Summary
Legionella pneumophila is a facultative intracellular pathogen capable of replicating in a wide spectrum of cells. Successful infection by Legionella requires the Dot/Icm type IV secretion system, which translocates a large number of effector proteins into infected cells. By co-opting numerous host cellular processes, these proteins function to establish a specialized organelle that allows bacterial survival and proliferation. Even within the vacuole, L. pneumophila triggers robust immune responses. Recent studies reveal that a subset of Legionella effectors directly target some basic components of the host innate immunity systems such as phagosome maturation. Others play essential roles in engaging the host innate immune surveillance system. This review will highlight recent progress in our understanding of these interactions and discuss implications for the study of the immune detection mechanisms.
Keywords: Type IV secretion, phagosome maturation, NFκB, Interferon Induction
Introduction
The innate immune system functions to provide the first line of defense against pathogen attack and to signal the adaptive immune system of the presence of potential infection. This system is composed of two major components. The first is the scavenging function by phagocytic cells such as macrophages that engulf and digest incoming pathogens. Phagocytosis and subsequent lysosomal digestion is one of the first challenges met by pathogens. The second component is signaling pathways consisting of germline-encoded receptors that are highly conserved across different kingdoms of organisms and, which sense pathogen-associated molecular patterns (PAMPs). These receptors, called pattern recognition receptors (PRRs), can be divided into two families: the Toll-like receptors (TLRs), which recognize ligands on the cell surface or those associated with the lysosome/endosome, and the Nod-like receptors (NLRs), which detect bacterial and viral molecules in the cytoplasm (Akira et al., 2006).
L. pneumophila was first recognized as a pathogen following an outbreak of pneumonia among attendees of the 1976 American Legion convention in Philadelphia (Fields et al., 2002). This bacterium is ubiquitous in aquatic environments as a parasite of freshwater protozoa. This habitat is believed to provide the evolutionary pressure for the acquisition of the traits necessary for survival and replication in phagocytes (Swanson et al., 2000). Development of this potentially fatal disease occurs when susceptible individuals inhale contaminated aerosols. Instead of being digested, L. pneumophila engulfed by alveolar macrophages undergoes robust intracellular growth, leading to tissue damage and subsequent development of the disease (Fields et al., 2002). Largely due to the progress in our understanding of the cell biological and biochemical basis for its intracellular life cycle (Isberg et al., 2009), L. pneumophila has become an attractive model in the study of bacterial pathogenesis as well as the mechanisms of pathogen detection by the immune system,.
The single most important pathogenic determinant of L. pneumophila is the Dot/Icm type IV secretion, which translocates effector proteins into host cells (Ensminger et al., 2009). These effectors have some highly intriguing features: First, the repertoire is extremely large and more than 270 experimentally verified substrates have been identified (Luo et al., 2004, Heidtman et al., 2008, Burstein et al., 2009, Huang et al., 2010, Zhu et al., 2011); this number is close to 10% of the predicted protein encoding genes in the genome of L. pneumophila (Cazalet et al., 2004, Chien et al., 2004). Second, the structures of these proteins are highly diverse. Whereas some proteins contain readily detectable signatures suggestive of potential biochemical activity (e.g. RalF (Nagai et al., 2002)), most proteins are unique to Legionella and do not show significant homology to other proteins in the current database (Zhu et al., 2011). Third, deletion of a single one of these genes rarely leads to defects in Legionella intracellular growth in standard experimental conditions (Isberg et al., 2009). Finally, host processes targeted by these proteins are very diverse and in some cases the potential benefit of such modulation is not yet understood (Dorer et al., 2006, Shin et al., 2008b). These effectors not only play essential roles in the biogenesis of the specialized replicative organelle but also function as messengers that trigger host immune responses. The latter activity has been exploited as a highly effective tool to dissect the mechanisms used by the host to distinguish pathogenic microorganisms from non-harmful ones.
Here, I will discuss the roles of Dot/Icm effectors directly engaging the host innate immune system. The immune responses mediated by TLRs in response to Legionella PAMP molecules such as LPS and the detection of Legionella flagellin by pathways involved the NLR family protein Naip5 and Ipaf have recently been reviewed (Neild et al., 2004, Vance, 2010) and will not be covered here.
Inhibition of phagosome maturation
After phagocytosis, the Legionella containing vacuole (LCV) bypasses the default phagosome maturation pathway. At least in the first 10 hours after uptake, LCVs maintain a neutral pH and are devoid of typical lysosomal proteins such as LAMP-1 (Sturgill-Koszycki et al., 2000). Lysosomal killing is one of the basic components of innate immunity and is the first challenge L. pneumophila faces after phagocytosis. As a parasite of freshwater amoeba, L. pneumophila has evolved sophisticated mechanisms to escape the delivery of its phagosome into the lysosomal network. Active remodeling of the phagosomal membranes to mimic those of resident organelles is a common strategy used by intravacuolar pathogens (Blander et al., 2006). Within minutes of its formation, membranes of the LCV are loaded with some resident proteins of the endoplasmic reticulum (ER), such as calnexin and Bip, indicating that the LCV interacts with the ER network (Swanson et al., 1995, Tilney et al., 2001). Further, inhibition of the activity of proteins such as the small GTPases Sar1 and Rab1, which regulate vesicle budding and trafficking from the ER leads to the arrest of intracellular bacterial growth (Kagan et al., 2002). Based on these observations, Roy and colleagues proposed that L. pneumophila redirects the trafficking of ER-originated vesicles to remodel the LCV membranes (Kagan et al., 2002). This model was supported by the fact that the LCV is surrounded by small smooth vesicles shortly after its formation (Tilney et al., 2001, Robinson et al., 2006), and was greatly substantiated by the discovery of Dot/Icm effectors that directly manipulate activities of Arf1 and Rab1, small GTPases essential for regulation of vesicle trafficking between the ER and the Golgi apparatus (Nagai et al., 2002, Machner et al., 2006, Murata et al., 2006).
Oscillating between an active GTP-bound form and an inactive GDP-bound form, Rab1 regulates several important steps of vesicle transport, most notably, the anterograde transport between the ER and the Golgi apparatus (Stenmark, 2009). L. pneumophila hijacks Rab1 activity with proteins that mimic the functions of its regulatory proteins. Among these, SidM/DrrA is a protein of multiple activities. Anchoring on the LCV by binding to phosphatidylinositol-4-phosphate (Brombacher et al., 2009), SidM/DrrA extracts Rab1 from the GDP association inhibitor (GDI) and recruits and activates it on the bacterial phagosome by its a guanine nucleotide exchange factor (GEF) activity (Murata et al., 2006, Machner et al., 2006, Ingmundson et al., 2007, Machner et al., 2007) (Fig. 1). The bacterial GTPase activation protein (GAP) LepB completes the activation cycle by inducing the GTPase activity of Rab1, leading to its subsequent removal from the LCVs (Ingmundson et al., 2007) (Table 1). Activated Rab1 is believed to facilitate the recruitment of membrane vesicles to the LCV, thus contributing to the membrane remodeling process (Machner et al., 2006).
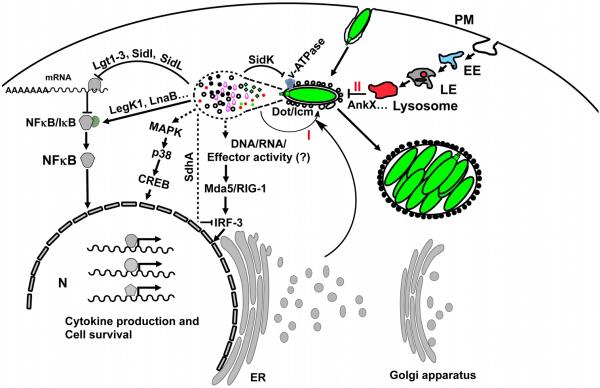
Figure 1. Innate immunity pathways modulated by Dot/Icm substrates.
L. pneumophila translocates a large number of effectors into host cytosol via the Dot/Icm type IV secretion system to facilitate its intracellular replication. Host protein synthesis inhibition by 5 effectors, including three glucosyltransferases (Lgt1-3), SidL and SidL led to inefficient resynthesis of the NF-κB inhibitor IκB and NFκB activation. LegK1 and LnaB activate NF-κB, most likely by initiating the kinase cascade that ultimately causes phosphorylation and subsequent degradation of IκB. Nuclear translocation of NF-κB leads to cytokine production and the induction of anti-apoptotic genes. A yet unidentified set of effectors induces the MAP kinase pathway, leading to similar effects. Undefined effectors or DNA/RNA released by the Dot/Icm transporter induce expression of type I interferon, a process which is inhibited by the effector SdhA. Membrane remodeling of the bacterial phagosome (I) is achieved by a set of effectors, some of which are well-characterized, including RalF, SidM/DrrA, LepB, SidD, AnkX, SidJ and LegK2. Inhibition of phagolysosomal fusion (II) probably is mediated by AnkX, and very likely other yet unidentified effectors. Dashed lines indicate undefined effectors involved in the specified pathway. For clarity, pyroptotic cell death caused by flagellin, which reaches the cytosol probably via the Dot/Icm transporter is not shown. CREB, cAMP response element-binding protein; EE, early endosome; LE, late endosome; ER, endoplasmic reticulum; MAPK, mitogen-activated protein (MAP) kinases; N, nucleus; PM, plasma membrane.
Table 1.
Dot/Icm effectors involved in the modulation of host innate immunity
| Effectors (Lpg number) |
synonym | Biochemical activity |
Targeted host function |
Role in innate immunity |
Reference |
|---|---|---|---|---|---|
| Lpg2464 | SidM/Drr A |
GEFa/GDF/AMPyl ator |
Vesicle trafficking |
Membrane remodeling |
Machner et al., 2006, Murata et al., 2006, Machner et al., 2007, Muller et al., 2010) |
| Lpg2464 | LepB | GAP | Vesicle trafficking |
Membrane remodeling |
(Ingmundson et al., 2007) |
| Lpg2465 | SidD | DeAMPylase | Vesicle trafficking |
Membrane remodeling |
(Neunuebel et al., 2011, Tan et al., 2011) |
| Lpg0695 | AnkX/Leg A8 |
PC transferase | Vesicle trafficking |
Membrane remodeling |
(Mukherjee et al., 2011) |
| Lpg1950 | RalF | GEF | Vesicle trafficking |
Membrane remodeling |
(Nagai et al., 2002) |
| Lpg2155 | SidJ | Unknown | Vesicle trafficking |
Membrane remodeling |
(Liu et al., 2005) |
| Lpl2066 | LegK2 | Protein kinase | Vesicle trafficking |
Membrane remodeling |
(Hervet et al., 2011) |
| Lpg0968 | SidK | Unknown | v-ATPase | Vacuolar acidification |
(Xu et al., 2010) |
| Lpg1483 | LegK1 | Protein kinase | NFκ-B activation |
Cell survival/immu ne response |
(Ge et al., 2009) |
| Lpg2527 | LnaB | Unknown | NFκ-B activation |
Cell survival/immu ne response |
(Losick et al., 2010) |
| Lpg0376 | SdhA | Unknown | Cell death and IFN induction |
Cell survival/immu ne response |
(Laguna et al., 2006, Monroe et al., 2009) |
| Lpg1368 | Lgt1 | glucosyltransferase | Protein translation |
immune response |
(Belyi et al., 2006, Fontana et al., 2011) |
| Lpg2862 | Lgt2 | glucosyltransferase | Protein translation |
immune response |
(Belyi et al., 2008, Fontana et al., 2011) |
| Lpg1488 | Lgt3 | glucosyltransferase | Protein translation |
immune response |
Belyi et al., 2008, Fontana et al., 2011) |
| Lpg2504 | SidI | Unknown | Protein translation |
immune response |
(Shen et al., 2009, Fontana et al., 2011) |
| Lpg0437 | SidL | Unknown | Protein translation |
immune response |
(Fontana et al., 2011) |
Note: GEF, guanosine exchange factor
GDF, GDI-displacement factor (GDF)
GAP
GTPase activation protein
It is intriguing to note that the activity of Rab1 recruited to the LCV is under another layer of regulation, which, strikingly, is imposed by SidM/DrrA. Using a catalytic mechanism similar to glutamine synthetase adenylyl transferase, SidM/DrrA posttranslationally modifies tyrosine 77 of Rab1 by AMPylation, locking the GTPase into its active form which cannot be induced by LepB (Muller et al., 2010). This paradox was recently resolved by the identification of the bacterial deAMPylase SidD, which reverses the modification, making Rab1 accessible to the GAP protein (Neunuebel et al., 2011, Tan et al., 2011). Despite the interesting mechanisms of regulation and the apparent importance of temporal control of Rab1 activity during L. pneumophila infection, it is not clear why it is necessary to lock this small GTPase in its active form during the several hours of its association with the LCVs. It is possible that AMPylation prevents the interference of Rab1 activity by host or bacterial factors. Remarkably, a recent study showed that Rab1 is phosphorylcholinated by AnkX at Ser76 (Mukherjee et al., 2011). Although the effect of this posttranslational modification on Rab1 is not clear, it is possible that AMPylation may prevent the potentially negative effect of AnkX on Rab1 at certain cellular location or phase of infection. In any case, the extensive and precise regulation of the activity of a host protein critical for vesicle transport highlights the importance of efficient membrane remodeling in meeting the challenges posed by the host innate immunity system.
It is worth noting that deletion of any of the genes involved in modulating Rab1 activity did not cause defects in L. pneumophila intracellular growth (Isberg et al., 2009), indicating that modulation of Rab1 activity does not entirely represent the factors involved in membrane remodeling of the LCV. In fact, Dot/Icm effectors such as SidJ and LegK2, which are important for efficient acquisition of ER specific proteins to the LCVs, have been identified (Liu et al., 2007, Hervet et al., 2011) (Table 1). However, their mechanisms of action are unknown. Similarly, many effectors capable of interfering with membrane trafficking in the model system yeast have been identified but the mechanism of action of these proteins remains to be determined (de Felipe et al., 2008, Heidtman et al., 2008). Detailed analysis of their functions will very likely reveal new layers of regulation in membrane trafficking and membrane remodeling of the LCV.
Nascent phagosomes mature by sequential fusion with endosomes and ultimately with lysosomes, which allows for terminal degradation of the engulfed particles or the killing of the phagocytosized microorganisms. A notable feature associated with matured phagosomes is low luminal pH (about 4.5), which is important for their digestive capacity because the optimal activity of many phagosomal hydrolytic enzymes requires an acidic environment (Greenberg et al., 2002). Regulation of phagosomal pH is mediated by the vacuolar ATPase (v-ATPase), a multi-component machinery responsible for proton transport across membranes in a process powered by ATP hydrolysis (Forgac, 2007). After phagocytosis, wild type L. pneumophila maintains a neutral phagosomal pH (Sturgill-Koszycki et al., 2000). Unexpectedly, given the absence of other endosomal markers, v-ATPase was found on membranes of LCVs (Urwyler et al., 2009), suggesting the necessity to inhibit the proton transporter. By screening for Legionella genes capable of causing the neutral pH-sensitive phenotype exhibited by yeast v-ATPase mutants, Xu et al identified SidK, a Dot/Icm substrate that specifically inhibits v-ATPase activity by binding to VatA, a component of the peripheral V1 domain involved in ATP hydrolysis (Xu et al., 2010) (Fig. 1). Macrophages loaded with SidK were impaired in the digestion of internalized nonpathogenic bacteria (Xu et al., 2010), highlighting the role of v-ATPase in lysosomal killing and the importance of SidK and other yet unidentified Legionella effectors for modulating the function of the proton transporter.
Regulatory factors such as members of the Rab small GTPase family and proteins involved in modulating lipid composition of the LCV membrane are believed to contribute to the prevention of the bacterial phagosome from entering the lysosomal pathway (Russell et al., 2006). For example, kinases involved in the metabolism of phosphatidylinositol-3-phosphate (PI3P) may be important because this lipid directly affects localization and function of proteins involved in phagosome maturation (Fratti et al., 2001). In fact, anchoring of two effectors, SidC and SidM to the LCV is mediated by binding to PI4P (Weber et al., 2006, Brombacher et al., 2009). However, effectors directly responsible for the presumed enrichment of phosphatidylinositol on the vacuole surface have not been reported. Interestingly, AnkX also targets Rab35, a small GTPase involved in endosomal trafficking (Mukherjee et al., 2011) (Fig. 1). In light of our current appreciation of the redundant mechanisms used by Legionella to regulate its membrane remodeling, it is attempting to predict the existence of more effectors involved in these processes. Future identification and functional characterization of these effectors will surely lead to better understanding of mechanisms of phagosome maturation and its contribution to host defense against intravacuolar pathogens.
Activation of the NFκ pathway
The second component of the innate immune response is mediated by the activation of specific signaling pathways at the transcriptional level, leading to the production of antimicrobial molecules such as cytokines and the activation of the adaptive immune system. Infection by Legionella triggers the classical TLRs-dependent immune responses (Archer et al., 2009). It also activates caspase 1 by the Naip5/Birc1e and Ipaf-dependent pathway involved in the detection of flagellin released into the host cytosol by the Dot/Icm transporter (Vance, 2010). Microarray analyses with bacterial flagellin mutants using macrophages derived from mice defective in the TLR pathways have revealed important roles for the Dot/Icm system in the induction of several innate immunity pathways, including the NFκB pathway, MAP kinase pathway and host stress response (Losick et al., 2006, Shin et al., 2008a, Fontana et al., 2011).
The master regulator NFκB plays an essential role in the mammalian innate immunity response by controlling the expression of a large number of genes (Hayden et al., 2006). Signals capable of activating NFκB are very diverse, ranging from classical PAMP molecules like LPS to cellular insults such as ER stress and disruption of the actin cytoskeleton (Akira et al., 2006, Bruno et al., 2009). In most cases, these molecules trigger a signaling cascade that causes the release of NFκB from members of the IκB inhibitor family and its subsequent nuclear translocation (Akira et al., 2006). Phosphorylation of IκB by the IκB kinases (IKKs) leads to K48–linked ubiquitination and degradation by the proteasome (Akira et al., 2006). Thus, any activity that leads to IκB phosphorylation will cause an NFκB-mediated response. The importance of the Dot/Icm system in the activation NFκB suggests the existence of substrates capable of causing IκB phosphorylation. Using NFκB-specific luciferase reporters, two Dot/Icm substrates, LegK1 and LnaB, which potently activate NFκB have been identified Ge et al., 2009, Losick et al., 2010) (Fig. 1). LegK1 is a Ser/Thr kinase, which in vitro directly phosphorylates some members of IκB family, including IκBα and p105, leading to the activation of the noncanonical NFκB pathway (Ge et al., 2009). However, the mechanism of action of LnaB is not known (Table 1). Further, the significance of LegK1 and LnaB in the activation of NFκB during L. pneumophila infection is unclear because mutants lacking either of these proteins exhibited very marginal defects, if any, in the activation of this pathway (Ge et al., 2009, Losick et al., 2010). Of note is that a number of Dot/Icm effectors that moderately activate the NFκB reporters were identified in these screenings (Ge et al., 2009, Losick et al., 2010; Haenssler et al., 2011). It is possible that the collective effects of some of these proteins contribute significantly to the NFκB activation during bacterial infection.
Fontana et al revealed a novel mechanism of immune regulation during L. pneumophila infection by demonstrating the requirement of five effectors involved in inhibiting host protein synthesis in the production of distinctive cytokines such as IL23a and Csf2, (Fontana et al., 2011). Compared to infections with the mutant lacking all five translation inhibitors (Table 1), a minimum of a 2 hr delay in IκB re-synthesis was observed in macrophages infected with the wild type strain (Fontana et al., 2011). Because of its labile property, delayed re-synthesis of IκB after its turnover can lead to prolonged NFκB activation, which induces the expression of a set of immune genes (Fig. 1). Thus, activity that prevents efficient production of IκB can activate NFκB. These results indicate that in addition to specific microbial molecules that activate TLRs or cytosolic pattern recognition receptors, a pathogen-encoded activity can be sensed by host immune surveillance mechanisms. It is important to note that mice infected by the Δ5 mutant were not defective in cytokine production nor did IL23a−/− mice exhibit detectably higher bacterial burden (Fontana et al., 2011). Such discrepancies may result from the many redundant pathways that recognize L. pneumophila in vivo (Fontana et al., 2011, Archer et al., 2010)
Because the death of a host cell will lead to premature termination of infection, induction of apoptotic pathways is considered a mechanism of innate immunity. In addition to the production of cytokines, NFκB activates the expression of many anti-apoptotic genes during L. pneumophila infection (Losick et al., 2006, Abu-Zant et al., 2007, Shin et al., 2008a). Two lines of evidence indicate that reprogramming of the host cell death pathway by NFκB activation is important for bacterial intracellular growth. First, in macrophages lacking the anti-apoptotic gene plasminogen activator inhibitor-2 (PAI-2),L. pneumophila infection caused more extensive cell death and a reduction in bacterial replication (Losick et al., 2006). Second, blocking NFκB nuclear translocation by genetic or pharmaceutical agents caused extensive cell death upon low dose bacterial challenge (Losick et al., 2006). The activities act in concert with effectors such as SidF and SdhA, which inhibit host cell death by distinctive mechanisms (Laguna et al., 2006, Banga et al., 2007).
Activation of the MAP kinase pathway
Infection by L. pneumophila also strongly induces the MAP kinase pathway, which is involved in innate immunity and regulation of cell death in parallel with the NFκB pathway (Haenssler et al., 2011). Unlike NFκB, which only exists in mammalian cells and is not believed to play a significant role in shaping the pathogenicity of L. pneumophila, the MAP kinase pathway is an important signaling conduit in its natural amoeba host. The NFκB and MAP kinase pathways, however share some common properties: First, in both cases, the stimulus is relayed by specific kinases, which form the step-wise signaling cascade. Second, like the NFκB pathway, stress response signals and stimuli engaged by TLRs or NLRs also induce the MAP kinase pathway (Huang et al., 2009, Ting et al., 2010). Regulation of MAPK signaling is mainly achieved by negative feedback loops via members of the dual specificity phosphatases (DUSPs) (Jeffrey et al., 2007). Analogous to the control of IκB expression by NFκB, transcription of DUSPs is directly regulated by MAPK signaling. Dot/Icm-dependent induction of the MAP kinase pathway occurs in both amoeba and mammalian hosts (Losick et al., 2006, Farbrother et al., 2006, Shin et al., 2008a). For example, an amoeba mutant lacking DupA failed to support maximal intracellular bacterial growth (Li et al., 2009). However, the lack of DupA caused constitutive activation of MAPK signaling, which led to significant changes in host gene expression, making it difficult to pinpoint the host proteins directly responsible for this phenotype (Li et al., 2009). The role of the MAPK signaling cascade in Legionella infection of mammalian hosts is unknown.
Although the involvement of the Dot/Icm transporter in the induction of the MAP kinase pathway is well established, effectors specifically targeting components of this pathway have not been identified. Under infection conditions, the putative kinases LegK1, LegK2 and LegK3 translocated by the Dot/Icm system do not appear to be important in MAPK signaling (Shin et al., 2008a). However, whether any one of these proteins is capable of activating this pathway when over-expressed is not known. Future studies using suitable MAPK-dependent reporters may lead to identification of such genes. It is also possible that the induction is initiated by indirect effects resulting from the perturbation of host processes by subsets of these effectors. Identification of L. pneumophila mutants unable to induce the MAP kinase pathway will provide excellent tools to probe its regulation. For example, if the mutant missing all five translation inhibitors no longer stimulates the induction, it would suggest the involvement of one or more labile proteins akin to IκB in the regulation of the MAPK signaling.
Whereas the role of the Dot/Icm transporter or its effectors in the induction of the NFκB pathway and the MAP kinase pathway has been established, it is important to note that in both cases, maximal and sustained induction of immune responses cannot be achieved by activity conferred by Dot/Icm alone. Instead, one or more pathways engaged by TLRs or NLRs are required for maximal immune responses. In fact, it has been suggested that NFκB activation during L. pneumophila infection is a two-phase process in which the classical TLR-dependent pathway occurs immediately after bacterial contact, whereas the Dot/Icm-dependent activation persists during the entire replication cycle (Bartfeld et al., 2009). For examples, maximal induction of IL23a and Csf2 by macrophages only occurs in the presence of protein synthesis agents such as ExoA and Pam3 CSK4, and ligands for TLR2 and/or Nod2 ligands (Fontana et al., 2011). Similarly, full induction of some interleukins such as Il1a, Il1b, Il6, and Il12, requires an active Dot/Icm system and the TLR pathway mediated by MyD88 and/or Rip2 (Shin et al., 2008a).
Modulation of type I Interferon induction
L. pneumophila infection leads to transcriptional activation of type I interferon (IFN) genes and restriction of bacterial replication. This observation is reminiscent to the role of type I Interferon in the innate immune response against viral infection (Akira et al., 2006). Several groups have contributed to our understanding of the mechanism of the induction of type I IFNs by L. pneumophila infected macrophages: First, this process is independent of the TLR pathway; second, a functional Dot/Icm type IV secretion system is indispensible for the induction; third, the response does not require the pathway involved the sensing of flagellin by Naip5/Nlrc4 (Opitz et al., 2006, Stetson et al., 2006, Monroe et al., 2009). However, the nature of the ligand and receptor involved in this signaling cascade is less clear. Based on the observation that IRF-3 or IPS-1 is essential for L. pneumophila-induced IFNβ expression, it has been suggested that bacterial DNA released by the Dot/Icm transporter is the signal sensed by the cell (Opitz et al., 2006, Stetson et al., 2006). On the other hand, another study showed that 5′-ppp RNA products generated by the host RNA polymerase III from bacterial DNA trigger IFNβ induction through the RIG-I pathway (Chiu et al., 2009). In contrast, more recent evidence indicates that in mouse macrophages, the RIG-I and Mda5-dependent type I IFN response occurs by sensing RNA molecules presumably directly released by the Dot/Icm transporter (Monroe et al., 2009). This conclusion is consistent with the fact that IPS-1/MAVS-KO mouse cells exhibited very marginal reduction in type I IFN response to the B form of DNA delivered by transfection (Kumar et al., 2006). Of greater interest, Monroe et al., found that the Dot/Icm substrate SdhA functions to suppress the type I IFN induction triggered by L. pneumophila infection (Monroe et al., 2009) (Fig. 1). SdhA also plays a role in protecting infected cells from cell death (Laguna et al., 2006), a function that appears to be independent of the suppression of type I IFN induction (Monroe et al., 2009). The suppression by SdhA protects the intracellular bacteria from the detrimental effect of type I IFN production.
The role of type I IFN induction in defense against L. pneumophila infection is a subject of debate. Whereas earlier studies suggested that mice deficient in type I interferon signaling alone are not more susceptible to Legionella infection (Ang et al., 2010, Monroe et al., 2009), a recent report showed that both type I and II IFNs pr tect mice during lung infections, because higher bacterial burdens were observed in mice deficient in receptors for IFNα(IFNAR−/−) or INFγIFNGR−/−) or both (Lippmann et al., 2011).
Dot/Icm-dependent transfer of DNA has been well established (Vogel et al., 1998), but the transfer of RNA by this transporter has not yet been documented. Thus, the potential discrepancy regarding the receptors involved in the type I interferon pathway may be due to the uncertainty of the ligands and the cell types used. It is possible that a subset of the many Dot/Icm effectors or the collective effects of subsets of them serve as the signal (Monroe et al., 2009). As exemplified by the activation of NFκB by bacterial proteins involved in inhibiting host protein synthesis (Fontana et al., 2011), such proteins may not directly target the RNA sensing pathways. Instead they may interfere with one or more as yet unrecognized host processes, leading to indirect activation of the pathway. In light of the emerging mechanisms for Dot/Icm effector-mediated regulation of the activity of other effectors (Kubori et al., 2010, Luo, 2011, Neunuebel et al., 2011, Tan et al., 2011), SdhA may function to countercheck the activity of one of more effectors directly involved in type I IFN induction. Regardless of their chemical nature, identification of the effectors responsible for the type I IFN induction will be instrumental in addressing these important questions.
Concluding Remarks
Study of the function of Dot/Icm effectors in the last decade has allowed us to better understand the mechanisms employed by L. pneumophila to manipulate host processes. To some extent, this pathogen can be considered a brilliant cell biologist with the tools necessary to exploit the intricacy of host cell biology. On the other hand, in contrast to highly adapted mammalian pathogens that have evolved sophisticated strategies to avoid acute immune responses from the host. L. pneumophila is “naïve” in its interactions with the immune system of higher organisms. The many “accidents” caused by Dot/Icm effectors in mammalian macrophages are invaluable tools for analyzing patterns of pathogenesis or activity-dependent immune surveillance mechanisms, which function in concert with the classical receptor-mediated recognition to distinguish pathogens from nonpathogens (Vance et al., 2009). Detailed analysis of the activity of the effectors involved and the cellular processes engaged by them should reveal unrecognized yet important interactions among signaling pathways that govern a variety of distinct cellular functions.
Acknowledgment
I thank Dr. Art Aronson for critical reading of the manuscript. I also acknowledge my colleagues who contributed to our study of Legionella and its effectors, including S. Banga, M. Fontana, Y. Liu, X. Shen, M. Swanson, Y. Tan, R. Vance, L. Xu and W. Zhu. I apologized to authors whose works cannot be cited here because of space limitation. Our work was supported by NIH-NIAID grants R01AI069344, K02AI085403 and R21AI092043.
Abbreviations
- RIG-I
retinoic-acid-inducible protein 1
- MDA-5
melanoma-differentiation-associated gene 5
- MAVS
mitochondrial antiviral signaling
- IPS-1
interferon-β promoter stimulator 1
- IRF
interferon regulatory factor
- IPS
IFN-β promoter stimulator
- RIP-2
receptor interacting protein 2
References
- Abu-Zant A, Jones S, Asare R, Suttles J, Price C, Graham J, Kwaik YA. Anti apoptotic signalling by the Dot/Icm secretion system of L. pneumophila. Cell Microbiol. 2007;9:246–264. doi: 10.1111/j.1462-5822.2006.00785.x. [DOI] [PubMed] [Google Scholar]
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Ang DK, Oates CV, Schuelein R, Kelly M, Sansom FM, Bourges D, et al. Cutting edge:pulmonary Legionella pneumophila is controlled by plasmacytoid dendritic cells but not type I IFN. J Immunol. 2010;184:5429–5433. doi: 10.4049/jimmunol.1000128. [DOI] [PubMed] [Google Scholar]
- Archer KA, Ader F, Kobayashi KS, Flavell RA, Roy CR. Cooperation between multiple microbial pattern recognition systems is important for host protection against the intracellular pathogen Legionella pneumophila. Infect Immun. 2010;78:2477–2487. doi: 10.1128/IAI.00243-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer KA, Alexopoulou L, Flavell RA, Roy CR. Multiple MyD88-dependent responses contribute to pulmonary clearance of Legionella pneumophila. Cell Microbiol. 2009;11:21–36. doi: 10.1111/j.1462-5822.2008.01234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banga S, Gao P, Shen X, Fiscus V, Zong WX, Chen L, Luo ZQ. Legionella pneumophila inhibits macrophage apoptosis by targeting pro-death members of the Bcl2 protein family. Proc Natl Acad Sci U S A. 2007;104:5121–5126. doi: 10.1073/pnas.0611030104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartfeld S, Engels C, Bauer B, Aurass P, Flieger A, Bruggemann H, Meyer TF. Temporal resolution of two-tracked NF-kappaB activation by Legionella pneumophila. Cell Microbiol. 2009;11:1638–1651. doi: 10.1111/j.1462-5822.2009.01354.x. [DOI] [PubMed] [Google Scholar]
- Belyi Y, Niggeweg R, Opitz B, Vogelsgesang M, Hippenstiel S, Wilm M, Aktories K. Legionella pneumophila glucosyltransferaseinhibits host elongation factor 1A. Proc Natl Acad Sci U S A. 2006;103:16953–16958. doi: 10.1073/pnas.0601562103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyi Y, Tabakova I, Stahl M, Aktories K. Lgt: a family of cytotoxic glucosyltransferases produced by Legionella pneumophila. J Bacteriol. 2008;190:3026–3035. doi: 10.1128/JB.01798-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blander JM, Medzhitov R. On regulation of phagosome maturation and antigen presentation. Nat Immunol. 2006;7:1029–1035. doi: 10.1038/ni1006-1029. [DOI] [PubMed] [Google Scholar]
- Brombacher E, Urwyler S, Ragaz C, Weber SS, Kami K, Overduin M, Hilbi H. Rab1 guanine nucleotide exchange factor SidM is a major phosphatidylinositol 4-phosphate-binding effector protein of Legionella pneumophila. J Biol Chem. 2009;284:4846–4856. doi: 10.1074/jbc.M807505200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno VM, Hannemann S, Lara-Tejero M, Flavell RA, Kleinstein SH, Galan JE. Salmonella Typhimurium type III secretion effectors stimulate innate immune responses in cultured epithelial cells. PLoS Pathog. 2009;5:e1000538. doi: 10.1371/journal.ppat.1000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein D, Zusman T, Degtyar E, Viner R, Segal G, Pupko T. Genome-scale identification of Legionella pneumophila effectors using a machine learning approach. PLoS Pathog. 2009;5:e1000508. doi: 10.1371/journal.ppat.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalet C, Rusniok C, Bruggemann H, Zidane N, Magnier A, Ma L, et al. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat Genet. 2004;36:1165–1173. doi: 10.1038/ng1447. [DOI] [PubMed] [Google Scholar]
- Chien M, Morozova I, Shi S, Sheng H, Chen J, Gomez SM, et al. The genomic sequence of the accidental pathogen Legionella pneumophila. Science. 2004;305:1966–1968. doi: 10.1126/science.1099776. [DOI] [PubMed] [Google Scholar]
- Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects 1 cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Felipe KS, Glover RT, Charpentier X, Anderson OR, Reyes M, Pericone CD, Shuman HA. Legionella eukaryotic-like type IV substrates interfere with organelle trafficking. PLoS Pathog. 2008;4:e1000117. doi: 10.1371/journal.ppat.1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorer MS, Kirton D, Bader JS, Isberg RR. RNA interference analysis of Legionella in Drosophila cells: exploitation of early secretory apparatus dynamics. PLoS Pathog. 2006;2:e34. doi: 10.1371/journal.ppat.0020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensminger AW, Isberg RR. Legionella pneumophila Dot/Icm translocated substrates: a sum of parts. Curr Opin Microbiol. 2009;12:67–73. doi: 10.1016/j.mib.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farbrother P, Wagner C, Na J, Tunggal B, Morio T, Urushihara H, et al. Dictyostelium transcriptional host cell response upon infection with Legionella. Cell Microbiol. 2006;8:438–456. doi: 10.1111/j.1462-5822.2005.00633.x. [DOI] [PubMed] [Google Scholar]
- Fields BS, Benson RF, Besser RE. Legionella and Legionnaires’ disease: 25 years of investigation. Clin Microbiol Rev. 2002;15:506–526. doi: 10.1128/CMR.15.3.506-526.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana MF, Banga S, Barry KC, Shen X, Tan Y, Luo ZQ, Vance RE. Secreted bacterial effectors that inhibit host protein synthesis are critical for induction of the innate immune response to virulent Legionella pneumophila. PLoS Pathog. 2011;7:e1001289. doi: 10.1371/journal.ppat.1001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol. 2007;8:917–929. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- Fratti RA, Backer JM, Gruenberg J, Corvera S, Deretic V. Role ophosphatidylinositol 3-kinase and Rab5 effectors in phagosomal biogenesis and mycobacterial phagosome maturation arrest. J Cell Biol. 2001;154:631–644. doi: 10.1083/jcb.200106049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge J, Xu H, Li T, Zhou Y, Zhang Z, Li S, et al. A Legionella type IV effector activates the NF-kappaB pathway by phosphorylating the IkappaB family of inhibitors. Proc Natl Acad Sci U S A. 2009;106:13725–13730. doi: 10.1073/pnas.0907200106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg S, Grinstein S. Phagocytosis and innate immunity. Curr Opin Immunol. 2002;14:136–145. doi: 10.1016/s0952-7915(01)00309-0. [DOI] [PubMed] [Google Scholar]
- Haenssler E, Isberg RR. Control of host cell phosphorylation by legionella pneumophila. Front Microbiol. 2011;2:64. doi: 10.3389/fmicb.2011.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden MS, West AP, Ghosh S. NF-kappaB and the immune response. Oncogene. 2006;25:6758–6780. doi: 10.1038/sj.onc.1209943. [DOI] [PubMed] [Google Scholar]
- Heidtman M, Chen EJ, Moy MY, Isberg RR. Large-scale identification of Legionella pneumophila Dot/Icm substrates that modulate host cell vesicle trafficking pathways. Cell Microbiol. 2008 doi: 10.1111/j.1462-5822.2008.01249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervet E, Charpentier X, Vianney A, Lazzaroni JC, Gilbert C, Atlan D, Doublet P. Protein kinase LegK2 is a type IV secretion system effector involved in endoplasmic reticulum recruitment and intracellular replication of Legionella pneumophila. Infect Immun. 2011;79:1936–1950. doi: 10.1128/IAI.00805-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Shi LZ, Chi H. Regulation of JNK and p38 MAPK in the immune system: signal integration, propagation and termination. Cytokine. 2009;48:161–169. doi: 10.1016/j.cyto.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Boyd D, Amyot WM, Hempstead AD, Luo ZQ, O’Connor TJ, et al. The E Block motif is associated with Legionella pneumophila translocated substrates. Cell Microbiol. 2010;13:227–245. doi: 10.1111/j.1462-5822.2010.01531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingmundson A, Delprato A, Lambright DG, Roy CR. Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature. 2007;450:365–369. doi: 10.1038/nature06336. [DOI] [PubMed] [Google Scholar]
- Isberg RR, O’Connor TJ, Heidtman M. The Legionella pneumophila 1 replication vacuole: making a cosy niche inside host cells. Nat Rev Microbiol. 2009;7:13–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey KL, Camps M, Rommel C, Mackay CR. Targeting dual-specificity phosphatases: manipulating MAP kinase signalling and immune responses. Nat Rev Drug Discov. 2007;6:391–403. doi: 10.1038/nrd2289. [DOI] [PubMed] [Google Scholar]
- Kagan JC, Roy CR. Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nat Cell Biol. 2002;4:945–954. doi: 10.1038/ncb883. [DOI] [PubMed] [Google Scholar]
- Kubori T, Shinzawa N, Kanuka H, Nagai H. Legionella metaeffector exploits host proteasome to temporally regulate cognate effector. PLoS Pathog. 2010;6:e1001216. doi: 10.1371/journal.ppat.1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar H, Kawai T, Kato H, Sato S, Takahashi K, Coban C, et al. Essential role of IPS-1 in innate immune responses against RNA viruses. J Exp Med. 2006;203:1795–1803. doi: 10.1084/jem.20060792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguna RK, Creasey EA, Li Z, Valtz N, Isberg RR. A Legionella pneumophila13 translocated substrate that is required for growth within macrophages and protection from host cell death. Proc Natl Acad Sci U S A. 2006;103:18745–18750. doi: 10.1073/pnas.0609012103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Dugan AS, Bloomfield G, Skelton J, Ivens A, Losick V, Isberg RR. The amoebal MAP kinase response to Legionella pneumophila is regulated by DupA. Cell Host Microbe. 2009;6:253–267. doi: 10.1016/j.chom.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippmann J, Muller HC, Naujoks J, Tabeling C, Shin S, Witzenrath M, et al. Dissection of a type I interferon pathway in controlling bacterial intracellular infection in mice. Cell Microbiol. 2011 doi: 10.1111/j.1462-5822.2011.01646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HY, Deng AM, Zhang J, Zhou Y, Yao DK, Tu XQ, et al. Correlation of Chlamydia pneumoniae infection with primary biliary cirrhosis. World J Gastroenterol. 2005;11:4108–4110. doi: 10.3748/wjg.v11.i26.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Luo ZQ. The Legionella pneumophila effector SidJ is required for efficient recruitment of endoplasmic reticulum proteins to the bacterial phagosome. Infect Immun. 2007;75:592–603. doi: 10.1128/IAI.01278-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick VP, Haenssler E, Moy MY, Isberg RR. LnaB: a Legionella pneumophila activator of NF-kappaB. Cell Microbiol. 2010;12:1083–1097. doi: 10.1111/j.1462-5822.2010.01452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick VP, Isberg RR. NF-kappaB translocation prevents host cell death after low-dose challenge by Legionella pneumophila. J Exp Med. 2006;203:2177–2189. doi: 10.1084/jem.20060766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo ZQ. Targeting One of its Own: Expanding Roles of Substrates of the Legionella Pneumophila Dot/Icm Type IV Secretion System. Front Microbiol. 2011;2:31. doi: 10.3389/fmicb.2011.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo ZQ, Isberg RR. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc Natl Acad Sci U S A. 2004;101:841–846. doi: 10.1073/pnas.0304916101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machner MP, Isberg RR. Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev Cell. 2006;11:47–56. doi: 10.1016/j.devcel.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Machner MP, Isberg RR. A bifunctional bacterial protein links GDI displacement to Rab1 activation. Science. 2007;318:974–977. doi: 10.1126/science.1149121. [DOI] [PubMed] [Google Scholar]
- Monroe KM, McWhirter SM, Vance RE. Identification of host cytosolic sensors and bacterial factors regulating the type I interferon response to Legionella pneumophila. PLoS Pathog. 2009;5:e1000665. doi: 10.1371/journal.ppat.1000665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Liu X, Arasaki K, McDonough J, Galan JE, Roy CR. Modulation of Rab GTPase function by a protein phosphocholintransferase. Nature. 2011;477:103–106. doi: 10.1038/nature10335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller MP, Peters H, Blumer J, Blankenfeldt W, Goody RS, Itzen A. The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science. 2010;329:946–949. doi: 10.1126/science.1192276. [DOI] [PubMed] [Google Scholar]
- Murata T, Delprato A, Ingmundson A, Toomre DK, Lambright DG, Roy CR. The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat Cell Biol. 2006;8:971–977. doi: 10.1038/ncb1463. [DOI] [PubMed] [Google Scholar]
- Nagai H, Kagan JC, Zhu X, Kahn RA, Roy CR. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science. 2002;295:679–682. doi: 10.1126/science.1067025. [DOI] [PubMed] [Google Scholar]
- Neild AL, Roy CR. Immunity to vacuolar pathogens: what can we learn from Legionella? Cell Microbiol. 2004;6:1011–1018. doi: 10.1111/j.1462-5822.2004.00450.x. [DOI] [PubMed] [Google Scholar]
- Neunuebel MR, Chen Y, Gaspar AH, Backlund PS, Jr., Yergey A, Machner MP. De-AMPylation of the small GTPase Rab1 by the pathogen Legionella pneumophila. Science. 2011;333:453–456. doi: 10.1126/science.1207193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opitz B, Vinzing M, van Laak V, Schmeck B, Heine G, Gunther S, et al. Legionella pneumophila induces IFNbeta in lung epithelial cells via IPS-1 and IRF3, which alsocontrol bacterial replication. J Biol Chem. 2006;281:36173–36179. doi: 10.1074/jbc.M604638200. [DOI] [PubMed] [Google Scholar]
- Robinson CG, Roy CR. Attachment and fusion of endoplasmic reticulum with vacuoles containing Legionella pneumophila. Cell Microbiol. 2006;8:793–805. doi: 10.1111/j.1462-5822.2005.00666.x. [DOI] [PubMed] [Google Scholar]
- Russell MR, Nickerson DP, Odorizzi G. Molecular mechanisms of late endosome morphology, identity and sorting. Curr Opin Cell Biol. 2006;18:422–428. doi: 10.1016/j.ceb.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Shen X, Banga S, Liu Y, Xu L, Gao P, Shamovsky I, et al. Targeting eEF1A by a Legionella pneumophila effector leads to inhibition of protein synthesis and induction of host stress response. Cell Microbiol. 2009;11:911–926. doi: 10.1111/j.1462-5822.2009.01301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S, Case CL, Archer KA, Nogueira CV, Kobayashi KS, Flavell RA, et al. Type IV secretion-dependent activation of hostMAP kinases induces an increased proinflammatory cytokine response to Legionella pneumophila. PLoS Pathog. 2008a;4:e1000220. doi: 10.1371/journal.ppat.1000220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S, Roy CR. Host cell processes that influence the intracellular survival of Legionella pneumophila. Cell Microbiol. 2008b;10:1209–1220. doi: 10.1111/j.1462-5822.2008.01145.x. [DOI] [PubMed] [Google Scholar]
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. NatRev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Sturgill-Koszycki S, Swanson MS. Legionella pneumophila replication vacuoles mature into acidic, endocytic organelles. J Exp Med. 2000;192:1261–1272. doi: 10.1084/jem.192.9.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson MS, Hammer BK. Legionella pneumophila pathogesesis: a fateful journey from amoebae to macrophages. Annu Rev Microbiol. 2000;54:567–613. doi: 10.1146/annurev.micro.54.1.567. [DOI] [PubMed] [Google Scholar]
- Swanson MS, Isberg RR. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect Immun. 1995;63:3609–3620. doi: 10.1128/iai.63.9.3609-3620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y, Luo ZQ. Legionella pneumophila SidD is a deAMPylase that modifies Rab1. Nature. 2011;475:506–509. doi: 10.1038/nature10307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney LG, Harb OS, Connelly PS, Robinson CG, Roy CR. How the parasitic bacterium Legionella pneumophila modifies its phagosome and transforms it into rough ER: implications for conversion of plasma membrane to the ER membrane. J Cell Sci. 2001;114:4637–4650. doi: 10.1242/jcs.114.24.4637. [DOI] [PubMed] [Google Scholar]
- Ting JP, Duncan JA, Lei Y. How the noninflammasome NLRs function in the innate immune system. Science. 2010;327:286–290. doi: 10.1126/science.1184004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urwyler S, Nyfeler Y, Ragaz C, Lee H, Mueller LN, Aebersold R, Hilbi H. Proteome analysis of Legionella vacuoles purified by magnetic immunoseparation reveals secretory and endosomal GTPases. Traffic. 2009;10:76–87. doi: 10.1111/j.1600-0854.2008.00851.x. [DOI] [PubMed] [Google Scholar]
- Vance RE. Immunology taught by bacteria. J Clin Immunol. 2010;30:507–511. doi: 10.1007/s10875-010-9389-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009;6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Andrews HL, Wong SK, Isberg RR. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 1998;279:873–876. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]
- Weber SS, Ragaz C, Reus K, Nyfeler Y, Hilbi H. Legionella pneumophila exploits PI(4)P to anchor secreted effector proteins to the replicative vacuole. PLoS Pathog. 2006;2:e46. doi: 10.1371/journal.ppat.0020046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Shen X, Bryan A, Banga S, Swanson MS, Luo ZQ. Inhibition of host vacuolar H+-ATPase activity by a Legionella pneumophila effector. PLoS Pathog. 2010;6:e1000822. doi: 10.1371/journal.ppat.1000822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Banga S, Tan Y, Zheng C, Stephenson R, Gately J, Luo ZQ. Comprehensive identification of protein substrates of the Dot/Icm type IV transporter of Legionella pneumophila. PLoS One. 2011;6:e17638. doi: 10.1371/journal.pone.0017638. [DOI] [PMC free article] [PubMed] [Google Scholar]