Abstract
PTEN is a well-defined tumor suppressor gene that antagonizes the PI3K/Akt pathway to regulate a multitude of cellular processes such as survival, growth, motility, invasiveness and angiogenesis. While the functions of PTEN have been studied extensively, the regulation of its activity during normal and disease conditions still remains incompletely understood. In this study, we identified the protein phosphatase-1 nuclear targeting subunit PNUTS (PPP1R10) as a PTEN associated protein. PNUTS directly interacted with the lipid-binding domain (C2 domain) of PTEN and sequestered it in the nucleus. Depletion of PNUTS leads to increased apoptosis and reduced cellular proliferation in a PTEN-dependent manner. PNUTS expression was elevated in certain cancers compared to matched normal tissues. Collectively, our studies reveal PNUTS as a novel PTEN regulator and a likely oncogene.
Introduction
PTEN is an important tumor suppressor, which has major roles in cell survival, proliferation, migration and cell death (1-3). PTEN was initially identified as a gene located in the chromosomal locus 10q23; one of the most frequently mutated or deleted loci in human cancers. Evidence for loss or mutations of the PTEN gene in many diverse tumor types (4) and high susceptibility of PTEN heterozygous mice to a wide range of tumors (5, 6) strongly support the status of PTEN as an important tumour suppressor for many types of cancers. Functionally, PTEN is a dual specific phosphatase that acts on both lipid and protein substrates (2). The tumor suppressor activity of PTEN is mostly mediated through its lipid substrates (7). One such crucial substrate is phosphatidylinositol 3,4,5-trisphosphate (PIP3), which is converted to phosphatidylinositol 4,5-bisphosphate (PIP2) by PTEN at the cellular membrane (8). PIP3 generated by PI3-Kinase is required for the downstream activation of AKT pathway, which further promotes cell growth and survival. Thus, PTEN keeps a check on tumorigenesis by negatively regulating AKT pathway through down regulating the cellular levels of PIP3. PTEN being a very crucial tumor suppressor, it is important to mechanistically understand its regulation during normal and disease conditions. Although PTEN functions were extensively studied, the regulation of PTEN is less understood.
To elucidate potential regulators of PTEN, we recently performed a tandem affinity purification using PTEN stable cell line and identified several PTEN associated proteins (9). We repeatedly found PNUTS as one of the potential PTEN associated proteins. PNUTS (Protein phosphatase-1 nuclear targeting subunit), also called PPP1R10, CAT53 and p99, was originally isolated as a nuclear protein that forms a stable complex with PP1α and PP1γ in mammalian cells (10, 11). PNUTS binds to PP1 and potently decreases the catalytic activity of PP1 towards exogenous substrates such as Retinoblastoma (Rb) protein in vitro, and reduced expression of PNUTS in mammalian cells affects cell viability (12, 13). However, the exact function of PNUTS in vivo remains to be elucidated.
Materials and Methods
Plasmids
Full length PNUTS and PTEN were cloned into mammalian expressing S-protein/FLAG/SBP (streptavidin binding protein) - triple tagged destination vector, and MYC-tagged destination vector using Gateway cloning system (Invitrogen). PNUTS domain deletions and PTEN domain deletions were cloned into S-protein/FLAG/SBP (Streptavidin binding protein) - triple tagged destination vector. Bacterially expressing GST-tagged PTEN, GST-tagged PNUTS, MBP-tagged PTEN and MBP-tagged PNUTS vectors were generated by transferring their coding sequences into destination vectors by using gateway cloning system.
Antibodies
Rabbit anti-PNUTS, anti-Foxo3a, anti-PNUTS (IHC specific) (Bethyl Laboratories), Monoclonal anti-PTEN clone 6H2.1 (cascade biosciences), Monoclonal anti-MBP (New England Bio Labs), anti-GST, anti-Myc clone 9E10, anti-p53, anti-HDAC2 (all from Santacruz Biotechnologies), Rabbit anti-pAkt (ser-473), anti-Akt, anti-PTEN, anti-pFoxo3a (ser-253), (all from Cell Signalling technology), anti-HDAC2 (Biomol, used in figure 2C), anti-GAPDH (Imgenex), anti-Rad51 (Calbiochem), anti-Flag, anti-Actin (Sigma), antibodies were used in this study.
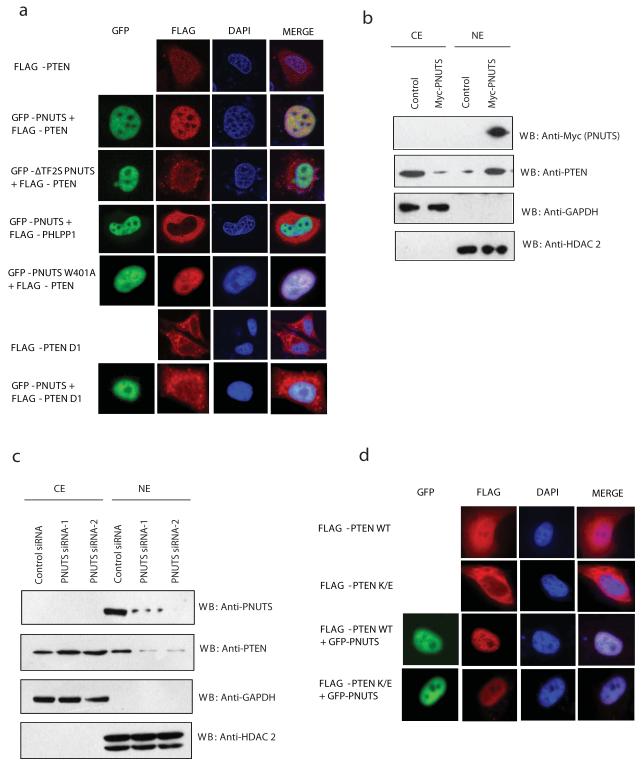
Figure 2. PNUTS sequesters PTEN in the nucleus.
(a) HeLa cells were transfected with Flag-PTEN alone, in combination with GFP-PNUTS, GFP-Delta TF2S PNUTS, GFP-PNUTS W401A mutant or PTEN D1 mutant alone and in combination with GFP-PNUTS. Flag-PHLPP1 together with GFP-PNUTS was used as a control. The localization of PTEN and PHLPP was detected by immunofluorescence staining using anti-FLAG antibody. (b) HeLa cells were transfected with Myc-tagged PNUTS and 24 hours post transfections the cytoplasmic (CE) and nuclear extracts (NE) were prepared. PTEN localization was detected by immunoblotting. GAPDH and HDAC2 were used as controls. (c) HeLa cells were transfected with either control siRNA or PNUTS siRNAs and PTEN was detected in cytoplasmic and nuclear extracts by immunoblotting. (d) Cells were transfected with the indicated constructs and the localization of PTEN was detected by immnoflourescence after staining the anti-Flag antibody.
Cell lines
293T, HeLa, K562, MDA-MB231, BPH1, DU145 and PC-3 cells were used in this study. All cell lines were obtained from American Type Culture Collection (ATCC), which were tested and authenticated by the cell bank using their standard STR (short tandem repeats) based techniques. Cells were also continuously monitored by microscopy to maintain their original morphology and also tested for mycoplasma contamination by using DAPI staining.
Cell transfections, Immunoprecipitation and Immunoblotting
Cells were transfected with various plasmids using Lipofectamine (Invitrogen) according to the manufacturer’s protocol. For immunoprecipitation assays, cells were lysed with NETN buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5 % Nonidet P-40) containing 50 mM β-glycerophosphate, 10 mM NaF, 1 μg/ml of each pepstatin A and aprotinin on ice for 30 minutes. The whole cell lysates obtained by centrifugation were incubated with 2 μg of specified antibody bound to either protein A or protein G sepharose beads (Amersham Biosciences) for 1 hour at 4 °C. The immunocomplexes were then washed with NETN buffer four times and applied to SDS-PAGE. Immunoblotting was performed following standard protocols.
Isolation of cytoplasmic and nuclear extracts
Cells grown in 100mm dish were resuspended in 400μl of Buffer A (1M HEPES pH 7.9, 2M Kcl, 0.5M EDTA pH 8.0, 0.1M EGTA pH 7.0 with protease inhibitors) and incubated in ice for 1 hour. Later 1% NP40 was added and the cytoplasmic extract (supernatant) was collected by centrifugation at 5,000rpm for 5 minutes. The pellet was resuspended in Buffer B (1M HEPES pH 7.9, 5M NaCl, 0.5M EDTA pH8.0, 0.1M EGTA pH 7.0 with protease inhibitors) and the nuclear extract was collected after incubation in ice for 45 minutes followed by centrifugation at 15,000rpm for 5 minutes.
GST pull-down and In vitro binding assays
Bacterially expressed GST-PTEN or control GST bound to Glutathione-Sepharose beads (Amersham) was incubated with 293T cell lysates for 1 hour at 4°C, and the washed complexes were eluted by boiling in SDS sample buffer, separated by SDS-PAGE, and the interactions were analyzed by Western blotting. Similarly, bacterially expressed GST-PNUTS or GST bound to Glutathione-Sepharose beads (Amersham) was incubated with cell lysates of bacteria in which MBP-PTEN is expressed.
RNA interference
Control siRNA and the on-target plus individual siRNAs against PNUTS (siRNA#1: ACAAUUGGCUGACGUAUUC, siRNA#2: GCAGACCCGUUCACCAGAA) were purchased from Dharmacon Inc. PTEN siRNA was purchased from Qiagen. Retroviral shRNA set for PNUTS was purchased from Open biosystems. Transfection was performed twice 30 hours apart with 20 nM of siRNA using Oligofectamine reagent (Invitrogen) according to the manufacturer’s protocol.
Retrovirus production and infection
Virus-containing supernatant was collected 48 and 72 hours after co-transfection of control shRNA or PNUTS shRNA vectors and pcl-ampho helper plasmid into BOSC23 packaging cells, and was used to infect DU-145 or PC-3 cells in the presence of polybrene. Two days later cells were transfered to puromycin containing selection medium. The clones stably expressing PNUTS shRNA were screened after 3 weeks and verified by western blotting using anti-PNUTS antibodies.
PTEN-Lipid binding assay
Phophatidylserine (PS) or phosphatidylethanolamine (PE) or phosphatidylcholine (PC) or PS: PE: PC mix (1:1:1) dissolved in 2:1:0.8 ratio of methanol/chloroform/water were spotted onto nitrocellulose membranes and dried at room temperature for 1 hour. After blocking (50mM Tris-HCl, pH 7.5, 150mM NaCl, 0.1% Tween, 2 mg/ml BSA) the membranes for 1 hour, they were incubated with GST-PTEN either in the presence or absence of PNUTS overnight at 4°C. The membranes were then washed 10 times for 5 minutes each with 1XTBS-T and incubated with HRP-conjugated anti-GST antibody. The binding of PTEN to the lipid was detected by enhanced chemiluminiscence.
Apoptosis assays
Cells were transfected with control, PNUTS, or PTEN and PNUTS siRNAs. After 72Hrs the cells were washed with PBS and stained with FITC-Annexin V and PI according to the manufacture’s protocol (BD Bioscience Annexin V Kit). Apoptotic cells (Annexin V positive, PI negative) were then determined by flow cytometry.
Soft-agar colony assays
Cells were resuspended in RPMI containing 10% FBS along with 0.5% low-melting agarose and seeded on a plate coated with 1% agarose in RPMI and 10% FBS. Viable colonies were scored after 3 weeks of incubation and the quantified data was presented from three independent experiments.
Quantitative RT- PCR
Tumor samples were collected following patient informed consent and approval from the ethics committee of each collaborating hospital. Total RNA was isolated using Trizol reagent (Invitrogen, USA) as per manufacturer’s instructions. 1μg of total RNA was reverse transcribed in the presence of anchored oligo dT (CyScribe Post labeling kit, Amersham, USA) using Superscript-II (Invitrogen, USA) as per manufacturer’s protocol. Quantitative PCR was then initiated using the SYBR Pre mix Ex Taq (perfect real time) kit (Takara Biotechnology, Japan) as per manufacturer’s protocol with 40 cycles of 95°C/30 seconds and 60°C/1 minute. The threshold cycle (Ct) values for PTEN and PNUTS were normalized to GAPDH for each sample and the ΔCt values thus obtained were plotted. Pearson’s correlation (r) between the genes was calculated using Microsoft Excel. Following Primers are used for Q-RT-PCR
| Gene/Exon | Primer sequence |
|---|---|
| PNUTS_F | CTGCCAGCTAAAGGTGAAGATA |
| PNUTS_R | AAGTGAAACATTTTGTCCTTTTT |
| PTEN_F | AAAACGAGCACAGAACCAA |
| PTEN_R | GGCTCCAAAGAGGCTGTAT |
| GAPDH_F | CAATGACCCCTTCATTGACC |
| GAPDH_R | GATCTCGCTCCTGGAAGATG |
Immunoflouroscence staining
Cells grown on coverslips were fixed with 3% paraformaldehyde solution in PBS containing 50 mM sucrose at room temperature for 15 minutes. After permeabilization with 0.5 % Triton X-100 buffer containing 20 mM HEPES pH7.4, 50 mM NaCl, 3mM MgCl2, and 300 mM sucrose at room temperature for 5 minutes, cells were incubated with a primary antibody at 37 °C for 20 minutes. After washing with PBS, cells were incubated with rhodamine-conjugated secondary antibody at 37 °C for 20 minutes. Nuclei were counterstained with 4′6-diamidino-2-phenylindole (DAPI). After a final wash with PBS, coverslips were mounted with glycerin containing paraphenylenediamine.
Immunohistochemsitry
Sections of matched tumor and normal tissues were placed on a single frosty coated slide (Fisher, USA). Antigen retrieval was performed using the pressure cooker method in citrate buffer (pH 6.0) and endogenous peroxidase was blocked using 2 % H2O2. The slides were incubated with PNUTS antibody (Bethyl, USA; 1:250 dilution) for 1 hour followed by incubation with Horse Radish Peroxidase (HRP) conjugated secondary antibody (DAKO, USA). Visualization was performed using Envision plus HRP kit using DAB substrate (DAKO, USA) as per manufacturer’s instructions. The slides were counterstained with hematoxylene and mounted. The IHC results were evaluated for both nuclear stain intensity (on a scale of 1 (negative stain) to 5 (very strong)) and for percentage positive epithelial cells. Overall Staining Intensity (OSI) was calculated as the product of intensity score and percentage of positive cells scored. The tumor/normal OSI fold change was converted to log2 scale and plotted.
Results
PNUTS is a novel PTEN associated protein
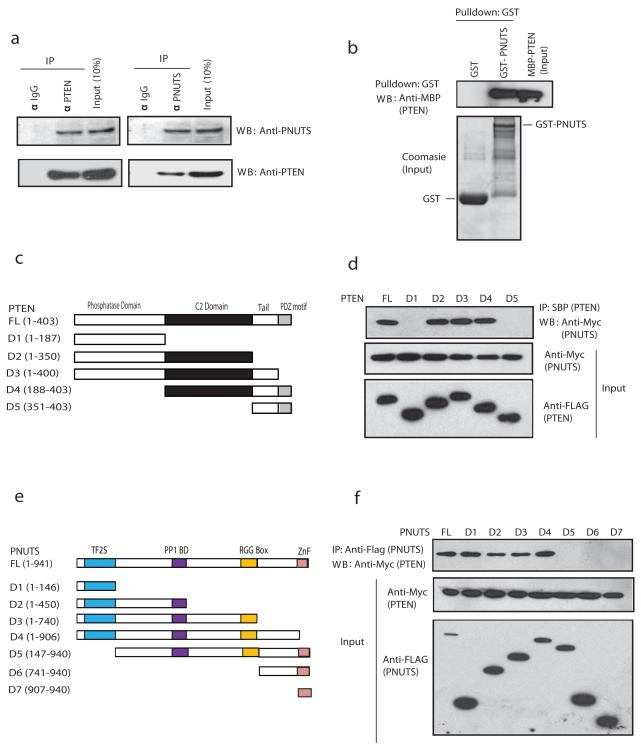
We demonstrated the interaction of endogenous PTEN with PNUTS by immunoprecipitating HEK-293T cell lysates using a PNUTS polyclonal antibody or a PTEN monoclonal antibody (Fig. 1a). By transiently expressing exogenous Flag-PTEN and Myc-PNUTS we further confirmed their interaction in HEK 293T cells (Supplementary Fig. 1a). In addition, an in vitro GST pull down assay with a bacterially expressed PTEN clearly shows the association of PNUTS with PTEN (Supplementary Fig. 1b). Importantly, bacterially purified recombinant GST-PNUTS and MBP-PTEN proteins interacted with each other suggesting a direct interaction between PTEN-PNUTS (Fig. 1b). To map the binding region of PTEN, we generated expression constructs for SFB-tagged PTEN and a series of domain deletion mutants (Fig. 1c). Our Immunoprecipitation results indicated that PNUTS interacts with the C2-domain of PTEN (Fig. 1d & Supplementary Fig. 1c). On the other hand, to map the PTEN binding region in PNUTS, we co-expressed SFB-tagged PNUTS constructs (Fig. 1e) along with Myc-tagged PTEN. Deletion of the N-terminal TF2S domain of PNUTS (ΔTF2S PNUTS) disrupted its binding to PTEN (Fig. 1f). Collectively, these interaction data suggests that PTEN C2 domain and PNUTS TF2S domain are both necessary and sufficient for the interaction between PTEN and PNUTS.
Figure 1. PNUTS is a novel PTEN associated protein.
(a) Co-immunoprecipitation of endogenous PTEN and PNUTS was carried out using extracts prepared from HEK 293T cells. The presence of PNUTS in PTEN complex and vice versa was evaluated by immunoblotting with the indicated antibodies. (b) Lysates from bacteria expressing MBP-PTEN were pulled down with GST alone or GST-PNUTS containing sepharose beads and immunoblotted with anti-MBP antibody. (c) Schematic representation of N-terminal SFB-tagged PTEN (FL), along with its various deletion mutants (D1-D5). (d) SFB-tagged PTEN Full Length and domain deletions were expressed in HEK 293T cells along with Full Length MYC-PNUTS, the cell lysates were pulled down with Streptavidin beads and the interaction of PNUTS was detected with anti-Myc antibody. The expression of PTEN and PNUTS was checked by immunoblotting with anti-Flag and anti-Myc antibodies respectively. (e) Schematic representation of N-terminal SFB-tagged PNUTS (FL), along with its deletion mutants (D1-D7). (f) SFB-tagged PNUTS constructs and Myc-PTEN were co-expressed in HEK293T cells, the cell lysates were pulled down with streptavidin beads and the interaction of PTEN was detected by immunoblotting with anti-Myc antibodies.
PNUTS sequesters PTEN in the nucleus
Previous studies have shown that PTEN is localized in cytoplasm and nucleus (14) whereas PNUTS is exclusively a nuclear protein (11). We further tested whether PTEN and PNUTS co-localize in the cell. PTEN is mainly localized in cytoplasm with sparse nuclear localization. Interestingly expression of full length PNUTS, but not ΔTF2S PNUTS leads to drastic re-localization of PTEN to the nucleus, where it co-localizes with PNUTS (Fig. 2a & supplementary Fig. 1d). On the other hand, PTEN mutant defective of PNUTS binding (PTEN D1) is exclusively localized in cytosol and overexpression of PNUTS has no effect on its localization. Further, expression of PNUTS mutant (W401A) that is defective in PP1 binding also relocalized PTEN to the nucleus indicating PP1 independent role of PNUTS in PTEN localization. Overexpression of PNUTS has no effect on PHLPP, another phosphatase in Akt pathway, indicating the specificity of PNUTS-PTEN interaction. We further demonstrated the positive role of PNUTS in relocalization of PTEN into the nucleus by extracting the cytoplasmic and nuclear fractions. Overexpression of PNUTS enhanced nuclear pool of PTEN (Fig. 2b) where as siRNA mediated knock down of PNUTS reduced nuclear PTEN levels (Fig. 2c) with no significant changes in the half-life of PTEN (Supplementary Fig. 1e). Previous studies have shown that ubiquitination and deubiquitination of PTEN plays a critical role in nuclear-cytoplasmic shuttling of PTEN (14, 15). We used PTEN K289E mutant that is defective in ubiquitination to further study the role of PNUTS in PTEN localization. PTEN K289E mutant is predominantly cytoplasmic but co-expression of PNUTS lead to its nuclear relocalization similar to wild type PTEN (Fig. 2d & Supplementary Fig. 1f), thus suggesting the ubiquitin independent PTEN nuclear localization by PNUTS. Collectively these data suggest the PNUTS sequesters PTEN in the nucleus by directly interacting with PTEN.
PNUTS is upregulated in human cancers and positively correlates with PTEN expression
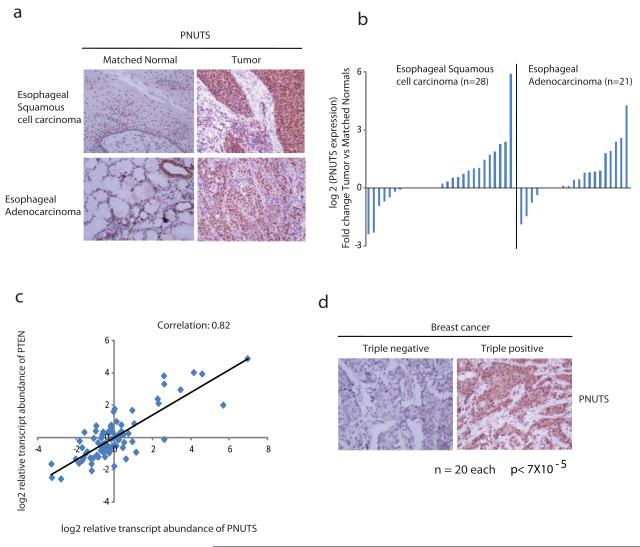
PNUTS is localized on chromosome 6p21.3, a region with frequent aberrations such as chromosome amplifications and increase in gene copy number reported in various human cancers (16, 17). In addition we have shown that PNUTS readily interacts with PTEN, a well known tumor suppressor thus we speculated that PNUTS may play a critical role in tumorigenic process. To test the role of PNUTS in tumorigenesis we have analysed the expression of PNUTS in tissues derived from various types of cancers. Our immunohistochemistry results clearly demonstrated that PNUTS expression is significantly upregulated in tumors derived from squamous cell carcinomas and adenocarcinomas of esophagus compared to their matched normal tissues (Fig. 3a & 3b). Additionally, gene expression data sets from publicly available cancer database (Oncomine) demonstrate increased expression of PNUTS in various tumors, including prostate, skin, renal and brain compared with that in normal tissues (Supplementary Fig. 2a). Interestingly, we observed a positive correlation between the expression of PTEN and PNUTS at the transcript level in esophageal, colon and pancreatic tumors along with normal tissues (Fig. 3c). In addition, PNUTS expression was significantly higher in triple positive breast cancer (with high PTEN levels) (18, 19) compared to the triple negative breast cancers (low PTEN) (Fig. 3d), which again supports a strong association of PNUTS and PTEN. Together these data suggest that PNUTS is concurrently expressed along with PTEN to regulate the survival and growth signalling in cells.
Figure 3. PNUTS is differentially expressed in normal and tumor tissues.
(a) Sections of matched tumor and normal tissues were stained with PNUTS antibody and the representative IHC images were shown. (b) Tissue samples obtained from esophageal squamous cell carcinoma and adenocarcinoma were subjected to immunohistochemistry with specific PNUTS antibody. Data represents the fold change in the overall staining density in tumor sample compared to matched normals. (c) A quantitative RT-PCR analysis was done after extracting RNA from tissues derived from esophageal, pancreatic and colon cancers (n=52) and normal tissues (n=32). Data represents the relative transcript abundance of PNUTS and PTEN normalized against GAPDH. The Pearson’s correlation coefficient is also shown. The trend line depicts the distribution of the data points with respect to perfect correlation (r=1). (d) Tissue samples obtained from triple positive (Er+, PR+, Her2/neu+) and triple negative (Er−, PR−, Her2/neu−) breast cancers (n=20 samples each) were subjected to immunohistochemistry with specific PNUTS antibody. Fisher exact test (two tailed) p value is also shown.
PNUTS control Akt activation and apoptosis in a PTEN dependent manner
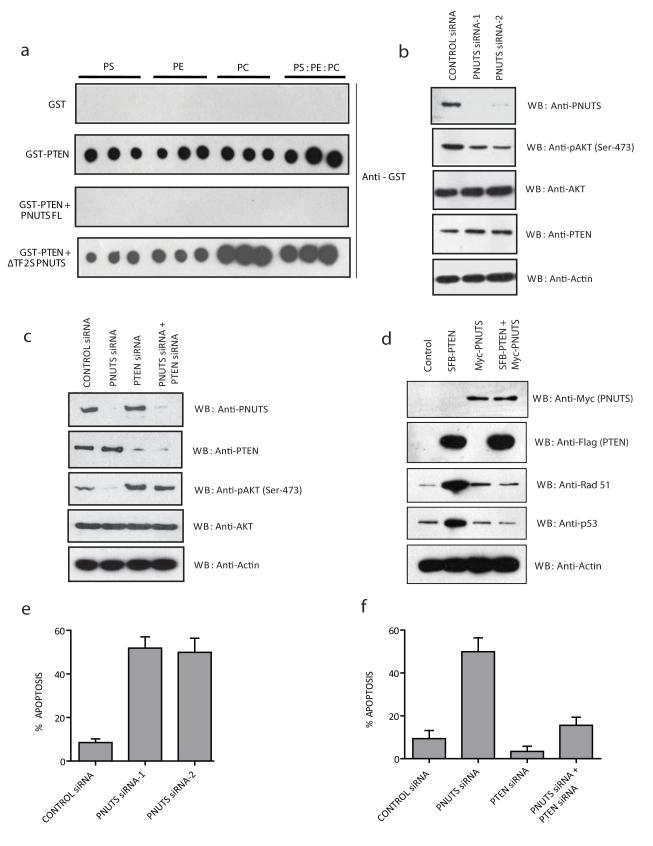
Since PNUTS binds the lipid binding domain (C2 domain) of PTEN, we investigated whether PNUTS interaction with PTEN blocks the association between PTEN and lipids. We performed an in vitro PTEN-lipid binding assay to test if PNUTS interferes with PTEN binding to lipids. As expected, GST-PTEN but not GST alone, strongly associated with the lipids. Interestingly addition of full length PNUTS dramatically reduced the association of PTEN with the lipids (Fig. 4a). But addition of ΔTF2S PNUTS showed modest effects on PTEN binding to lipids. To further explore the effect of PNUTS on the PTEN access to lipids at the membrane, we tested whether Akt signalling downstream of PTEN is affected when PNUTS levels were modulated in cells. Indeed, knockdown of PNUTS by specific siRNAs in HeLa cells resulted in decreased Akt phosphorylation (Fig. 4b) with no significant effect on PTEN and total Akt levels (Fig. 4b). PNUTS regulates Akt activation in a PTEN dependent manner, because a simultaneous depletion of PNUTS along with PTEN by siRNA reversed PNUTS effect on Akt phosphorylation (Fig. 4c). As PNUTS relocalized PTEN to the nucleus we further analyzed the effect of PNUTS on the nuclear functions of PTEN. Expression of PTEN in PTEN null PC-3 prostate cancer cells enhanced rad51 and p53 protein levels but surprisingly co-expression of PNUTS along with PTEN reduced rad51 and p53 to normal levels (Fig. 4d). These results suggest that PNUTS sequesters PTEN in an inactive state in the nucleus. It is well known that PTEN promotes apoptosis (20), and since PNUTS acts as a negative regulator of PTEN function, we hypothesized that depletion of PNUTS might induce apoptosis. To test this hypothesis, we depleted PNUTS by siRNA in HeLa cells and checked for cell death. Knockdown of PNUTS by siRNA readily induced cellular apoptosis (Fig. 4e), but a simultaneous depletion of PNUTS and PTEN rescued cells from apoptosis when compared to PNUTS depletion alone (Fig. 4f). Similar results were observed in K562 myeloid leukemic cells (Supplementary fig. 3a-c), MDA-MB231 breast cancer cells (Supplementary fig. 4a-c) and BPH1 prostate epithelial cells (Supplementary fig. 4d-f) depleted with PNUTS and PTEN. In addition to reduced Akt phosphorylation, depletion of PNUTS also affects Foxo3A phosphorylation, a downstream substrate of Akt (Supplementary Fig. 4a & 4d). Taken together, these results indicate that PNUTS negatively regulates apoptosis in a PTEN dependent manner.
Figure 4. PNUTS control Akt activation and apoptosis in a PTEN dependent manner.
(a) Nitrocellulose membranes spotted with phophatidylserine (PS) or phosphatidylethanolamine (PE) or phosphatidylcholine (PC) or PS: PE: PC mix (1:1:1) in triplicate was incubated with indicated recombinant proteins. Bound PTEN was detected with anti-GST antibody. (b) HeLa cells were transfected with either control siRNA or two different PNUTS siRNAs. After 72hrs of transfection, cells were lysed and cell lysates were blotted with the indicated antibodies. (c) HeLa Cells were treated with control siRNA, PNUTS siRNA, PTEN siRNA, or a combination of PTEN siRNA and PNUTS siRNA, and the cell lysates were immunoblotted with indicated antibodies. (d) PC-3 cells were transfected with SFB-PTEN alone or in combination with Myc-PNUTS and the expression of Rad51 and p53 was detected by immunoblotting with specific antibodies. (e) HeLa cells were transfected with the indicated siRNAs and the percentage of apoptosis was measured by using Annexin-V staining. Error bar indicates standard deviation (n=3), P<0.01; students t-test. (f) HeLa cells were transfected with the indicated siRNAs and the percentage of apoptosis was measured by using Annexin-V staining. Error bar indicates standard deviation (n=3), P<0.01; students t-test.
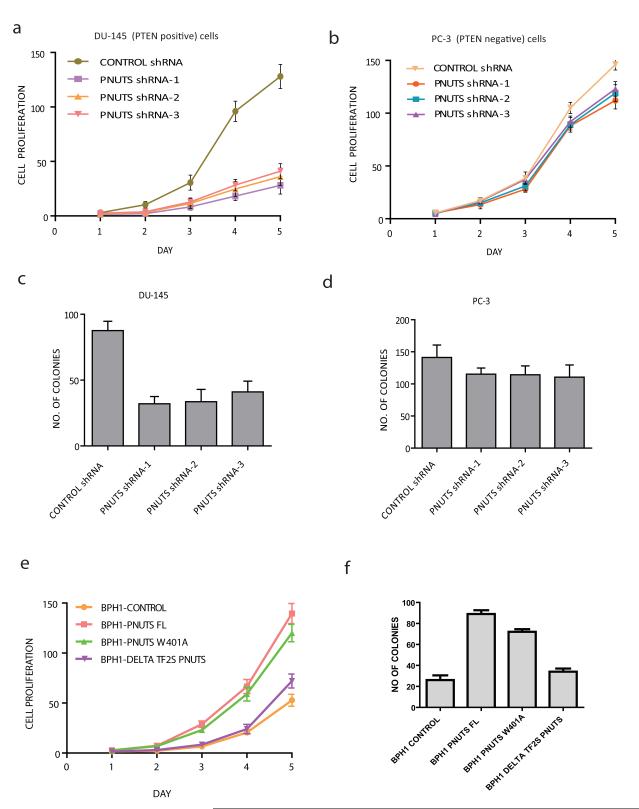
PNUTS is required for tumorigenicity of cells
To further test whether endogenous PNUTS influences cellular survival and growth and to establish its role as a proto-oncogene, we stably depleted PNUTS with short hairpin RNA (shRNA) in a PTEN–positive prostate cancer cell line (DU145) and a PTEN negative prostate cancer cell line (PC3). DU-145 PNUTS shRNA expressing cells showed decreased phosphorylation of Akt and Foxo3a, increased p27 levels, (Supplementary Fig. 5a & 5b) and reduced cell proliferation (Fig. 5a) when compared with control shRNA expressing cells. In contrast, levels of phosphorylated Akt (pAkt) and extent of cell proliferation were not significantly changed in PC-3 PNUTS shRNA expressing cells compared with control shRNA (Fig. 5b & Supplementary Fig. 5c & 5d). In addition, we analysed the cell transforming ability of PNUTS by carrying out soft agar colony formation assays. Depletion of PNUTS significantly reduced the transforming capability of DU-145 cancer cells when compared with control shRNA (Fig. 5c), whereas the transforming capability of PC-3 cancer cells was unaffected with depletion of PNUTS (Fig. 5d). Conversely, stable expression of full length PNUTS but not ΔTF2S PNUTS in normal BPH1 prostate epithelial cells (Supplementary Fig. 5e) enhanced the proliferation (Fig. 5e) and the transforming ability of cells (Fig. 5f). Interestingly, the expression of PNUTS W401A mutant that is defective in binding to PP1 also shows increased proliferation and transformation although less compared to full length PNUTS suggesting that PNUTS mediates its cellular functions mainly through PTEN inhibition and to some extent via PP1 inhibition. Collectively these results indicate that PNUTS acts as a potential oncogene by sequestering PTEN.
Figure 5. PNUTS is required for tumorigenicity of cells.
(a) DU-145 cells stably expressing control shRNA or PNUTS shRNA were seeded and cell proliferation was measured by tryphan blue exclusion for 5 days. Error bar indicates standard deviation (n=3), P<0.01 for all three sets of PNUTS shRNA; students t-test. (b) PC-3 cells stably expressing control shRNA or PNUTS shRNA were analysed for proliferation in a similar way that was described in figure 5a. Error bar indicates standard deviation (n=3), P>0.05 for all three sets of PNUTS shRNA; students t-test. (c) Control shRNA or PNUTS shRNA expressing DU-145 stable cell lines were tested for anchorage independent growth in a soft agar colony assay. Viable colonies were counted after 3 weeks. Error bar indicates standard deviation (n=3), P<0.01for all three sets of PNUTS shRNA; students t-test. (d) PC-3 cells stably expressing control shRNA or PNUTS shRNA were tested for anchorage independent growth in a soft agar colony assay. Viable colonies were counted after 3 weeks. Error bar indicates standard deviation (n=3), P>0.05 for all three sets of PNUTS shRNA; students t-test. (e) A prostate epithelial BPH1 parental cell line along with stably expressing BPH1-PNUTS wt or PNUTS W401A mutant or Delta TF2S PNUTS cells (1×104 cells) were seeded in 100mm dishes. Cells were trypsinized, and viable cells were counted following staining by tryphan blue exclusion method at the indicated times. (f) BPH1 cell line along with BPH1-PNUTS wt or PNUTS W401A mutant or Delta TF2S PNUTS expressing cells were tested for their anchorage independent growth in a soft agar colony assay. Viable colonies after 3 weeks were counted and the data was presented from three independent experiments. Error bar indicates standard deviation (n=3), P<0.01; students t-test.
Discussion
Since PTEN is a very critical tumor suppressor and trivial changes in the PTEN levels lead to susceptibility to malignancy, it is essential to define the regulatory mechanisms that control PTEN functions. Several studies have indicated that PTEN is regulated by multiple mechanisms either at the transcriptional or post translational level. PTEN expression is regulated by transcription factors such as p53, PPARγ and egr-1 at transcription level (21-23), whereas PTEN ubiquitination and phosphorylation regulate its protein levels and activity at the posttranslational level (9, 24). In this study, we demonstrate that PTEN activity can be regulated through a direct protein-protein interaction.
PNUTS tightly associates with PTEN and block the lipid access at the membrane, since PNUTS via its N-terminal TF2S domain binds directly to the lipid binding C2 domain of PTEN. In fact we have shown that expression of PNUTS sequesters PTEN in the nucleus compared to non-expressing cells, where PTEN is mainly cytoplasmic and membrane associated. Thus PNUTS might affect the PIP3 metabolism at the membrane by nuclear sequestration of PTEN. In addition our data also suggested that PNUTS negatively regulates nuclear PTEN induced expression of rad51 and p53. These results suggest that PNUTS sequesters PTEN in an inactive state in the nucleus.
Our studies also suggested that PNUTS might be acting as a potential oncogene by positively regulating the Akt pathway, promoting cell growth and proliferation and suppressing cellular apoptotic potential in a PTEN dependent manner. Importantly, we demonstrated that PNUTS expression is significantly upregulated in tumors derived from squamous cell carcinomas and adenocarcinomas of esophagus compared to their matched normal tissues. Further we observed a positive correlation between the expression of PTEN and PNUTS at the transcript level both in tumors (esophageal, colon and pancreas) and normal tissues, thus suggesting a tight association of PNUTS with PTEN in regulating its function. PTEN is known to be down-regulated by different mechanisms such as mutations or deletions in various types of cancers. Here we are proposing an additional mechanism of PTEN inactivation via PNUTS overexpression in at least some cancers such as oesophageal cancers where mutations and/or deletions of PTEN are rarely reported. Therefore the role of PNUTS may be expected to be important in those tumors where PTEN in not inactivated by mutations, deletions etc. Earlier studies have shown that depletion of PNUTS increased apoptosis in PP1 and Retinoblastoma dependent manner (12). Our results suggest that it would be possible that PNUTS may exert its cellular functions via acting through multiple pathways. In addition to its role in human cancers, PI3Kinase-Akt-PTEN axis has multitude of functions in regulating various pathological conditions such as diabetes, neurological disorders, aging and heart diseases (25). Hence the identification of PNUTS as a critical regulator of PTEN through our studies has provided an additional line of potential therapeutic intervention for human diseases.
Supplementary Material
Acknowledgements
This work was supported in part by a Wellcome Trust/DBT India Alliance grant (to S.M; 500230/Z/11/Z), a grant from Department of Biotechnology, Ministry of Science and Technology, India (to S.M; BT/PR13134/GBD/27/202/2009), and CDFD core funds. S.M is a recipient of Department of Biotechnology’s IYBA award (BT/01/IYBA/2009). SRS is supported by a fellowship from UGC, India. We thank Nanci Rani for providing technical assistance, Pandilla Ramaswamy for processing esophageal tissues for IHC and real time analysis, and Leena Bashyam for her assistance in Quantitative RT-PCR. The authors thank Dr Anirban Maitra, Johns Hopkins University school of Medicine, USA for providing pancreatic cancer xenograft samples.
Footnotes
Conflicts of interest: The authors declare that they have no conflict of interest
References
- 1.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science (New York, NY. 1997;275:1943–7. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 2.Myers MP, Stolarov JP, Eng C, Li J, Wang SI, Wigler MH, et al. P-TEN, the tumor suppressor from human chromosome 10q23, is a dual-specificity phosphatase. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:9052–7. doi: 10.1073/pnas.94.17.9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nature genetics. 1997;15:356–62. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 4.Ali IU, Schriml LM, Dean M. Mutational spectra of PTEN/MMAC1 gene: a tumor suppressor with lipid phosphatase activity. Journal of the National Cancer Institute. 1999;91:1922–32. doi: 10.1093/jnci/91.22.1922. [DOI] [PubMed] [Google Scholar]
- 5.Kwabi-Addo B, Giri D, Schmidt K, Podsypanina K, Parsons R, Greenberg N, et al. Haploinsufficiency of the Pten tumor suppressor gene promotes prostate cancer progression. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:11563–8. doi: 10.1073/pnas.201167798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang SI, Parsons R, Ittmann M. Homozygous deletion of the PTEN tumor suppressor gene in a subset of prostate adenocarcinomas. Clin Cancer Res. 1998;4:811–5. [PubMed] [Google Scholar]
- 7.Myers MP, Pass I, Batty IH, Van der Kaay J, Stolarov JP, Hemmings BA, et al. The lipid phosphatase activity of PTEN is critical for its tumor supressor function. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:13513–8. doi: 10.1073/pnas.95.23.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. The Journal of biological chemistry. 1998;273:13375–8. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 9.Maddika S, Kavela S, Rani N, Palicharla VR, Pokorny JL, Sarkaria JN, et al. WWP2 is an E3 ubiquitin ligase for PTEN. Nature cell biology. 2011;13:730–5. doi: 10.1038/ncb2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allen PB, Kwon YG, Nairn AC, Greengard P. Isolation and characterization of PNUTS, a putative protein phosphatase 1 nuclear targeting subunit. The Journal of biological chemistry. 1998;273:4089–95. doi: 10.1074/jbc.273.7.4089. [DOI] [PubMed] [Google Scholar]
- 11.Kim YM, Watanabe T, Allen PB, Kim YM, Lee SJ, Greengard P, et al. PNUTS, a protein phosphatase 1 (PP1) nuclear targeting subunit. Characterization of its PP1- and RNA-binding domains and regulation by phosphorylation. The Journal of biological chemistry. 2003;278:13819–28. doi: 10.1074/jbc.M209621200. [DOI] [PubMed] [Google Scholar]
- 12.Grana X. Downregulation of the phosphatase nuclear targeting subunit (PNUTS) triggers pRB dephosphorylation and apoptosis in pRB positive tumor cell lines. Cancer biology & therapy. 2008;7:842–4. doi: 10.4161/cbt.7.6.6298. [DOI] [PubMed] [Google Scholar]
- 13.Udho E, Tedesco VC, Zygmunt A, Krucher NA. PNUTS (phosphatase nuclear targeting subunit) inhibits retinoblastoma-directed PP1 activity. Biochemical and biophysical research communications. 2002;297:463–7. doi: 10.1016/s0006-291x(02)02236-2. [DOI] [PubMed] [Google Scholar]
- 14.Trotman LC, Wang X, Alimonti A, Chen Z, Teruya-Feldstein J, Yang H, et al. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell. 2007;128:141–56. doi: 10.1016/j.cell.2006.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song MS, Salmena L, Carracedo A, Egia A, Lo-Coco F, Teruya-Feldstein J, et al. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature. 2008;455:813–7. doi: 10.1038/nature07290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santos GC, Zielenska M, Prasad M, Squire JA. Chromosome 6p amplification and cancer progression. Journal of clinical pathology. 2007;60:1–7. doi: 10.1136/jcp.2005.034389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bashyam MD, Bair R, Kim YH, Wang P, Hernandez-Boussard T, Karikari CA, et al. Array-based comparative genomic hybridization identifies localized DNA amplifications and homozygous deletions in pancreatic cancer. Neoplasia (New York, NY. 2005;7:556–62. doi: 10.1593/neo.04586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marty B, Maire V, Gravier E, Rigaill G, Vincent-Salomon A, Kappler M, et al. Frequent PTEN genomic alterations and activated phosphatidylinositol 3-kinase pathway in basal-like breast cancer cells. Breast Cancer Res. 2008;10:R101. doi: 10.1186/bcr2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saal LH, Johansson P, Holm K, Gruvberger-Saal SK, She QB, Maurer M, et al. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:7564–9. doi: 10.1073/pnas.0702507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stambolic V, Suzuki A, de la Pompa JL, Brothers GM, Mirtsos C, Sasaki T, et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 21.Patel L, Pass I, Coxon P, Downes CP, Smith SA, Macphee CH. Tumor suppressor and anti-inflammatory actions of PPARgamma agonists are mediated via upregulation of PTEN. Curr Biol. 2001;11:764–8. doi: 10.1016/s0960-9822(01)00225-1. [DOI] [PubMed] [Google Scholar]
- 22.Stambolic V, MacPherson D, Sas D, Lin Y, Snow B, Jang Y, et al. Regulation of PTEN transcription by p53. Molecular cell. 2001;8:317–25. doi: 10.1016/s1097-2765(01)00323-9. [DOI] [PubMed] [Google Scholar]
- 23.Virolle T, Adamson ED, Baron V, Birle D, Mercola D, Mustelin T, et al. The Egr-1 transcription factor directly activates PTEN during irradiation-induced signalling. Nature cell biology. 2001;3:1124–8. doi: 10.1038/ncb1201-1124. [DOI] [PubMed] [Google Scholar]
- 24.Wang X, Jiang X. Post-translational regulation of PTEN. Oncogene. 2008;27:5454–63. doi: 10.1038/onc.2008.242. [DOI] [PubMed] [Google Scholar]
- 25.Tamguney T, Stokoe D. New insights into PTEN. Journal of cell science. 2007;120:4071–9. doi: 10.1242/jcs.015230. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.