Abstract
Background
Blood vessel formation is important for many physiological and pathological processes, and is therefore a critical target for drug development. Inhibiting angiogenesis to starve a tumor or promoting “normalization” of tumor blood vessels in order to facilitate delivery of anticancer drugs are both areas of active research. Recapitulation of vessel formation by human cells in vitro allows investigating cell-cell and cell-matrix interactions in a controlled environment, and is thereby a crucial step in developing high content (HC) and high throughput (HT) screening assays to search for modulators of blood vessel formation. Human umbilical vein endothelial cells (HUVECs) exemplify primary cells used in angiogenesis assays. However, primary cells have significant limitations that include phenotypic decay and/or senescence by 6–8 passages in culture, making stable integration of fluorescent markers and large-scale expansion for high throughput screening problematic. To overcome these limitations for HTS, we developed a novel angiogenic model system that employs stable fluorescent endothelial cell lines based on immortalized human microvascular endothelial cells (HMEC-1, hereafter HMECs). We then evaluated HMEC cultures, both alone and co-cultured with an epicardial mesothelial cell (EMC) line that contributes vascular smooth muscle cells, to determine suitability for HTS or HCS.
Results
The endothelial and epicardial lines were engineered to express a panel of nuclear- and cytoplasm-localized fluorescent proteins to be mixed and matched to suit particular experimental goals. HMECs retained their angiogenic potential and stably expressed fluorescent proteins for at least 13 passages after transduction. Within 8 hours upon plating on Matrigel, the cells migrated and coalesced into networks of vessel-like structures. If co-cultured with EMCs, the branches formed cylindrical-shaped structures of HMECs surrounded by EMC-derivatives reminiscent of vessels. Network formation measurements revealed responsiveness to media composition and control compounds.
Conclusions
HMEC-based lines retain most of the angiogenic features of primary endothelial cells, yet possess long-term stability and ease of culture, making them intriguing candidates for large-scale primary HC and HT screening (of ~10,000–1,000,000 molecules). Furthermore, inclusion of EMCs demonstrates the feasibility of using epicardial-derived cells, which normally contribute to smooth muscle, to model large vessel formation. In summary, the immortalized fluorescent HMEC and EMC lines and straightforward culture conditions will enable assay development for HCS of angiogenesis.
Keywords: Angiogenesis, microvascular, fluorescent proteins, time-lapse microscopy of live cells in culture
INTRODUCTION
Blood vessels are critical for embryonic development, tumor growth and metastasis, as well as inflammation-related disorders, including those associated with obesity, diabetes, and ischemia/reperfusion injuries (reviewed in (Carmeliet, 2005) and (Carmeliet, 2003). However, current methods of modulating angiogenesis are not meeting clinical goals, reinforcing the need for additional drug targets to inhibit or create functional vessels. For example, inhibiting angiogenesis to starve a tumor with drugs that target VEGF (Vascular Endothelial Growth Factor) or its receptors does not provide long term improvement for cancer patients, and only mildly prolongs survival, even when used in combination with chemotherapy (reviewed in (Loges et al., 2009), and consideration has recently been given to the idea that strong inhibition of angiogenesis might hinder drug delivery and promote cancer cell selection towards higher malignancy and metastatic potential (Paez-Ribes et al., 2009). Consequently, novel combination therapies of existing drugs are being explored (e.g. (Bergers et al., 1999; Jendreyko et al., 2005). However, the discovery of new drugs with varied modes of action and the development of novel therapies to promote or inhibit blood vessel formation is severely limited by the lack of quantifiable, high throughput assays.
Increasingly successful generation of vessel-like structures in vitro for models of angiogenesis and advances in high content screening, wherein evolving automated microscopy and computer vision are enabling assays of even more complex cellular models and phenotypic screens, led us to ask if primary screening of blood vessel formation is possible. Since preliminary approaches to automating vessel formation measurements have appeared promising, the major limitation in developing angiogenesis assays for large-scale, high throughput applications has been the reliance on primary cells such as HUVECs that exhibit the needed phenotypic characteristics. Primary cells have a limited replicative capacity and lose their phenotypic potential for vessel formation after several passages in culture. Limited life span precludes making stable cell lines engineered with stable fluorescent reporters for assays, and the alternative use of chemical dyes causes significant photochemical damage, thus limiting most screens to end-point assays where the cells are fixed and stained at the end of the experiment. For analysis, fluorescence labeling has been required thus far and even if vessel formation measurements of bright field images becomes possible, corresponding specific labeling of key vessel-formation components for revealing mechanistic components do not exist. Alternatively, transient transduction of primary cells with viral-based vectors may be possible (Evensen et al., 2010), but is cumbersome for HTS and may increase assay to assay variation.
Here we describe development of a novel immortalized cell-based model system and evaluation of its suitability for future scale-up for high throughput analysis of angiogenesis to screen for modulators of vessel formation and function. Human microvascular endothelial cells (HMEC-1) (Ades et al., 1992) were stably labeled with nuclear fluorescent proteins. The fluorescent HMECs retain most of the angiogenic features of primary endothelial cells, yet are stable in culture. Co-culture with epicardial mesothelial cells (EMCs), which can differentiate into smooth muscle cells and pericytes (Eid et al., 1992), create realistic in vitro vessel-like structures, including inner endothelial and outer smooth muscle layers that resemble the intima and media of developing vessels.
RESULTS
Expression of endothelial cells receptors and response to TNFα in HMEC-1 cells
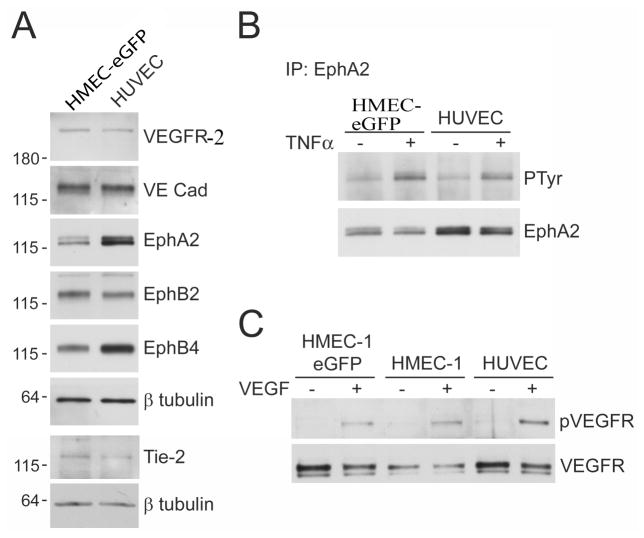
The immortalized HMEC-1 cells have been extensively characterized previously (Ades et al., 1992; Xu et al., 1994; Bouis et al., 2001; Unger et al., 2002; Bender et al., 2008 and references therein). We also analyzed the expression of key markers in our HMEC lines expressing eGFP to ensure that the fluorescently labeled cells still possessed endothelial characteristics after being transduced. We compared HMEC-eGFP cells to HUVECs, the cell type most commonly used in angiogenesis assays, for expression of proteins that play important roles in the development and function of blood vessels, such as endothelial growth factor receptor (VEGFR-2), vascular endothelial (VE) cadherin, Tie-2 and the Eph receptors (EphA2, EphB2, and EphB4), (Yancopoulos et al., 2000; Harris and Nelson, 2010). We found that VEGFR-2, VE cadherin and EphB2 were expressed at similar levels in HMEC-eGFP cells and HUVECs, while expression of EphA2 and EphB4 was slightly lower and Tie-2 expression slightly higher in HMEC-eGFP cells compared to HUVECs (Fig. 1A). Importantly, in HMEC-eGFP cells the EphA2 receptor retains the ability to become phosphorylated (Fig. 1B) on tyrosine residues (i.e. activated) after stimulation of the cells with tumor necrosis factor α (TNFα), an angiogenic factor that is known to cause upregulation of the ephrin-A1 ligand for EphA2 in HUVEC cells (Pandey et al., 1995). In addition, VEGFR-2 becomes similarly phosphorylated in HMECs, both in the non-fluorescent parental and in the eGFP expressing cell lines, as compared to HUVECs after VEGF stimulation (Fig. 1C). These results suggest that HMECs have properties comparable to primary endothelial cells, making them a suitable model cell line for angiogenesis studies.
Figure 1.
Expression of endothelial cells receptors and response to TNFa and VEGF in HMECs and HUVECs. (A) VEGFR-2, VE cadherin, Tie-2, EphA2, EphB2, and EphB4 were detected by immunoblotting with specific antibodies in HMEC-eGFP and HUVEC cell lysates. Reprobing for β-tubulin verified that similar amounts of protein were loaded for the two cell types. (B) HMEC-eGFP cells and HUVECs were left unstimulated or stimulated with TNFα for 2 hours. EphA2 immunoprecipitates were probed with anti-phosphotyrosine antibody (PTyr) and reprobed for EphA2. As estimated from the relative band intensities, the stimulation with TNFα resulted in 2.8 and 2.7 fold increase of PTyr labeling in HMECs and HUVECs, respectively. (C) Parental HMECs, eGFP expressing HMECs and HUVECs were left unstimulated or stimulated with VEGF for 15 min. Cell lysates were probed with anti-phospho-VEGF receptor and reprobed for VEGFR-2.
Characterization of vessel-like network formation by HMECs in comparison to HUVECs
We studied the timeline of network formation exhibited by HMECs and HUVECs in prolonged Matrigel based assays in 12-hour experiments (Fig. 2). Within the first few hours after plating, single scattered cells began to converge into clusters, and by 5 hours they formed proto-networks with many protrusions connecting the cell groupings (Figs. 2A, B). Time-lapse movies revealed active dynamics within the networks, with branches thickening and condensing and new connections continuing to appear (Supplemental Movie 1). During the next few hours, both cell types demonstrated network remodeling. This involved condensing/merging of protrusions between the cellular groupings as cells organized into fewer, thicker branches and appearance of distinct nodes (points where cellular branches converge/diverge) in both HUVECs and HMECs. Finally, after 9–12 hours in culture network remodeling slowed down and minimal changes were observed within the networks (Figs. 2A, B). For the purposes of this study we called these quiescent and stable networks “mature networks”. Networks formed by both cell types typically began deteriorating approximately 20–24 hours post plating (data not shown).
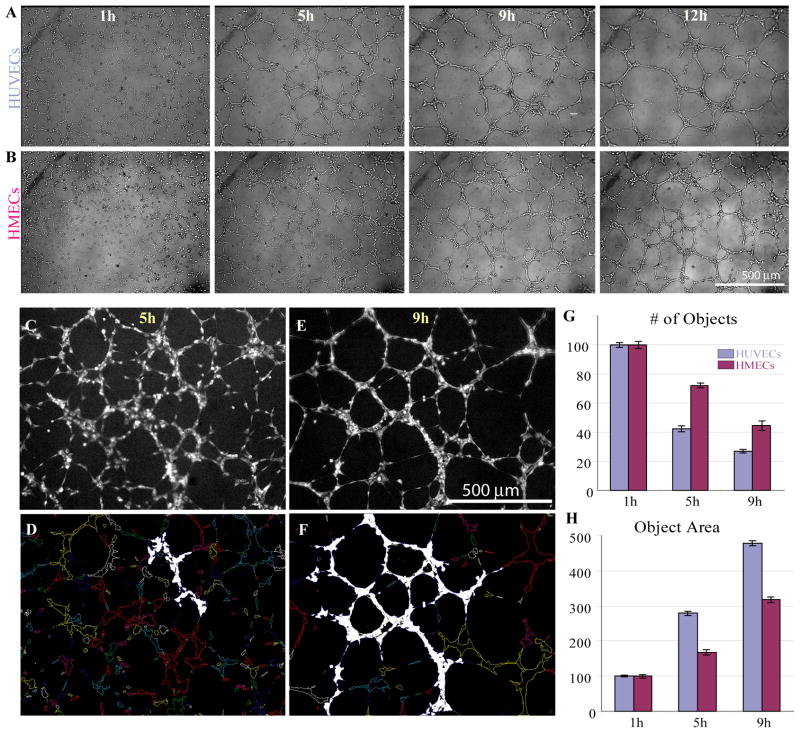
Figure 2.
Networks formed by HMECs and HUVECs in culture are morphologically similar. Brightfield images (corresponding to 1h, 5h, 9h and 12h time points) taken from time-lapse movies of HUVECs (A) and HMECs (B) forming vascular networks on Matrigel. Sample fluorescent images of CellTracker Red-labeled HMEC cells at 5h (C) and 9h (E) are shown along with the corresponding object outlines (D, F, respectively), generated by MetaMorph. The largest “objects” (1 per image) are highlighted in white. (G, H) Quantification of network parameters using MetaMorph Integrated Morphometry Analysis module. For this analysis, number of objects and their average area were quantified, for each field of view (corresponding to the central region of the well), averaged for at least 16 wells and normalized to the 1h timepoints, separately for HMECs and HUVECs. Data are presented as mean +/− SEM.
We quantified network formation (Figs. 2C-H) using the MetaMorph Integrated Morphometry Analysis module, which segments and characterizes individual objects within fields of view (Figs. 2D, F). As cells formed clusters that eventually developed into connected networks, the number of individual objects decreased (Fig. 2G) while the size of each object increased (Fig. 2H). We also manually estimated the degree of network formation in Figs. 2C, D to show that it matched the trends revealed by the MetaMorph analysis (Supplemental Table 1). Although both cell types demonstrated similar dynamics and vascularization trends, they were not identical: HUVECs tended to form networks of fewer, thicker branches with slightly larger mesh size, while HMECs had a higher concentration of vascular nodes (resulting in more cellular branches) and slightly thinner branches that were more difficult to segment as one connected object (Figs. 2G, H). These differences in network formation might be due to the intrinsic differences of each cell type. HUVECs are derived from large umbilical veins, while HMECs are of microvascular origin. Nevertheless, HMECs and HUVECs exhibited similar network formation timeline and the networks formed by both cell types reached the maximal maturation between 8–12 hours with the typical connected object area for both cell types ranging between 300–600 μm2.
Modulation of network formation dynamics by medium composition
We then investigated the effects of medium composition on network formation by HMECs and found substantial differences between control morphogenesis and that occurring in the absence of growth factors or in the presence of drugs commonly used in angiogenic research (Fig. 3). The most interconnected and robust networks formed in complete EGM2-MV medium, which is specially formulated for microvascular cells. To compare data obtained in different experiments, we normalized the number of contiguous objects (as defined in Fig. 2) to that formed in EGM2-MV wells on each particular plate. Thus, the greater the number of objects, the more broken and discontinuous is the network. EGM2 (a complete medium formulated for macrovascular cells such as HUVECs) also supported good network formation. However, the networks formed in EGM2-MV matured earlier (at around 4 hours) and persisted later, until 8–12 hours, whereas networks in EGM2 matured at around 8 hours and began to deteriorate soon afterwards (data not shown). Thus, EGM2-MV is the preferred medium for HMECs because it stimulates the best network formation, both in terms of timing and morphological characteristics.
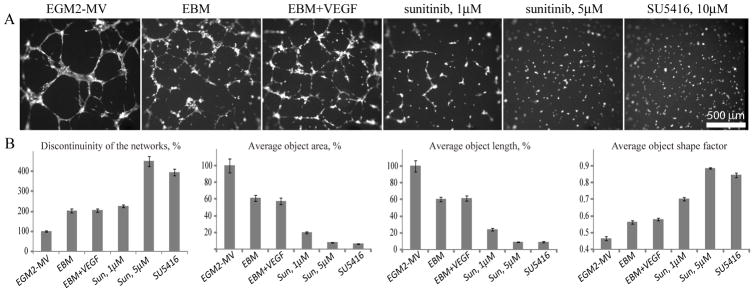
Figure 3.
Network formation by HMECs depends on VEGF signaling. (A) Representative pictures of networks formed by HMECs in multiple wells of a 384 well plate in various media (as indicated) for approximately 8–9 hours. (B) Quantitation of the network formation using MetaMorph Integrated Morphometry Analysis. Discontinuinity of the networks (expressed as number of continuous objects), average object area, length and shape factor are all normalized to EGM2-MV control condition and presented as average +/−SEM (N=27 wells per condition).
Network formation by HMECs depends on VEGF signaling
In order to investigate the VEGF requirements for network formation, we first established that the endothelial basal medium (EBM) not supplemented with extracellular factors supported only the initial formation of networks that never fully developed and remained mostly as uncondensed, disconnected branching clusters (Fig. 3). Analysis of the time-lapse movies revealed that these immature networks usually start breaking down by 12 hours in culture, thus confirming the need for extracellular factors (Supplemental Movie 2). Such a basal medium would be useful for assays of network promoting activity.
Supplementing EBM with VEGF at concentrations up to 240 ng/ml did not result in significant improvement of the network formation, as judged by visual observation and a number of network quantitation parameters (Fig 3B). HMECs have previously been shown to express low levels of VEGF (Frick et al., 2003; Loboda et al., 2006) and, therefore, their inability to complete network formation in the basal medium even with supplementary VEGF, could reflect their dependence on other factors present in the complete medium (such as FGF, IGF, EGF, ascorbic acid and FBS).
Although HMECs may not need exogenous VEGF, we found (Fig. 3) that network formation was strongly inhibited when VEGF signaling was perturbed by either receptor tyrosine kinase inhibitor sunitinib (reviewed by Chow and Eckhardt, 2007) or SU5416, specific VEGFR inhibitor (Fong et al., 1999). Thus, network formation in HMECs strongly depends on VEGF signaling.
Network formation assay can discriminate among pharmacologically diverse compounds
The ability to discriminate among structurally diverse compounds is a requisite for a screening assay. With EBM and additions of sunitinib and SU5416 to EGM2-MV having demonstrated network formation inhibition, we tested several other conditions. DMEM supplemented with 10% FBS did not support network formation and, instead, resulted in large numbers of small disconnected clusters of cells (Figs. 4A, B). The alkaloid vinblastine, which inhibits assembly of microtubules (Rai and Wolff, 1996), also inhibited network formation at a concentration of 50 nM, probably as a consequence of reduced migration (Figs. 4A, B). Similarly, 30 nM suramin, which inhibits angiogenesis by blocking the binding of growth factors (such as PDGF, FGF and VEGF) to their receptors (Waltenberger et al., 1996; Gagliardi et al., 1998a) and through other less characterized mechanisms (Gagliardi et al., 1998b; McCain et al., 2004), also inhibited branching morphogenesis in the HMEC assay, causing the formation of multicellular clusters that often merged together in the middle of the well forming one or several large aggregates (Fig. 4C, Supplemental Movie 3). Since the decrease in number of objects would not be a good metric of network formation when large cell aggregates form, we quantified suramin-induced inhibition using the combination of MetaMorph’s Angiogenesis Tube module and Integrated Morphometry analysis tool, reporting the shape factor, length and outer radius of the objects (Fig. 4D).
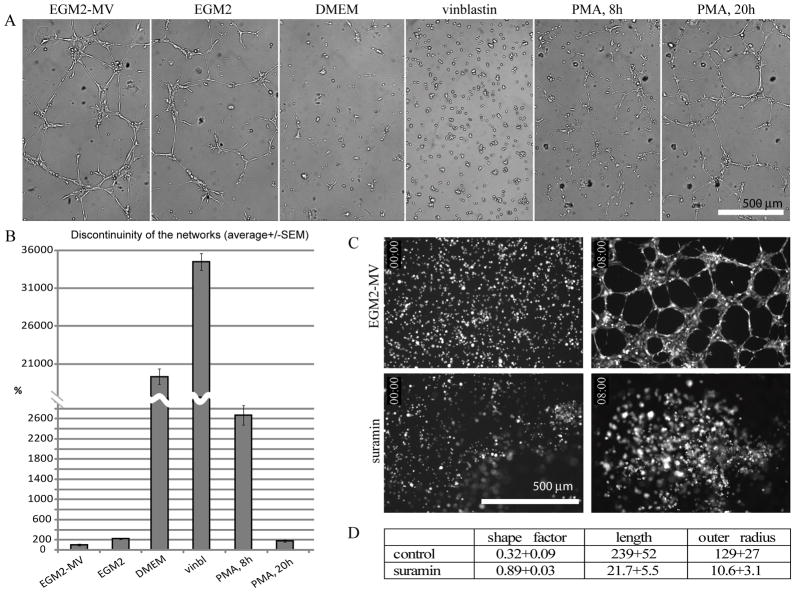
Figure 4.
Media effects on network formation by HMEC cells. (A) Representative pictures of networks formed by HMECs in multiple wells of a 96 well plate in various media for approximately 8–9 hours (except for the right-most PMA image which was taken at 20h post-plating). (B) Quantitation of the network formation using MetaMorph Integrated Morphometry Analysis. Discontinuinity of the networks is expressed as number of continuous objects normalized to EGM2-MV positive control condition and presented as average +/−SEM, minimum of 16 wells were analyzed per condition. Well formed, mature networks consist of small number of large objects, while disrupted networks and cell clusters are characterized by a larger number of disconnected smaller objects. (C, D) Quantitation of suramin effects on network formation. (C) Control cells (top panels) and cells treated with suramin (bottom panels), time points 0 and 8 hours are shown. (D) The parameters of cell structures formed after 8 hours were quantified using MetaMorph Integrated Morphology Analysis Module; comparisons of shape factor, length (μm) and outer radius (μm) for control and suramin are presented.
Phorbol-12-myristate-13-acetate (PMA), which activates certain protein kinase C isoforms (Goel et al., 2007), delayed formation of the networks (at 100 ng/ml in EGM2-MV) and thus caused network discontinuity at 8 hours (i.e., the cells were still in discontinuous branched clusters), which resolved at 20 hours (Fig. 4B). The networks matured more slowly – typically by 12–14 hours, but persisted longer – for over 30 hours post-plating (~10 hours after control networks begin deteriorating).
To evaluate the robustness of the effects of inhibitors and suitability of our assay for HCS applications, we calculated Z′ values, which range from -∞ to 1.0 and for which Z′ > 0.2 is considered sufficient statistical significance for HTS (Zhang et al., 1999). For control versus DMEM, vinblastine and suramin in 96-well plates, networks parameters generated 0.35 ≤ Z′ ≤ 0.67 (Table 1, best Z′ for each condition). With sufficient statistical significances for HTS in 96-well plates, we miniaturized the angiogenic assay to the 384-well format for the well-defined anti-angiogenic compounds sunitinib and SU5416 and obtained 0.61 ≤ Z′ ≤ 0.87 (Table 1). These Z′ results provide strong evidence for the suitability of this assay for scaling up to HTS.
Table 1.
Z′ data for HCS assay validation. The table presents sample Z′ values obtained from several representative experiments (minimum of 16 wells in a 96 well plate and minimum of 27 wells in a 384 well plate per condition) using complete EGM2-MV medium as the positive control and DMEM or EBM (basal) media, or EGM2-MV medium supplemented with vinblastine, suramin, sunitinib or SU5416 as negative controls. The images were analyzed using MetaMorph Integrated Morphometry Analysis and Angiogenesis Tube Formation modules. Shaded cells, data not available.
| format | treatment | # of objects | Object area | Object length | Shape factor | Total tube length | Total tube area | Outer radius |
|---|---|---|---|---|---|---|---|---|
| 96 well plate | DMEM | 0.35 | ||||||
| Vinblastine | 0.41 | 0.67 | ||||||
| Suramin | 0.37 | 0.31 | 0.44 | 0.25 | ||||
| 384 well plate | EBM | 0.61 | 0.07 | 0.4 | 0.29 | 0.45 | ||
| Sunitinib | 0.76 | 0.73 | 0.85 | 0.87 | 0.85 | |||
| SU5416 | 0.79 | 0.73 | 0.84 | 0.79 | 0.85 |
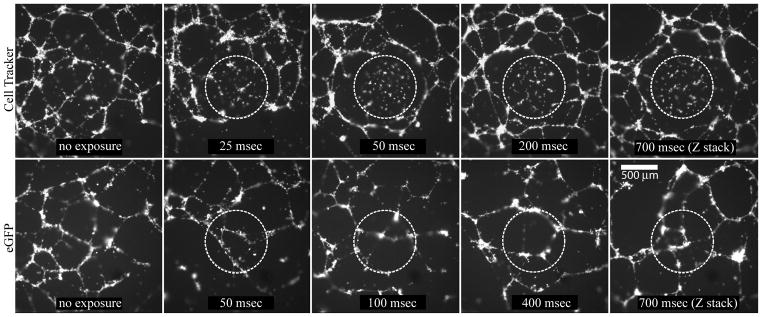
HMEC cells stably expressing fluorescent proteins exhibit less phototoxicity compared to the paternal HMECs labeled with CellTracker dye
To investigate if expression of fluorescent proteins confers an advantage over live cell dyes in time lapse imaging assays, we performed a time lapse experiment to compare HMEC-eGFP cells with CellTracker green-labeled HMEC cells in a 21 hour Matrigel assay in a 384 well plate format. We used a 10x objective and a partially closed fluorescence illumination field diaphragm to selectively illuminate a small region in the center of a well for different exposure settings in different wells (ranging from 25 to 400 msec). For some wells 4D images were acquired (z-stack over time) with 700-msec combined exposure (seven z-planes at 100-msec each) for each time point. At the end of the experiment the fluorescence illumination field diaphragm was opened and images of the whole well were taken with a 4x objective. The results (Fig. 5) revealed that Cell Tracker labeled cells exhibited substantial phototoxicity even with the lowest exposure setting, as evidenced from the dead cells and perturbed network formation in the illuminated areas, whereas perfect networks formed in the non-illuminated areas. In contrast, HMEC-eGFP cells underwent normal morphogenesis under all settings in both illuminated and non-illuminated regions of the well.
Figure 5.
CellTracker labeled HMECs (top panel) exhibit higher photoxicity compared to the eGFP-expressing HMECs (lower panel). The images represent projections from z-stacks acquired with a 4x objective at the end of a 21 hour, 10-minute interval time lapse experiment set up using a 10x objective and a partially closed diaphragm. The illuminated areas in the center of each well are outlined by dotted lines and the exposure times are indicated. For the right-most images, a 4D acquisition (z-stack over time) was set up, resulting in 700 msec combined exposure of the central areas.
Organization of networks formed by HMECs co-cultured with EMCs
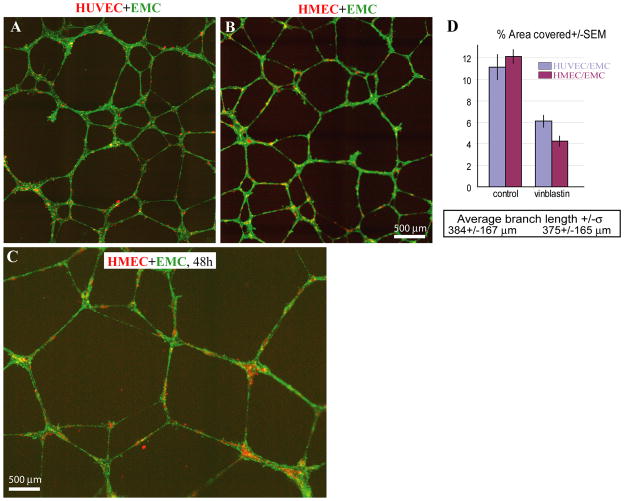
In order to examine whether HMECs have the potential to associate with smooth muscle cells and form structures resembling larger blood vessels, we co-cultured endothelial cells with EMCs, which have been shown to differentiate into smooth muscle cells and pericytes (Eid et al., 1992) and characterized the branching morphogenesis in a Matrigel assay. HMECs and HUVECs formed networks with EMCs with comparable dynamics, such that by 8 hours the networks were completely formed. However, the presence of EMCs caused the networks to persist longer, as demonstrated by characteristic branched network morphologies after 48 hours in culture (Figs. 6 A-C). The % area coverage and average branch length of the networks formed by HUVECs and HMECs in co-culture with EMCs were comparable (Fig. 6D).
Figure 6.
2D organization of networks formed by HMECs co-cultured with EMCs. (A, B) Representative images of CellTracker-labeled HUVECs and HMECs (pseudocolored red) and EMCs (pseudocolored green) co-cultured on Matrigel for about 20 hours. (C) Representative image of an HMEC/EMC network at 48 hours post-plating. (D) Quantification of the field of view occupied by tube structures as opposed to vacant space (% Area Covered) in control and vinblastin-treated samples. Comparison of images in A and B, as well as morphometric parameters such as average branch length and % area covered, indicate a great degree of similarity between the networks.
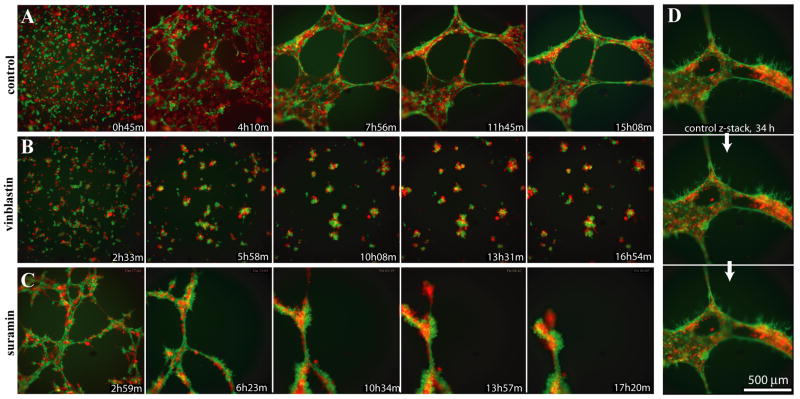
To further characterize the organization of the HMEC/EMC networks we used mCherry and GFP-expressing HMEC and EMC stable lines, respectively. Importantly, we found that cells, initially distributed randomly, gradually organized themselves so that the endothelial HMECs were primarily found inside the vessel-like structures while EMC mostly lined the outside (Fig. 7A, D, Supplemental Movie 4). We also noticed that after the networks matured, the EMCs started forming thin filopodia-like protrusions from the branches into the medium (Fig. 7D), and this morphology became more prominent over time.
Figure 7.
3D organization of networks formed by HMECs co-cultured with EMCs. Montages of the best focal planes taken from time lapse 4D movies of HMEC-mCherry cells grown in co-culture with EMC-eGFP cells in control media (A), in the presence of vinblastin (B), or in the presence of low concentration of suramin (C). The timepoints are not absolutely consistent between the different conditions because the cells were plated in 40 different wells of a 96-well plate and each time point constituted a z-stack of 31 focal planes, in two colors, that took about 5 minutes to acquire. (D) Z-stack showing correct relative orientation of HMEC-mCherry (red, inside) and EMC-eGFP (green, outside) cells in control culture approximately 34 h post plating. Z-planes are 10 mm apart. Note the filopodia-like projections extended from the green EMC-eGFP cells.
Addition of 50 nM vinblastine to the HMEC/EMC co-cultures altered the relative disposition of HMECs and EMCs, such that not only network formation was inhibited, but the two cell types were randomly mixed in clusters (Figs. 7B), consistent with a requirement for normal microtubule dynamics in vessel organization.
The assay was able to discern a dose-dependent effect of suramin. 30 μM suramin caused the cells to aggregate into large multicellular clusters. In contrast, abnormal transient networks were able to form in 15 μM suramin, but they were highly contractile and quickly collapsed into rods and clusters (Fig. 7C). In striking contrast to normal cultures, the branches of the collapsing networks (at ~10 hours post plating) formed green and red (HMECs and EMCs, respectively) rope-like structures running parallel, loosely wrapped around each other. These results demonstrate vessel-like morphological characteristics comparable to HMECs and HUVECs, adding further support for the suitability for HTS for angiogenesis modulators.
DISCUSSION
We have developed an in vitro angiogenesis system using immortalized endothelial cell lines that form branched networks of vessel-like structures that assume a normal inside-outside orientation in co-culture with EMCs, which form smooth muscle and pericytes, and appears suitable for scale-up to HTS. The cells retain their vessel-forming potential over many passages (>30). Moreover, both cell types were engineered to express a panel of nuclear- or cytoplasm-localized green (eGFP) or red (mCherry) fluorescent proteins that greatly facilitate time-lapse imaging and analysis to monitor cellular behavior during vessel formation. The characteristics of branched networks were quantified using MetaMorph, revealing that the in vitro assay can discriminate the effects of culture supplements and bioactive small molecules. Most assays for vessel formation use primary cells that are poorly suited for high throughput applications, and below we discuss the biological similarities of our system to primary cell assays and the added utility for HTS.
The HMEC-1 line, that we used for creating our fluorescent lines, was originally derived from human dermal microvascular endothelial cells and has already been extensively characterized and compared to both microvascular and macrovascular primary cells (Ades et al., 1992; Xu et al., 1994; Bouis et al., 2001; Unger et al., 2002; Bender et al., 2008) and references therein). In previous studies HMECs were concluded to be one of the best characterized and also one of the most physiologically preserved (i.e. similar to primary endothelia) microvascular lines currently available (Bouis et al., 2001; Unger et al., 2002; van, Jr. et al., 2008). Consistent with this conclusion, HMEC-1 line has been widely used in numerous assays related to microvascular endothelia functions with many of the results later confirmed in vivo, thus validating HMECs as an appropriate endothelial cell model (see the specific references in Supplemental Note 1).
In our hands, fluorescent HMECs were found to express a repertoire of endothelial cells receptors similar to primary endothelial cells (HUVECs), such as VEGFR-2, VE-cadherin, Tie-2 and several Eph receptors (Fig. 1). Moreover, we have shown that HMECs respond to proangiogenic factors, as exemplified by the ability of VEGF and EphA2 receptors to become tyrosine phosphorylated (activated) in response to VEGF and TNFα treatment, respectively, consistent with the formation of branched networks in our and prior studies (Ades et al., 1992; Meade-Tollin and Van Noorden, 2000). The similar levels of VEGFR-2 activation obtained in parental and eGFP expressing HMECs, together with the robust network formation we have observed with the fluorescent HMEC-based stable lines suggest that the expression of fluorescent proteins does not alter their angiogenic properties.
We found that stably transduced HMECs expressing fluorescent proteins showed far less toxicity than CellTracker dye labeling over the time course of our studies (Fig. 5), and are therefore a convenient source of labeled cells for HCS. Similarly, fluorescent protein-labeled EMCs did not show significant toxicity or impair vessel network formation. Other endothelial lines might be similarly engineered, including three human microvascular endothelial cell lines that have been immortalized using human telomerase catalytic protein (Yang et al., 1999; Venetsanakos et al., 2002; Shao and Guo, 2004). Differences between the endothelial cell lines might be useful to generalize and validate results obtained from one line, or pinpoint compounds that affect particular vascular beds.
Importantly, the vessel formation assay can detect activating and inhibiting factors. Maintenance of the HMEC vessel-like network depends on the presence of factors in the complete medium (that contains FGF, VEGF, IGF, EGF, ascorbic acid and FBS), and the immature networks quickly deteriorate in incomplete medium (Fig. 3). In addition to indicating a dependence on characterized angiogenic factors, this result also suggests that the basal system can be used to study angiogenic factors. In this regard, it is interesting that a low dose of PMA delayed network formation, while also prolonging the persistence of endothelial networks (Fig. 4B).
Conversely, sunitinib, SU5416, suramin, and vinblastine inhibited vessel network formation. Taken together, our results indicate that the normal responsiveness of the assay to angiogenic factors, plus the ability to use high content imaging to dynamically monitor vessel formation for over 24 hours, rather than a single endpoint, in high throughput should enable screens to discern complex effects on angiogenesis. Indeed, the success of our semi-automated preliminary quantification of network parameters formed in 96 and 384-well formats yielded Z′ values between 0.3 and 0.9, indicating the high dynamic range needed for minimizing false positive and negative results with only one experiment (or well) per condition, as needed for large scale HCS. This evidence of suitability for scale-up has also led us to begin developing more robust algorithms to segment and automatically evaluate network morphogenesis and the “correctness” of the inside-outside cellular orientation.
HMEC-1 cells have previously been described to migrate and form branching structures when grown on Matrigel (Ades et al., 1992; Meade-Tollin and Van Noorden, 2000). However, here we present quantitative analyses of the dynamics and morphological parameters of networks formed by HMECs alone, as well as (for the first time) in co-culture with EMCs. Although co-culture of HUVECs and smooth muscle cells have already been proposed as a model for high content screening (Evensen et al., 2010), we are the first to demonstrate that microvascular HMECs are not only capable of network formation in conjunction with EMCs, but also assume correct orientation within differentiated branches.
Inclusion of EMCs resulted in branching structures with normal inside-outside disposition (Figs. 6–7), similar to normal vessels and primary assay co-cultures (Evensen et al., 2010), and this should facilitate the automated study of factors that influence the formation of intermediate and large vessels. HMEC-EMC vessel-like structures (Fig. 7) sprouted spike-like projections from the EMCs into the extracellular matrix. Similar spikes also form in HUVEC/pericyte co-cultures, and in vivo such projections are not found on the normal vessels, but are associated with immature, leaky vessels (M. Komatsu, personal communication). Thus, while the co-culture model does not fully recapitulate the formation of normal vasculature, it might be useful to study vessel normalization, or screen and evaluate molecules that exert a normalizing effect on defective vasculature analogous to that associated with tumors. Specifically, defective vasculature has been considered a potential reason for poor delivery of anti-tumor drugs to within solid tumors, and inhibition of angiogenesis has been suggested to hinder drug delivery and promote cancer cell selection towards higher malignancy and metastatic potential (Paez-Ribes et al., 2009). Thus, the approach of promoting or normalizing vasculature, instead of inhibiting tumor vasculogenesis, may prove to be a more helpful strategy by allowing more efficient delivery of chemotherapy drugs (reviewed in (Jain, 2005). Thus, our experimental results should enable HCS assays based on functional vessel formation that may lead to drugs that normalize defective vasculature.
In conclusion, we have shown that our engineered HMEC fluorescent lines retain sufficient features of the angiogenesis phenotype to serve as a model of blood vessel formation that should enable large scale HCS. These features include retention of angiogenic potential and phenotypic stability, relative ease of maintenance, and suitability for creation of stable fluorescent protein cell lines that facilitate image analysis. Nuclear localized fluorescent proteins will enable future cellular tracking of each cell type, while cytoplasm-localized fluorescent facilitates image segmentation and analysis necessary for accurate quantification of dynamic aspects of network morphogenesis, and both will facilitate automated subcellular localization of mechanistically important signaling proteins. Use of a model characterized by formation of vessel-like structures will be an exciting advance over the migration-only surrogates that have been used thus far in large-scale screens. While it’s not necessary for an initial HC/HT screen to recapitulate normal biology perfectly because each can be followed by secondary screens using more temperamental primary cell models, a better initial screen is much more likely to generate pertinent hits. The novel features of our model enable both large scale primary screening and fluorescent time-lapse imaging, the latter of which will be critical for identifying the key time point for each mechanistic/assay goal and for identifying differences in the behavioral/phenotypic actions of molecular hits in follow-up studies without the need for fixing the cells at many different time points.
MATERIAL AND METHODS
Cells and Cell culture
EMCs were a kind gift from Dr. Hoda Eid. HUVECs were obtained from Lonza (Walkersville, MD) or Cascade Biologics (Portland, OR). The original HMEC-1 cell line was obtained from ATCC (cat # CRL-10636). HMECs and HUVECs were routinely propagated in complete endothelial growth media EGM2 or EGM2-MV (Lonza, Walkersville, MD). EMCs were grown in 10% FBS DMEM supplemented with Pen/Strep antibiotics.
All cells were grown at 37°C in humidified atmosphere of 5% CO2 to 80–90% confluence before passing them onto 25 or 75 cm2 tissue culture flasks. HMEC an HUVEC cells were usually split at a ratio of 1:3 or 1:4, while EMCs were split at a ratio of 1:5. Cells were harvested by washing with warm, sterile PBS followed by brief incubation in trypsin 0.25%/EDTA solution (1 ml per 100 cm2 tissue culture surface area). Care should be taken not to over-trypsinize the cells.
For experiments, non-fluorescent HMECs or HUVECs were labeled with green or orange CellTracker (Molecular Probes, Eugene, OR) according to manufacturer’s instructions and plated on top of Matrigel layer (see below) in coverslip-thick-glass-bottom 8-chamber slides, 96- or 384-well plates. Alternatively, HMEC lines stably expressing eGFP or mCherry fluorescent proteins were used. For all experiments HUVECs were used between 3rd and 8th passages. HMECs were used between passages #25 and 32.
Generation of fluorescently labeled stable cell lines
Lentivirus constructs expressing eGFP, mCherry, H2B-eGFP and H2B-mCherry (Kita-Matsuo et al., 2009) were applied to the cells at MOI 5 for 24 hours. Cells were then transferred to 96 well plates at approximately 0.5 cells per well, and single colonies with appropriate fluorescence levels, cell morphology, and growth rate were selected and expanded. Five passages were made to ensure that the expression of the fluorescent protein was stable.
Immunoblotting and immunoprecipitations
HMECs and HUVECs were lysed in modified RIPA buffer (1% Triton X-100, 1% Na deoxycholate; 0.1% SDS; 20 mM Tris; 150 mM NaCl; 1 mM EDTA) containing 10 μM NaF, 1μM sodium pervanadate and protease inhibitors. Equal amounts of cell lysates were probed by immunoblotting with anti-VEGFR-2 (Cell Signaling, Danvers, MA), anti-VE-cadherin (Santa Cruz Biotechnology, Santa Cruz, CA), anti Tie-2 (Santa-Cruz biotechnology, Santa Cruz, CA), anti EphA2 (Millipore-Upstate, Inc, Temecula, CA), anti-EphB2, anti-EphB4 or anti β-tubulin antibodies. The EphB2 and EphB4 antibodies were made to a GST fusion protein of the carboxy-terminal region (~100 amino acids) of the receptors (Holash and Pasquale, 1995;Noren et al., 2004). The protein concentration was calculated using the BCA protein assay kit (Pierce Biotecnology, Rockford, IL).
To assess whether EphA2 is tyrosine phosphorylated in response to stimulation with tumor necrosis factor alpha (TNFα), which upregulates the ligand ephrin-A1, HMECs and HUVECs were starved for 2 hours in serum free medium before adding 20 nM TNFα for 2 hours. The cells were then lysed in modified RIPA buffer (see above) and incubated with 2 μg anti-EphA2 antibody (Millipore-Upstate, Inc, Temecula, CA). The amount of lysates used for HMEC-1 immunoprecipitations was 1.5 times higher than that used for HUVECs. Immunoprecipitates were probed by immunoblotting with anti-phosphotyrosine antibody (Millipore, Inc, Temecula, CA) and reprobed with a mouse anti-EphA2 antibody (Invitrogen/Zymed Laboratories, San Francisco, CA) followed by a secondary anti-IgG peroxidase-conjugated antibody (GE Healthcare, UK). To assess VEGFR-2 activation HMECs and HUVECs were starved for 2 hours in serum free medium before addind 100 ng/ml VEGF for 15 min. Cell lysates were probed by immunoblotting with anti-phospho-VEGFR (Cell signaling, Boston, MA) and reprobed with a mouse anti-EphA2 antibody (Invitrogen/Zymed Laboratories, San Francisco, CA).
Network formation angiogenic assays
Angiogenic assays were typically performed on glass-bottom 96-well plates or 384-well plates (Greiner Bio-One, Monroe, NC). A few hours before plating the cells, 7–8 mg/ml solution of Matrigel (BD Biosciences, San Jose, CA) was added at 40 μl per well for a 96-well plate, or at 10 μl per well for a 384-well plate. To reduce menisci and eliminate bubbles within the Matrigel layers, prechilled plates were coated with Matrigel on ice and then immediately centrifuged at 1800 rcf at 4°C for one minute. The plates were then transferred to a tissue culture incubator, and Matrigel was allowed to polymerize for 1 hour before appropriate culture media was added to the wells. Endothelial cells (HUVECs or HMECs) were plated at a concentration of 2.5×104 cells/well in a 96 well plate or 4×103 cells/well in a 384 well plate. For co-cultures, endothelial cells and mesothelial cells were plated at a one-to-one to one-to-two ratio, respectively, maintaining a total cell concentration of 7.5×104 cells per 1 cm2 of surface area. Typically, EGM2 medium (for HUVECs) and EGM2-MV medium (for HMECs) were used as complete media in single or co-culture angiogenic assays. Most assays were performed on HMECs stably expressing mCherry or eGFP fluorescent proteins in the cytoplasm and involved epi-fluorescent time lapse imaging. When original HMECs were used, they were typically prelabeled with CellTracker Green or Red (Molecular Probes, Eugene, OR) according to manufacturer instructions.
Experimental Conditions
Various media and drug compounds were tested for their effects on angiogenic network formation. Cell cultures were incubated in either complete media (see above), EBM2 (serum-free, growth factor-free basal media), 10% FBS DMEM or EGM2-MV medium containing various compounds. Typically, 190 μl of media with compound solution was prepared and added to each well in a 96-well plate format prior to addition of 10 μl of concentrated cell suspension to each well. As such, compound concentrations in the 190 μl were scaled up to account for a final volume of 240 μl (200 μl media and cells plus 40 μl Matrigel). Compounds tested included VEGF, suramin, vinblastine and SU5416 (Calbiochem, San Diego, CA), sunitinib (BioVision, San Diego, CA), as well as PMA (US Biological, Swampscott, MA). Working concentrations for each compound were as follows: VEGF, 80 or 240 ng/ml; vinblastine, 50 nM; suramin, 15 or 30 μM; SU5416, 10 μM; SU5416, 1 or 5 μM; PMA, 100 ng/ml.
Imaging
For image acquisition we used an inverted Nikon TE2000 PFS-equipped fully automated epi-fluorescence microscope controlled by MetaMorph software (Molecular Devices, USA) and enclosed in a custom made incubator to keep cells at 37°C with 5% CO2. Images were acquired with either CoolSnap (Roper Scientific/Photometrics, Tucson, AZ ) or an Orca II (Hamamatsu, Japan) CCD cameras using a 4x 0.2 numerical aperture (NA), 10x 0.3NA or 20x 0.5NA objectives.
Image analysis
Most measurements, including quantitation of Western blots, as well as semi-automated image analyses and cell tracking were done using MetaMorph (Molecular Devices, USA). For analysis of angiogenic network features, we used MetaMorph Integrated Morphology and Angio Tubes modules, as described in the text. All measurements were then exported into Excel, which was used for analysis and plotting.
Z′ values were calculated as described (Zhang et al., 1999). Briefly, Z′ is a function of the ratio of the sum of standard deviations (σ) of the positive and negative control values and the difference between their means (μ).
Supplementary Material
Acknowledgments
This work was supported by the following grants: Mathers Charitable Foundation to MM and JHP; National Institute of Health 5U54HG005033-02, R01CA116099 and P01CA138390 to JHP; R37HL059502 to MM, R01HL065484 and R01HL086879 to PR-L, R01CA116099 and P01CA138390 to EBP, and the California Institute for Regenerative Medicine RC1-000132 to MM. KW is a California Institute for Regenerative Medicine Postdoctoral Fellow. Authors would like to thank Dr. Hoda Eid for kindly providing EMC cell line.
Abbreviations
- HUVECs
Human Umbilical Vein Endothelial Cells
- HMECs
Human Microvascular Endothelial Cells
- EMCs
Epicardial Mesothelial Cells
- VEGF
Vascular Endotelial Growth Factor
- VEGFR
Vascular Endotelial Growth Factor Receptor
- VE cadherin
Vascular Endothelial Cadherin
- HCS
High Content Screening
- HTS
high throughput screening
- H2B
Histon-2B
Footnotes
AUTHOR CONTRIBUTION
Natalie L. Prigozhina supervised the project, designed most of the experiments, analyzed and interpreted the data, and was responsible for writing the manuscript. Andrew Heisel and Jordan R. Seldeen performed the experiments and assisted in data analysis. Ke Wei created the stable cell lines and provided technical expertise. Roberta Noberini performed experiments shown in Figure 1. Edward A. Hunter advised on the quantification aspect of the research and is developing the automated analysis software. Diego Calzolari assisted with image preparation and data management. Pilar Ruiz-Lozano, Elena B. Pasquale, Mark Mercola and Jeffrey H. Price directed and obtained funding for this project, as well as provided expertise, advice and editing of the manuscript.
References
- Ades EW, Candal FJ, Swerlick RA, George VG, Summers S, Bosse DC, Lawley TJ. HMEC-1: establishment of an immortalized human microvascular endothelial cell line. J Invest Dermatol. 1992;99:683–690. doi: 10.1111/1523-1747.ep12613748. [DOI] [PubMed] [Google Scholar]
- Bender A, Zapolanski T, Watkins S, Khosraviani A, Seiffert K, Ding W, Wagner JA, Granstein RD. Tetracycline suppresses ATP gamma S-induced CXCL8 and CXCL1 production by the human dermal microvascular endothelial cell-1 (HMEC-1) cell line and primary human dermal microvascular endothelial cells. Exp Dermatol. 2008;17:752–760. doi: 10.1111/j.1600-0625.2008.00716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergers G, Javaherian K, Lo KM, Folkman J, Hanahan D. Effects of angiogenesis inhibitors on multistage carcinogenesis in mice. Science. 1999;284:808–812. doi: 10.1126/science.284.5415.808. [DOI] [PubMed] [Google Scholar]
- Bouis D, Hospers GA, Meijer C, Molema G, Mulder NH. Endothelium in vitro: a review of human vascular endothelial cell lines for blood vessel-related research. Angiogenesis. 2001;4:91–102. doi: 10.1023/a:1012259529167. [DOI] [PubMed] [Google Scholar]
- Carmeliet P. Angiogenesis in health and disease. Nat Med. 2003;9:653–660. doi: 10.1038/nm0603-653. [DOI] [PubMed] [Google Scholar]
- Carmeliet P. Angiogenesis in life, disease and medicine. Nature. 2005;438:932–936. doi: 10.1038/nature04478. [DOI] [PubMed] [Google Scholar]
- Chow LQ, Eckhardt SG. Sunitinib: from rational design to clinical efficacy. J Clin Oncol. 2007;25:884–896. doi: 10.1200/JCO.2006.06.3602. [DOI] [PubMed] [Google Scholar]
- Eid H, Larson DM, Springhorn JP, Attawia MA, Nayak RC, Smith TW, Kelly RA. Role of epicardial mesothelial cells in the modification of phenotype and function of adult rat ventricular myocytes in primary coculture. Circ Res. 1992;71:40–50. doi: 10.1161/01.res.71.1.40. [DOI] [PubMed] [Google Scholar]
- Evensen L, Micklem DR, Link W, Lorens JB. A novel imaging-based high-throughput screening approach to anti-angiogenic drug discovery. Cytometry A. 2010;77:41–51. doi: 10.1002/cyto.a.20808. [DOI] [PubMed] [Google Scholar]
- Fong TA, Shawver LK, Sun L, Tang C, App H, Powell TJ, Kim YH, Schreck R, Wang X, Risau W, Ullrich A, Hirth KP, McMahon G. SU5416 is a potent and selective inhibitor of the vascular endothelial growth factor receptor (Flk-1/KDR) that inhibits tyrosine kinase catalysis, tumor vascularization, and growth of multiple tumor types. Cancer Res. 1999;59:99–106. [PubMed] [Google Scholar]
- Frick M, Dulak J, Cisowski J, Jozkowicz A, Zwick R, Alber H, Dichtl W, Schwarzacher SP, Pachinger O, Weidinger F. Statins differentially regulate vascular endothelial growth factor synthesis in endothelial and vascular smooth muscle cells. Atherosclerosis. 2003;170:229–236. doi: 10.1016/s0021-9150(03)00299-5. [DOI] [PubMed] [Google Scholar]
- Gagliardi AR, Kassack M, Kreimeyer A, Muller G, Nickel P, Collins DC. Antiangiogenic and antiproliferative activity of suramin analogues. Cancer Chemother Pharmacol. 1998;41:117–124. doi: 10.1007/s002800050717. [DOI] [PubMed] [Google Scholar]
- Gagliardi AR, Taylor MF, Collins DC. Uptake of suramin by human microvascular endothelial cells. Cancer Lett. 1998;125:97–102. doi: 10.1016/s0304-3835(97)00496-5. [DOI] [PubMed] [Google Scholar]
- Goel G, Makkar HP, Francis G, Becker K. Phorbol esters: structure, biological activity, and toxicity in animals. Int J Toxicol. 2007;26:279–288. doi: 10.1080/10915810701464641. [DOI] [PubMed] [Google Scholar]
- Harris ES, Nelson WJ. VE-cadherin: at the front, center, and sides of endothelial cell organization and function. Curr Opin Cell Biol. 2010;22:651–658. doi: 10.1016/j.ceb.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holash JA, Pasquale EB. Polarized expression of the receptor protein tyrosine kinase Cek5 in the developing avian visual system. Dev Biol. 1995;172:683–693. doi: 10.1006/dbio.1995.8039. [DOI] [PubMed] [Google Scholar]
- Jain RK. Normalization of tumor vasculature: an emerging concept in antiangiogenic therapy. Science. 2005;307:58–62. doi: 10.1126/science.1104819. [DOI] [PubMed] [Google Scholar]
- Jendreyko N, Popkov M, Rader C, Barbas CF., III Phenotypic knockout of VEGF-R2 and Tie-2 with an intradiabody reduces tumor growth and angiogenesis in vivo. Proc Natl Acad Sci USA. 2005;102:8293–8298. doi: 10.1073/pnas.0503168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita-Matsuo H, Barcova M, Prigozhina N, Salomonis N, Wei K, Jacot JG, Nelson B, Spiering S, Haverslag R, Kim C, Talantova M, Bajpai R, Calzolari D, Terskikh A, McCulloch AD, Price JH, Conklin BR, Chen HS, Mercola M. Lentiviral vectors and protocols for creation of stable hESC lines for fluorescent tracking and drug resistance selection of cardiomyocytes. PLoS One. 2009;4:e5046. doi: 10.1371/journal.pone.0005046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loboda A, Jazwa A, Jozkowicz A, Molema G, Dulak J. Angiogenic transcriptome of human microvascular endothelial cells: Effect of hypoxia, modulation by atorvastatin. Vascul Pharmacol. 2006;44:206–214. doi: 10.1016/j.vph.2005.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loges S, Schmidt T, Carmeliet P. “Antimyeloangiogenic” therapy for cancer by inhibiting PlGF. Clin Cancer Res. 2009;15:3648–3653. doi: 10.1158/1078-0432.CCR-08-2276. [DOI] [PubMed] [Google Scholar]
- McCain DF, Wu L, Nickel P, Kassack MU, Kreimeyer A, Gagliardi A, Collins DC, Zhang ZY. Suramin derivatives as inhibitors and activators of protein-tyrosine phosphatases. J Biol Chem. 2004;279:14713–14725. doi: 10.1074/jbc.M312488200. [DOI] [PubMed] [Google Scholar]
- Meade-Tollin LC, Van Noorden CJ. Time lapse phase contrast video microscopy of directed migration of human microvascular endothelial cells on matrigel. Acta Histochem. 2000;102:299–307. doi: 10.1078/S0065-1281(04)70037-9. [DOI] [PubMed] [Google Scholar]
- Njauw CN, Yuan H, Zheng L, Yao M, Martins-Green M. Origin of periendothelial cells in microvessels derived from human microvascular endothelial cells. Int J Biochem Cell Biol. 2008;40:710–720. doi: 10.1016/j.biocel.2007.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noren NK, Lu M, Freeman AL, Koolpe M, Pasquale EB. Interplay between EphB4 on tumor cells and vascular ephrin-B2 regulates tumor growth. Proc Natl Acad Sci USA. 2004;101:5583–5588. doi: 10.1073/pnas.0401381101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez-Ribes M, Allen E, Hudock J, Takeda T, Okuyama H, Vinals F, Inoue M, Bergers G, Hanahan D, Casanovas O. Antiangiogenic therapy elicits malignant progression of tumors to increased local invasion and distant metastasis. Cancer Cell. 2009;15:220–231. doi: 10.1016/j.ccr.2009.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey A, Shao H, Marks RM, Polverini PJ, Dixit VM. Role of B61, the ligand for the Eck receptor tyrosine kinase, in TNF-alpha-induced angiogenesis. Science. 1995;268:567–569. doi: 10.1126/science.7536959. [DOI] [PubMed] [Google Scholar]
- Rai SS, Wolff J. Localization of the vinblastine-binding site on beta-tubulin. J Biol Chem. 1996;271:14707–14711. doi: 10.1074/jbc.271.25.14707. [DOI] [PubMed] [Google Scholar]
- Shao R, Guo X. Human microvascular endothelial cells immortalized with human telomerase catalytic protein: a model for the study of in vitro angiogenesis. Biochem Biophys Res Commun. 2004;321:788–794. doi: 10.1016/j.bbrc.2004.07.033. [DOI] [PubMed] [Google Scholar]
- Unger RE, Krump-Konvalinkova V, Peters K, Kirkpatrick CJ. In vitro expression of the endothelial phenotype: comparative study of primary isolated cells and cell lines, including the novel cell line HPMEC-ST1.6R. Microvasc Res. 2002;64:384–397. doi: 10.1006/mvre.2002.2434. [DOI] [PubMed] [Google Scholar]
- van B, Jr, van der LE, Griffioen AW. Angiogenic profiling and comparison of immortalized endothelial cells for functional genomics. Exp Cell Res. 2008;314:264–272. doi: 10.1016/j.yexcr.2007.08.013. [DOI] [PubMed] [Google Scholar]
- Venetsanakos E, Mirza A, Fanton C, Romanov SR, Tlsty T, McMahon M. Induction of tubulogenesis in telomerase-immortalized human microvascular endothelial cells by glioblastoma cells. Exp Cell Res. 2002;273:21–33. doi: 10.1006/excr.2001.5424. [DOI] [PubMed] [Google Scholar]
- Waltenberger J, Mayr U, Frank H, Hombach V. Suramin is a potent inhibitor of vascular endothelial growth factor. A contribution to the molecular basis of its antiangiogenic action. J Mol Cell Cardiol. 1996;28:1523–1529. doi: 10.1006/jmcc.1996.0142. [DOI] [PubMed] [Google Scholar]
- Xu Y, Swerlick RA, Sepp N, Bosse D, Ades EW, Lawley TJ. Characterization of expression and modulation of cell adhesion molecules on an immortalized human dermal microvascular endothelial cell line (HMEC-1) J Invest Dermatol. 1994;102:833–837. doi: 10.1111/1523-1747.ep12382086. [DOI] [PubMed] [Google Scholar]
- Yancopoulos GD, Davis S, Gale NW, Rudge JS, Wiegand SJ, Holash J. Vascular-specific growth factors and blood vessel formation. Nature. 2000;407:242–248. doi: 10.1038/35025215. [DOI] [PubMed] [Google Scholar]
- Yang J, Chang E, Cherry AM, Bangs CD, Oei Y, Bodnar A, Bronstein A, Chiu CP, Herron GS. Human endothelial cell life extension by telomerase expression. J Biol Chem. 1999;274:26141–26148. doi: 10.1074/jbc.274.37.26141. [DOI] [PubMed] [Google Scholar]
- Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.