Summary
Background
Multimarker quantitative real-time polymerase chain reaction (qRT-PCR) represents an effective method for detecting circulating tumour cells in the peripheral blood of patients with melanoma.
Objectives
To investigate whether the phenotype of circulating melanoma cells represents a useful indicator of disease stage, recurrence and treatment efficacy.
Methods
Peripheral blood was collected from 230 patients with melanoma and 152 healthy controls over a period of 3 years and 9 months. Clinical data and blood samples were collected from patients with primary melanoma (early stages, 0–II, n = 154) and metastatic melanoma (late stages, III–IV, n = 76). Each specimen was examined by qRT-PCR analysis for the expression of five markers: MLANA, ABCB5, TGFβ2, PAX3d and MCAM.
Results
In total, 212 of the patients with melanoma (92%) expressed markers in their peripheral blood. Two markers, MLANA and ABCB5, had the greatest prognostic value, and were identified as statistically significant among patients who experienced disease recurrence within our study period, being expressed in 45% (MLANA) and 49% (ABCB5) of patients with recurrence (P = 0·001 and P = 0·031, respectively). For patients administered nonsurgical treatments, MCAM expression correlated with poor treatment outcome.
Conclusions
Circulating tumour cells were detectable at all stages of disease and long after surgical treatment, even when patients were considered disease free. Specifically, expression of ABCB5 and MLANA had significant prognostic value in inferring disease recurrence, while MCAM expression was associated with poor patient outcome after treatment, confirming multimarker qRT-PCR as a potential technique for monitoring disease status.
Cutaneous melanoma is a tumour originating from normal skin melanocytes. Global incidence of melanoma is increasing, 1 with an estimated 70 230 new cases annually in the U.S.A.2 Melanoma has a high metastatic potential,3 and metastatic melanoma is difficult to treat. It is important therefore to find methods of detecting melanoma spread at early stages for more accurate treatment and prediction of prognosis.
A staging system was devised by the American Joint Committee on Cancer (AJCC), based on histological features of the primary lesion, and the presence of nodal and/or systemic metastases in advanced disease.3 Due to increased awareness, most patients present with thin, localized melanoma, which is usually curable by surgical resection. However, approximately 5% of patients with lesions < 1 mm thick, 25–40% of patients with 2–4 mm lesions, and 50–75% of patients with lesions > 4 mm thick develop metastatic disease and die within 10 years.3 Patients diagnosed with regional and distant metastases have 10-year survival rates of 64% and 16%, respectively.4
Patients may suffer relapse following disease-free periods of 10 years or more.5,6 Evidently then, diagnosing patients as clinically disease free after surgical removal of early-stage melanomas may be inaccurate, as tumour cells dispersed into the bloodstream may remain dormant, prior to formation of distant metastases.7
Considerable research into the detection of circulating tumour cells in the blood of patients with breast, colorectal and prostate cancer has shown that the presence of these cells correlates with both disease progression and poor outcome.8–16 For melanoma, circulating cell analysis has not been successfully utilized clinically. Although several articles have indicated their usefulness,17–22 markers and methodology remain controversial. Moreover, it is unclear whether circulating melanoma stem cells are prognostic23 or whether all circulating melanoma cells have metastatic potential.24
Here we used multimarker quantitative real-time polymerase chain reaction (qRT-PCR) to investigate the phenotype of circulating cells in patients with melanoma. Given the sensitivity of qRT-PCR, it is suitable for assessment of rare circulating melanoma cells,25 and multiple markers are essential given the heterogeneous nature of melanoma cells.18,26 Our markers included melanocyte (PAX3d27 and MLANA),28 tumour (TGFβ229 and MCAM)30 and stem cell (ABCB5)31 markers. PAX3 regulates melanocytic development,32–34 and the PAX3d isoform is highly expressed in melanoma.35 ABCB5, a stem cell marker, marks a subset of rare, chemoresistent melanoma stem cells,24,31,36 while MCAM is a marker of melanoma tumour progression.34
We show that circulating tumour cells are detectable at all stages of disease and persist long after treatment. Certain circulating cells, specifically those marked by MCAM expression, correlated with a poor treatment outcome, while ABCB5 and MLANA correlated with disease progression and could aid in monitoring disease status.
Patients and methods
Patient cohort
For this study, 230 patients and 152 healthy control participants provided written consent. Patients were recruited from the Medical Oncology Department of Sir Charles Gairdner Hospital and the Perth Melanoma Clinic at Hollywood Hospital (Perth, Western Australia) and consisted of 81 women and 149 men aged 24–96 years (mean 69 years), while the aged-matched, healthy cohort, recruited from the general population, contained 86 women and 66 men aged 18–99 years (mean 51 years). The Human Research Ethics Committees of Edith Cowan University (No. 2932) and Sir Charles Gairdner Hospital (No. 2007-123) approved the study.
The study included 154 patients with primary cutaneous melanoma and 76 with metastatic melanoma. AJCC clinical staging categorized 16·5% of our patients as stage 0, 33·5% as stage I, 17% as stage II, 13% as stage III and 20% as stage IV (Table 1). Patients were sampled only once and their clinical history was recorded; however, a subset of patients (n = 140) was sampled serially throughout the study. Tumour burden was calculated from pathology reports for a subset of 32 patients by calculating the sum of maximal tumour diameters for all tumours within the patient, at the time of sampling.
Table 1.
Quantitative real-time polymerase chain reaction marker detection in the peripheral blood of healthy controls and in patients with melanoma, categorized by American Joint Committee on Cancer (AJCC) stage
| Marker | AJCC stage of patients with melanoma % (n)
|
|||||
|---|---|---|---|---|---|---|
| 0 | I | II | III | IV | Healthy controls | |
| MLANA | 26 (10) | 25 (19) | 18 (7) | 40 (12) | 43 (20) | 13 (19) |
| ABCB5 | 45 (17) | 27 (21) | 33 (13) | 50 (15) | 52 (24) | 22 (33) |
| PAX3d | 47 (18) | 30 (23) | 36 (14) | 50 (15) | 46 (21) | 14 (21) |
| MCAM | 42 (16) | 49 (38) | 46 (18) | 53 (16) | 43 (20) | 14 (21) |
| TGFβ2 | 63 (24) | 64 (49) | 74 (29) | 77 (23) | 59 (27) | 22 (33) |
| N | 38 | 77 | 39 | 30 | 46 | 152 |
Melanoma cell lines
Human melanoma cell line A2058 cells (EACC: 91100402), were grown in Dulbecco’s modified Eagle’s medium media containing 10% heat-inactivated fetal bovine serum (Invitrogen, Grand Island, NY, U.S.A.) in a T75 cm2 flask (Greiner, Monroe, NC, U.S.A.) until cells were ~70% confluent.
Blood collection, RNA isolation and amplification protocols
At venipuncture, the first 4 mL of blood was discarded to avoid epithelial contamination.37 A 2·5 mL sample of whole blood was then collected into a PAXgene Blood RNA Tube (PreAnalytiX, Hombrechtikon, Switzerland), containing RNA stabilizers.38
Total RNA was isolated using the PAXgene Blood RNA Kit (Qiagen, Hilden, Germany) and DNase treated (DNA-free Kit, Ambion, Carlsbad, CA, U.S.A.). RNA was quantified and the integrity was qualified (Agilent 2100 Bioanalyser, Agilent Technologies, Santa Clara, CA, U.S.A.). Samples with insufficient RNA levels (RNA integrity number < 8) were excluded.
Total RNA (250 ng) was converted to cDNA (Omniscript Reverse Transcriptase, Qiagen). Each reaction (20 μL) contained 2 μL of RT Buffer (1 ×), 2 μL of deoxyribonucleotide triphosphates (dNTPs, 5 mmol L−1), 2 μL of oligo (dT) primer (0·4 μg μL−1), 1 μL of RNase inhibitor (10 U μL−1, Invitrogen) and 1 μL of Omniscript Reverse Transcriptase (4 U μL−1). Incubation conditions were 37 °C for 1 h; then 5 min at 95 °C, and included a no template control (NTC). qRT-PCR assays assessed the number of mRNA transcripts (level of expression) for six genes: MLANA, ABCB5, TGFB2, PAX3, MCAM and GAPDH. Each 15 μL reaction contained 1 μL of cDNA template, 1 × SYBR GreenER qPCR SuperMix (Invitrogen) and 200 nmol L−1 of primer (Table S1; see Supporting Information). Using an iCycler iQ5 Real-Time Thermocycler (Bio-Rad, Hercules, CA, U.S.A.), incubation was as per the manufacturer’s instructions.
To prevent contamination, all PCR reactions contained uracil-N-glycosylase to prevent reamplification of carryover PCR products,39 and different steps were performed in separate ultraviolet-treatable areas. Melting point determination and gel electrophoresis confirmed the expected size and identity of PCR products.
Glyceraldehyde 3-phosphate dehydrogenase (GAPDH), our internal reference for reverse transcription efficiency, is not upregulated in melanoma tissues or cultured cells relative to normal samples;40 samples with insufficient levels were excluded. Every assay included a standard curve, negative controls (NTC and reverse transcription control), positive control (A2058 cell line) and cDNA from a single healthy control.
Standard curve
A standard curve, to quantify mRNA copy number, was constructed using larger PCR products that included the target sequence used in qRT-PCR assays. For each standard, an end-point PCR primer set was designed (Table S2; see Supporting Information). A2058 cDNA, synthesized from 250 ng of RNA, was used as a template. Using a Taq DNA polymerase kit (Qiagen), each 20 μL reaction consisted of 1 × Coral Load buffer, 0·2 mmol L−1 of each dNTP, 2·5 U of Taq DNA polymerase, 1 μL cDNA and 200 nmol L−1 of primer. Incubation conditions were 94 °C for 5 min, then 45 cycles of 94 °C for 30 s, then 55 °C for 30 s and 72 °C for 1 min. PCR products were purified after agarose gel electrophoresis using the freeze–squeeze method.41 Resulting stock template solutions were quantified by spectrophotometric analysis (Nanodrop, Thermo Scientific, Waltham, MA, U.S.A.) and diluted into eight tenfold serial dilutions (101–108 copies μL−1), from which a standard curve for each target was generated on the iCycler iQ5 Real-Time Thermocycler. The amplification efficiencies of our target genes were between 96·4% and 100%. Samples were analysed in duplicate with averaged Ct values used for relative quantification. Relative mRNA copies for each transcript were calculated by dividing absolute mRNA copies of the target by that of GAPDH.
Statistical analysis
The Pearson χ2-test and Fisher’s exact test were used to examine the statistical significance of marker expression between patient groups and controls. The Cochrane–Armitage trend test examined the correlation between clinical stage, Clark level and/or expression level. Positive and negative predictive values were calculated to examine the sensitivity of our test.
The relationship between level of marker expression and stage or presence of disease was investigated by the Mann–Whitney U-test and Kruskal–Wallis H-test. Generalized linear modelling was used to examine marker consistency. SPSS (SPSS Inc., Chicago, IL, U.S.A.) was used for analysis, and differences of P < 0·05 were considered statistically significant.
Results
Multimarker qRT-PCR analysis was used to estimate the phenotype and quantity of circulating cells in patient blood. Marker positivity and level of expression were analysed for correlation with disease presence, stage and recurrence, and treatment outcome.
Marker expression in patient blood relative to controls
Patients with melanoma were significantly more likely to express a melanoma cell marker in their blood (92%, n = 212) than were controls (59%, n = 89; P = 0·000, odds ratio 4·206, 95% confidence interval 2·305–7·676). The number of markers expressed by participants was important, with 83% (n = 126) of healthy controls expressing either no marker or just one marker, in contrast to patients with melanoma, who were highly likely to express two or more markers (72%, n = 165; P = 0·012) (Table 2). The positive and negative predictive values of expressing a marker were 70% and 78%, respectively. Expression of two or more melanoma markers improved the positive predictive value to 86%, with a negative predictive value of 66%.
Table 2.
Number of markers detected in the peripheral blood of patients with cutaneous malignant melanoma (CMM) and healthy controls
| Number of markers | 0 | 1 | 2 | 3 | 4 | 5 | n |
|---|---|---|---|---|---|---|---|
| Patients with CMM % (n) | 8 (18) | 20 (47) | 34 (79) | 23 (54) | 10 (24) | 3 (8) | 230 |
| Healthy controls % (n) | 41 (63) | 41 (63) | 11 (17) | 4 (6) | 2 (3) | 0 | 152 |
To identify the most informative markers, we analysed the expression incidence of each marker in the patients. MLANA, ABCB5, MCAM, PAX3d, and TGFβ2 were detected in 30% (n = 68), 40% (n = 90), 47% (n = 108), 40% (n = 91) and 66% (n = 152) of patients with melanoma, respectively.
Marker expression in patients relative to clinical stage
To assess marker utility, we analysed marker expression relative to clinical stage. MLANA and ABCB5 were significantly more likely to be expressed in late-stage patients (stages III–IV; 42% and 51%, respectively) than in early-stage patients (stages 0–II; 23% and 33%; P = 0·003 and P = 0·006, respectively). No other markers appeared to be stage related (Table 1).
Clinical factors including primary location, Clark level, Breslow thickness and ulceration did not correlate with marker expression (Table 3).
Table 3.
Characteristics of the patients with melanoma
| Prognostic factor | Number of patients | |
|---|---|---|
| Sex | Male | 81 |
| Female | 149 | |
| American Joint Committee on Cancer stage | 0 | 38 |
| I | 77 | |
| II | 39 | |
| III | 30 | |
| IV | 46 | |
| Primary site | Face | 36 |
| Torso | 78 | |
| Arms | 31 | |
| Legs | 37 | |
| Scalp | 14 | |
| No primary lesion or unknown | 34 | |
| Breslow thickness (primary) mm | < 1 mm | 103 |
| 1–4 mm | 82 | |
| > 4 mm | 20 | |
| Unknown | 25 | |
| Clark level | I | 43 |
| II | 30 | |
| III | 20 | |
| IV | 81 | |
| V | 15 | |
| Unknown | 41 | |
| Ulceration | No | 178 |
| Yes | 34 | |
| Unknown | 18 |
The Pearson χ2-test was used to analyse whether the incidence of marker expression in patient blood samples taken within 1 month of clinical diagnosis differed from samples taken at times > 1 month following diagnosis. For most markers, expression did not differ between samples taken at times less than and greater than 1 month following diagnosis. However, TGFβ2 was expressed slightly more frequently in samples obtained more than 1 month following diagnosis (47%, n = 187) relative to samples collected within the first month following diagnosis (32%, n = 50; P = 0·001). Samples taken at times > 1 month after diagnosis were as likely to express a marker (79%, n = 311), than samples taken within the first month following diagnosis (71%, n = 110).
Marker expression correlates with disease recurrence
To investigate the prognostic value of our markers, the relationship between marker expression and disease recurrence was analysed. During our study, 73 patients (32%) experienced recurrence, and in these patients both MLANA and ABCB5 were expressed significantly more frequently (45%, n = 33 and 49%, n = 36, respectively) than in patients without recurrence (23%, n = 35 and 34%, n = 54; P = 0·001 and P = 0·031, respectively) (Table 4). Patients with recurrence expressed five markers more commonly (8%, n = 6) than patients who experienced no recurrence during our study (1%, n = 2; P = 0·013). Patients with recurrence during our study were more likely to express both ABCB5 and MLANA (26%, n = 19) than patients without recurrence during the study period (10%, n = 15; P = 0·002). Coexpression of MLANA and ABCB5 produced a positive predictive value of 56% and a negative predictive value of 72%.
Table 4.
Quantitative real-time polymerase chain reaction marker detection in the peripheral blood of patients with melanoma, categorized by recurrence and time since diagnosis
| Marker | Patients with recurrence during study period % (n)
|
Time since diagnosis in relation to sampling in patients with recurrence % (n)
|
||||
|---|---|---|---|---|---|---|
| Recurrence | No recurrence | < 1 month | < 1 year | < 5 years | > 5 years | |
| MLANA | 45 (33)a | 23 (35) | 33 (2) | 18 (7) | 26 (32) | 22 (18) |
| ABCB5 | 49 (36)a | 34 (54) | 0 | 18 (7) | 22 (27) | 32 (26) |
| PAX3d | 41 (30) | 39 (61) | 33 (2) | 21 (8) | 22 (27) | 15 (12) |
| MCAM | 49 (36) | 46 (72) | 0 | 21 (8) | 21 (26) | 25 (25) |
| TGFβ2 | 63 (46) | 68 (106) | 50 (3) | 29 (11) | 34 (42) | 34 (34) |
| N | 73 | 157 | 6 | 38 | 124 | 81 |
P < 0·01 by Pearson χ2-test.
When divided into stages, 48% of stage III–IV patients with recurrence (n = 29) expressed MLANA compared with only 19% of stage III–IV patients without recurrence (n = 3; P = 0·033). There was no correlation between MLANA expression and recurrence in early-stage (0–II) patients. Analysis of the presence and level of markers relative to time since diagnosis, in patients who developed recurrence during our study, revealed that markers were present in patient blood at all stages, including many years after diagnosis or surgical removal of the primary tumour, even when there were no clinical signs of disease (Table 4).
Marker expression in relation to tumour burden
To examine whether marker expression correlated with tumour burden, stage III–IV patients were divided into those with one metastasis (n = 16, mean tumour diameter, 19·8 ± 13·2 mm) or multiple metastases (n = 16, mean of sum of tumour diameter, 76·5 ± 36·06 mm) at the time of sampling. Only MCAM expression was significantly related to tumour burden, with the level of expression greater among stage III–IV patients with multiple metastases (1·23 ± 1·66, n = 4) than in those with only a single metastasis (0·67 ± 0·49, n = 8; P = 0·027).
Among stage III–IV patients, the surgical removal of a clinically measureable metastatic tumour also influenced marker expression. MLANA and PAX3d were expressed less frequently in patients following the surgical removal of a clinically measureable metastatic tumour (18%, n = 29; 19%, n = 30, respectively), compared with patients with an existing tumour burden (48%, n = 16 and 42%, n = 14; P = 0·000 and P = 0·003, respectively). A significant reduction in the level of ABCB5 expression was observed in stage III–IV samples corresponding to the removal of a clinically measureable metastasis (0·19 ± 0·79), compared with samples with an existing tumour burden (0·22 ± 0·42; P = 0·011).
Marker expression in relation to clinical treatment
To analyse the effect of nonsurgical treatment on circulating tumour cell phenotypes, marker expression was investigated in 34 patients (15%) who received nonsurgical treatment. Treatments included radiotherapy (n = 29), interferon alfa 2b (n = 5), limb infusion (n = 1), vaccine (n = 4), chemotherapy (n = 9) and radio frequency ablation (RFA, n = 1).
Three patients were sampled both before and after their nonsurgical treatment, whereas the majority of nonsurgically treated patients were sampled during or following treatment. The condition of two patients deteriorated post-treatment, while the third remained stable (Fig. 1). Patient 1 was sampled at diagnosis of a stage III melanoma, 2 months prior to radiotherapy treatment of the left inguinal node, and expressed only ABCB5. After 17 months, with their left axilla node identified as positive, the patient gained MLANA and MCAM expression and the level of ABCB5 expression was maintained. No markers were detected in patient 2, 3 months prior to RFA treatment for a lung metastasis. Sampled 3 months after treatment, MLANA, ABCB5 and PAX3d were detected; the patient died 3 months later. Patient 3 had positive lymph nodes and expressed no markers the day before interferon alfa 2b treatment. Serial blood collections were then made at 6, 12, 21 and 26 months post-treatment. TGFβ2 was detected at 6 and 12 months, and TGFβ2, MCAM and ABCB5 were detected by 26 months with a fivefold decrease in MCAM between the last two serial samples.
Fig 1.
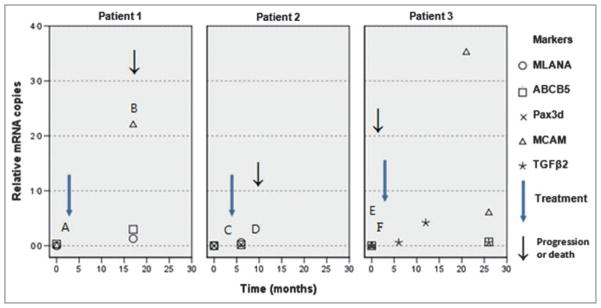
Variations in the level of cell marker expression (relative mRNA copies) in patients 1, 2 and 3 relative to time (months). Significant clinical events for each patient are indicated by capitalized letters. Patient 1: A, radiotherapy treatment to the left inguinal node; B, left axilla node positive. Patient 2: C, radio frequency ablation treatment to a lung metastasis; D, patient deceased. Patient 3: E, positive lymph nodes; F, interferon alfa 2b treatment.
The majority of the nonsurgically treated stage IV samples were collected after nonsurgical treatment (n = 26). To examine whether the phenotype of circulating melanoma cells reflects the effectiveness of treatment, samples were divided into those from patients who had a negative treatment outcome (n = 40, progression and/or death) and those with a positive outcome (n = 22, disease remained stable or tumour size reduced). The expression of MCAM was significantly more common in patients with a negative treatment outcome, with 43% expressing MCAM (n = 17), while only 9% of samples from patients with a positive outcome expressed this marker (n = 2; P = 0·006) (Table 5). The positive predictive value for MCAM expression as a marker of poor clinical outcome in patients with stage IV melanoma is 89·5%, while the negative predictive value is 46·5%.
Table 5.
MCAM expression in the peripheral blood of patients with stage IV melanoma in relation to clinical outcome
| Clinical outcome | MCAM expression % (n)
|
N | |
|---|---|---|---|
| + | − | ||
| Negative (disease progression and/or death) | 42 (17) | 58 (23) | 40 |
| Positive (disease remained stable or tumour size reduced) | 9 (2) | 91 (20) | 22 |
Marker consistency and serial blood collection
To determine consistency, 21·7% of all acquired blood samples had a technical repeat (second PAXgene Blood RNA Tube) taken on the same day. Expression in these samples was highly consistent, with 70% of duplicate samples having identical marker incidence and expression levels (n = 273).
Generalized linear modelling was employed to examine serially collected samples (n = 140), acquired for each patient at intervals of 6 months over a period of 2 years. For the majority of our markers, time was not a significant variable, with marker expression remaining consistent, regardless of sampling time and regardless of stage (Table S3, see Supporting Information). However, for ABCB5 expression, the incidence significantly reduced over time (Wald χ2 = 16·209, P = 0·000) in stage IV patients (odds ratio 0·2518, P = 0·000, 95% confidence interval 0·131–0·483), which may represent the effect of nonsurgical treatment on ABCB5 expression in stage IV patients.
Discussion
This study showed that a multimarker qRT-PCR test for melanoma circulating cells correlated with melanoma diagnosis. Very few patients with melanoma expressed no markers at all, only 8%. However, a significant number of healthy controls also expressed one marker. Therefore, patients and controls are best differentiated by the number of markers expressed, i.e. by a multimarker test.
Multiple markers were detected at all disease stages, even in early-stage patients; however, the incidence was lower in these patients than in later-stage patients. In contrast to previous papers,18,42 the levels and numbers of markers were not stage related, possibly because of the use of ABCB5 and TGFβ2, both previously unused markers of circulating tumour cells. TGFβ2 was the most common marker in controls, so cannot be recommended as a single marker for patient analysis.
By contrast, ABCB5, described by Frank and colleagues in 2003,31 was a highly informative marker.23,24,36 ABCB5 reportedly represents a subset of melanoma cells with a stem cell phenotype and is thought here to represent a subset of heterogeneous circulating melanoma cells present at all disease stages. In this study it was identified as a potential predictor of active recurrence/progression events, with its expression in blood correlating with disease recurrence and progression in patients, a finding that warrants further testing of this marker. This marker was detected among patients regardless of whether or not they were considered clinically disease free, indicating that these stemlike circulating cells remain for long periods in the blood. A role for ABCB5 cells in disease spread has been demonstrated in mouse-to-mouse transplantation experiments, where circulating ABCB5-expressing tumour cells were shown to be capable of initiating metastatic disease after transplantation into donor mice.23
Besides ABCB5, MLANA expression was also shown to be a potential predictor of recurrence events in our patient cohort. As ABCB5-positive cells do not coexpress MLANA,24 circulating cells must therefore include both stem and nonstem cell phenotypes during recurrence. This may reflect a change in stem cells to a nonstem cell phenotype during formation of the metastatic tumour.43 Previous studies described MLANA expression as correlating with disease stage,25 and in this study both MLANA and ABCB5 expression correlated with disease stage, with expression more common in advanced (stage III–IV) than in early-stage (stage 0–II) patients. In fact, removal of a clinically measurable metastasis significantly reduced the expression of PAX3d, ABCB5 and MLANA in stage III–IV patients, signifying the importance of metastases in maintaining the haematogenous spread of tumour cells.7,44
Utilizing these markers, circulating tumour cells were detected at all disease stages long after surgical treatment, even when a patient was considered clinically disease free. Thus, as found in previous studies of circulating tumour cells,42,45 the mere presence of circulating tumour cells does not always correlate with disease stage or metastatic recurrence, and additional markers are required to stratify those early-stage patients likely to have disease progression.
In our study, MCAM expression correlated with increased tumour burden, a result consistent with recent in vivo studies with nude mice that revealed a significant correlation between MCAM expression and the formation of metastases.46 MCAM was also the most suitable marker for monitoring response to therapy, and indicates, presumably, ineffective eradication of circulating melanoma cells, evidenced here by an increase in the incidence of MCAM expression in patients with a negative treatment outcome. MCAM expression could therefore be used to monitor treatment resistance or risk of relapse. Given that MCAM expression is thought to play a role in cell–cell and cell–matrix interactions during formation of metastases, the expression of MCAM in circulating cells may aid in the spread of malignant melanoma cells throughout the body.7,44 A previous study found MCAM to be a predictor of poor patient survival and disease prognosis.43 Overall, 68% of nonsurgically treated patients had a negative treatment outcome, highlighting the poor response rate of patients to nonsurgical treatments, and reflected here by 64% of these patients expressing MCAM. Essentially, MCAM expression may aid in identifying a subset of patients with metastatic disease who respond poorly to conventional systemic treatments and may benefit from an alternative treatment regime.
Some P-values were achieved by considering several different predictors. However, corrections were not made for multiple comparisons because the markers were considered to be independent. Moreover, even when we applied Bonferroni corrections to secondary hypotheses for five comparisons, the majority of hypotheses remained significant (P < 0·01). For example, comparisons between early- vs. late-disease stage (MLANA, P = 0·003 and ABCB5, P = 0·006), disease recurrence (MLANA, P = 0·001) and negative treatment outcome (MCAM, P = 0·006) remained significant. Only the correlation of MCAM expression and tumour burden (MCAM, P = 0·027), and ABCB5 expression in relation to recurrence (ABCB5, P = 0·031), and the surgical removal of a clinically measurable lesion (ABCB5, P = 0·011) were no longer significant. Despite this, MCAM and ABCB5 remain valuable markers of clinical outcome and disease recurrence, especially ABCB5 expression in combination with MLANA.
It is clear that circulating tumour cells are detectable at all stages of disease, and the specific detection of ABCB5-, MLANA- and MCAM-expressing tumour cells in the blood may be of prognostic value. Importantly, we demonstrated that a qRT-PCR multimarker blood test provides a suitable method from which to develop further a reliable test for monitoring disease status without the need for specific isolation of circulating tumour cells.
Supplementary Material
What’s already known about this topic?
Considerable effort has been devoted to developing an effective noninvasive method for monitoring disease progression and outcome among patients with cancer. One method is multimarker quantitative real-time polymerase chain reaction (qRT-PCR), used to detect circulating tumour cells in patient blood.
In breast, colorectal and prostate cancer, circulating tumour cells correlate with both disease progression and poor treatment outcome.
Circulating cell analysis in melanoma has had varied success; despite the capacity to detect cells, the appropriate markers remain undefined.
What does this study add?
This is the first application of ABCB5 as a marker of circulating melanoma cells, incorporated into a multimarker qRT-PCR test of blood in patients with melanoma.
ABCB5 is expressed by a subset of melanoma cells with a stem cell phenotype, and its detection in this study, along with MLANA, a melanocyte marker, correlated with disease recurrence, suggesting both stem and nonstem cell phenotypes are present in patient peripheral blood.
MCAM expression is shown for the first time to correlate with negative treatment outcomes.
Acknowledgments
The authors thank Liz Watson, Jenny Fairweather and Katrina Burton for their assistance with this study.
Funding sources
NHMRC application number 1013349; Cancer and Palliative Care Research and Evaluation Unit WAPCN Small Grants 2010/11; and Cancer Council of WA Research Grant (to M.Z and S.M.). Grant support MHF: National Institutes of Health (U.S.A.) grant numbers 5R01CA113796, 1R01CA158467, 1R01CA138231 and 2P50CA093683 (to M.H.F).
Footnotes
Conflicts of interest
None declared.
Additional Supporting Information may be found in the online version of the article:
Table S1. Quantitative real-time polymerase chain reaction primer sequences (sense and antisense) (Geneworks).
Table S2. Endpoint polymerase chain reaction primer sequences (sense and antisense) (Geneworks).
Table S3. Generalized linear model analysis examining the effect of time on individual marker expression in serial blood collections.
References
- 1.Tucker MA. Melanoma Epidemiology. Hematol Oncol Clin North Am. 2009;23:383–95. doi: 10.1016/j.hoc.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–36. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 3.Balch CM, Buzaid AC, Soong SJ, et al. Final version of the American Joint Committee on Cancer staging system for cutaneous melanoma. J Clin Oncol. 2001;19:3635–48. doi: 10.1200/JCO.2001.19.16.3635. [DOI] [PubMed] [Google Scholar]
- 4.Pollack AL, Li J, Berkowitz Z, et al. Melanoma survival in the United States, 1992 to 2005. J Am Acad Dermatol. 2011;65:S78–86. doi: 10.1016/j.jaad.2011.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsao H, Cosimi AB, Sober AJ. Ultra-late recurrence (15 years or longer) of cutaneous melanoma. Cancer. 1997;79:2361–70. [PubMed] [Google Scholar]
- 6.Shen P, Guenther JM, Wanek LA, Morton DL. Can elective lymph node dissection decrease the frequency and mortality rate of late melanoma recurrences? Ann Surg Oncol. 2000;7:114–19. doi: 10.1007/s10434-000-0114-x. [DOI] [PubMed] [Google Scholar]
- 7.Stoecklein N, Hosch S, Bezler M, et al. Direct genetic analysis of single disseminated cancer cells for prediction of outcome and therapy selection in esophageal cancer. Cancer Cell. 2008;13:441–53. doi: 10.1016/j.ccr.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 8.Sastre J, Maestro ML, Puente J, et al. Circulating tumour cells in colorectal cancer: correlation with clinical and pathological variables. Ann Oncol. 2008;19:935–8. doi: 10.1093/annonc/mdm583. [DOI] [PubMed] [Google Scholar]
- 9.Gaforio JJ, Serrano MJ, Sanchez-Rovira P, et al. Detection of breast cancer cells in the peripheral blood is positively correlated with estrogen-receptor status and predicts for poor prognosis. Int J Cancer. 2003;107:984–90. doi: 10.1002/ijc.11479. [DOI] [PubMed] [Google Scholar]
- 10.Shariat SF, Kattan MW, Song W, et al. Early postoperative peripheral blood reverse transcription PCR assay for prostate-specific antigen is associated with prostate cancer progression in patients undergoing radical prostatectomy. Cancer Res. 2003;63:5874–8. [PubMed] [Google Scholar]
- 11.Müller V, Stahmann N, Riethdorf S, et al. Circulating tumor cells in breast cancer: correlation to bone marrow micrometastases, heterogeneous response to systemic therapy and low proliferative activity. Clin Cancer Res. 2005;11:3678–85. doi: 10.1158/1078-0432.CCR-04-2469. [DOI] [PubMed] [Google Scholar]
- 12.Benoy IH, Elst H, Philips M, et al. Real-time RT-PCR detection of disseminated tumour cells in bone marrow has superior prognostic significance in comparison with circulating tumour cells in patients with breast cancer. Br J Cancer. 2006;94:672–80. doi: 10.1038/sj.bjc.6602985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lembessis P, Msaouel P, Halapas A, et al. Combined androgen blockade therapy can convert RT-PCR detection of prostate-specific antigen (PSA) and prostate-specific membrane antigen (PSMA) transcripts from positive to negative in the peripheral blood of patients with clinically localized prostate cancer and increase biochemical failure-free survival after curative therapy. Clin Chem Lab Med. 2007;45:1488–94. doi: 10.1515/CCLM.2007.301. [DOI] [PubMed] [Google Scholar]
- 14.Pfitzenmaier J, Ellis WJ, Hawley S, et al. The detection and isolation of viable prostate-specific antigen positive epithelial cells by enrichment: a comparison to standard prostate-specific antigen reverse transcriptase polymerase chain reaction and its clinical relevance in prostate cancer. Urol Oncol. 2007;25:214–20. doi: 10.1016/j.urolonc.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 15.Wong SC, Chan CM, Ma BB, et al. Clinical significance of cytokeratin 20-positive circulating tumor cells detected by a refined immunomagnetic enrichment assay in colorectal cancer patients. Clin Cancer Res. 2009;15:1005–12. doi: 10.1158/1078-0432.CCR-08-1515. [DOI] [PubMed] [Google Scholar]
- 16.Allen JE, El-Deiry WS. Circulating tumor cells and colorectal cancer. Curr Colorectal Cancer Rep. 2010;6:212–20. doi: 10.1007/s11888-010-0069-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith B, Selby P, Southgate J, et al. Detection of melanoma cells in peripheral blood by means of reverse transcriptase and polymerase chain reaction. Lancet. 1991;338:1227–9. doi: 10.1016/0140-6736(91)92100-g. [DOI] [PubMed] [Google Scholar]
- 18.Hoon DS, Wang Y, Dale PS, et al. Detection of occult melanoma cells in blood with a multiple-marker polymerase chain reaction assay. J Clin Oncol. 1995;13:2109–16. doi: 10.1200/JCO.1995.13.8.2109. [DOI] [PubMed] [Google Scholar]
- 19.Brownbridge GG, Gold J, Edward M, MacKie RM. Evaluation of the use of tyrosinase-specific and melanA/MART-1-specific reverse transcriptase-coupled polymerase chain reaction to detect melanoma cells in peripheral blood samples from 299 patients with malignant melanoma. Br J Dermatol. 2001;144:279–87. doi: 10.1046/j.1365-2133.2001.04015.x. [DOI] [PubMed] [Google Scholar]
- 20.Reynolds SR, Albrecht J, Shapiro RL, et al. Changes in the presence of multiple markers of circulating melanoma cells correlate with clinical outcome in patients with melanoma. Clin Cancer Res. 2003;9:1497–502. [PubMed] [Google Scholar]
- 21.Gkalpakiotis S, Arenberger P, Kremen J, Arenbergerova M. Quantitative detection of melanoma-associated antigens by multimarker real-time RT-PCR for molecular staging: results of a 5 years study. Exp Dermatol. 2010;19:994–9. doi: 10.1111/j.1600-0625.2010.01123.x. [DOI] [PubMed] [Google Scholar]
- 22.Rao C, Bui T, Connelly M, et al. Circulating melanoma cells and survival in metastatic melanoma. Int J Oncol. 2011;38:755–60. doi: 10.3892/ijo.2011.896. [DOI] [PubMed] [Google Scholar]
- 23.Ma J, Frank MH. Tumor initiation in human malignant melanoma and potential cancer therapies. Anticancer Agents Med Chem. 2010;10:131–6. doi: 10.2174/187152010790909254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schatton T, Frank MH. Antitumor immunity and cancer stem cells. Ann N Y Acad Sci. 2009;1176:154–69. doi: 10.1111/j.1749-6632.2009.04568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koyanagi K, Kuo C, Nakagawa T, et al. Multimarker quantitative real-time PCR detection of circulating melanoma cells in peripheral blood: relation to disease stage in melanoma patients. Clin Chem. 2005;51:981–8. doi: 10.1373/clinchem.2004.045096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parker KA, Glaysher S, Polak M, et al. The molecular basis of the chemosensitivity of metastatic cutaneous melanoma to chemotherapy. J Clin Pathol. 2010;63:1012–20. doi: 10.1136/jcp.2010.080119. [DOI] [PubMed] [Google Scholar]
- 27.Vachtenheim J, Novotná H. Expression of genes for microphthalmia isoforms, Pax3 and MSG1, in human melanomas. Cell Mol Biol. 1999;45:1075–82. [PubMed] [Google Scholar]
- 28.Coulie PG, Brichard V, Van Pel A, et al. A new gene coding for a differentiation antigen recognized by autologous cytolytic T lymphocytes on HLA-A2 melanomas. J Exp Med. 1994;180:35–42. doi: 10.1084/jem.180.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kokkinakis DM, Liu X, Chada S, et al. Modulation of gene expression in human central nervous system tumors under methionine deprivation-induced stress. Cancer Res. 2004;64:7513–25. doi: 10.1158/0008-5472.CAN-04-0592. [DOI] [PubMed] [Google Scholar]
- 30.Bardin N, Francès V, Lesaule G, et al. Identification of the S-Endo 1 endothelial-associated antigen. Biochem Biophys Res Commun. 1996;218:210–16. doi: 10.1006/bbrc.1996.0037. [DOI] [PubMed] [Google Scholar]
- 31.Frank NY, Pendse SS, Lapchak PH, et al. Regulation of progenitor cell fusion by ABCB5 P-glycoprotein, a novel human ATP-binding cassette transporter. J Biol Chem. 2003;278:47156–65. doi: 10.1074/jbc.M308700200. [DOI] [PubMed] [Google Scholar]
- 32.Medic S, Rizos H, Ziman M. Differential PAX3 functions in normal skin melanocytes and melanoma cells. Biochem Biophys Res Commun. 2011;411:832–7. doi: 10.1016/j.bbrc.2011.07.053. [DOI] [PubMed] [Google Scholar]
- 33.Medic S, Ziman M. PAX3 across the spectrum: from melanoblast to melanoma. Crit Rev Biochem Mol Biol. 2009;44:85–97. doi: 10.1080/10409230902755056. [DOI] [PubMed] [Google Scholar]
- 34.Medic S, Ziman M. PAX3 expression in normal skin melanocytes and melanocytic lesions (naevi and melanomas) PLoS ONE. 2010;5:e9977. doi: 10.1371/journal.pone.0009977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuzaki Y, Hashimoto S, Fujita T, et al. Systematic identification of human melanoma antigens using serial analysis of gene expression (SAGE) J Immunother. 2005;28:10–19. doi: 10.1097/00002371-200501000-00002. [DOI] [PubMed] [Google Scholar]
- 36.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–11. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 37.Koyanagi K, O’Day SJ, Boasberg P, et al. Serial monitoring of circulating tumor cells predicts outcome of induction biochemotherapy plus maintenance biotherapy for metastatic melanoma. Clin Cancer Res. 2010;16:2402–8. doi: 10.1158/1078-0432.CCR-10-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rainen L, Oelmueller U, Jurgensen S, et al. Stabilization of mRNA expression in whole blood samples. Clin Chem. 2002;48:1883–90. [PubMed] [Google Scholar]
- 39.Longo MC, Berninger MS, Hartley JL. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene. 1990;93:125–8. doi: 10.1016/0378-1119(90)90145-h. [DOI] [PubMed] [Google Scholar]
- 40.Giricz O, Lauer-Fields JL, Fields GB. The normalization of gene expression data in melanoma: investigating the use of glyceraldehyde 3-phosphate dehydrogenase and 18S ribosomal RNA as internal reference genes for quantitative real-time PCR. Anal Biochem. 2008;380:137–9. doi: 10.1016/j.ab.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tautz D, Renz M. An optimized freeze-squeeze method for the recovery of DNA fragments from agarose gels. Anal Biochem. 1983;132:14–19. doi: 10.1016/0003-2697(83)90419-0. [DOI] [PubMed] [Google Scholar]
- 42.Curry BJ, Myers K, Hersey P. Polymerase chain reaction detection of melanoma cells in the circulation: relation to clinical stage, surgical treatment, and recurrence from melanoma. J Clin Oncol. 1998;16:1760–9. doi: 10.1200/JCO.1998.16.5.1760. [DOI] [PubMed] [Google Scholar]
- 43.Hoek K, Eichhoff O, Schlegel N, et al. In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res. 2008;68:650–6. doi: 10.1158/0008-5472.CAN-07-2491. [DOI] [PubMed] [Google Scholar]
- 44.Hüsemann Y, Geigl JB, Schubert F, et al. Systemic spread is an early step in breast cancer. Cancer Cell. 2008;13:58–68. doi: 10.1016/j.ccr.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 45.Aubin F, Chtourou M, Teyssier JR, et al. The detection of tyrosinase mRNA in the peripheral blood of stage I melanoma patients is not of clinical relevance in predicting metastasis risk and survival. Melanoma Res. 2000;10:113–18. [PubMed] [Google Scholar]
- 46.Luca M, Hunt B, Bucana CD, et al. Direct correlation between MUC18 expression and metastatic potential of human melanoma cells. Melanoma Res. 1993;3:35–41. doi: 10.1097/00008390-199304000-00006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


