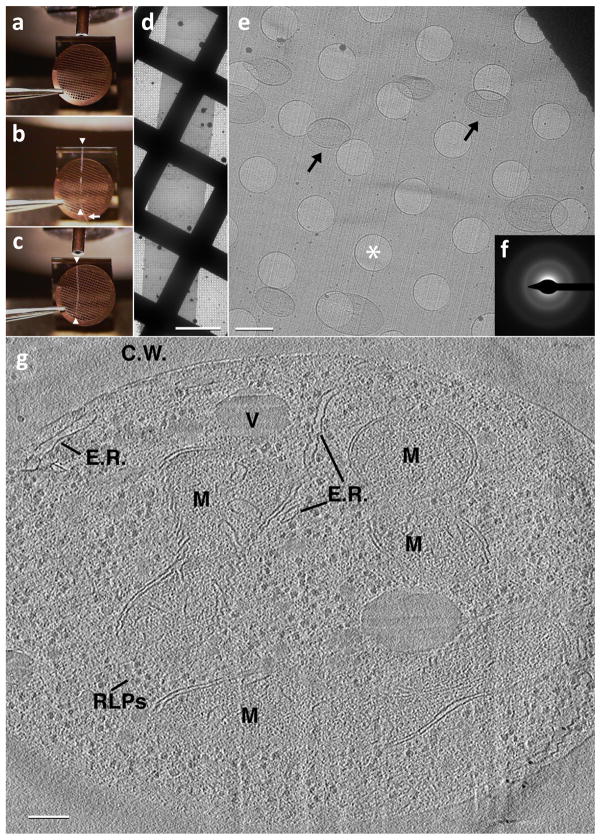
Other applications of cryo-electron tomography have included imaging of thin regions of mammalian cells [45] and investigation of a variety of filamentous and cytoskeletal assemblies [46–48] and of thin vitreous sections prepared from mammalian cells and tissues [38, 49], leading to significant new information on sub-cellular structures [1, 8, 50]. As cellular cryo-electron tomography progresses towards charting macromolecular landscapes [51–54], tomography of vitreous cryo-sections (reviewed in [55]) is emerging as a particularly important tool for the investigation of large cells and tissues. Historically, this technique has been hindered by problems with creating proper attachment of the ribbons of vitreous sections to the support film of the electron microscopic grid. Recently, electrostatic charging has been used to successfully attach vitreous sections to the EM support film [56]. Figure 3 shows a relatively long ribbon of vitreous cryo-sections pulled from the edge of the microtome blade over the EM grid (Figure 3a-c). It is then attached using electrostatic charge to the EM support film. S. cerevisiae cells can be seen scattered throughout the vitreous section, shown at higher magnification (Figure 3e). Another major limitation has been artifacts that are introduced during sectioning, evident in most images [57, 58], although the structures of large macromolecular complexes such as 80S ribosomes remain unaltered by section-induced compression (Figure 3g) [49]. Nevertheless, electron diffraction patterns from high pressure-frozen lysozyme crystals subjected to vitreous cryo-sectioning can extend to 7.9 Å [59], and there is hope that the throughput of many of the steps can be improved, suggesting that vitreous cryo-sectioning could become a routine method for observing macromolecular structures in their native cellular environment.
Figure 3.
Imaging of vitreous sections by cryo-electron microscopy. (a) An EM grid is placed at the diamond knife-edge using tweezers with a bent tip. (b) A ribbon of vitreous sections (white arrowheads) is guided over the EM grid using an eyelash (white arrow) attached to a wooden dowel. (c) Once the ribbon of vitreous sections is of suitable length (white arrowheads), approximately the diameter of the EM grid, an electrostatic generator is switched from the discharge mode to the charge mode, causing the ribbon to attach to the EM grid. (d) A low magnification image showing the ribbon of vitreous sections after electrostatic charging. Scale bar: 50 μm. (e) A medium magnification micrograph showing a vitreous section from within the ribbon. Note that the section is smooth and relatively flat with no apparent mechanically induced damage as is often seen with stamping/pressing techniques. S. cerevisiae cells can be seen scattered throughout the vitreous section (black arrows). Holes within the C-Flat EM grid can be seen and one such is denoted with a white asterisk. Scale bar: 2 μm. (f) A selected area diffraction pattern confirming vitreous ice. (g) A 50 Å thick slice through a reconstructed tilt series from a 500 Å thick vitreous section of an S. cerevisiae cell: C.W. – Cell Wall, V – Vacuole, M – Mitochondrion, E.R. – pieces of Endoplasmic reticulum, RLPs – Ribosome-like particles. Scale bar: 1000 Å.
As discussed above, tomography is emerging as a powerful method to visualize one-of-a-kind, structurally heterogenous entities at resolutions between ~ 100 Å and ~ 50 Å. These resolutions are two orders of magnitude better than can be achieved by visualizing subcellular detail using optical microscopy, filling a critical gap in cell biology. In addition, cryo-electron tomography is a unique approach for imaging all of the cellular components simultaneously in unstained cells, as the image contrast arises from the intrinsic scattering of electrons by the protein and nucleic acid components. The development of clonable labels for use in electron microscopy [60, 61], as well as of correlative electron-microscopic/light-microscopic techniques [62, 63], provides new avenues to determine the localization of individual biological complexes in situ in cells when specific labeling is desired. While the attainable spatial resolution of fluorescently-labeled proteins within a cell can reach resolutions better than ~1000 Å, unlabeled cellular protein complexes are not visualized by light microscopy. Identification of fluorescently labeled entities at low resolution combined with correlative electron microscopy could thus be highly useful for describing subcellular architecture, while simultaneously targeting specific molecules of interest.
While cryo-electron tomography provides a unique opportunity to explore the organization of protein complexes and cellular components in situ in intact viruses, cells and tissues, individual tomograms from protein complexes are at resolutions that are often not useful for describing molecular structures. However, in cases where there are multiple copies of the structure of interest, it is possible to improve the final resolution by averaging. This procedure, in which multiple copies of sub-volumes containing the assembly of interest are extracted from tomograms, aligned, classified and averaged in 3D, is emerging as a powerful approach at the interface between structural and cell biology [35, 64–69].
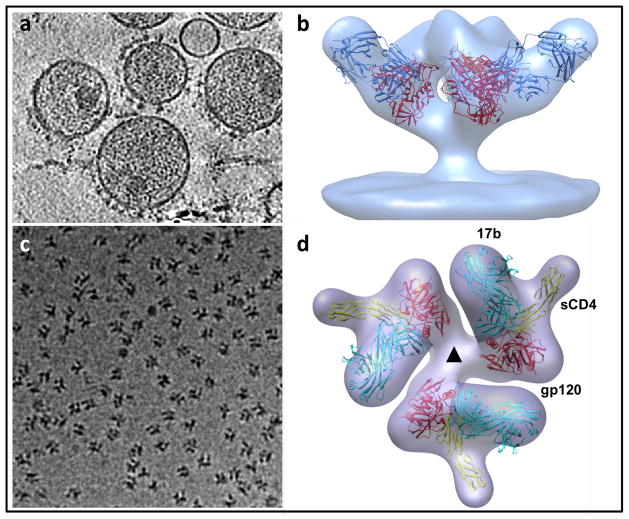
One area in which sub-volume averaging has been particularly useful is in structural studies of viral surface proteins, where crystallographic structural information can be difficult to obtain from proteins in their native state. For example, sub-volume averaging approaches have yielded numerous 3D structures, at ~ 20 Å resolution, of trimeric HIV (human immunodeficiency virus) and SIV (simian immunodeficiency virus) envelope glycoproteins in complex with neutralizing antibodies (Figure 4a, 4b) [70–73], providing new and fundamental insights into the mechanism of viral entry. Sub-volume averaging can also be applied to soluble protein complexes where there is excellent contrast between the protein assembly and its surrounding milieu (Figure 4c, 4d) [35, 74, 75]. Subvolume averaging has also led to structural information on complex systems as yet intractable to X-ray crystallography; examples of this include bacterial membrane protein assemblies [76, 77] and myosin V assemblies [78]. The resolutions that have been achieved with this approach are typically in the range of ~ 30 Å to ~ 20 Å. When unambiguous fitting constraints are available, such as the presence of one or more antibody Fab fragments bound to specific sites, individual sub-components whose structures have been determined by X-ray crystallography and NMR can be fitted reliably into the tomographic density maps. The accuracy of this type of fitting is only likely to get better with improvement in the resolutions obtained with tomographic density maps [79] using methods such as “constrained single-particle tomography” that have been developed recently [80]. In this approach, the accuracy of determining molecular orientations is greatly improved by exploiting the known angular constraints present in tomographic tilt series. The finding that resolutions as high as ~ 8 Å can be obtained with this strategy for averaging and 3D reconstruction is thus an exciting development that could have broad applicability for structural analysis of molecular conplexes using cryo-electron tomography.
Figure 4.
Structural analysis of membrane protein complexes using cryo-electron tomography combined with sub-volume averaging. (a) Tomographic slice through a field of HIV recorded from a specimen grid that was plunge-frozen and stored at liquid nitrogen temperatures. In this image, the viral membrane is decorated with trimeric envelope glycoproteins, which are required for viral entry into target cells. (b) Density map at ~ 20 Å resolution of the trimeric envelope glycoproteins complexed with the neutralizing antibody VRC01. The map was obtained by missing wedge-corrected, subvolume averaging of cryo-electron tomographic images. The map was then fitted with three copies of the X-ray crystallographically determined structure for the complex of monomeric gp120, a portion of the HIV envelope glycoprotein, complexed with VRC01. (c) Projection image of individual molecular complexes of soluble trimeric envelope glycoproteins from human immunodeficiency virus (HIV; strain KNH1144). (d) Density map at ~ 20 Å resolution of the complex of HIV envelope glycoproteins (molecular weight of polypeptide portion ~ 240 kDa) with soluble CD4 (molecular weight ~ 24 kDa) and Fab fragment (molecular weight ~ 50 kDa). The map is fitted with three copies of the structure of the ternary complex of monomeric gp120, sCD4 and 17b Fab determined by X-ray crystallography. Figure panels adapted from[73] and [74].
Single-particle cryo-electron microscopy
Perhaps the most commonly used variant of cryo-electron microscopy is single-particle analysis. In this technique, data from a large number of 2D projection images, featuring identical copies of a protein complex in different orientations, are combined to generate a 3D reconstruction of the structure. When atomic models are available for some or all of the sub-components of the complex, they can be placed or fitted into the density map to provide pseudo-atomic models, considerably extending the information obtained by electron microscopy (reviewed in [10, 81]).
As in most other cryo-EM applications, the first step in single-particle imaging involves spreading soluble complexes across a hole in a carbon film. The specimen is then plunge-frozen, creating a thin layer of vitreous ice that ideally contains identical copies of the complex present in different orientations. The thickness of the ice layer can vary from a few hundred to a few thousand Ångstroms, and is influenced both by the dimensions of the particle and the composition of the buffer. Starting with images of fields containing many molecular complexes, individual particles are selected by hand or by automated algorithms. Once selected, statistical methods, such as principal component analysis, multivariate analysis or covariance analysis, can be used to sort images based on variations in their structural features. A number of computer programs such as EMAN [82], SPIDER [83] and IMAGIC [84], which have been available for many years, can carry out these strategies for image processing. The degree of relatedness between individual “particle” images is used to identify clusters of similar images within the data set. Related images are then averaged to obtain characteristic projection views of the complex at much higher signal-to-noise ratios than in the original images. Iteration of this classification process, using characteristic views of the newly generated class averages as alignment references, improves the accuracy of alignment and permits visualization of finer structural features.
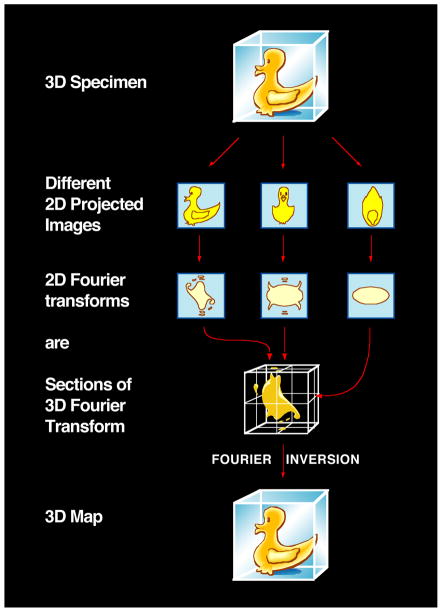
Generation of a 3D reconstruction from the 2D electron microscopic projection views of the molecular complex is dependent upon knowing the relative orientations of all of the particles. The steps involved are mathematically complex, and take advantage of the central projection theorem, which states that, for a 3D object, the Fourier transform of each 2D projection is a central slice through the 3D Fourier transform of the object (Figure 5). Thus, by acquiring a sufficiently large number of molecular images that encompass a wide range of orientations relative to the electron beam, one can build up the 3D reconstructions one image at a time. The 2D Fourier transform of each image contributes a single slice to the 3D Fourier transform. Relative orientations of a pair of images can be derived from the fact that the Fourier transforms of two projections share a common line where they intersect in 3D Fourier space. Angular assignment by identifying common lines can be conducted in Fourier space, as typified by the methodology developed originally for icosahedral virus reconstructions [85], or in real space by obtaining a set of 1D line projections from each pair of 2D class average images and assigning relative orientations based on identifying the line common to both sets [86]. Pair-wise comparisons of at least three class averages to generate three common lines is necessary to assign the relative angular orientations of each projection. Following angular determination, an initial 3D reconstruction can be obtained using real-space reconstruction methods or Fourier-space-based methods. Many variants of these general strategies exist, including those tailored for highly symmetric samples or those with helical symmetry [10, 87]. Initial maps can be refined to higher resolution using projection-matching refinement strategies, which compare individual particle images to uniformly sampled re-projections of the 3D map, performed in real space (using programs such as IMAGIC or SPIDER) or in Fourier space (using programs such as FREALIGN). If appropriate, available atomic coordinates of corresponding protein components derived from X-ray or NMR analyses can be then fitted into the final structure using programs (such as UCSF Chimera) designed for rigid-body or flexible fitting [10, 88, 89].
Figure 5.
Principle of reconstruction of 3D structure by Fourier inversion. Using the “Fourier duck” as the prototypical biological specimen, the schematic illustrates that projection images of the object, each with a different orientation, have 2D Fourier transforms that correspond to sections (indicated by red arrows) through the 3D Fourier transform of the original object. Thus, once the 3D Fourier transform is built up from a collection of 2D images spanning a complete range of orientations, Fourier inversion enables recovery of the 3D structure.
Distinct single-particle strategies that are not reliant on common lines approaches are employed to better handle commonly encountered situations such as the presence of preferred molecular orientations on the grid or of multiple molecular conformations/states. This latter problem arises when different conformations of a homogeneous sample, different ligand occupancy of a complex, or different oligomerization states of a complex coexist within the sample, leading to size variability or multiple symmetries. The use of geometric constraints combined with computational approaches is particularly useful in samples where the particles present on the grid display a preferred orientation relative to the incident electron beam. For example, in random conical tilt imaging [90], two images of each grid region are recorded, the first with the specimen stage tilted 50–60 degrees and the second after the stage position is returned to 0 degrees, and the information is combined. This strategy is also useful when there is conformational heterogeneity because the untilted molecular images can be classified into distinct groups of 2D projection images, which can then be used in conjunction with the corresponding tilted images to obtain multiple 3D reconstructions. Although random conical tilt effectively samples a large swath of Fourier space, this approach still results in a conical region of missing data in 3D space (“missing cone”), given the practical limitation to the highest tilt angles that can be used to obtain useful images. A recently proposed orthogonal tilt reconstruction scheme [91] circumvents the missing cone problem by collecting +/− 45 degree tilts of a large population of randomly oriented particles; here the two views of each molecule are related by a 90 degree rotation. One tilt set is sorted to generate class averages while the corresponding orthogonal images of each class average sample an array of equatorial views around a sphere, with specific positions dictated via angular information obtained during the initial alignment and classification step. Initial 3D reconstructions of each class can then be determined and compared to identify regions of conformational heterogeneity.
The highest resolutions achieved using single-particle approaches have been found when imaging icosahedral viruses. Two factors account for this: (1) the presence of high symmetry (60-fold or higher) confers a powerful advantage for averaging within each virion, and (2) the large size of the virions (tens of megadaltons in most instances) provides excellent contrast for accurately determining particle orientations in projection images. Specialized software that is targeted to automatically solving icosahedrally symmetric structures works by taking advantage of the special symmetry features of these viruses [92]. The ordered packing arrangement also lowers conformational heterogeneity of viral protein components. Because of these features, structures of numerous icosahedral viruses have now been determined to resolutions better than 4 Å (reviewed in [11]), and approaching 3 Å in selected cases (Figure 7) [19]. Other recent highlights include reconstructions, at resolutions better than ~ 10 Å, of dynamic protein complexes with much lower levels of symmetry than icosahedral viruses, such as chaperones [93], and complexes with no internal symmetry, such as ribosomes [94]. Notably, several large and symmetric membrane protein complexes have been successfully reconstructed to sub-nanometer resolutions [95, 96], indicating that this is a promising strategy for rapidly obtaining medium-resolution structures of protein complexes.
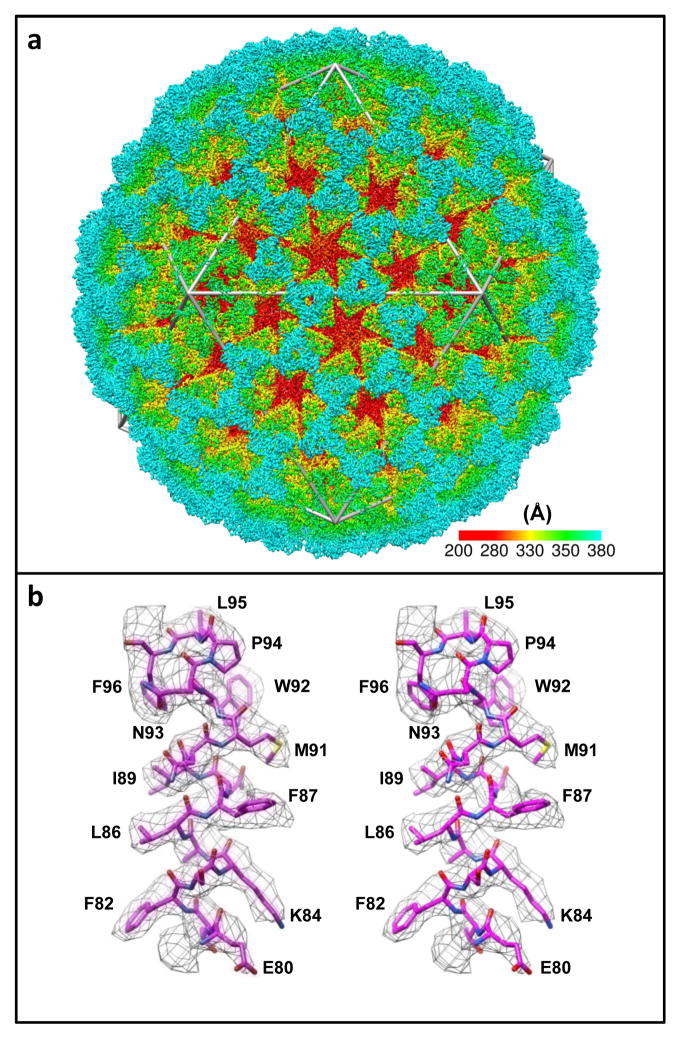
Figure 7.
Determination of structure of a non-enveloped icosahedral virus using cryo-electron microscopy. Visualization of (a) entire structure and (b) selected region demonstrating that near-atomic resolution maps can be obtained for highly ordered assemblies such as icosahedral viruses using advanced image processing methods. Figure adapted from [19].
Progress is also being made in technology development for electron microscopy and computational aspects of data processing, leading to extraordinary increases in speed and automation. An example of this is presented in Figure 6, where we show that the 3D structure of GroEL can be determined to a resolution of ~ 7 Å using automated strategies for data acquisition, automated particle selection, and automated 3D reconstruction. In addition to the single-particle methods developed to analyze conformationally heterogeneous specimens [97], experimental and computational strategies that combine full tomographic sub-volume averaging to generate one or more initial 3D reconstructions at low resolution, with single-particle cryo-EM refinement of the structure(s) at higher resolution, can be integrated into a seamless pipeline. Improvements in both computational sorting aspects and increased tomographic resolution [80] promise to make this an exceptionally powerful, streamlined strategy for 3D structure determination.
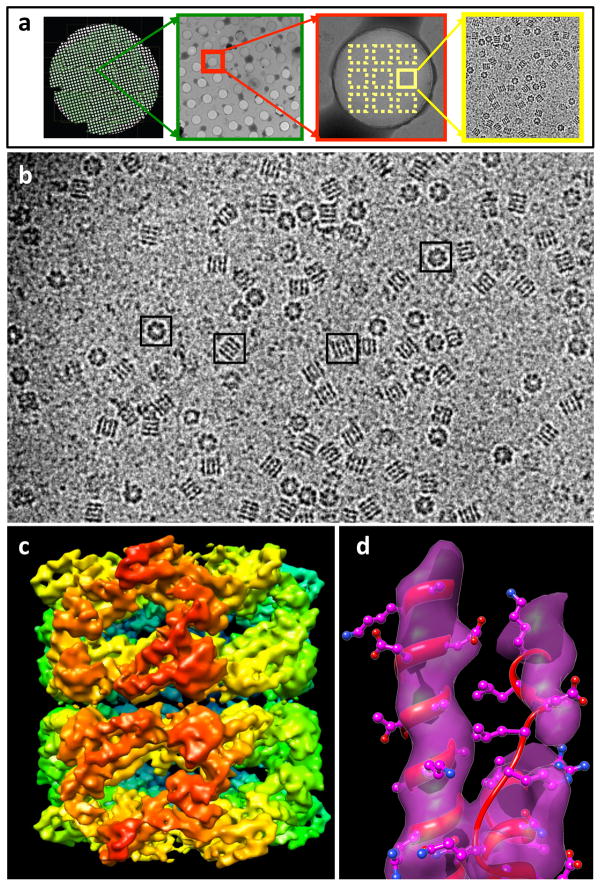
Figure 6.
Automation of macromolecular assembly structure determination by cryo-EM single-particle analysis. (a) Images of a cryo-EM grid at sequentially higher magnification, beginning (left) with an image of the entire grid and concluding with an image of individual structures (right). (b) Representative projection image from a frozen-hydrated specimen of purified GroEL protein complexes. Complexes with distinct orientations relative to the electron beam can be discerned as indicated in the boxed examples. (c) 3D reconstruction using ~ 28,000 individual projection images such as those boxed in panel (b) to generate a density map of the complex at ~ 7 Å resolution. The initial 3D reconstruction was derived by sub-volume averaging using ~ 2000 GroEL particles. Refinement of the initial reconstruction was carried out using almost completely automated procedures as implemented in the software package FREALIGN [111] to refine the structure to ~ 7 Å resolution. (d) Demonstration that the resolution achieved is adequate to visualize α-helices, illustrated by the superposition of a density map of a region of the polypeptide with the corresponding region of a GroEL structure determined by X-ray crystallography (PDB ID: 3E76). Figure panels (b–d) adapted from Bartesaghi et al. (manuscript in preparation).
Cryo-electron microscopy of ordered assemblies
Because 2D projection images can contain information at very high resolutions, in some cases exceeding 3Å, it is possible, in principle, for structures determined by cryo-electron microscopy to achieve near-atomic resolution [98], even in the absence of high symmetry. However, the intrinsic conformational heterogeneity of most isolated macromolecular assemblies poses a major limitation to reaching this goal [99]. One solution to this challenge is to find experimental conditions that induce the formation of helical or crystalline assemblies of the protein of interest. In cases when this is successful, the ordered packing ensures a high degree of conformational homogeneity, while knowledge of the geometry of the packing arrangement provides a highly accurate determination of the relative alignment of the repeating units. This strategy has been extremely effective with membrane proteins that form two-dimensional crystals in the plane of the membrane, allowing structure determination at resolutions approaching ~ 3 Å, as in the case of bacteriorhodopsin (Figure 8) [16]. In addition to two-dimensional crystals, other types of ordered assemblies such as tubular crystals and helical assemblies have also been extremely valuable for structural analysis of conformational changes at resolutions in the 7 Å to 4 Å range. One of the factors limiting resolution even in these specialized cases is the steep fall-off in image amplitudes at high resolution due to factors such as beam-induced specimen movement, charging from incident electrons, and mechanical instabilities in the microscope. In the case of two-dimensional crystals, this problem can be partially offset with the use of electron diffraction to obtain more accurate amplitudes. When this is possible, it is the best of both worlds, enabling structure determination that combines accurate phases derived from the images with accurate amplitudes derived from the diffraction patterns. Resolutions better than ~ 2 Å have been achieved using electron crystallography [18], making this a powerful approach, although restricted to the few special cases when proteins can be induced to form ordered assemblies such as helical or two-dimensional crystals.
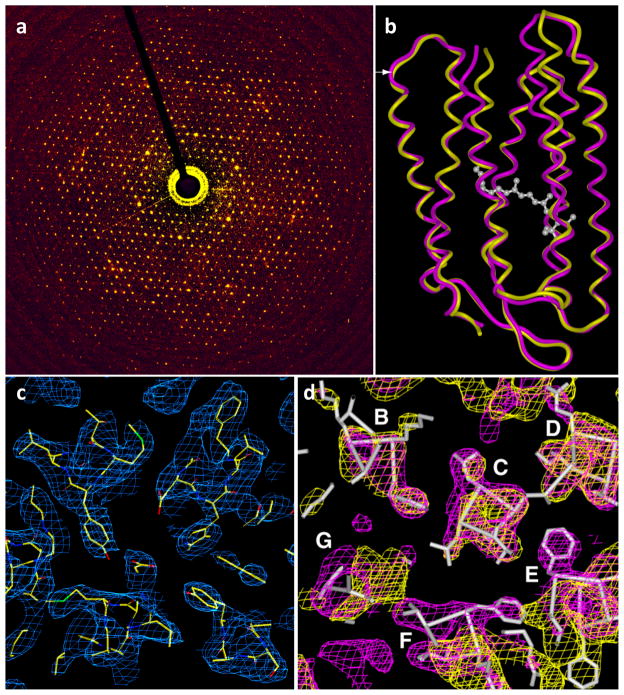
Figure 8.
Determination of membrane protein structure using electron crystallography of 2D crystals. (a) Electron diffraction pattern from bacteriorhodopsin crystals, with reflections extending to ~ 2 Å. (b) Structures of bacteriorhodopsin in native and intermediate conformations at 3.2 Å resolution were obtained by combining phase information present in images of 2D crystals with amplitude information obtained from electron diffraction patterns. (c) σA-weighted density (2FO-FC) map of the open intermediate of bacteriorhodopsin in the center of a lipid bilayer. The map is fitted with the refined atomic model (PDB ID: 1FBK). (d) Sections of bacteriorhodopsin in wild-type (purple) and open intermediate (yellow) conformations, showing the helix movements (from magenta to yellow coordinates) at the cytoplasmic ends of transmembrane helices F and G. The location of the section is marked by the white arrow at the left edge of (b)). The maps are superimposed on the structure of wild-type bacteriorhodopsin, derived by cryo-electron microscopy at 3.2 Å resolution. Figure adapted from [16].
New technology developments and automation
One hurdle that has historically slowed progress in the cryo-EM field has been the time- and personnel-intensive nature of the work. Collecting images for structure determination at the highest resolutions requires precise imaging conditions and specimen placement within the electron microscope. In modern microscopes, the alignments and calibrations involved in imaging tend to be very stable, and are usually performed once for every multi-day session of data collection. However, maintaining and compensating for specimen placement, particularly in single-particle analysis and tomography, presents a significant challenge. The position and orientation of the specimen is usually determined with micrometer precision by a mechanized stage. As the stage is adjusted during data acquisition, the movement of this stage leads to continual small changes in the coordinates of the specimen. This can be compensated for by a combination of beam shift and image shift (for the X and Y axes) and defocus (for the Z axis). Using image and beam shift, several adjacent images can also be collected from a single hole. In tomographic data collection, recording tilt series while maintaining the same region in the field of view can pose a significant obstacle because tilting the specimen holder leads to changes in the Z axis, requiring appropriate adjustment of defocus. These challenges have been addressed successfully with the advent of computerized stages, which together with optically stable microscopes have allowed the development of expert systems capable of managing automated data collection for tomography. Robust data collection packages are now available both as freely distributed (SerialEM [100]) and as commercially available programs (Xplor3D) that effectively automate the tedious task of adjusting image position and defocus for multiple exposures on a single region of interest. Additionally, these programs collect data in a “batch” mode, where the microscopes can continue collecting data from different regions of the specimen for periods exceeding 24 hours without manual intervention.
Automation and improvements in many of the steps in the workflow for cryo-electron microscopy have not only led to dramatic increases in the speed and ease of use, but have also substantially improved data quality. The use of robotics in preparation of frozen-hydrated samples has increased specimen reproducibility and identification of optimal conditions for specimen preparation. Automation in data acquisition, environmental enclosures, the use of patterned grids, and the implementation of remote operation schemes, together with hardware improvements such as automated liquid nitrogen refilling systems, have reduced the need for personnel intervention during data acquisition. The commercial and freely available software routines, which allow automated data collection for single-particle microscopy as well as tomography, have also had a major impact on the development of the cryo-EM field.
One important technical development on the horizon is the use of direct electron detectors [101], which adds to the repertoire of devices currently used (photographic film and charge-coupled device (CCD) cameras) for detection. Two parameters that are important in evaluating detector performance are the modulation transfer function (MTF) and the detective quantum efficiency (DQE). The MTF represents the response of the detector at different spatial frequencies, while the DQE reports on the quality with which incident electrons are recorded at each frequency. Thus the MTF essentially describes the resolution of the detector, while the DQE describes how strong the signal is relative to the noise level. Having the highest possible MTF across the spectrum enables obtaining the best DQE across all frequencies. Although photographic film has remained the standard so far for optimal detector performance, and CCDs have remained the choice for ease of use and automation, the new generation of direct electron detectors appear poised to provide performance that exceeds that of film, while preserving all of the convenience of CCD detectors for automation.
Converting 2D projection images into 3D reconstructions is a process requiring both significant computational power and well-designed image processing algorithms. The broad availability of Graphics Processing Units (GPUs) and large computer clusters over the last decade has been critical for reducing the amount of time required to turn raw data into a 3D reconstruction [102–104]. In addition, image-processing routines that can bypass the need for computationally intensive search strategies for orientation assignment have contributed to additional computational savings [64, 105, 106]. Other important advances in image processing include the development of refinement protocols that require limited user involvement and improved classification strategies for sorting out variability within cryo-EM images [75, 97, 107, 108]. Together, these improvements will enable the analysis of larger and more complex cryo-EM datasets at increasingly higher resolutions.
An emerging paradigm for integrated structural biology
Since the first genomes were published more than a decade ago, genomic and proteomic studies have revealed vast networks of protein-protein and protein-nucleic acid interactions in the cell [109]. This explosion in the genomic and proteomic fields highlights the need for a similar growth in the structural biology field. Already, the application of X-ray crystallographic and NMR spectroscopic approaches have yielded >15,000 unique structures that have been deposited in the Protein Data Bank (PDB). These have included entries from single chain proteins as well as multi-protein complexes, and span the range from small complexes with sizes < 50 kDa that are amenable to analysis by NMR spectroscopy to larger complexes that are amenable to crystallization and analysis by X-ray crystallography, providing a wealth of structural insights into biological mechanisms. However, key gaps remain, especially in the analysis of small, dynamic protein assemblies that are not tractable to analysis by either NMR or X-ray crystallographic methods, but may be uniquely suited for study by cryo-EM. Examples are beginning to emerge where the information from very low resolution cryo-EM maps can nevertheless provide useful information when used in conjunction with NMR spectroscopic information [90].
A glimpse of where the greatest challenges and rewards lie in this emerging discipline can be gleaned by inspection of the data deposited with the EM data bank (EMDB), a world-wide repository for the data generated by all forms of cryo-electron microscopy. EMDB was founded at the European Bioinformatics Institute (EBI) in 2002 and since 2007 has been managed by three partnering entities: the Protein Data Bank in Europe (PDBe), the Research Collaboratory for Structural Bioinformatics (RCSB PDB), and the National Center for Macromolecular Imaging (NCMI). In addition to maintaining the archive and developing services for it, the EMDB plays a key role in leading community-wide initiatives for developing standards and improved practices in the field. EMDB has grown to hold over 1300 released maps, and based on the current growth trend, the holdings are expected to grow 5–10 fold by 2020. A unique bird’seye view of the state of the EM field and current trends can be obtained by analyzing the contents of the EMDB. Two useful tools for this purpose are EMStats (pdbe.org/emstats), which is a web service that presents dynamically generated, up-to-date charts of trends and distributions derived from data mined from the archive, and EMSearch (pdbe.org/emsearch), which is an advanced web search service.
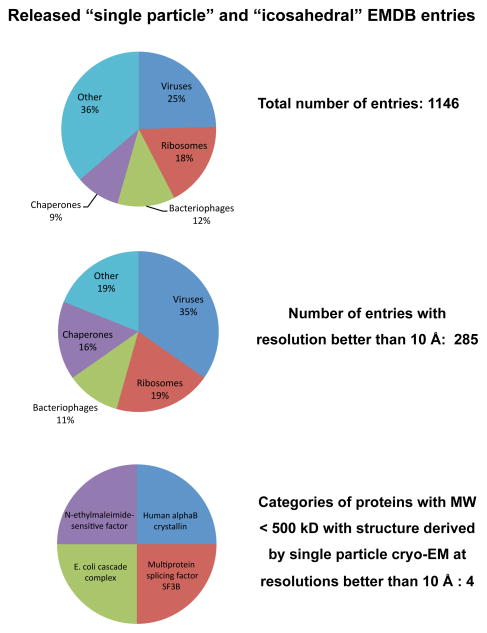
Inspection of the distribution of the released entries belonging to the “single-particle” and “icosahedral” aggregation state categories reveals that icosahedral viruses and ribosomes make up nearly half of the entries, with the rest dominated by assemblies such as chaperones that display a high level of symmetry (Figure 9). Only a quarter of the entries are at resolutions better than 10 Å, which is the resolution at which α-helical regions in the structure can begin to be discerned. Remarkably, the vast majority of these structures at 10 Å or better resolution are from large protein complexes, with only six examples (representing four distinct protein complexes) below 500 kDa, reflecting smaller, potentially dynamic protein complexes (Figure 9). These types of complexes are at the heart of cell function, and understanding how they work will generate fundamental insights that will be key to the development of biomedical therapeutics in the coming decades. It is safe to predict that addressing this critical gap in the landscape of structural biology is where we may expect the greatest future impact of cryo-EM.
Figure 9.
Distribution of released entries belonging to the “single-particle” and “icosahedral” categories in the Electron Microscopy Data Bank (emdb.org) as of June 2012. Structures from ribosomes and icosahedral viruses dominate the deposited entries, both in the entire data set, and even more strikingly, in the subset of entries with resolutions better than 10 Å. Further inspection shows that there are only four distinct protein complexes with molecular masses < 500 kDa (Multiprotein splicing factor SF3B: EMD-1043; DegQ: EMD-5290; E. coli cascade complex: EMD-5314 and EMD-5315; N-ethylmaleimide-sensitive factor: EMD-5370 and EMD-5371), thus identifying a key gap in structural biology that can potentially be filled by taking advantage of improvements in microscope hardware and algorithms for image processing.
Acknowledgments
Work in our laboratory at NIH is supported by the Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD. We thank Hong Zhou for permission to use the images presented in Figure 7. More information on our research program is available at http://electron.nci.nih.gov and at http://livinglab.nih.gov.
References
- 1.Hoenger A, Bouchet-Marquis C. Cellular tomography. Adv Protein Chem Struct Biol. 2011;82:67–90. doi: 10.1016/B978-0-12-386507-6.00003-8. [DOI] [PubMed] [Google Scholar]
- 2.Hurbain I, Sachse M. The future is cold: cryo-preparation methods for transmission electron microscopy of cells. Biol Cell. 2011;103:405–20. doi: 10.1042/BC20110015. [DOI] [PubMed] [Google Scholar]
- 3.Milne JL, Subramaniam S. Cryo-electron tomography of bacteria: progress, challenges and future prospects. Nat Rev Microbiol. 2009;7:666–75. doi: 10.1038/nrmicro2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tocheva EI, Li Z, Jensen GJ. Electron cryotomography. Cold Spring Harb Perspect Biol. 2010;2:a003442. doi: 10.1101/cshperspect.a003442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pierson J, Vos M, McIntosh JR, Peters PJ. Perspectives on electron cryotomography of vitreous cryo-sections. J Electron Microsc (Tokyo) 2011;60(Suppl 1):S93–100. doi: 10.1093/jmicro/dfr014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang J, Liu X, Rochat RH, Baker ML, Chiu W. Reconstructing virus structures from nanometer to near-atomic resolutions with cryo-electron microscopy and tomography. Adv Exp Med Biol. 2012;726:49–90. doi: 10.1007/978-1-4614-0980-9_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Subramaniam S, Bartesaghi A, Liu J, Bennett AE, Sougrat R. Electron tomography of viruses. Curr Opin Struct Biol. 2007;17:596–602. doi: 10.1016/j.sbi.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yahav T, Maimon T, Grossman E, Dahan I, Medalia O. Cryo-electron tomography: gaining insight into cellular processes by structural approaches. Curr Opin Struct Biol. 2011;21:670–7. doi: 10.1016/j.sbi.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 9.Wisedchaisri G, Reichow SL, Gonen T. Advances in structural and functional analysis of membrane proteins by electron crystallography. Structure. 2011;19:1381–93. doi: 10.1016/j.str.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orlova EV, Saibil HR. Structural analysis of macromolecular assemblies by electron microscopy. Chem Rev. 2011;111:7710–48. doi: 10.1021/cr100353t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grigorieff N, Harrison SC. Near-atomic resolution reconstructions of icosahedral viruses from electron cryo-microscopy. Curr Opin Struct Biol. 2011;21:265–73. doi: 10.1016/j.sbi.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ubarretxena-Belandia I, Stokes DL. Present and future of membrane protein structure determination by electron crystallography. Adv Protein Chem Struct Biol. 2010;81:33–60. doi: 10.1016/B978-0-12-381357-2.00002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pilhofer M, Ladinsky MS, McDowall AW, Jensen GJ. Bacterial TEM: new insights from cryo-microscopy. Methods Cell Biol. 2010;96:21–45. doi: 10.1016/S0091-679X(10)96002-0. [DOI] [PubMed] [Google Scholar]
- 14.Frank J. Single-particle reconstruction of biological macromolecules in electron microscopy--30 years. Q Rev Biophys. 2009;42:139–58. doi: 10.1017/S0033583509990059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henderson R, Unwin PN. Three-dimensional model of purple membrane obtained by electron microscopy. Nature. 1975;257:28–32. doi: 10.1038/257028a0. [DOI] [PubMed] [Google Scholar]
- 16.Subramaniam S, Henderson R. Molecular mechanism of vectorial proton translocation by bacteriorhodopsin. Nature. 2000;406:653–7. doi: 10.1038/35020614. [DOI] [PubMed] [Google Scholar]
- 17.Henderson R, Baldwin JM, Ceska TA, Zemlin F, Beckmann E, Downing KH. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J Mol Biol. 1990;213:899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- 18.Gonen T, Cheng Y, Sliz P, Hiroaki Y, Fujiyoshi Y, Harrison SC, Walz T. Lipid-protein interactions in double-layered two-dimensional AQP0 crystals. Nature. 2005;438:633–8. doi: 10.1038/nature04321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X, Jin L, Fang Q, Hui WH, Zhou ZH. 3.3 A cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry. Cell. 2010;141:472–82. doi: 10.1016/j.cell.2010.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scott MC, Chen CC, Mecklenburg M, Zhu C, Xu R, Ercius P, Dahmen U, Regan BC, Miao J. Electron tomography at 2.4-angstrom resolution. Nature. 2012;483:444–7. doi: 10.1038/nature10934. [DOI] [PubMed] [Google Scholar]
- 21.Glaeser RM. Limitations to significant information in biological electron microscopy as a result of radiation damage. J Ultrastruct Res. 1971;36:466–82. doi: 10.1016/s0022-5320(71)80118-1. [DOI] [PubMed] [Google Scholar]
- 22.Glaeser RM, Cosslett VE, Valdre U. Low temperature electron microscopy: Radiation damage in crystalline biological materials. J Microsc. 1971;12:133–38. [Google Scholar]
- 23.Egerton RF, Li P, Malac M. Radiation damage in the TEM and SEM. Micron. 2004;35:399–409. doi: 10.1016/j.micron.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 24.Ohi M, Li Y, Cheng Y, Walz T. Negative Staining and Image Classification - Powerful Tools in Modern Electron Microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goringer HU, Stark H, Bohm C, Sander B, Golas MM. Three-dimensional reconstruction of Trypanosoma brucei editosomes using single-particle electron microscopy. Methods Mol Biol. 2011;718:3–22. doi: 10.1007/978-1-61779-018-8_1. [DOI] [PubMed] [Google Scholar]
- 26.Chiu PL, Kelly DF, Walz T. The use of trehalose in the preparation of specimens for molecular electron microscopy. Micron. 2011;42:762–72. doi: 10.1016/j.micron.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor KA, Glaeser RM. Electron diffraction of frozen, hydrated protein crystals. Science. 1974;186:1036–7. doi: 10.1126/science.186.4168.1036. [DOI] [PubMed] [Google Scholar]
- 28.Dubochet J, Adrian M, Chang JJ, Homo JC, Lepault J, McDowall AW, Schultz P. Cryo-electron microscopy of vitrified specimens. Q Rev Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 29.Saibil HR. Macromolecular structure determination by cryo-electron microscopy. Acta Crystallogr D Biol Crystallogr. 2000;56:1215–22. doi: 10.1107/s0907444900010787. [DOI] [PubMed] [Google Scholar]
- 30.DeRosier DJ, Klug A. Reconstruction of three-dimensional structures from electron micrographs. Nature. 1968;217:130–34. doi: 10.1038/217130a0. [DOI] [PubMed] [Google Scholar]
- 31.Unwin PN, Henderson R. Molecular structure determination by electron microscopy of unstained crystalline specimens. J Mol Biol. 1975;94:425–40. doi: 10.1016/0022-2836(75)90212-0. [DOI] [PubMed] [Google Scholar]
- 32.Bammes BE, Jakana J, Schmid MF, Chiu W. Radiation damage effects at four specimen temperatures from 4 to 100 K. J Struct Biol. 2010;169:331–41. doi: 10.1016/j.jsb.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitsuoka K. Obtaining high-resolution images of biological macromolecules by using a cryo-electron microscope with a liquid-helium cooled stage. Micron. 2011;42:100–6. doi: 10.1016/j.micron.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 34.Koster AJ, Grimm R, Typke D, Hegerl R, Stoschek A, Walz J, Baumeister W. Perspectives of molecular and cellular electron tomography. J Struct Biol. 1997;120:276–308. doi: 10.1006/jsbi.1997.3933. [DOI] [PubMed] [Google Scholar]
- 35.Schmid MF. Single-particle electron cryotomography (cryoET) Adv Protein Chem Struct Biol. 2011;82:37–65. doi: 10.1016/B978-0-12-386507-6.00002-6. [DOI] [PubMed] [Google Scholar]
- 36.Marko M, Hsieh C, Schalek R, Frank J, Mannella C. Focused-ion-beam thinning of frozen-hydrated biological specimens for cryo-electron microscopy. Nat Methods. 2007;4:215–217. doi: 10.1038/nmeth1014. [DOI] [PubMed] [Google Scholar]
- 37.Strunk KM, Wang K, Ke D, Gray JL, Zhang P. Thinning of large mammalian cells for cryo-TEM characterization by cryo-FIB milling. J Microsc. 2012;247:220–7. doi: 10.1111/j.1365-2818.2012.03635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rigort A, Bauerlein FJ, Villa E, Eibauer M, Laugks T, Baumeister W, Plitzko JM. Focused ion beam micromachining of eukaryotic cells for cryoelectron tomography. Proc Natl Acad Sci U S A. 2012;109:4449–54. doi: 10.1073/pnas.1201333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Z, Trimble MJ, Brun YV, Jensen GJ. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 2007;26:4694–708. doi: 10.1038/sj.emboj.7601895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raddi G, Morado DR, Yan J, Haake DA, Yang XF, Liu J. Three-dimensional structures of pathogenic and saprophytic Leptospira species revealed by cryo-electron tomography. J Bacteriol. 2012;194:1299–306. doi: 10.1128/JB.06474-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang P, Khursigara CM, Hartnell LM, Subramaniam S. Direct visualization of Escherichia coli chemotaxis receptor arrays using cryo-electron microscopy. Proc Natl Acad Sci U S A. 2007;104:3777–81. doi: 10.1073/pnas.0610106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu X, Walter MH, Paredes A, Morais MC, Liu J. The mechanism of DNA ejection in the Bacillus anthracis spore-binding phage 8a revealed by cryo-electron tomography. Virology. 2011;421:141–8. doi: 10.1016/j.virol.2011.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kudryashev M, Cyrklaff M, Alex B, Lemgruber L, Baumeister W, Wallich R, Frischknecht F. Evidence of direct cell-cell fusion in Borrelia by cryogenic electron tomography. Cell Microbiol. 2011;13:731–41. doi: 10.1111/j.1462-5822.2011.01571.x. [DOI] [PubMed] [Google Scholar]
- 44.Butan C, Hartnell LM, Fenton AK, Bliss D, Sockett RE, Subramaniam S, Milne JL. Spiral architecture of the nucleoid in Bdellovibrio bacteriovorus. J Bacteriol. 2011;193:1341–50. doi: 10.1128/JB.01061-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lucic V, Leis A, Baumeister W. Cryo-electron tomography of cells: connecting structure and function. Histochem Cell Biol. 2008;130:185–96. doi: 10.1007/s00418-008-0459-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hanein D. Tomography of actin cytoskeletal networks. Methods Enzymol. 2010;483:203–14. doi: 10.1016/S0076-6879(10)83010-1. [DOI] [PubMed] [Google Scholar]
- 47.Li S, Fernandez JJ, Marshall WF, Agard DA. Three-dimensional structure of basal body triplet revealed by electron cryo-tomography. EMBO J. 2012;31:552–62. doi: 10.1038/emboj.2011.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nans A, Mohandas N, Stokes DL. Native ultrastructure of the red cell cytoskeleton by cryo-electron tomography. Biophys J. 2011;101:2341–50. doi: 10.1016/j.bpj.2011.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pierson J, Ziese U, Sani M, Peters PJ. Exploring vitreous cryo-section-induced compression at the macromolecular level using electron cryo-tomography; 80S yeast ribosomes appear unaffected. J Struct Biol. 2011;173:345–9. doi: 10.1016/j.jsb.2010.09.017. [DOI] [PubMed] [Google Scholar]
- 50.Davies KM, Anselmi C, Wittig I, Faraldo-Gomez JD, Kuhlbrandt W. Structure of the yeast F1Fo-ATP synthase dimer and its role in shaping the mitochondrial cristae. Proc Natl Acad Sci U S A. 2012;109:13602–7. doi: 10.1073/pnas.1204593109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grunewald K, Medalia O, Gross A, Steven AC, Baumeister W. Prospects of electron cryotomography to visualize macromolecular complexes inside cellular compartments: implications of crowding. Biophys Chem. 2003;100:577–91. doi: 10.1016/s0301-4622(02)00307-1. [DOI] [PubMed] [Google Scholar]
- 52.Ortiz JO, Forster F, Kurner J, Linaroudis AA, Baumeister W. Mapping 70S ribosomes in intact cells by cryoelectron tomography and pattern recognition. J Struct Biol. 2006;156:334–41. doi: 10.1016/j.jsb.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 53.Beck M, Malmstrom JA, Lange V, Schmidt A, Deutsch EW, Aebersold R. Visual proteomics of the human pathogen Leptospira interrogans. Nat Methods. 2009;6:817–23. doi: 10.1038/nmeth.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuhner S, van Noort V, Betts MJ, Leo-Macias A, Batisse C, Rode M, Yamada T, Maier T, Bader S, Beltran-Alvarez P, Castano-Diez D, Chen WH, Devos D, Guell M, Norambuena T, Racke I, Rybin V, Schmidt A, Yus E, Aebersold R, Herrmann R, Bottcher B, Frangakis AS, Russell RB, Serrano L, Bork P, Gavin AC. Proteome organization in a genome-reduced bacterium. Science. 2009;326:1235–40. doi: 10.1126/science.1176343. [DOI] [PubMed] [Google Scholar]
- 55.Al-Amoudi A, Chang JJ, Leforestier A, McDowall A, Salamin LM, Norlén LPO, Richter K, Sartori Blanc N, Studer D, Dubochet J. Cryo-electron microscopy of vitreous sections. EMBO J. 2004;23:3583–3588. doi: 10.1038/sj.emboj.7600366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pierson J, Fernandez JJ, Bos E, Amini S, Gnaegi H, Vos M, Bel B, Adolfsen F, Carrascosa JL, Peters PJ. Improving the technique of vitreous cryo-sectioning for cryo-electron tomography: electrostatic charging for section attachment and implementation of an anti-contamination glove box. J Struct Biol. 2010;169:219–25. doi: 10.1016/j.jsb.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 57.Zhang P, Bos E, Heymann J, Gnaegi H, Kessel M, Peters PJ, Subramaniam S. Direct visualization of receptor arrays in frozen-hydrated sections and plunge-frozen specimens of E. coli engineered to overproduce the chemotaxis receptor Tsr. J Microsc. 2004;216:76–83. doi: 10.1111/j.0022-2720.2004.01395.x. [DOI] [PubMed] [Google Scholar]
- 58.Al-Amoudi A, Studer D, Dubochet J. Cutting artefacts and cutting process in vitreous sections for cryo-electron microscopy. J Struct Biol. 2005;150:109–21. doi: 10.1016/j.jsb.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 59.Sader K, Studer D, Zuber B, Gnaegi H, Trinick J. Preservation of high resolution protein structure by cryo-electron microscopy of vitreous sections. Ultramicroscopy. 2009;110:43–7. doi: 10.1016/j.ultramic.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Q, Mercogliano CP, Lowe J. A ferritin-based label for cellular electron cryotomography. Structure. 2011;19:147–54. doi: 10.1016/j.str.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 61.Bouchet-Marquis C, Pagratis M, Kirmse R, Hoenger A. Metallothionein as a clonable high-density marker for cryo-electron microscopy. J Struct Biol. 2012;177:119–27. doi: 10.1016/j.jsb.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Driel LF, Valentijn JA, Valentijn KM, Koning RI, Koster AJ. Tools for correlative cryo-fluorescence microscopy and cryo-electron tomography applied to whole mitochondria in human endothelial cells. Eur J Cell Biol. 2009;88:669–84. doi: 10.1016/j.ejcb.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 63.Jun S, Ke D, Debiec K, Zhao G, Meng X, Ambrose Z, Gibson GA, Watkins SC, Zhang P. Direct visualization of HIV-1 with correlative live-cell microscopy and cryo-electron tomography. Structure. 2011;19:1573–81. doi: 10.1016/j.str.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bartesaghi A, Sprechmann P, Liu J, Randall G, Sapiro G, Subramaniam S. Classification and 3D averaging with missing wedge correction in biological electron tomography. J Struct Biol. 2008;162:436–50. doi: 10.1016/j.jsb.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bartesaghi A, Subramaniam S. Membrane protein structure determination using cryo-electron tomography and 3D image averaging. Curr Opin Struct Biol. 2009;19:402–7. doi: 10.1016/j.sbi.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Forster F, Pruggnaller S, Seybert A, Frangakis AS. Classification of cryo-electron sub-tomograms using constrained correlation. J Struct Biol. 2008;161:276–86. doi: 10.1016/j.jsb.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 67.Heumann JM, Hoenger A, Mastronarde DN. Clustering and variance maps for cryo-electron tomography using wedge-masked differences. J Struct Biol. 2011;175:288–99. doi: 10.1016/j.jsb.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu Z, Frangakis AS. Classification of electron sub-tomograms with neural networks and its application to template-matching. J Struct Biol. 2011;174:494–504. doi: 10.1016/j.jsb.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 69.Stolken M, Beck F, Haller T, Hegerl R, Gutsche I, Carazo JM, Baumeister W, Scheres SH, Nickell S. Maximum likelihood based classification of electron tomographic data. J Struct Biol. 2011;173:77–85. doi: 10.1016/j.jsb.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 70.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–13. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.White TA, Bartesaghi A, Borgnia MJ, de la Cruz MJ, Nandwani R, Hoxie JA, Bess JW, Lifson JD, Milne JL, Subramaniam S. Three-dimensional structures of soluble CD4-bound states of trimeric simian immunodeficiency virus envelope glycoproteins determined by using cryo-electron tomography. J Virol. 2011;85:12114–23. doi: 10.1128/JVI.05297-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.White TA, Bartesaghi A, Borgnia MJ, Meyerson JR, de la Cruz MJ, Bess JW, Nandwani R, Hoxie JA, Lifson JD, Milne JL, Subramaniam S. Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: strain-dependent variation in quaternary structure. PLoS Pathog. 2010;6:e1001249. doi: 10.1371/journal.ppat.1001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tran EEH, Borgnia MJ, Kuybeda O, Schauder DM, Bartesaghi A, Frank GA, Sapiro G, Milne JLS, Subramaniam S. Structural Mechanism of Trimeric HIV-1 Envelope Glycoprotein Activation. PLoS Pathog. 2012;8:e1002797. doi: 10.1371/journal.ppat.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Harris A, Borgnia MJ, Shi D, Bartesaghi A, He H, Pejchal R, Kang YK, Depetris R, Marozsan AJ, Sanders RW, Klasse PJ, Milne JL, Wilson IA, Olson WC, Moore JP, Subramaniam S. Trimeric HIV-1 glycoprotein gp140 immunogens and native HIV-1 envelope glycoproteins display the same closed and open quaternary molecular architectures. Proc Natl Acad Sci U S A. 2011;108:11440–5. doi: 10.1073/pnas.1101414108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Frank GA, Bartesaghi A, Kuybeda O, Borgnia MJ, White TA, Sapiro G, Subramaniam S. Computational separation of conformational heterogeneity using cryo-electron tomography and 3D sub-volume averaging. J Struct Biol. 2012;178:165–76. doi: 10.1016/j.jsb.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Khursigara CM, Wu X, Zhang P, Lefman J, Subramaniam S. Role of HAMP domains in chemotaxis signaling by bacterial chemoreceptors. Proc Natl Acad Sci U S A. 2008;105:16555–60. doi: 10.1073/pnas.0806401105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu J, Hu B, Morado DR, Jani S, Manson MD, Margolin W. Molecular architecture of chemoreceptor arrays revealed by cryoelectron tomography of Escherichia coli minicells. Proc Natl Acad Sci U S A. 2012;109:E1481–8. doi: 10.1073/pnas.1200781109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Winkler H, Zhu P, Liu J, Ye F, Roux KH, Taylor KA. Tomographic subvolume alignment and subvolume classification applied to myosin V and SIV envelope spikes. J Struct Biol. 2009;165:64–77. doi: 10.1016/j.jsb.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Eibauer M, Hoffmann C, Plitzko JM, Baumeister W, Nickell S, Engelhardt H. Unraveling the structure of membrane proteins in situ by transfer function corrected cryo-electron tomography. J Struct Biol. 2012 doi: 10.1016/j.jsb.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 80.Bartesaghi A, Lecumberry F, Sapiro G, Subramaniam S. Protein secondary structure determination by constrained single particle cryo-electron tomography. Structure. 2012 doi: 10.1016/j.str.2012.10.016. (In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Frank J. Three-dimensional Electron Microscopy of Macromolecular Assemblies: Visualization of Biological Molecules in Their Native State. Oxford University Press; 2006. [Google Scholar]
- 82.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 83.Shaikh TR, Gao H, Baxter WT, Asturias FJ, Boisset N, Leith A, Frank J. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat Protoc. 2008;3:1941–74. doi: 10.1038/nprot.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 85.Crowther RA, Amos LA, Finch JT, De Rosier DJ, Klug A. Three dimensional reconstructions of spherical viruses by fourier synthesis from electron micrographs. Nature. 1970;226:421–5. doi: 10.1038/226421a0. [DOI] [PubMed] [Google Scholar]
- 86.Van Heel M. Angular reconstitution: a posteriori assignment of projection directions for 3D reconstruction. Ultramicroscopy. 1987;21:111–23. doi: 10.1016/0304-3991(87)90078-7. [DOI] [PubMed] [Google Scholar]
- 87.Penczek PA. Fundamentals of three-dimensional reconstruction from projections. Methods Enzymol. 2010;482:1–33. doi: 10.1016/S0076-6879(10)82001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ludtke SJ, Lawson CL, Kleywegt GJ, Berman H, Chiu W. The 2010 cryo-em modeling challenge. Biopolymers. 2012;97:651–4. doi: 10.1002/bip.22081. [DOI] [PubMed] [Google Scholar]
- 89.Ahmed A, Whitford PC, Sanbonmatsu KY, Tama F. Consensus among flexible fitting approaches improves the interpretation of cryo-EM data. J Struct Biol. 2012;177:561–70. doi: 10.1016/j.jsb.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Radermacher M, Wagenknecht T, Verschoor A, Frank J. Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J Microsc. 1987;146:113–36. doi: 10.1111/j.1365-2818.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- 91.Leschziner A. The orthogonal tilt reconstruction method. Methods Enzymol. 2010;482:237–62. doi: 10.1016/S0076-6879(10)82010-5. [DOI] [PubMed] [Google Scholar]
- 92.Yan X, Sinkovits RS, Baker TS. AUTO3DEM--an automated and high throughput program for image reconstruction of icosahedral particles. J Struct Biol. 2007;157:73–82. doi: 10.1016/j.jsb.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cong Y, Baker ML, Jakana J, Woolford D, Miller EJ, Reissmann S, Kumar RN, Redding-Johanson AM, Batth TS, Mukhopadhyay A, Ludtke SJ, Frydman J, Chiu W. 4.0-A resolution cryo-EM structure of the mammalian chaperonin TRiC/CCT reveals its unique subunit arrangement. Proc Natl Acad Sci U S A. 2010;107:4967–72. doi: 10.1073/pnas.0913774107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Penczek PA, Kimmel M, Spahn CM. Identifying conformational states of macromolecules by eigen-analysis of resampled cryo-EM images. Structure. 2011;19:1582–90. doi: 10.1016/j.str.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ludtke SJ, Tran TP, Ngo QT, Moiseenkova-Bell VY, Chiu W, Serysheva II. Flexible architecture of IP3R1 by Cryo-EM. Structure. 2011;19:1192–9. doi: 10.1016/j.str.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lau WC, Rubinstein JL. Subnanometre-resolution structure of the intact Thermus thermophilus H+-driven ATP synthase. Nature. 2012;481:214–8. doi: 10.1038/nature10699. [DOI] [PubMed] [Google Scholar]
- 97.Scheres SH, Gao H, Valle M, Herman GT, Eggermont PP, Frank J, Carazo JM. Disentangling conformational states of macromolecules in 3D-EM through likelihood optimization. Nat Methods. 2007;4:27–9. doi: 10.1038/nmeth992. [DOI] [PubMed] [Google Scholar]
- 98.Henderson R. The potential and limitations of neutrons, electrons and X-rays for atomic resolution microscopy of unstained biological molecules. Q Rev Biophys. 1995;28:171–93. doi: 10.1017/s003358350000305x. [DOI] [PubMed] [Google Scholar]
- 99.Henderson R, Chen S, Chen JZ, Grigorieff N, Passmore LA, Ciccarelli L, Rubinstein JL, Crowther RA, Stewart PL, Rosenthal PB. Tilt-pair analysis of images from a range of different specimens in single-particle electron cryomicroscopy. J Mol Biol. 2011;413:1028–46. doi: 10.1016/j.jmb.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 101.Faruqi AR, McMullan G. Electronic detectors for electron microscopy. Q Rev Biophys. 2011;44:357–90. doi: 10.1017/S0033583511000035. [DOI] [PubMed] [Google Scholar]
- 102.Castano-Diez D, Kudryashev M, Arheit M, Stahlberg H. Dynamo: a flexible, user-friendly development tool for subtomogram averaging of cryo-EM data in high-performance computing environments. J Struct Biol. 2012;178:139–51. doi: 10.1016/j.jsb.2011.12.017. [DOI] [PubMed] [Google Scholar]
- 103.Li X, Grigorieff N, Cheng Y. GPU-enabled FREALIGN: accelerating single particle 3D reconstruction and refinement in Fourier space on graphics processors. J Struct Biol. 2010;172:407–12. doi: 10.1016/j.jsb.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schmeisser M, Heisen BC, Luettich M, Busche B, Hauer F, Koske T, Knauber KH, Stark H. Parallel, distributed and GPU computing technologies in single-particle electron microscopy. Acta Crystallogr D Biol Crystallogr. 2009;65:659–71. doi: 10.1107/S0907444909011433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cong Y, Zhang Q, Woolford D, Schweikardt T, Khant H, Dougherty M, Ludtke SJ, Chiu W, Decker H. Structural mechanism of SDS-induced enzyme activity of scorpion hemocyanin revealed by electron cryomicroscopy. Structure. 2009;17:749–58. doi: 10.1016/j.str.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xu M, Beck M, Alber F. High-throughput subtomogram alignment and classification by Fourier space constrained fast volumetric matching. J Struct Biol. 2012;178:152–64. doi: 10.1016/j.jsb.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sander B, Golas MM, Luhrmann R, Stark H. An approach for de novo structure determination of dynamic molecular assemblies by electron cryomicroscopy. Structure. 2010;18:667–76. doi: 10.1016/j.str.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 108.Yang Z, Fang J, Chittuluru J, Asturias FJ, Penczek PA. Iterative stable alignment and clustering of 2D transmission electron microscope images. Structure. 2012;20:237–47. doi: 10.1016/j.str.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T, Gnau V, Bauch A, Bastuck S, Huhse B, Leutwein C, Heurtier MA, Copley RR, Edelmann A, Querfurth E, Rybin V, Drewes G, Raida M, Bouwmeester T, Bork P, Seraphin B, Kuster B, Neubauer G, Superti-Furga G. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–7. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 110.Subramaniam S. The SIV surface spike imaged by electron tomography: one leg or three? PLoS Pathog. 2006;2:e91. doi: 10.1371/journal.ppat.0020091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Grigorieff N. Three-dimensional structure of bovine NADH:ubiquinone oxidoreductase (complex I) at 22 A in ice. J Mol Biol. 1998;277:1033–46. doi: 10.1006/jmbi.1998.1668. [DOI] [PubMed] [Google Scholar]