Abstract
Centromeres are chromosomal loci that direct segregation of the genome during cell division. The histone H3 variant CENP-A (also known as CenH3) defines centromeres in monocentric organisms, which confine centromere activity to a discrete chromosomal region, and holocentric organisms, which distribute centromere activity along the chromosome length1–3. Because the highly repetitive DNA found at most centromeres is neither necessary nor sufficient for centromere function, stable inheritance of CENP-A nucleosomal chromatin is postulated to epigenetically propagate centromere identity4. Here, we show that in the holocentric nematode Caenorhabditis elegans pre-existing CENP-A nucleosomes are not necessary to guide recruitment of new CENP-A nucleosomes. This is indicated by lack of CENP-A transmission by sperm during fertilization and by removal and subsequent reloading of CENP-A during oogenic meiotic prophase. Genome-wide mapping of CENP-A location in embryos and quantification of CENP-A molecules in nuclei revealed that CENP-A is incorporated at low density in domains that cumulatively encompass half the genome. Embryonic CENP-A domains are established in a pattern inverse to regions that are transcribed in the germline and early embryo, and ectopic transcription of genes in a mutant germline altered the pattern of CENP-A incorporation in embryos. Furthermore, regions transcribed in the germline but not embryos fail to incorporate CENP-A throughout embryogenesis. We propose that germline transcription defines genomic regions that exclude CENP-A incorporation in progeny, and that zygotic transcription during early embryogenesis remodels and reinforces this basal pattern. These findings link centromere identity to transcription and shed light on the evolutionary plasticity of centromeres.
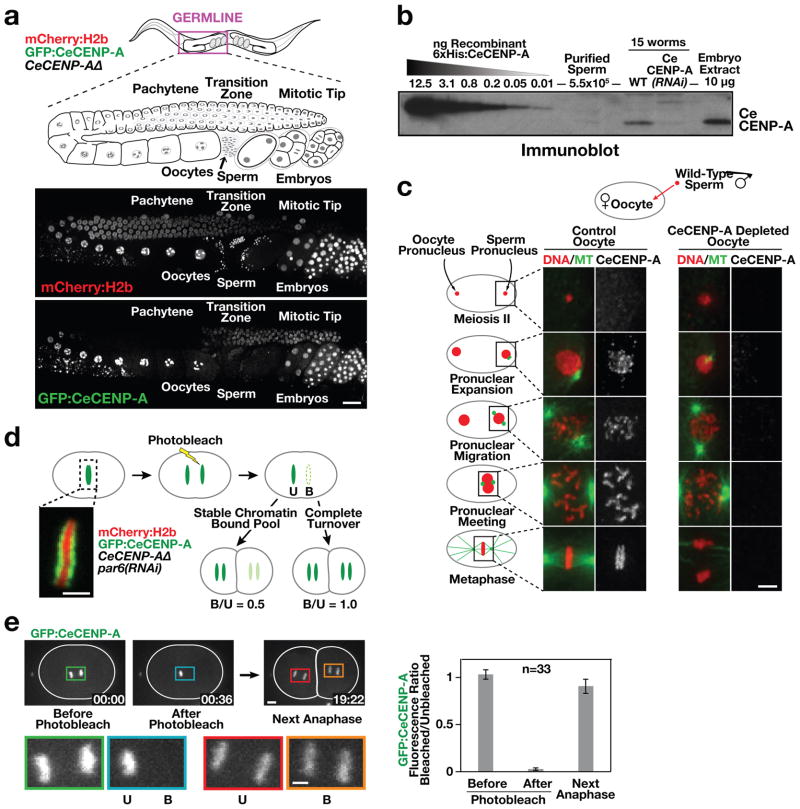
To characterize CENP-A localization dynamics in C. elegans (CeCENP-A; Supplementary Figs 1 and 2), we generated a strain in which the only source of CENP-A is a single copy green fluorescent protein (GFP)-conjugated transgene encoding GFP–CeCENP-A (Supplementary Fig. 1b). Imaging in adult hermaphrodites revealed that, in the maternal germline, CeCENP-A is removed from chromosomes as they enter the pachytene stage of meiotic prophase, and is reloaded when nuclei progress into diplotene (Fig. 1a). CeCENP-A was not detected in the nuclei of mature sperm (Supplementary Fig. 2a, b), and quantitative immunoblotting indicated that sperm have fewer than the detection limit of 300 CeCENP-A molecules (Fig. 1b and Supplementary Fig. 2c, d). To test for sperm-derived CeCENP-A in embryos, we fertilized CeCENP-A-depleted oocytes with wild-type sperm. In control embryos, CeCENP-A localized to both sperm and oocyte chromatin during chromosome condensation, pronuclear migration and mitosis (Fig. 1c). As reported in other systems5, this recruitment was independent of DNA replication (Supplementary Fig. 3). After fertilization of CeCENP-A-depleted oocytes with wild-type sperm, no CeCENP-A signal was detected on sperm or oocyte chromatin throughout the cell cycle (Fig. 1c). Thus, sperm chromatin does not retain CeCENP-A to propagate centromere identity through fertilization in C. elegans.
Figure 1. CeCENP-A dynamics in meiotic prophase, at fertilization and across embryonic divisions.
a, Gonad region of an adult hermaphrodite co-expressing GFP–CeCENP-A and mCherry–histone H2b in a CeCENP-AΔ (hcp-3(ok1892; see also Supplementary Figs 1 and 2) background (see also Supplementary Figs 1 and 2). Scale bar, 20 μm. b, Quantitative immunoblot showing that sperm lack a significant pool of CeCENP-A (see also Supplementary Fig. 2c, d). c, Fertilized one-cell control or CeCENP-A-depleted embryos at different stages of the first mitotic division were immunostained for CeCENP-A and α-tubulin (MT). Wild-type (N2) males were mated to fem-1 mutant worms to ensure all embryos were cross-progeny. Scale bar, 5 μm. d, Schematic of photobleaching experiment to assay CeCENP-A inheritance across early embryonic divisions. par-6 RNA interference (RNAi) abolishes developmental asynchrony in the two-cell embryo. Unbleached (U) and bleached (B) chromatid sets are indicated. Scale bar, 2 μm. e, Representative images and quantification of the photobleaching experiment. Higher magnification views highlight bleached and unbleached chromatid sets. Error bars are 95% confidence intervals for the means. Scale bars, 5 μm.
Pulse-chase experiments in human cells have suggested that stable inheritance of CENP-A on chromatin propagates centromere identity through cell division6,7. To test whether CeCENP-A is stably inherited on chromatin in embryos, we photobleached one set of GFP–CeCENP-A-labelled chromatids after separation from their sisters at anaphase onset in the one-cell embryo. GFP–CeCENP-A signals were then compared in the next round of division between cells inheriting bleached or unbleached chromatid sets. Stable inheritance of CeCENP-A on chromatin predicts that 50% of CeCENP-A is old and the other 50% is new, resulting in a bleached/unbleached ratio of 0.5. In contrast, if CeCENP-A is not stably inherited on chromatin between the two rounds of division, the bleached/unbleached ratio should be 1.0, which is close to the observed value (Fig. 1d–f and Supplementary Fig. 4). Thus, despite the short division time (only ~15 min between consecutive metaphases), CeCENP-A is nearly completely turned over on chromatin during embryonic cell divisions.
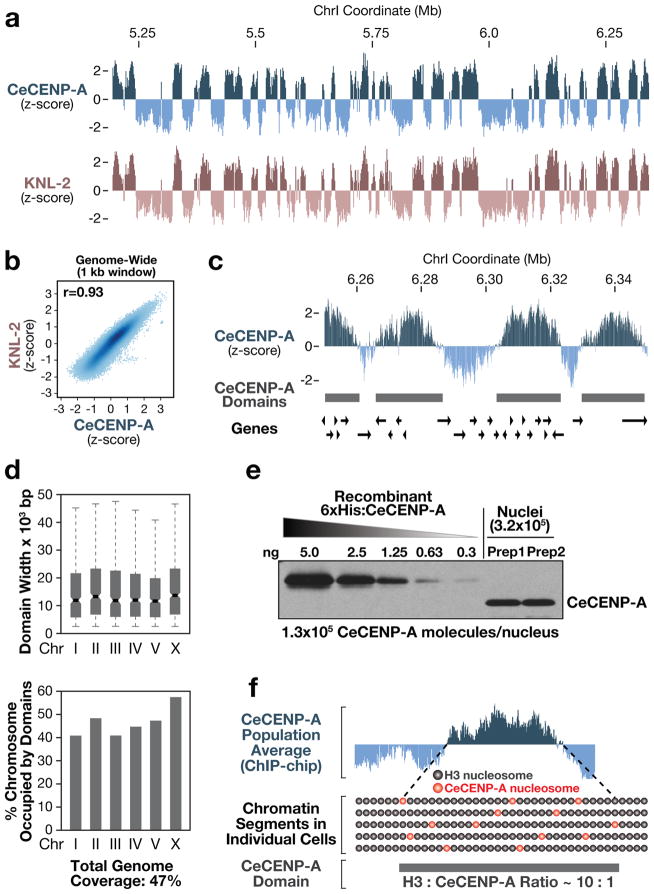
The above results indicate that pre-existing CeCENP-A nucleosomes may not be the cue that targets new CeCENP-A nucleosomes. To define the unknown guiding cue(s), we analysed the genome-wide distribution of CeCENP-A in embryos using chromatin immunoprecipitation with a CeCENP-A-specific antibody8 followed by hybridization to a tiling microarray (ChIP-chip). As CENP-A chromatin is characterized by highly repetitive DNA in most higher eukaryotes, this offered the opportunity to define the distribution of CENP-A in an organism naturally lacking large stretches of repeats. Our ChIP-chip analysis revealed regions of CeCENP-A enrichment along the entire length of chromosomes, as predicted for holocentric chromosome architecture (Fig. 2a and Supplementary Fig. 5a–c). The genome-wide distribution of the conserved CENP-A-specific loading factor KNL-2 was indistinguishable from that of CeCENP-A, indicating that the CeCENP-A distribution reflects specific incorporation (Fig. 2a, b). A sliding-window-based domain definition algorithm revealed that CeCENP-A domains vary considerably in size (median 10–12 kilobase), cover both genic and intergenic regions, are distributed evenly throughout the genome, and do not correlate with repeat density (Fig. 2c–e and Supplementary Fig. 5d–f). Although nearly half the genome is occupied by CeCENP-A domains (Fig. 2d), quantification of CeCENP-A molecules in purified embryonic nuclei showed that there is only enough CeCENP-A to occupy at most 4% of the genome (Fig. 2e and Supplementary Fig. 6a–c). Therefore, the domains enriched for CeCENP-A identified by ChIP-chip must be comprised primarily of H3 nucleosomes (Fig. 2f). Consistent with this, histone H3 ChIP-chip analysis does not show depletion in regions enriched for CeCENP-A9 (Supplementary Fig. 6d, e). Thus, the holocentric architecture of C. elegans chromosomes arises from reproducible definition of domains that are permissive for low-density CeCENP-A incorporation.
Figure 2. Genome-wide mapping of CeCENP-A-enriched chromatin.
a, Regions enriched for CeCENP-A and its loading factor KNL-2 in a representative portion of chromosome I. For each track, the average z-score probe signal of two independent biological replicates is plotted. b, Genome-wide correlation plot of CeCENP-A and KNL-2 occupancy. The correlation coefficient (r) is in the upper left corner. c, Regions enriched for CeCENP-A with the positions of annotated genes. CeCENP-A domains were defined by a sliding window algorithm. d, Features of CeCENP-A domains for individual chromosomes. Boxplots: boxes indicate 25th to 75th percentile, whiskers 2.5th to 97.5th percentile. Wedges around the medians indicate 95% confidence intervals for the medians (see also Supplementary Fig. 5d–f). e, Two independent nuclei preparations (Prep) from early embryos (<100 nuclei) were blotted alongside a purified CeCENP-A standard (see also Supplementary Figs 2c, d and 6a–c). f, Hypothetical model for CeCENP-A permissive domain.
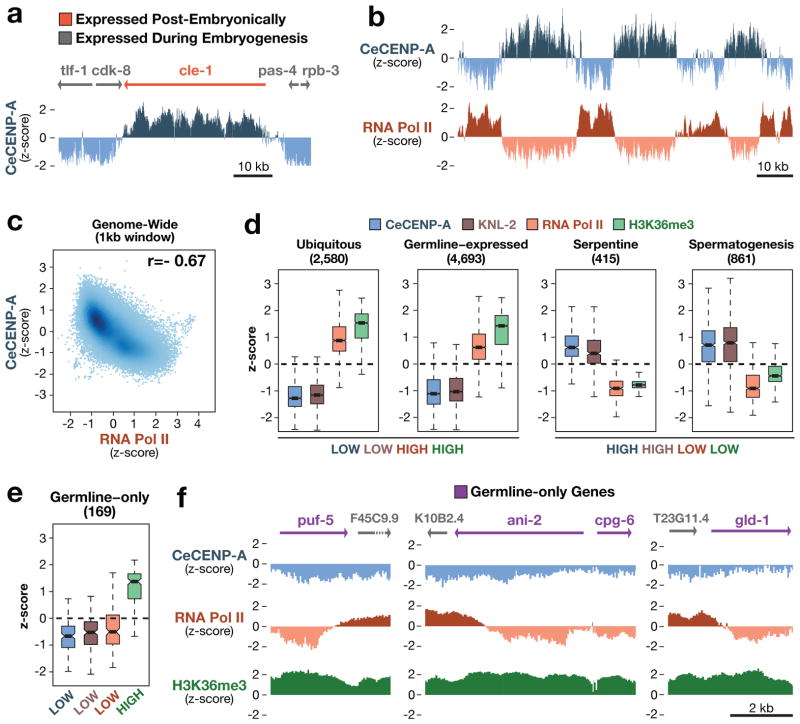
The abundance of genomic regions permissive for CeCENP-A incorporation makes it unlikely that they are defined by a specific DNA sequence. Instead, a correlation emerged with transcriptional status: genes transcribed in embryos were refractory to CeCENP-A incorporation, whereas genes that are silent in embryos (but transcribed in post-embryonic tissues) were permissive for CeCENP-A incorporation (Fig. 3a). ChIP-chip analysis of RNA polymerase II (Pol II) revealed an inverse correlation with CeCENP-A that extended genome-wide (Fig. 3b, c and Supplementary Fig. 7).
Figure 3. Relationship between CeCENP-A and gene expression.
a, Chromosomal region containing the cle-1 gene, which is expressed in neurons, and flanking genes that are expressed during embryogenesis. b, Genome browser view showing inverse correlation between CeCENP-A and RNA Pol II occupancy. c, Genome-wide correlation plot of CeCENP-A and RNA Pol II occupancy. The correlation coefficient (r) is in the upper right corner. d, CeCENP-A, KNL-2, RNA Pol II and H3K36me3 occupancy for various gene sets defined on the basis of expression data. The number of genes in each set is shown in parentheses. Boxplots (as in Fig. 2d) show the range of z-scores averaged over gene bodies. e, CeCENP-A, KNL-2, RNA Pol II and H3K36me3 occupancy for the germline-only gene set. f, Genome browser views of CeCENP-A, RNA Pol II, and H3K36me3 occupancy on germline-only genes, flanked by genes expressed in embryos.
The inverse correlation between transcription and CeCENP-A incorporation was puzzling, given that there is no significant RNA Pol II-dependent transcription during the first two rounds of embryonic division, and transcriptional activity remains low until the 30-cell stage10–12. In addition, inhibition of transcription using α-amanitin did not cause defects in chromosome segregation in early embryos (Supplementary Fig. 8). We analysed the CeCENP-A and RNA Pol II distribution in four populations of embryos that formed a developmentally timed series from very early (73% of embryos with ≤8 nuclei) to old (67% of embryos with > 200 nuclei) (Supplementary Fig. 9a, b). The CeCENP-A distribution remained constant across this series, despite the activation or repression of genes (Supplementary Figs 9b–d and 10a, b). Thus, CeCENP-A incorporation in embryos may not be dictated simply by active transcription.
To better assess the relationship between transcription and CeCENP-A incorporation, we analysed CeCENP-A and RNA Pol II enrichment in different gene classes defined in previous work by their expression profiles13. The inverse correlation between CeCENP-A and RNA Pol II held true for four of five gene classes (Fig. 3d). However, the ‘germline-only’ class, which is comprised of genes transcribed in the maternal germline but not in embryos, did not show an inverse correlation. Instead, both CeCENP-A and embryonic RNA Pol II levels were low (Fig. 3e), which was confirmed by individual analysis of well-characterized genes transcribed exclusively in the germline (Fig. 3f). In addition, ‘germline-only’ genes failed to incorporate CeCENP-A throughout embryogenesis, despite the persistent absence of RNA Pol II (Supplementary Fig. 10a, b). Germline-only genes are enriched for histone H3 lysine 36 methylation (H3K36me), indicating that the absence of CeCENP-A and RNA Pol II signals is not a false negative13 (Fig. 3d–f). Thus, genes transcribed in the maternal germline are refractory to CeCENP-A incorporation, even if they are transcriptionally silent in embryos. This result, together with the paucity of transcription in early embryos and the consistent CeCENP-A distribution throughout embryogenesis, indicates that transcriptional activity in the maternal germline may render genomic regions refractory to CeCENP-A incorporation. Activation of transcription during early embryogenesis probably remodels and reinforces this basal CeCENP-A pattern. In support of this, genes transcribed in early embryos, but lacking signatures of germline transcription, also show low CeCENP-A occupancy (Supplementary Fig. 10c).
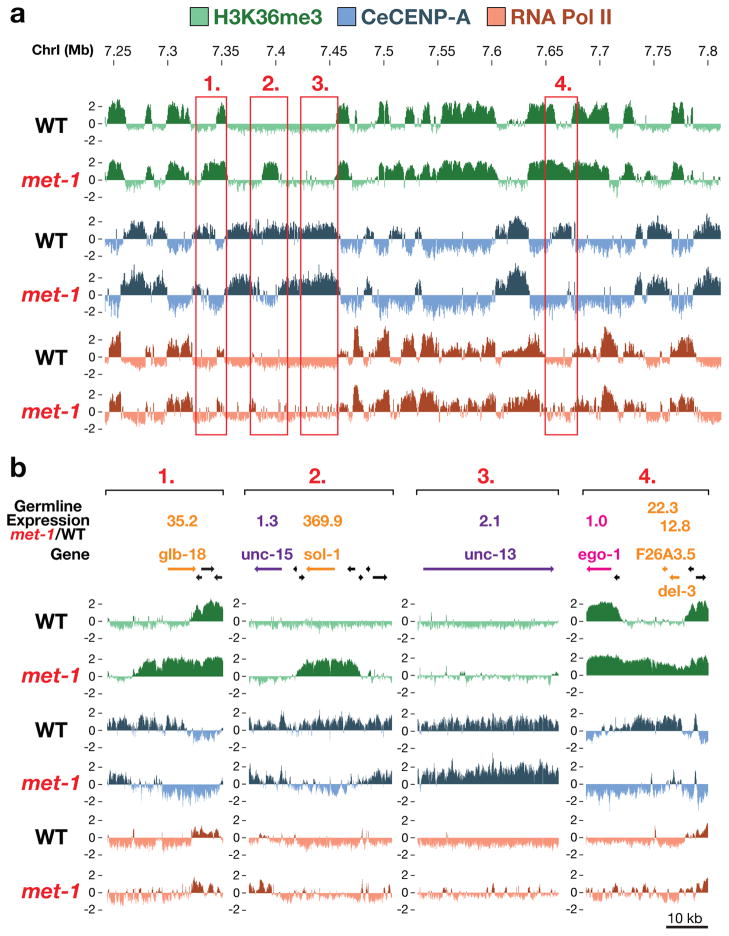
If germline transcriptional activity influences CeCENP-A incorporation in the progeny, changes in germline transcription should alter the CeCENP-A distribution in embryos. An unexpected observation in embryos derived from a null mutant of met-1, which encodes one of two C. elegans H3K36 methyltransferases14, enabled us to test this prediction. The met-1 mutant is viable and fertile, indicating that transcription is not globally misregulated in this mutant. Consistent with this, the genome-wide distributions of H3K36me3, RNA Pol II and CeCENP-A were similar in met-1 and wild-type embryos (Fig. 4a; genome-wide correlation coefficients of 0.88, 0.9 and 0.9, respectively). However, we observed rare regions of ectopic H3K36me3 enrichment in the met-1 mutant (Fig. 4 and Supplementary Figures 11 and 12). Out of 132 regions > 5 kb in size that acquired an ectopic H3K36me3 signal in met-1 mutant embryos, 75 did not show significant RNA Pol II occupancy in embryos, indicating that these regions are mistranscribed in mutant germlines but not in embryos. CeCENP-A was depleted from these regions in embryos (Fig. 4a, b and Supplementary Figs 11 and 12a). To test if acquisition of H3K36me3 and loss of CeCENP-A in met-1 mutant embryos is associated with ectopic germline gene transcription, we hand-dissected germlines from adult wild-type and met-1 mutant worms and measured messenger RNA levels by quantitative PCR for nine genes in ectopic H3K36me3 regions. As controls, we used eight genes located in regions that did not show a change in H3K36me3 signal. Genes in regions with ectopic H3K36me3 signal indeed showed significantly elevated RNA levels in met-1 mutant germlines compared to wild type (Fig. 4b and Supplementary Fig. 12b). Thus, the data obtained with the met-1 mutant indicate that ectopic transcription in the germline converts regions from permissive to non-permissive for CeCENP-A incorporation in the embryo progeny.
Figure 4. Germline expression controls CeCENP-A occupancy in the progeny embryos.
a, Portion of chromosome I featuring specific regions with ectopic H3K36me3 signal in the met-1 mutant (see also Supplementary Figs 11 and 12). b, Screen shots of the regions boxed in (a). Real-time quantitative reverse transcription PCR was performed on hand-dissected wild-type and met-1 mutant gonads. Mean met-1:wild-type expression ratio (four independent biological replicates each) is listed above genes (see table in Supplementary Fig. 12b for all genes analysed).
While the genome-wide inverse relationship to embryonic transcription suggests the simple model that CeCENP-A deposition is random and antagonized by active transcription, such a model fails to explain restriction of de-novo-deposited CeCENP-A on transcriptionally silent sperm chromatin (Fig. 1d), the low CeCENP-A occupancy on the 169 ‘germline-only’ genes throughout embryogenesis, and the results from the met-1 mutant analysis (Supplementary Discussion). Thus, we favour the model that transcription in the germline makes regions non-permissive for CeCENP-A incorporation in the progeny (Supplementary Fig. 12c), and the onset of transcriptional activity in embryos reinforces and remodels this pattern.
In C. elegans, H3K36 methylation and the Argonaute CSR-1, which binds short 22G-RNAs (named for their 5′ Guanosine residue and 22 nucleotide length) derived from germline transcripts, are candidate mechanisms for transmitting memory of germline transcription to early embryos13,15. Both H3K36 methylation (Fig. 3d–f and Supplementary Fig. 7c) and CSR-1 22G-RNA targets (Supplementary Fig. 13) are inversely correlated with CeCENP-A occupancy, and inhibition of CSR-1 and its co-factors leads to early embryonic chromosome segregation defects15,16.
The results here demonstrate that cues unrelated to pre-existing CENP-A nucleosomes can dictate the incorporation of new CENP-A nucleosomes, challenging the view that centromeres are patterned by stable inheritance of CENP-A domains with the key mark for centromere identity being CENP-A itself. Discontinuity of chromatin-localized CENP-A in the germline, similar to the one we describe here for C. elegans, has recently been proposed to also occur in plants17, suggesting that removal and reloading of CENP-A during every generation may be more common than is currently appreciated. In addition, the processes of neocentromerization and centromere repositioning, which occur with appreciable frequency in humans and are observed frequently during evolution18–20, may be guided by cues that are linked to transcription.
METHODS SUMMARY
For ChIP-chip, chitinase-treated embryos were fixed with 1% formaldehyde in PBS for 10 min, suspended in ChIP buffer (50 mM HEPES-KOH pH 7.6, 140 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.5% NP-40, 0.1% deoxycholate, 1% sarkosyl), and sonicated with a Branson sonifier microtip. Antibody (5 μg) pre-bound to 50 μl Dynabeads (Dynal Biotech) were incubated for 4 h with embryo extract (3 mg total protein). Beads were washed and eluted, and the purified DNA amplified as described13 before hybridization at the Roche Nimblegen Service Laboratory (2.1M probe tiling arrays with 50-bp probes; WormBase version WS170). Genome-wide scatter plots and Pearson correlations were obtained using log2 z-scores after median smoothing over 1-kb windows. GFP–CeCENP-A images were acquired with a CSU10 spinning disk confocal head (Yokogawa) and a CCD camera (iXon DV887; Andor Technology) mounted on a Nikon TE2000-E inverted microscope equipped with a solid state laser combiner (ALC) 491 nm and 561 nm lines. For quantification of CeCENP-A in nuclei, embryos were treated with chitinase and lysed by douncing in nuclei buffer (10 mM Tris-HCl pH 8, 80 mM KCl, 2 mM K-EDTA, 0.75 mM spermidine, 0.3 mM spermine, 0.1% digitonin). Nuclei were separated from debris by low-speed centrifugation steps. For germline expression analysis, total RNA from 50–100 dissected gonads was isolated using TRIzol (Invitrogen), and complementary DNA was synthesized with Superscript III reverse transcriptase (Invitrogen). Quantitative real-time PCR was performed with iQ SYBR Green Supermix (Bio-Rad) in the iQ5 cycler (Bio-Rad) using standard protocols.
METHODS
Antibodies
ChIP for HCP-3/CeCENP-A was performed with OD79, an affinity-purified rabbit polyclonal antibody raised against amino acids 3–183 of CeCENP-A8. A second antibody (SDQ0804) raised against amino acids 40–118 of CeCENP-A confirmed the ChIP-chip pattern observed with antibody OD79 (genome-wide correlation coefficient of 0.92). ChIP for KNL-2 was performed with two polyclonal rabbit antibodies (SDQ08003, SDQ08010) raised against amino acids 207–306. ChIP for RNA Pol II was performed using a monoclonal antibody against the CTD repeat YSPTSPS of RNA Pol II (8WG16; abcam ab817mod). The antibody for H3K36me3 ChIP was described previously13. Other antibodies used for immunoblotting and immunofluorescence were: α-tubulin (DM1α; Sigma-Aldrich), NPP-112 and SQV-8 (gifts from J. Audhya), BUB-1 (ref. 21) and KNL-2 (ref. 22).
Worm strains
For monitoring GFP–CeCENP-A and GFP–CPAR-1 localization in live adult gonads, sperm and embryos (Fig. 1 and Supplementary Figs 1 and 2), and for the photobleaching experiments of GFP–CeCENP-A in Fig. 1, worm strains expressing transgenes from a single-copy locus that includes endogenous 5′ and 3′ regions were generated using the MosSCI technique23, as outlined in Supplementary Fig. 1b. The GFP–CeCENP-A strain OD347 fully rescued the CeCENP-A/hcp-3 deletion allele ok1892, and all experiments involving GFP–CeCENP-A were performed in the ok1892 deletion background. For imaging, the GFP–CeCENP-A (OD347) and GFP–CPAR-1 (OD588) strains were crossed with a strain expressing mCherry–histone H2b (OD56) to generate strains OD421 and OD416, respectively.
For the mating-based analysis of CeCENP-A inheritance through fertilization (Fig. 1c), wild-type (N2) males were crossed to strain BA17, which harbours a temperature-sensitive mutation that abrogates sperm production. Use of the fem-1 strain ensured that the embryos analysed by immunofluorescence were cross-progeny. Injection of double-stranded RNA targeting CeCENP-A and mating with males was performed as previously described24.
For the transcription inhibition assay (Supplementary Fig. 8), GFP–CeCENP-A-expressing hermaphrodites (OD347) were mated with his-72p:GFP–H3.3 and end-3p:mCherry–H1 co-expressing males (RW10007).
For the photobleaching analysis in Supplementary Fig. 4, a strain expressing GFP–CeCENP-A was generated by bombarding plasmid pJM12 into unc-119(de3) worms. In pJM12, a GFP–CeCENP-A transgene is expressed under control of the pie-1 promoter and 3′ untranslated region (UTR). The GFP is inserted at amino acid 174 between the amino-terminal tail and histone core (Supplementary Fig. 4a). Coding sequence and introns preceding amino acid 134 were altered to preserve coding information, but make the transgene-encoded mRNA resistant to RNAi. The two introns in this part of the CeCENP-A locus were replaced with introns from SPhased GFP (Fire lab 2005 vector kit). Ballistic bombardment of pJM12 generated strain OD136. The transgene insertion in OD136 cannot be homozygosed—there is a low amount of embryonic lethality (12–14%) and Unc progeny. This is probably due to the transgene insertion site, as both of these phenotypes segregated with the GFP fluorescence through multiple outcrosses. A dsRNA to the re-encoded region was used to selectively deplete endogenous CeCENP-A and assess functional rescue by the transgene (Supplementary Fig. 4b). OD136 was used to generate strain OD265, where one copy of the endogenous CeCENP-A locus is deleted and GFP–CeCENP-A as well as mCherry–H2b are co-expressed. Photobleaching experiments in Supplementary Fig. 4d, e were performed using OD265. Strain genotypes are listed in Supplementary Table 1.
RNA interference
L4 worms were injected with dsRNA prepared as described previously21 and incubated for 48 h at 20 °C, except for the mating experiment using the fem-1 mutant (see above).
N2 genomic DNA was used as a template to generate PCR products for dsRNA production. The dsRNA for depletion of CeCENP-A in the inheritance experiment of Fig. 1c was described previously8.
Oligonucleotides for production of dsRNA against the re-encoded sequence of CeCENP-A used in the rescue experiments with strain OD136 (Supplementary Fig. 4a, b): oOD1887, 5′-AATTAACCCTCACTAAAGGgccgatgacaccccaattat-3′; oOD1888, 5′-TAATACGACTCACTATAGGccgtgggagtaatcgacaag-3′. Oligonucleotides for dsRNA against GFP: oOD2423, 5′-TAATACGACTCACTATAGGgtcagtggagagggtggaaggtg-3′; oOD2424, 5′-AATTAACCCTCACTAAAGGcatgccatgtgtaatcccagcagc-3′.
For replication inhibition (Supplementary Fig. 3), dsRNAs targeting cdc-6 and cdt-1 were mixed to obtain equal concentrations. Oligonucleotides used for dsRNA against cdc-6: oOD1265, 5′-AATTAACCCTCACTAAAGGCAAATTCCTGCTGCTCCAAT-3′; oOD1266, 5′-TAATACGACTCACTATAGGCGGTCGAACCTCAAGTTCAT-3′. Oligonucleotides used for dsRNA against cdt-1: oOD801, 5′-AATTAACCCTCACTAAAGGCAAAAACAACGAAGCGTGTG-3′; oOD802, 5′-TAATACGACTCACTATAGGCCTCGTTTTTCATTTTATCATTCA-3′.
Immunofluorescence and immunoblotting
Embryos were fixed and processed for immunofluorescence as described previously21,25. Antibodies directly labelled with fluorescent dyes (Cy2, Cy3 or Cy5; Amersham Biosciences) were used at 1 μg ml−1. Images were recorded on a DeltaVision microscope at 1 × 1 binning with a ×100 numerical aperture (NA) 1.3 U-planApo objective (Olympus). Z-stacks (0.2-μm sections) were deconvolved using softWoRx (Applied Precision), and maximum intensity projections were imported into Adobe Photoshop CS4 for further processing.
Immunoblotting was performed using standard methods. For CeCENP-A immunoblots (Figs 1b and 2e), proteins were transferred for 5 h at 30 V in 25 mM Tris-HCl pH 8.3, 192 mM glycine, 20% methanol. These blotting conditions were optimized to result in quantitative transfer of CeCENP-A onto the membrane.
Live imaging and photobleaching
All live imaging was performed at 20 °C. For images of adult hermaphrodite gonads (Fig. 1a and Supplementary Fig. 2a, b) worms were anesthetized with a mixture of 1 mg ml−1 ethyl 3-aminobenzoate methanesulphonate and 0.1 mg ml−1 of tetramisole hydrochloride in M9 for 15–30 min before transferring them to an 2% agarose pad under a coverslip. Images of gonad regions were acquired with a ×40–1.3 NA PlanFluor objective by collecting an 80 × 0.5 μm Z- series of GFP and mCherry images for every Z-plane. The whole gonad views shown in Fig. 1a and Supplementary Fig. 2a were stitched together from three individual, overlapping images. Embryos (Supplementary Fig. 1c) and -1 oocytes/spermatheca (Supplementary Fig. 2b) were imaged using a ×100–1.4 NA PlanApochromat objective. Images were acquired with 1 × 1 binning on a spinning disk confocal setup mounted on a Nikon TE2000-E inverted microscope equipped with a solid-state laser combiner (ALC) (491 nm and 561 nm lines0, a Yokogawa CSU10 head and a CCD camera (iXon DV887; Andor Technology). Acquisition parameters, shutters and focus were controlled by iQ 1.10.0 software (Andor Technology). Images were processed with Fiji 1.0 and Adobe Photoshop CS4.
For the transcription inhibition assay (Supplementary Fig. 8), cross-progeny embryos from GFP–CeCENP-A expressing hermaphrodites mated with his-72p:GFP–H3.3 and end-3p:mCherry–H1 co-expressing males were dissected in L-15 blastomere culture medium26 containing 200 μg ml−1 α-amanitin. GFP and mCherry Z-stacks of permeable and impermeable embryos in the same field of view were acquired at 1–4 min intervals with a ×60–1.4 NA PlanApochromat objective until embryos contained more than 50 cells.
The photobleaching experiments in Fig. 1d, e were performed with the FRAPPA unit (Andor Technology) using a ×60–1.4 NA PlanApochromat objective. Thirteen Z-sections were acquired with a spacing of 1 μm before and after bleaching in the first embryonic division and at 1-min intervals thereafter until anaphase of the second division. Maximum intensity projections were generated for each Z-stack. For each image sequence, an identical sized rectangle (R1) was drawn around each anaphase chromatid set before and after photobleaching in the first division and around anaphase chromatid sets in the second division. A larger rectangle (Rb) was drawn around each rectangle R1 and the area between the two rectangles served as a measure of background intensity. The average intensity (Avg. Int.) of the GFP signal in each R1 was measured using the formula: , and the ratio of average intensities on anaphase chromatid sets before/after bleaching and in the subsequent anaphase were calculated and averaged.
For the photobleaching experiments in Supplementary Fig. 4d, e, the microscope setup differed from the one used for the experiments in Fig. 1 as follows: the microscope was equipped with a krypton – argon 2.5 W water-cooled laser (Spectra-Physics), acquisition parameters, shutters and focus were controlled by MetaMorph software (MDS Analytical Technologies), and the 488 nm laser line for photobleaching was steered into a custom-modified epifluorescence port. GFP–CeCENP-A intensity ratios were calculated as described above, except that anaphase chromatid sets before/after bleaching in the first division were compared with metaphase plates in the second division.
Expression analysis on dissected germlines by quantitative PCR
Worms containing one or two embryos were dissected with 30-gauge needles in Egg buffer (25 mM HEPES-KOH pH 7.6, 118 mM NaCl, 48 mM KCl, 2 mM CaCl2, 2 mM MgCl2,) containing 1 mM levamisole and 0.5% Tween-20. Total RNA from 50–100 gonads was isolated using TRIzol (Invitrogen), and cDNA was synthesized using Superscript III reverse transcriptase (Invitrogen). Quantitative real-time PCR was performed with iQ SYBR Green Supermix (Bio-Rad) in the iQ5 cycler (Bio-Rad) using standard protocols. The average amplification efficiency (E) of primer pairs (Supplementary Table 2) was calculated from two standard curves (tenfold dilution series of cDNA prepared from mixed-stage N2 worms). The relative transcript abundance (RTA) of target genes in met-1 mutant (met-1) versus wild-type (WT) germlines was calculated after normalization to the reference gene act-2 (actin homologue), using the formula: , where Ct denotes the threshold cycle. The assay was performed in duplicate on four biological replicates each for wild-type and met-1 mutant germlines.
Quantification of CeCENP-A molecules in purified nuclei and sperm
6xHis–CeCENP-A was expressed in bacteria, purified by nickel affinity chromatography under denaturing conditions followed by electroelution using the Bio-Rad Electro-Eluter. The concentration of the purified protein was measured relative to BSA on a gel. Purified sperm were a gift of S. Ward. Sperm concentration was measured by microscopy following 4′,6-diamidino-2-phenylindole (DAPI) staining.
For isolation of nuclei, early embryos (<100 cells) were harvested from synchronized adult worms and treated with chitinase as described below for embryo extract preparation. Packed embryos (1 ml) were washed with 2 × 50 ml chilled Egg buffer (25 mM HEPES-KOH pH 7.6, 118 mM NaCl, 48 mM KCl, 2 mM CaCl2, 2 mM MgCl2), hypotonically swollen for 15 min in 10 ml of 0.5 × Nuclei buffer (5 mM Tris-HCl pH 8, 40 mM KCl, 1 mM K-EDTA, 0.375 mM spermidine, 0.15 mM spermine), then washed into 10 ml of 1 × Nuclei buffer (10 mM Tris-HCl pH 8, 80 mM KCl, 2 mM K-EDTA, 0.75 mM spermidine, 0.3 mM spermine) supplemented with 0.1% digitonin (Sigma-Aldrich) and protease inhibitors, and immediately dounced with about 50 strokes in a 15-ml Wheaton Dounce homogenizer using pestle B. Large debris was pelleted at 100g for 3 min and re-dounced once as above. Supernatants containing the nuclei were combined and spun at 2,000g for 15 min. The nuclei pellet was suspended in 1 × Nuclei buffer supplemented with 0.1% digitonin and protease inhibitors and layered onto a 30% (w/v in Nuclei buffer + 0.1% digitonin) sucrose cushion. Nuclei were recovered in the pellet after spinning at 2,000g for 15 min.
Embryo isolation, fixation and extract preparation
N2 adult worms were grown from synchronized L1 larvae in S-basal medium. Batches of 500 ml in 2.8-l Fernbach flasks shaking at 230 r.p.m. were incubated at 17 °C (early embryos) or 19 °C (late embryos) for approximately 65 h. The exact time of harvest was determined by checking embryo production under a microscope (for early embryos, this was five embryos per worm or less). Gravid adults were separated from debris by sucrose floating, and embryos were recovered by dissolving adults with a bleach/NaOH solution. 10 μl and 2 μl of packed embryos were set aside for expression profiling and staging by fluorescence microscopy after DAPI staining, respectively. The remainder was suspended in 2 volumes of Egg buffer (25 mM HEPES-KOH pH 7.6, 118 mM NaCl, 48 mM KCl, 2 mM CaCl2, 2 mM MgCl2) and incubated with 0.15 units ml−1 chitinase (Sigma-Aldrich) until visible disintegration of the eggshell. Embryos were washed with 2 × 50 ml chilled phosphate-buffered saline (PBS) and suspended in 40 ml PBS. Fixation was performed for 10 min on ice after adding 4 ml of cross-linking solution (11% formaldehyde, 50 mM HEPES-KOH pH 8, 0.1 M NaCl, 1 mM Na-EDTA, 0.5 mM Na-EGTA), and excess formaldehyde was quenched with 120 mM glycine. Fixed embryos were washed with 3 × 50 ml PBS and suspended in five pellet volumes of ChIP buffer (50 mM HEPES-KOH pH 7.6, 140 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.5% NP-40, protease inhibitors). Sonication was performed using a Branson sonifier microtip in cycles of 10 s duration (0.9 s on, 0.1 s off) with the power setting at 25% (2 cycles), 30% (2 cycles), 35% (10 cycles), 40% (2 cycles), and 45% (2 cycles), and a pause of 1 min between cycles. Crude extracts were spun for 20 min at 10,000g, the supernatant was removed and glycerol added to 10%. Protein concentration was determined by the Bradford method and aliquots of 3 mg protein were flash-frozen in liquid nitrogen.
Chromatin immunoprecipitation (ChIP)
Extract corresponding to 3 mg protein was diluted to 900 μl with ChIP buffer. After addition of sarcosyl to 1%, Na-deoxycholate to 0.1%, and PMSF to 1 mM, the extract was spun for 10 min at maximum speed in a tabletop centrifuge and the supernatant was used for ChIP. 50 μl was removed for preparation of input DNA. To the rest, 50 μl of Dynabead Sheep anti-Rabbit or anti-Mouse IgG suspension (Dynal Biotech), pre-bound to 5 μg of target antibody, were added, and the mixture was incubated at 4 °C for 4 h or overnight (<16 h). Beads were recovered with a Dynal Magnetic Particle Concentrator (Invitrogen) and washed 2 × 5 min with buffer FA (50 mM HEPES-KOH pH 7.6, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate), 10 min with FA-1000 (50 mM HEPES-KOH pH 7.6, 1 M NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate), 10 min with FA-500 (50 mM HEPES-KOH pH 7.6, 500 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate) in a new tube, 10 min with TEL buffer (10 mM Tris-HCl pH 8.0, 0.25 M LiCl, 1 mM EDTA, 1% NP-40, 1% Na-deoxycholate), and once briefly with TE (10 mM Tris pH 8, 1 mM EDTA). Antibody-bound chromatin was recovered in 50 μl Elution buffer (10 mM Tris-Cl pH 8.0, 1 mM EDTA, 250 mM NaCl, 1% SDS) by shaking at 67 °C for 15 min. Input and ChIP chromatin samples were subsequently processed in parallel. Cross-links were reversed over night at 65 °C in Elution buffer, proteins were digested with 0.45 mg ml−1 proteinase K for 2 h at 37 °C, and nucleic acids were recovered by phenol/chloroform extraction and precipitation with ethanol. RNA was digested with 0.3 mg ml−1 RNase A in TE at 37 °C for 2 h, and DNA purified with a column (PCR Purification Kit, Qiagen).
The ChIP procedure for the met-1 mutant analysis (Fig. 4 and Supplementary Figs 11 and 12) differs slightly from the above and was described in detail previously13.
Ligation-mediated PCR
DNA ends were blunted with 5 units ml−1 T4 DNA polymerase (New England Biolabs) at 12 °C for 20 min and the DNA recovered by phenol/chloroform extraction and ethanol precipitation. Annealed oligomer adaptors (oligo1: 5′-GCGGTGACCCGGGAGATCTGAATTC-3′; oligo2: 5′-GAATTCAGATC-3′) were ligated to blunt DNA ends at 2 μM with 4,000 units ml−1 T4 DNA ligase (New England Biolabs) overnight at 16 °C, and DNA was precipitated with ethanol. DNA fragments were amplified for 22 cycles (55 °C, 2 min; 72 °C, 5 min; 95 °C, 2 min; 95 °C, 1 min; 60 °C, 1 min; 72 °C, 2 min; start cycle again at step 4) using 1 μM oligo 1 and a Taq DNA polymerase (100 units ml−1)/Pfu DNA polymerase (0.5 units ml−1) mix. Amplified DNA was purified with a column (PCR Purification Kit, Qiagen).
Microarray hybridizations, data analysis and display
Amplified ChIP DNA was labelled and hybridized by the Roche Nimblegen Service Laboratory. 2.1M probe tiling arrays, with 50-bp probes, designed against WormBase version WS170 (ce4) were used for all experiments. ChIP samples were labelled with Cy5 and their input reference with Cy3. One ChIP was dye-swapped, which resulted in the same pattern (not shown). For each probe, the intensity from the sample channel was divided by the reference channel and transformed to log2. The enrichment scores for each replicate were calculated by standardizing the log ratios to mean zero and standard deviation one (z-score). Genome-wide scatter plots and Pearson correlations between all ChIP targets and replicates were obtained using all probe z-scores after median smoothing over 1-kb windows.
The average z-score of two replicates was used for all analyses, except in Supplementary Figs 9, 10 and 13c, where individual data sets from extracts with distinct age distributions of embryos are compared. Accession numbers for data sets used in this study are listed in Supplementary Table 3. Scatter plots and boxplots for genes were generated by averaging z-scores of probes located completely within the transcript start site (TSS) and end site (TES). TSS and TES coordinates were obtained from WormBase (WS170).
Gene body profile plots (Supplementary Fig. 7b) were generated by aligning genes of length greater than 2 kb at their TSS and TES. The genomic regions 1.5 kb upstream to 1 kb downstream from TSS and 1 kb upstream to 1 kb downstream from TES were divided into 50-bp bins, and probes were assigned to the nearest bin. Gene group profiles were generated by averaging probe z-scores within each bin across genes in the group.
Definition of CeCENP-A-enriched domains
CeCENP-A signal was averaged over 2-kb windows, every 50 bp. A random distribution for the window averages was obtained by randomly sampling and assigning CeCENP-A values for each chromosome. The resulting random CeCENP-A tracks were also averaged over 2-kb windows. A cutoff was selected so that the number of random window averages above the cutoff was less than 3% of the number of non-randomized windows above the cutoff, effectively providing a 3% false positive rate with respect to the random window averages. Overlapping windows above the cutoff were combined into domains. Domains with gaps smaller than 2 kb were merged, and domains smaller than 2.5 kb were excluded.
Definition of gene classes based on expression profiling data sets
Gene classes were defined on the basis of expression data, as described previously13. In brief, ‘Ubiquitous’ or housekeeping have transcripts present in muscle, gut, neuron and adult germline SAGE (serial analysis of gene expression) data sets27,28; ‘Germline-expressed’ genes have transcripts present in the dissected adult hermaphrodite germ line SAGE data set27; ‘Serpentine’ receptor genes are expressed in mature neurons and silent in embryos29; ‘Spermatogenesis’ genes are classified as expressed during sperm production on the basis of comparative microarray analysis30; ‘Germline-only’ genes are expressed exclusively in the maternal germ line, as their transcripts are enriched in the germline30, maternally loaded into embryos11, and absent from muscle, gut and neuron SAGE data sets27,28; transcripts of ‘Embryo-expressed’ genes are not maternally provided and increase in level during embryogenesis11.
Criteria for identifying genes with maximal changes in RNA Pol II levels
Genes with maximal changes in RNA Pol II levels between the two averaged early embryo (EE) and late embryo (LE) extracts (Supplementary Fig. 10a, b), were identified by applying a moderated t-test31 and requiring a false discovery rate smaller than 5% (ref. 32). In addition, RNA Pol II levels for those genes were required to show at least a twofold change in RNA Pol II ChIP-chip hybridization signal between the averaged EE and LE extracts.
Transcriptional profiling of embryos
RNA was isolated from 10 μl of packed embryos using TRIzol (Invitrogen) and the RNeasy kit (Qiagen). RNA (20 μg) was hybridized to a single-colour 4-plex Nimblegen expression array with 72,000 probes (three 60-mer oligo probes per gene). Quantile normalization33 and the robust multichip average (RMA) algorithm34 were used to normalize and summarize the multiple probe values per gene to obtain one expression value per gene and sample. The expression values per gene were averaged across samples as indicated in the figure legends.
Supplementary Material
Acknowledgments
We thank S. Ward for the purified sperm sample, M. Gupta for help with analysis, J. Ahringer for advice on fixation and ChIP procedures, and A. Dernburg and other members of the Lieb modENCODE group for helpful discussions. This work was supported by a modENCODE grant (U01 HG004270), and by grants from NIH to A.D. (GM074215 and ARRA supplement) and S.S. (GM34059). R.G. was supported by a fellowship from the National Science Foundation of Switzerland. L.G. was supported by NIH T32 GM008646. A.D. and K.O. receive salary and other support from the Ludwig Institute for Cancer Research.
Footnotes
Author Contributions R.G., A.M. and T.E. performed ChIP experiments; A.R. performed analysis of all ChIP-chip datasets with advice from S.S.; K.W.Y. performed the photobleaching and α-amanitin experiments; F.B. and P.M. performed the mating inheritance experiment, analysed replication-independence and measured CeCENP-A levels in sperm; R.G. performed GFP CeCENP-A localization analysis, quantified CeCENP-A levels in nuclei with K.W.Y., and performed qPCR on germline RNA provided by L.G.; A.E., A.M. and J.M. generated GFP CeCENP-A strains; S.E. and J.D.L. helped initiate ChIP analysis of CeCENP-A; K.O. and A.D. made initial observations that established the project; A.D., R.G., A.R., K.W.Y. and S.S. prepared the figures and wrote the paper with advice from J.D.L. and K.O.; A.D. and S.S. supervised the project.
Author Information Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Malik HS, Henikoff S. Major evolutionary transitions in centromere complexity. Cell. 2009;138:1067–1082. doi: 10.1016/j.cell.2009.08.036. [DOI] [PubMed] [Google Scholar]
- 2.Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nature Rev Genet. 2008;9:923–937. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choo K. The Centromere. Ocford Univ. Press; 1997. [Google Scholar]
- 4.Sullivan KF. A solid foundation: functional specialization of centromeric chromatin. Curr Opin Genet Dev. 2001;11:182–188. doi: 10.1016/s0959-437x(00)00177-5. [DOI] [PubMed] [Google Scholar]
- 5.Shelby RD, Monier K, Sullivan KF. Chromatin assembly at kinetochores is uncoupled from DNA replication. J Cell Biol. 2000;151:1113–1118. doi: 10.1083/jcb.151.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jansen LET, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schuh M, Lehner CF, Heidmann S. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr Biol. 2007;17:237–243. doi: 10.1016/j.cub.2006.11.051. [DOI] [PubMed] [Google Scholar]
- 8.Monen J, Maddox PS, Hyndman F, Oegema K, Desai A. Differential role of CENP-A in the segregation of holocentric C. elegans chromosomes during meiosis and mitosis. Nature Cell Biol. 2005;7:1248–1255. doi: 10.1038/ncb1331. [DOI] [PubMed] [Google Scholar]
- 9.Liu T, et al. Broad chromosomal domains of histone modification patterns in C. elegans. Genome Res. 2011;21:227–236. doi: 10.1101/gr.115519.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seydoux G, Dunn MA. Transcriptionally repressed germ cells lack a subpopulation of phosphorylated RNA polymerase II in early embryos of Caenorhabditis elegans and Drosophila melanogaster. Development. 1997;124:2191–2201. doi: 10.1242/dev.124.11.2191. [DOI] [PubMed] [Google Scholar]
- 11.Baugh LR, Hill AA, Slonim DK, Brown EL, Hunter CP. Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development. 2003;130:889–900. doi: 10.1242/dev.00302. [DOI] [PubMed] [Google Scholar]
- 12.Edgar LG, Wolf N, Wood WB. Early transcription in Caenorhabditis elegans embryos. Development. 1994;120:443–451. doi: 10.1242/dev.120.2.443. [DOI] [PubMed] [Google Scholar]
- 13.Rechtsteiner A, et al. The histone H3K36 methyltransferase MES-4 acts epigenetically to transmit the memory of germline gene expression to progeny. PLoS Genet. 2010;6:e1001091. doi: 10.1371/journal.pgen.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andersen EC, Horvitz HR. Two C. elegans histone methyltransferases repress lin-3 EGF transcription to inhibit vulval development. Development. 2007;134:2991–2999. doi: 10.1242/dev.009373. [DOI] [PubMed] [Google Scholar]
- 15.Claycomb JM, et al. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell. 2009;139:123–134. doi: 10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Wolfswinkel JC, et al. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell. 2009;139:135–148. doi: 10.1016/j.cell.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 17.Ravi M, et al. Meiosis-specific loading of the centromere-specific histone CENH3 in Arabidopsis thaliana. PLoS Genet. 2011;7:e1002121. doi: 10.1371/journal.pgen.1002121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lomiento M, Jiang Z, D’Addabbo P, Eichler EE, Rocchi M. Evolutionary-new centromeres preferentially emerge within gene deserts. Genome Biol. 2008;9:R173. doi: 10.1186/gb-2008-9-12-r173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piras FM, et al. Uncoupling of satellite DNA and centromeric function in the genus Equus. PLoS Genet. 2010;6:e1000845. doi: 10.1371/journal.pgen.1000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warburton PE. Chromosomal dynamics of human neocentromere formation. Chromosome Res. 2004;12:617–626. doi: 10.1023/B:CHRO.0000036585.44138.4b. [DOI] [PubMed] [Google Scholar]
- 21.Oegema K, Desai A, Rybina S, Kirkham M, Hyman AA. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J Cell Biol. 2001;153:1209–1226. doi: 10.1083/jcb.153.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maddox PS, Hyndman F, Monen J, Oegema K, Desai A. Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J Cell Biol. 2007;176:757–763. doi: 10.1083/jcb.200701065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frøkjær-Jensen C, et al. Single-copy insertion of transgenes in Caenorhabditis elegans. Nature Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dammermann A, et al. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev Cell. 2004;7:815–829. doi: 10.1016/j.devcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 25.Cheeseman IM, et al. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 2004;18:2255–2268. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edgar LG. Blastomere culture and analysis. Methods Cell Biol. 1995;48:303–321. doi: 10.1016/s0091-679x(08)61393-x. [DOI] [PubMed] [Google Scholar]
- 27.Wang X, et al. Identification of genes expressed in the hermaphrodite germ line of C. elegans using SAGE. BMC Genomics. 2009;10:213. doi: 10.1186/1471-2164-10-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meissner B, et al. An integrated strategy to study muscle development and myofilament structure in Caenorhabditis elegans. PLoS Genet. 2009;5:e1000537. doi: 10.1371/journal.pgen.1000537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolasinska-Zwierz P, et al. Differential chromatin marking of introns and expressed exons by H3K36me3. Nature Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reinke V, Gil IS, Ward S, Kazmer K. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development. 2004;131:311–323. doi: 10.1242/dev.00914. [DOI] [PubMed] [Google Scholar]
- 31.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 32.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 34.Irizarry RA, et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.