Summary
DNA ligase IV (LigIV) and Artemis are central components of the Non-Homologous End Joining (NHEJ) machinery that is required for V(D)J recombination and the maintenance of genomic integrity in mammalian cells. We report here crystal structures of the LigIV DNA binding domain (DBD) in both its apo form and in complex with a peptide derived from the Artemis C-terminal region. We show that LigIV interacts with Artemis through an extended hydrophobic surface. In particular, we find that helix α2 in LigIV DBD is longer than in other mammalian ligases and presents residues which specifically interact with the Artemis peptide that adopts a beta-helix conformation on binding. Mutations of key residues on the LigIV DBD hydrophobic surface abolish the interaction. Together, our results provide structural insights into the specificity of LigIV-Artemis interaction and how the enzymatic activities of the two proteins may be coordinated during NHEJ.
Introduction
The adaptive immune system gains its ability to recognize a plethora of antigens by the generation of an extensive repertoire of antigen receptors on B and T lymphocytes. These numerous receptors are created by V(D)J recombination, a process during which DNA double-strand breaks (DSBs) are repaired by the Non-Homologous End Joining Pathway (NHEJ) (Lieber, 2010). NHEJ is one of the major pathways for the repair of DSBs in mammalian cells, playing not only a central role in V(D)J recombination and class switch recombination (CSR), but also in the repair of pathological DSBs generated by ionizing radiation and other chemical and enzymatic damage. This pathway works throughout the cell cycle and does not require the presence of a homologous sister chromatid (Lieber, 2010).
DNA ligase IV (LigIV) is an essential factor for NHEJ (Grawunder et al., 1998). Mutations in LigIV lead to a rare disease called the Lig4 syndrome, which is characterized by microcephaly, radiosensitivity, growth and developmental delay, and immunodeficiency (Chistiakov, 2010). Moreover, it has been shown that in mice the absence of LigIV increases tumor generation in non-lymphoid tissues (Sharpless et al., 2001).
LigIV is unique among other mammalian ligases in its ability to perform nuclear DNA DSB repair (Simsek et al., 2011). It exhibits a high degree of similarity with LigI and LigIII, particularly at the level of domain organization of the catalytic core (Ellenberger and Tomkinson, 2008). Thus, its unique ability to repair DSB might be partly delineated through its interacting partners. LigIV directly interacts with XRCC4 (Critchlow et al., 1997; Grawunder et al., 1997) and Ku (Chen et al., 2000; Nick McElhinny et al., 2000), and indirectly with DNA-PKcs (Drouet et al., 2005), Cernunnos-XLF (Ahnesorg et al., 2006) and the MRN complex (Chen et al., 2001), all which are necessary for NHEJ in mammalian cells. Recently, we have identified Artemis as a new interaction partner of LigIV (Malu et al., 2012). Artemis, in complex with DNA-PKcs, has a diverse array of nuclease activities and is a necessary factor for the processing of DSBs during NHEJ (Ma et al., 2002; Ma et al., 2005b). Association-binding experiments indicate that the C-terminal region (C-ter) of Artemis directly interacts with the DNA binding domain of LigIV (LigIV-DBD). Moreover, deletion analysis of Artemis C-ter identified an eleven amino acid peptide (485-DVPQWEVFFKR-495) that contains the minimal interacting region. Aromatic residues in this region are crucial for the interaction and are important for efficient V(D)J recombination (Malu et al., 2012). However, the corresponding interacting surface on LigIV-DBD remains uncharacterized due to the limited structural information for LigIV. As such, it is unclear how the interaction between Artemis C-ter and LigIV is mediated and how it facilitates NHEJ in mammalian cells.
We report here crystal structures of the LigIV-DBD in both its apo form and in complex with a peptide derived from the Artemis C-ter. We show that LigIV-DBD interacts with Artemis through a hydrophobic surface that is much more extended than in other mammalian DNA ligases, such as LigI and LigIII. Based on the structure, the interaction surface was further evaluated by isothermal titration calorimetry (ITC) as well as by mutational analysis of critical residues. Together, these results offer for the first time structural insights into the specificity of interaction between LigIV and Artemis, important for development of the immune system and genomic stability.
Results
Structure of apo LigIV-DBD
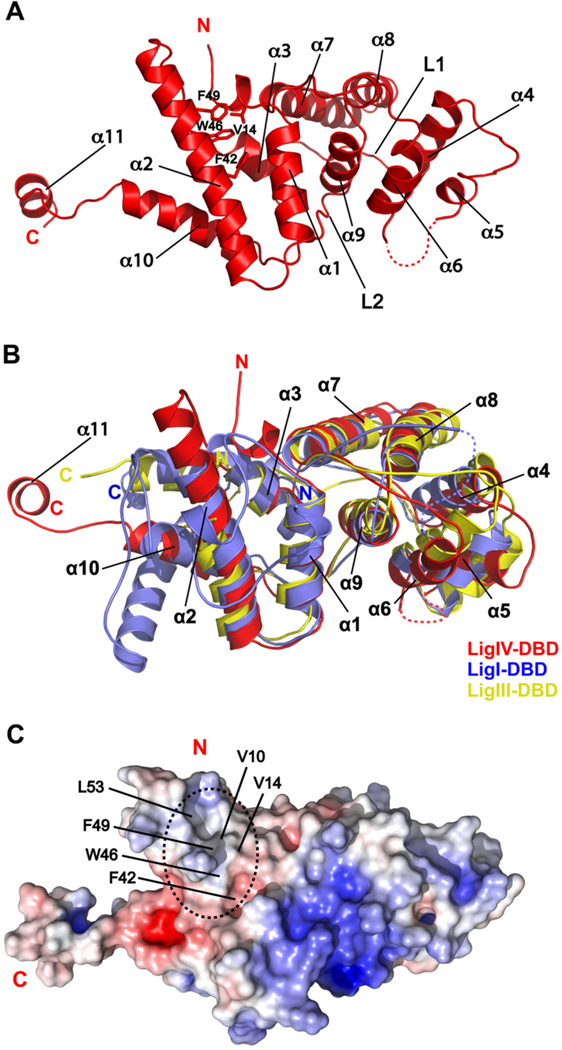
The crystal structure of LigIV-DBD, determined at 2.8 Å resolution (Figure S1), is composed of two helical subdomains, related by an approximate two fold axis (Figure 1A). The first subdomain is composed of helices α1, α2, α3, α10, and α11 while the second subdomain consists of helices α4, α5, α6, α7, α8, and α9, and the two are connected by long loops, L1 (71–86) and L2 (191–201). This type of pseudo-symmetry is also observed in the DBDs of other mammalian DNA ligases (Ellenberger and Tomkinson, 2008), as well as in the C-terminal helix-hairpin-helix (HhH) domain of E. coli LigA (Lee et al., 2000), which is functionally similar to the DBD of human DNA LigIV. As with LigI and LigIII, the helical subdomains in LigIV DBD lend to both a polar and non-polar surfaces (Figure S2). A superimposition of LigIV-DBD onto LigI/DNA and LigIII/DNA complex structures is consistent with DNA binding to the LigIV DBD on its positively charged surface (Figure S2) (Cotner-Gohara et al., 2010; Pascal et al., 2004). In all, even though the LigIV-DBD shares low sequence similarity with the LigI (13% sequence identity) and LigIII (15% sequence identity) DBDs (Figure S3), the structure is broadly similar, superimposing with rmsds of 4.1 Å (101 Cαs) and 2.5 Å (157 Cαs), respectively (Figure 1B).
Figure 1. Crystal structure of LigIV-DBD apo form.
(A) Ribbon diagram of LigIV-DBD. Secondary structure elements and important amino acids side chains are labeled. (B) Structural alignment of human Lig-DBDs. LigIV-DBD superimposed on LigI-DBD (PDB ID 1X9N, residues 284 to 531) and LigIII-DBD (PDB ID 3L2P, residues 168 to 375). (C) Electrostatic surface representation of LigIV-DBD colored, with saturating blue and red at + 7kT/e and −7 kT/e, respectively. A relatively uncharged and exposed surface area on LigIV-DBD is highlighted by a dashed oval. Important amino acids are labeled. See also Figure S1
The main difference between the LigIV DBD and that of LigI and LigIII is a substantially longer helix α2 (Figure 1B). The helix is longer by about two turns compared to LigI and by three full-turns when compared to LigIII (Figure 1B). This extended portion of helix α2 supports an extended hydrophobic surface characterized by partially solvent exposed residues on both helices α2 (F42, W46, F49, and L53) and α1 (V10 and V14) (Figure 1C). The presence of this hydrophobic surface and the extension of helix α2 suggested that they could play a role in the interaction with Artemis.
Structure of LigIV-DBD/cArt-Pep complex
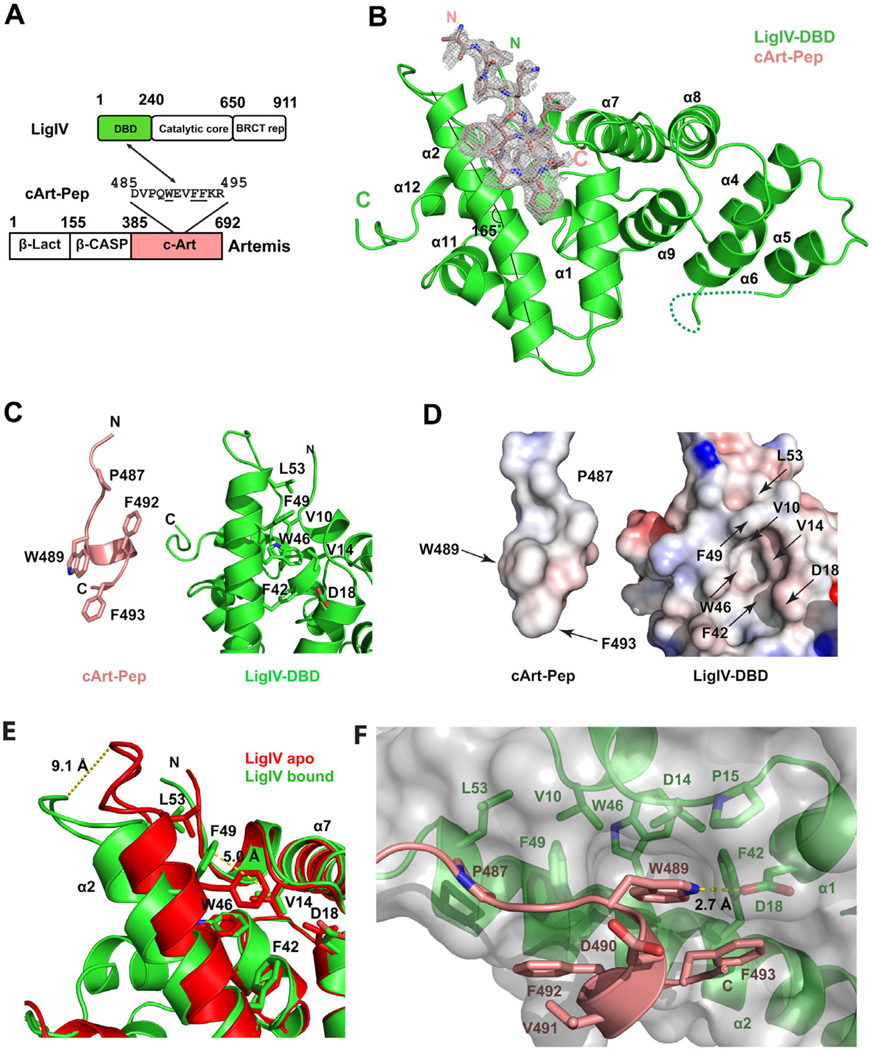
To understand the basis of specificity between LigIV and Artemis, we determined the crystal structure of LigIV DBD in complex with a peptide derived from the C-ter of Artemis (cArt-Pep) (Figures 2A and 2B). The structure, determined at 2.25 Å resolution, contains two complexes (A and B) in the asymmetric unit, which superimpose with an RMSD of 0.54 Å (for 218 Cαs). The cArt-Pep (485-DVPQWEVFFKR-495) is well defined in complex A, with ten of the eleven residues built into the electron density map (Figure 2B). The cArt-Pep is less defined in complex B, with three residues traced in the electron density map. We describe below the structure of complex A.
Figure 2. Crystal structure of LigIV-DBD/cArt-Pep complex.
(A) A schematic of LigIV and Artemis domain organization and of regions that interact. (B) Overall structure of the LigIV-DBD/cArt-Pep complex. LigIV-DBD (green) is represented in ribbon and cArt-Pep (pink) is represented in stick. Simulated annealing 2Fo-Fc omit electron density is also shown for cArt-Pep, contoured at σ=1.0. The axis of helix α2 is marked by two straight lines to indicate the bending. (C) LigIV-DBD and cArt-Pep displayed separately to highlight the important amino acids. (D) Electrostatic potential surface of LigIV-DBD and cArt-Pep when displayed separately. (E) Superposition of apo (red) and peptide bound (green) forms of LigIV-DBD to show the movements in helix α2 and residue F49. The peptide was omitted for clarity. (F) Detailed view of LigIV-DBD/cArt-Pep interaction surface. cArt-Pep and LigIV-DBD are shown in pink and green, respectively. The molecular surface of LigIV-DBD is presented in grey. Hydrogen bonds are depicted in dashed yellow lines. See also Figure S2.
The C-terminal portion of cArt-Pep adopts a helical conformation, supported by two intra-chain hydrogen bonds (N488(O)-V491(N)) and W489(O)-F493(N)). This helical portion of the peptide fits into a shallow hydrophobic pocket on the surface of LigIV DBD, bordered by helices α2 and α1 (Figures 2C and 2D). The N-terminal region of the peptide (residues 485–488) adopts an extended beta-chain conformation that tracks the lengthened portion of helix α2.
The hydrophobic pocket on LigIV DBD is created by an outward movement of helix α2, including a bending of the helical axis at L43 by ~20° (Figure 2B). This results in an outward displacement of the helix α2 C-terminus by ~9 Å, when compared to the apo structure, and creates the space necessary to bind cArt-Pep (Figure 2E). In addition to this gross movement of helix α2, the side chain of F49 moves and reorients by several Angstroms to create the cavity to accommodate W489 of cArt-Pep (Figure 2E). Indeed, W489 is the most deeply buried residue of cArt-Pep and it occupies roughly the position that F49 occupied in the DBD apoenzyme. In addition to interactions with F49, the indole ring of W489 is involved in extensive hydrophobic interactions with V10, V14, F42, and W46 (Figure 2D and 2F). The F42, W46 and F49 aromatic rings pack in a herring-bone pattern against W489. V10 and V14 stem from helix α1 and they complete the hydrophobic cage around W489 of cArt-Pep. A hydrogen bond (2.74 Å) is also observed between the nitrogen of W489 indole ring and the carbonyl group of D18 of LigIV-DBD (Figure 2F). F492 and F493 are on the same side of the helical portion of cArt-Pep as W489, and they too interact with the hydrophobic pocket on the surface of LigIV DBD; in particular, with residues F42 and F49 on helix α2. In addition, the main chain amide of F492 makes a direct hydrogen bond with the side chain of S45. Supplementing these interactions between W489, F492 and F493 on the helical portion of cArt-Pep and the LigIV DBD are van der Waals contacts between P487 on the N-terminal extension of cArt-Pep and L53 from the extended portion of helix α2 of LigIV DBD (Figure 2F).
Taken together, LigIV and Artemis interact via an induced-fit mechanism, wherein helix α2 in LigIV DBD rotates outward to create a hydrophobic pocket for binding a partially helical peptide from the Artemis C-ter. The mainly hydrophobic nature of the interaction is consistent with the stability of the complex at high salt concentrations, up to 500 mM NaCl (data not shown).
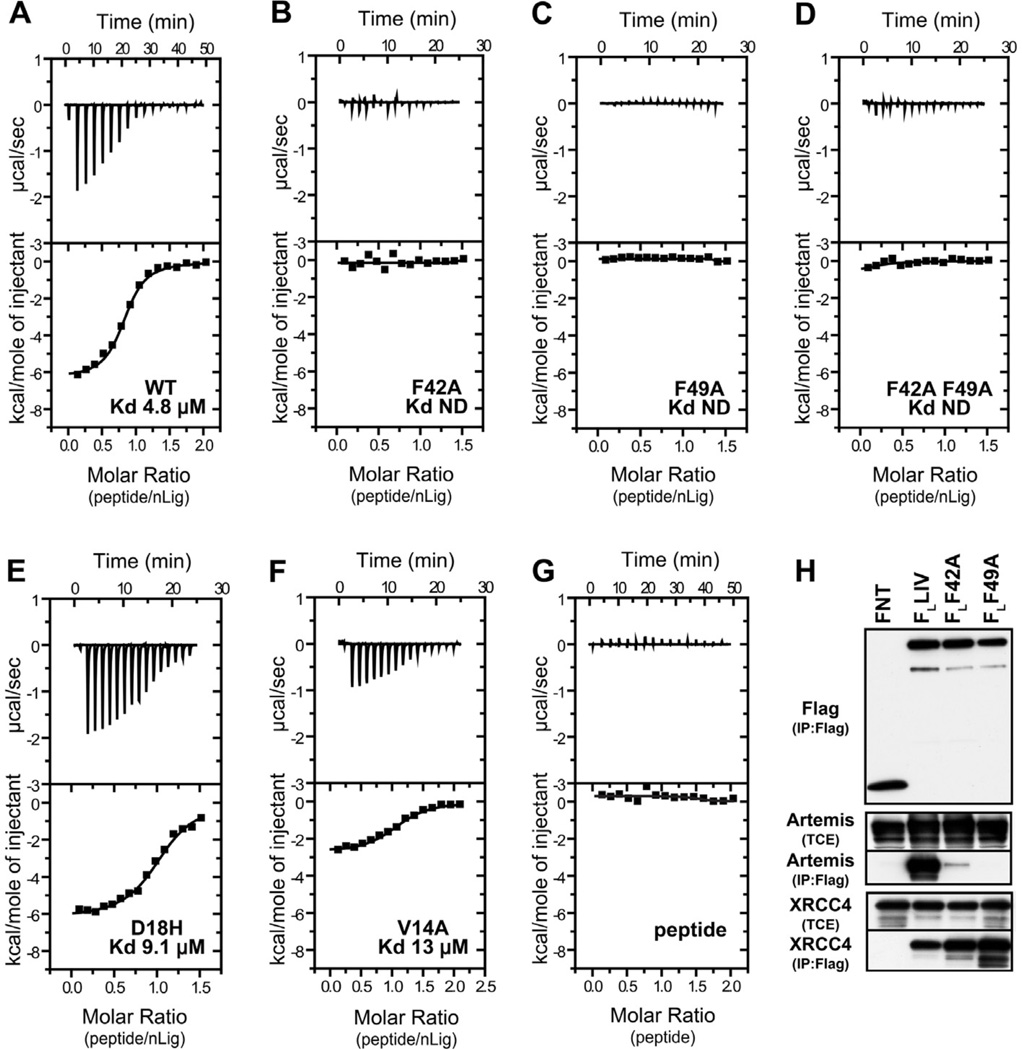
Structural analysis of LigIV-DBD/cArt-Pep complex shows that approximately 400 Å2 of the LigIV-DBD surface area is buried at the interface. To test the interface in solution, we used isothermal titration calorimetry (ITC). Consistent with the crystal structure, wild-type LigIV-DBD binds the cArt-Pep with a Kd of 4.8 µM and a binding stoichiometry of 1:1 (n =1.0 ± 0.1) (Figure 3A). Mutations of F49 and F42 to alanine result in a complete loss of peptide binding (Figure 3B and 3C), consistent with the central role of these phenylalanines on helix α2 in binding to the aromatic residues on cArt-Pep. By contrast, a modest effect on binding is incurred when D18 and V14 on helix α1 are mutated. D18 makes a hydrogen bond with the indole nitrogen of W489 and a D18H mutation increases the Kd to 9.1 µM (Figure 3E), while a V14A mutation increases the Kd to 13.64 µM (Figure 3F). Taken together, the ITC measurements are consistent with the crystal structure in showing the dominant role of F49 and F42 in binding the cArt-Pep.
Figure 3. LigIV-Artemis interaction.
ITC titration profile of cArt-Pep with (A) LigIV-DBD WT, (B) LigIV-DBD F42A, (C) LigIV-DBD F49A, (D) LigIV-DBD F42AF49A, (E) LigIV-DBD D18H, and (F) and, LigIV-DBD V14A. (G) ITC profile for cArt-Pep dilution. (H) Co-IP experiments with full-length and mutant LigIV. Flag IP was performed on lysates of 293T cells transfected with FNT (Flag-NLS-Thioredoxin), Flag Ligase IV (FLLIV) or its point mutants FL42A and FL49A with untagged Artemis. Western blot analysis for Flag, Artemis and endogenous XRCC4 was performed. Levels of Artemis and XRCC4 in TCE (Total Cell Extract) are shown as controls. See also Figure S3.
To examine the role of F42 and F49 amino acids in context of full-length proteins in vivo, we tested the ability of F42A and F49A mutants to interact with Artemis by co-immunoprecipitation. Consistent with the crystal structure and ITC measurements, only wild-type LigIV interacted with Artemis. However, both the wild type and mutant LigIV interacted equally well with XRCC4 (Figure 3H). Together, these data provide further evidence that LigIV interacts with Artemis and XRCC4 via mutually exclusive surfaces: the DBD at the N-terminus interacting with Artemis and the BRCT repeats at the C-terminus engaging XRCC4 (Wu et al., 2009).
Comparison to LigI and LigIII
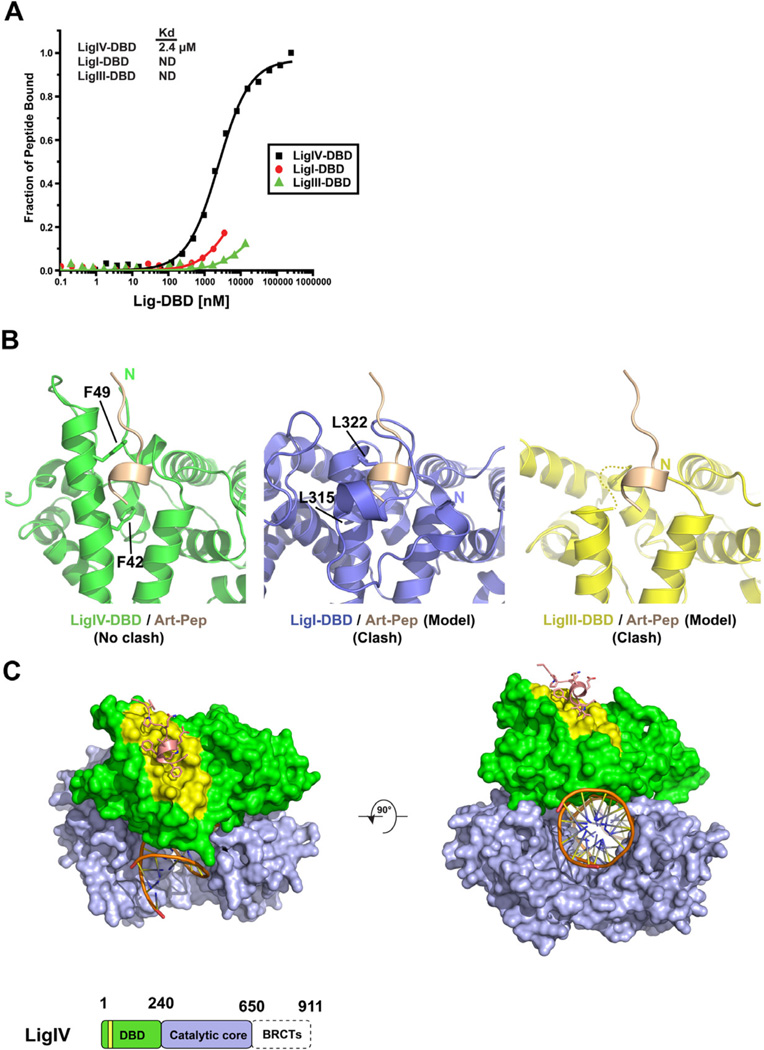
Figure 1B shows the structure of LigIV-DBD superimposed on the structures of LigI-DBD and LigIII-DBD. To examine whether the LigI and LigIII DBDs can also bind cArt-Pep, we performed Florescence Anisotropy (FA) peptide-binding experiments with purified DBDs. As shown in Figure 4A, LigI-DBD and LigIII-DBD bind cArt-Pep with substantially lower affinity than LigIV. Together, these in vitro results are in agreement with the co-immunoprecipitation experiments (Malu et al., 2012), wherein only LigIV-DBD was found to interact with Artemis. Figure 4B shows the cArt-Pep modeled on the LigI-DBD and the LigIII-DBD. In contrast to the LigIV-DBD, there is substantial steric overlap between cArt-Pep and the LigI-DBD and LigIII-DBD. Strikingly, the putative binding site in LigI-DBD is blocked by its unique N-terminal “extension”, which coils around the potential binding site (Pascal et al., 2004). Also, as noted above, helix α2 is shorter by about two turns in LigI-DBD, which decreases the potential binding surface for cArt-Pep by ~60 Å2. Finally, residues F49 and F42 that are essential for LigIV-DBD-cArt-Pep interaction are substituted by smaller L315 and L322 in LigI DBD (Figure S3), which likely diminishes the van der Waals interactions with W489 of cArt-Pep. In LigIII, since helix α2 is shorter by three full-turns compared to LigIV there is no residue equivalent to F49. Indeed, this region of LigIII is disordered in the crystal structure (Figure 4B) (Cotner-Gohara et al., 2010). Taken together, the LigIV DBD differs from the LigI and LigIII DBDs in presenting a hydrophobic surface that is adapted to bind the C-terminal region of Artemis. Both the length of helix α2 and the identity of specific residues on helix α2 contribute to the specificity of LigIV-DBD-cArt-Pep interaction.
Figure 4. Comparison to LigI and LigIII.
(A) Fluorescent anisotropy isothermal binding curves for cArt-Pep against LigIV-DBD, LigI-DBD, and LigIII-DBD. cArt-Pep binds specifically to LigIV-DBD. Because of low affinity, LigI-DBD and LigIII-DBD could not be fully titrated against cArt-Pep and the Kds were not determined (ND) (B) Close up views of LigIV-DBD/cArt-Pep binding (left panel) and models of cArt-Pep docked against LigI-DBD (center panel) and LigIII–DBD (right panel). Note the steric overlap between cArt-Pep and the N-terminal region of LigI-DBD. (C) A model of LigIV catalytic core bound to cArt-Pep (pink) and DNA substrate (orange). LigIV-DBD (green) and catalytic core (NTase and OBD) (light purple) are represented as surfaces. The peptide binding site (yellow) and the DNA binding site are on opposite surfaces. A schematic of LigIV domain organization is shown at the bottom. See also Figure S4.
Discussion
We present here the first molecular details of how LigIV and Artemis interact. We show that LigIV recruits Artemis via a hydrophobic surface that is more extended than in other mammalian DNA ligases. Specifically, a helix (α2) in LigIV DBD is longer than in other mammalian ligases and presents a set of hydrophobic residues that interact selectively with the C-terminal region of Artemis.
Various models for NHEJ have been proposed in recent years, from the sequential recruitment of factors to DSB to the simultaneous recruitment of factors (Dobbs et al., 2010; Ma et al., 2005a; Yano et al., 2009). The structure we present is consistent with the idea that, in presence of Artemis, NHEJ factors are recruited simultaneously to a DSB, leading to the formation of a large complex. Both Artemis and LigIV contain multiple domains and bind other NHEJ factors. For example, in addition of LigIV, the C-terminal region of Artemis also interacts with the kinase DNA-PKcs, leading to both phosphorylation of Artemis and autophosphorylation of the kinase itself (Goodarzi et al., 2006), which promotes DNA-PKcs release from the DNA (Hammel et al., 2010a). The N-terminal region of Artemis is composed of a metallo-β-lactamase fold and forms the catalytic core for its nuclease activity (Callebaut et al., 2002). In LigIV, the DBD at the N-terminus is followed by the catalytic adenylation domain and an oligo-binding (OB) domain, while the C-terminus contains two BRCT repeats. The LigIV’s BRCT repeats are required for interaction with XRCC4 (Sibanda et al, 2001; Wu et al., 2009), which in turn recruits XLF (Cernunnos); a complex with a stoichiometry of 1 BRCTs, 2 XRCC4 and 2 XLF can be isolated by gel filtration (Hammel et al., 2010b). Intriguingly, from our structure, the Artemis C-terminal region (or cArt-Pep) binds to LigIV DBD on the opposite face as the DNA, leaving the N-terminal nuclease domain of Artemis free to engage the DNA substrate in conjunction with the catalytic domain of LigIV (Figure 4C). Such coordination of the nuclease and ligation activities may help to increase the efficiency of the NHEJ reaction, though this remains to be fully tested. XRCC4 and XLF lack enzymatic activity but increase the efficiency of the ligation reaction (Grawunder et al., 1997; Gu et al., 2007). From recent SAXS and crystallographic studies, XRCC4 and XLF can form long filaments and may serve as “holders” of the DNA for the ligation reaction (Hammel et al., 2010b; Hammel et al., 2011; Ropars et al., 2011). Although, a clearer picture of how the Artemis and LigIV enzymatic activities are coordinated will emerge from a structure of the complex with intact proteins, based on the current structure one can begin to construct an initial model of how Artemis and LigIV are recruited together to a DSB (Figure S4).
Along with other damage tolerance pathways, the NHEJ machinery is potentially an important therapeutic target for sensitizing tumor cells to chemotherapy and radiation treatment (Lord and Ashworth, 2012). The LigIV-Artemis structure will aid in the design of small molecule and peptidomimetic inhibitors for increasing the vulnerability of tumor cells to chemotherapy and radiation efficacy. The limited size of the LigIV-Artemis interface and the fact that it is largely a solvent exposed hydrophobic surface, in principle, should facilitate the design of chemical inhibitors for the LigIV-Artemis interaction, analogous to PARP inhibitors for the treatment of certain cancers (Hakimelahi et al., 2001; Lord and Ashworth, 2012).
In conclusion, despite the central role of LigIV in V(D)J recombination and DSB repair, there is limited structural information on this key enzyme. The only atomic structures currently available for LigIV are for BRCTs domains and a subdomain of the nucleotidyltransferase domain (Wu et al., 2009; Ochi et al., 2012), with low resolution EM and SAXS studies also suggesting a degree of heterogeneity and/or flexibility when intact LigIV interacts with XRCC4 (Recuero-Checa et al., 2009; Hammel et al., 2010). The atomic structures we present here are the first for LigIV DBD, revealing a unique DBD that has evolved to bind both DNA and Artemis in a mutually exclusive manner.
Experimental Procedures
Protein and peptide production
The native and Selenomethionine (SeMet) DNA binding domain of LigIV (LigIV-DBD; residues 1–240) were expressed in E. coli. The proteins were purified by nickel affinity chromatography, cation exchange chromatography, and size exclusion chromatography. The DBDs of the human Ligase I (LigI, residues 260 – 535) and the human Ligase III (LigIII, residues 1 – 388) were expressed and purified in the same manner as LigIV-DBD. cArt-Pep encompassing the LigIV interaction motif (DVPQWEVFFKR) was synthesized by Bio-Synthesis Inc (Lewisville, Texas).
Crystallization and Data collection
LigIV-DBD was crystallized from solutions containing 16 % PEG 3350 and 200 mM (NH4)3PO4, at 20°C. SeMet LigIV-DBD crystals were obtained under similar conditions as the native protein except that (NH4)2SO4 replaced NaCl in the crystallization mix. The LigIV-DBD/cArt-Pep co-crystals were obtained from solutions containing 18% PEG 1000 and 200 mM Tris-HCl pH 8.0. All crystals were cryo-protected in paratone-N oil (Hampton research), and flash cooled to 90° K. All data sets were measured at the Brookhaven National Laboratory (BNL; beamline X6A) and the Advanced Photon Source (APS; beamline 24-ID). Due to significant anisotropy in LigIV-DBD/cArt-Pep diffraction, the data were truncated and anisotropic scaled by the Diffraction Anisotropy Server (Strong et al., 2006).
Structure determination and refinement
LigIV-DBD apo structure was solved by MAD phasing, followed by density modification. The initial model was refined in program Phenix with strict geometric and B-factor restraints (Adams et al., 2009), followed by several rounds of refinement that included simulated annealing, energy minimization, geometry optimization, TLS refinement, and manual rebuilding as needed. The final apo LigIV-DBD model contains 212 amino acids and 2 phosphate ions with an Rfactor of 21.7% and Rfree of 25.6% at 2.8 Å resolution (Table S1).
The structure of LigIV-DBD/cArtPep complex was solved by molecular replacement (MR) by the program PHASER (McCoy et al., 2007), using the apo LigIV-DBD as a search model. The structure was refined in the same manner as the apo LigIV-DBD. The final model contains 2 LigIV-DBD/cArt-Pep complexes in the asymmetric unit, complex A (aa 7-112 and 122-237 of LigIV DBD and aa 485-494 of Artemis) and complex B (aa 8–113 and 123–237 of LigIV DBD and 487–489 of Artemis).
Interestingly, because of crystal contacts, helices α10 and α11 at the C-terminus take different paths in the apo LigIV DBD and the LigIV DBD/cArt-Pep structures. The topology α10 and α11 in apo LigIV DBD differs from that observed in the LigI/DNA, LigIII/DNA, and LigIV/cArt-Pep crystal structures due to the crystal contacts these helices in apo LigIV DBD structure make with symmetry related molecules.
Peptide binding experiments
Peptide binding experiments were performed by mixing 6-FAM (6-Caboxyfluorescein) labeled cArt-Pep (5 nM final concentration) with increasing concentrations of Lig-DBDs (0.1 nM – 300 µM) in a peptide dilution buffer.
Isothermal titration calorimetry (ITC) measurements
Calorimetric measurements were performed with the MicroCal ITC200 instrument (GE healthcare) in the temperature range of 5 – 25°C.
Transfections and flag immunoprecipitation from 293T cells
HEK 293T cells were transfected using the Calcium Phosphate method. Cellular extracts were incubated with 200 µg/ml EtBr at 4°C. The cell lysate (supernatant) obtained after centrifugation was used for Flag IP using anti Flag M2-agarose beads (Sigma) and proteins were eluted with 0.2 µg/ml Flag peptide (Sigma) in Buffer C (150 mM KCl) (Ma et al., 2002). Western blot analysis was performed at least two times from independent transfections and representative results are shown.
Supplementary Material
First crystal structure of Ligase-IV-Artemis complex
Ligase IV DNA binding domain contains a unique hydrophobic cavity
The structure shows why only Ligase IV can recruit Artemis
Insights into how activities of Ligase IV and Artemis may be coordinated in NHEJ
Acknowledgements
We thank the staff at BNL (beamline X6A) and APS (beamline 24-ID) for facilitating X-ray data collection. We thank W. Hendrickson and F. Ferrage for discussions, and C. Escalante for help with the ITC experiments. Work in the P. Cortes laboratory is supported in part by grants R01 AI080755 and R01 AI070532.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
The Protein Data Bank accession numbers for the atomic coordinates and structure factors are 4HTO (LigIV DBD) and 4HTP (LigIV DBD/Artemis).
Supplemental Information
Supplemental information includes extended experimental procedure, four figures, and a table.
References
- Adams P, Afonine P, Bunkoczi G, Chen V, Davis I, Echols N, Headd J, Hung L, Kapral G, Grosse-Kunstleve R, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2009;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- Callebaut I, Moshous D, Mornon J-P, de Villartay J-P. Metallo-beta-lactamase fold within nucleic acids processing enzymes: the beta-CASP family. Nucleic Acids Res. 2002;30:3592–3601. doi: 10.1093/nar/gkf470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Trujillo K, Ramos W, Sung P, Tomkinson AE. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Molecular Cell. 2001;8:1105–1115. doi: 10.1016/s1097-2765(01)00388-4. [DOI] [PubMed] [Google Scholar]
- Chen L, Trujillo K, Sung P, Tomkinson AE. Interactions of the DNA ligase IV-XRCC4 complex with DNA ends and the DNA-dependent protein kinase. The J Biol Chem. 2000;275:26196–26205. doi: 10.1074/jbc.M000491200. [DOI] [PubMed] [Google Scholar]
- Chistiakov DA. Ligase IV syndrome. Adv Exp Med Biol. 2010;685:175–185. doi: 10.1007/978-1-4419-6448-9_16. [DOI] [PubMed] [Google Scholar]
- Cotner-Gohara E, Kim I-K, Hammel M, Tainer JA, Tomkinson AE, Ellenberger T. Human DNA ligase III recognizes DNA ends by dynamic switching between two DNA-bound states. Biochemistry. 2010;49:6165–6176. doi: 10.1021/bi100503w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Critchlow SE, Bowater RP, Jackson SP. Mammalian DNA double-strand break repair protein XRCC4 interacts with DNA ligase IV. Current biology : CB. 1997;7:588–598. doi: 10.1016/s0960-9822(06)00258-2. [DOI] [PubMed] [Google Scholar]
- Dobbs TA, Tainer JA, Lees-Miller SP. A structural model for regulation of NHEJ by DNA-PKcs autophosphorylation. DNA Repair (Amst) 2010;9:1307–1314. doi: 10.1016/j.dnarep.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouet J, Delteil C, Lefrancois J, Concannon P, Salles B, Calsou P. DNA-dependent protein kinase and XRCC4-DNA ligase IV mobilization in the cell in response to DNA double strand breaks. J Biol Chem. 2005;280:7060–7069. doi: 10.1074/jbc.M410746200. [DOI] [PubMed] [Google Scholar]
- Ellenberger T, Tomkinson AE. Eukaryotic DNA ligases: structural and functional insights. Annu Rev Biochem. 2008;77:313–338. doi: 10.1146/annurev.biochem.77.061306.123941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi AA, Yu Y, Riballo E, Douglas P, Walker SA, Ye R, Härer C, Marchetti C, Morrice N, Jeggo PA, et al. DNA-PK autophosphorylation facilitates Artemis endonuclease activity. EMBO J. 2006;25:3880–3889. doi: 10.1038/sj.emboj.7601255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grawunder U, Wilm M, Wu X, Kulesza P, Wilson TE, Mann M, Lieber MR. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- Grawunder U, Zimmer D, Fugmann S, Schwarz K, Lieber MR. DNA ligase IV is essential for V(D)J recombination and DNA double-strand break repair in human precursor lymphocytes. Molecular Cell. 1998;2:477–484. doi: 10.1016/s1097-2765(00)80147-1. [DOI] [PubMed] [Google Scholar]
- Gu J, Lu H, Tsai AG, Schwarz K, Lieber MR. Single-stranded DNA ligation and XLF-stimulated incompatible DNA end ligation by the XRCC4-DNA ligase IV complex: influence of terminal DNA sequence. Nucleic Acids Res. 2007;35:5755–5762. doi: 10.1093/nar/gkm579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakimelahi GH, Ly TW, Moosavi-Movahedi AA, Jain ML, Zakerinia M, Davari H, Mei HC, Sambaiah T, Moshfegh AA, Hakimelahi S. Design, synthesis, and biological evaluation of novel nucleoside and nucleotide analogues as agents against DNA viruses and/or retroviruses. J Med Chem. 2001;44:3710–3720. doi: 10.1021/jm010216r. [DOI] [PubMed] [Google Scholar]
- Hammel M, Yu Y, Mahaney BL, Cai B, Ye R, Phipps BM, Rambo RP, Hura GL, Pelikan M, So S, et al. Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. J. Biol. Chem. 2010;285:1414–1423. doi: 10.1074/jbc.M109.065615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammel M, Yu Y, Fang S, Lees-Miller SP, Tainer JA. XLF Regulates Filament Architecture of the XRCC4·Ligase IV Complex. Structure. 2010;18:1431–1442. doi: 10.1016/j.str.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammel M, Rey M, Yu Y, Mani RS, Classen S, Liu M, Pique ME, Fang S, Mahaney BL, Weinfeld M, et al. XRCC4 protein interactions with XRCC4-like factor (XLF) create an extended grooved scaffold for DNA ligation and double strand break repair. J Biol. Chem. 2011;16:32638–32650. doi: 10.1074/jbc.M111.272641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Chang C, Song HK, Moon J, Yang JK, Kim HK, Kwon ST, Suh SW. Crystal structure of NAD(+)-dependent DNA ligase: modular architecture and functional implications. EMBO J. 2000;19:1119–1129. doi: 10.1093/emboj/19.5.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR. The Mechanism of Double-Strand DNA Break Repair by the Nonhomologous DNA End-Joining Pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481:287–294. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
- Ma Y, Lu H, Schwarz K, Lieber MR. Repair of double-strand DNA breaks by the human nonhomologous DNA end joining pathway: the iterative processing model. Cell Cycle. 2005a;4:1193–1200. doi: 10.4161/cc.4.9.1977. [DOI] [PubMed] [Google Scholar]
- Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- Ma Y, Schwarz K, Lieber MR. The Artemis:DNA-PKcs endonuclease cleaves DNA loops, flaps, and gaps. DNA Repair (Amst) 2005b;4:845–851. doi: 10.1016/j.dnarep.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Malu S, De Ioannes P, Kozlov M, Greene M, Francis D, Hanna M, Pena J, Escalante CR, Kurosawa A, Erdjument-Bromage H, et al. Artemis C-terminal region facilitates V(D)J recombination through its interactions with DNA Ligase IV and DNA-PKcs. J Exp Med. 2012;209:955–963. doi: 10.1084/jem.20111437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A, Grosse-Kunstleve R, Adams P, Winn M, Storoni L, Read R. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Snowden CM, McCarville J, Ramsden DA. Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol Cell Biol. 2000;20:2996–3003. doi: 10.1128/mcb.20.9.2996-3003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochi T, Wu Q, Chirgadze DY, Grossmann JG, Bolanos-Garcia VM, Blundell TL. Structural Insights into the Role of Domain Flexibility in Human DNA Ligase IV. Structure. 2012;20:1212–1222. doi: 10.1016/j.str.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal JM, O'Brien PJ, Tomkinson AE, Ellenberger T. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature. 2004;432:473–478. doi: 10.1038/nature03082. [DOI] [PubMed] [Google Scholar]
- Ropars V, Drevet P, Legrand P, Baconais S, Amram J, Faure G, Marquez JA, Pietrement O, Guerois R, Callebaut I, et al. Structural characterization of filamnets formed by human Xrcc4-Cernunnos/XLF complex involved in nonhomologous DNA end joining. Proc. Natl. Acad. Sci. 2011;108:12663–12668. doi: 10.1073/pnas.1100758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rucero-Checa MA, Sore AS, Arias-Palomo E, Rivera-Cazada A, Scheres SHW, Maman JD, Pearl LH, Llorca O. Electron microscopy of Xrcc4 and the DNA ligase-Xrcc4 DNA repair complex. DNA Repair. 2009;8:1380–1389. doi: 10.1016/j.dnarep.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Sibanda BL, Critchlow SE, Begun J, Pei XY, Jackson SP, Blundell TL, Pellegrini L. Crystal structure of an Xrcc4-DNA ligase IV complex. Nature Struct. Biol. 2001;8:1015–1019. doi: 10.1038/nsb725. [DOI] [PubMed] [Google Scholar]
- Sharpless NE, Ferguson DO, O'Hagan RC, Castrillon DH, Lee C, Farazi PA, Alson S, Fleming J, Morton CC, Frank K, et al. Impaired nonhomologous end-joining provokes soft tissue sarcomas harboring chromosomal translocations, amplifications, and deletions. Molecular Cell. 2001;8:1187–1196. doi: 10.1016/s1097-2765(01)00425-7. [DOI] [PubMed] [Google Scholar]
- Simsek D, Furda A, Gao Y, Artus J, Brunet E, Hadjantonakis A-K, Van Houten B, Shuman S, McKinnon PJ, Jasin M. Crucial role for DNA ligase III in mitochondria but not in Xrcc1-dependent repair. Nature. 2011;471:245–248. doi: 10.1038/nature09794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong M, Sawaya MR, Wang S, Phillips M, Cascio D, Eisenberg D. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P-Y, Frit P, Meesala S, Dauvillier S, Modesti M, Andres SN, Huang Y, Sekiguchi J, Calsou P, Salles B, et al. Structural and functional interaction between the human DNA repair proteins DNA ligase IV and XRCC4. Mol Cell Biol. 2009;29:3163–3172. doi: 10.1128/MCB.01895-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K, Morotomi-Yano K, Adachi N, Akiyama H. Molecular mechanism of protein assembly on DNA double-strand breaks in the non-homologous end-joining pathway. J Radiat Res (Tokyo) 2009;50:97–108. doi: 10.1269/jrr.08119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.