Abstract
Background
Epidemiologic studies have reported that frequent consumption of quercetin-rich foods is inversely associated with lung cancer incidence. A quercetin-rich diet might modulate microRNA (miR) expression; however, this mechanism has not been fully examined.
Methods
miR expression data were measured by a custom-made array in formalin-fixed paraffin-embedded tissue samples from 264 lung cancer cases (144 adenocarcinomas and 120 squamous cell carcinomas). Intake of quercetin-rich foods was derived from a food-frequency questionnaire. In individual-miR-based analyses, we compared the expression of miRs (n=198) between lung cancer cases consuming high-versus-low quercetin-rich food intake using multivariate ANOVA tests. In family-miR-based analyses, we used Functional Class Scoring (FCS) to assess differential effect on biologically functional miRs families. We accounted for multiple testing using 10,000 global permutations (significance at p-valueglobal <0.10). All multivariate analyses were conducted separately by histology and by smoking status (former and current smokers).
Results
Family-based analyses showed that a quercetin-rich diet differentiated miR expression profiles of the tumor suppressor let-7 family among adenocarcinomas (p-valueFCS<0.001). Other significantly differentiated miR families included carcinogenesis-related miR-146, miR-26, and miR-17 (p-valuesFCS<0.05). In individual-based analyses, we found that among former and current smokers with adenocarcinoma, 33 miRs were observed to be differentiated between highest-and-lowest quercetin-rich food consumers (23 expected by chance; p-valueglobal = 0.047).
Conclusions
We observed differential expression of key biologically functional miRNAs between high-versus-low consumers of quercetin-rich foods in adenocarcinoma cases.
Impact
Our findings provide preliminary evidence on the mechanism underlying quercetin-related lung carcinogenesis.
Introduction
Quercetin is a polyphenol ubiquitously present in certain fruits (e.g., apples and grapes) and vegetables (e.g., onions, kale, broccoli, lettuce, and tomatoes) and has been found to possess anticarcinogenic properties (1). We (2) and others (3–5) previously observed that a quercetin-rich diet was associated with lower risk of lung cancer in epidemiologic studies. Quercetin and quercetin-rich foods may prevent carcinogenesis via several mechanisms, including free radical scavenging, pro-apoptotic and anti-proliferation pathway mediation, modification of anti-inflammatory responses, and activation of detoxifying Phase II enzymes (6–9).
New findings suggested that polyphenol compounds like quercetin are more likely to interact with cellular signaling cascades that regulate transcription factors (10). More specifically, in vivo and in vitro studies showed that they modulate a wide range of miR expressions and may consequently influence carcinogenesis (11–13). MiRs are short, non-coding, single-stranded RNAs involved in gene expression of multiple target mRNAs (14). Mis-regulated miRs have been implicated in many cancers where they act to promote over-expression of oncogenes and under-expression of tumor suppressor genes (14, 15). For example, the let-7 class of miRs function as tumor suppressors by repressing cell proliferation and regulating both RAS and c-myc oncogenes (16). In lung cancer, we previously showed that the let-7 family is differentially expressed by histology and is associated with survival in the Environment And Genetics in Lung Cancer Etiology (EAGLE) study (17).
Polyphenolic compounds, including quercetin, have been shown in experimental studies to alter expression of several cancer-related miRs, including the cancer-associated let-7 family (18–23). Quercetin metabolites were observed to modulate miR-155 in murine macrophages (18) and miR-146a in colon cancer cells (21). Another polyphenol, Epigallocatechin, has been shown to up-regulate miR-16 in human hepatocellular cells (23). Additionally, differential expression of the let-7 family and other miRs was observed in human hepatocellular cancer cells exposed to Ellagitannin (22).
The emerging evidence from in vitro and in vivo investigations provides biological rationale to examine the influence of quercetin on miR expression in lung carcinogenesis in the present epidemiologic study. As a follow-up study to our observation that a quercetin-rich diet was inversely associated with lung cancer in EAGLE participants (2), we investigated the influence of quercetin-rich food consumption on miR expression signatures in lung tissues of EAGLE lung cancer patients. Given the importance of let-7 in lung carcinogenesis (24, 25) and their association with polyphenols (20), we specifically focused on several members of the let-7 family as a priori candidates for quercetin modification. To our knowledge, this is the first mechanistic investigation of this nature using human tissues in relation to dietary quercetin-rich food consumption.
Materials and Methods
Study population
The present study is based on 144 adenocarcinoma (AD) and 120 squamous cell carcinoma (SQ) subjects from the EAGLE case-control study. The EAGLE study design has been previously described (26). Briefly, EAGLE is a population-based case-control study of lung cancer conducted in the Lombardy region of Italy between 2002 and 2005. Primary, incident, lung cancer cases (N=2100) were recruited from 13 hospitals that examined ~80% of all cases within the catchment area, which included 5 cities (Milan, Monza, Brescia, Pavia, and Varese), surrounding towns, and villages. The majority of cases (95%) were confirmed by pathology reports and the remaining cases by imaging and documentation of clinical history. Histologic type was recorded for all cases.
miR expression data
We previously described the miR expression data from EAGLE (17). Briefly, the miR expression data were derived from formalin-fixed paraffin-embedded (FFPE) tissue samples in 144 lung adenocarcinoma and 120 squamous cell carcinoma cases from EAGLE. The 264 individuals included in the current study were a subgroup with both dietary quercetin information and miR expression data. These individuals did not differ markedly in relevant characteristics (e.g. age, sex, body mass index, smoking, and alcohol consumption, Supplemental Table S1) from EAGLE lung cancer cases that were excluded due to lack of data on quercetin and/or miR expression data.
The miRs were analyzed using a custom-made, two-channel oligo-array using one EBV cell line as the reference sample. The array included a total of 713 human, mammalian, and viral mature antisense miRs plus 2 internal controls with 7 serial dilutions. Intensities for duplicate spots of each miR were averaged. Individual miRs with low overall signal intensity (<100) in both channels and/or low signal/noise ratio were excluded. A spot size smaller than 25 pixels and miRs with >50% missing data were additionally filtered out. Global median normalization was utilized as the most robust method with median-normalization calculated by subtracting out the median log-ratio for each array. Of the 440 human miRs, a total of 198 miRs were retained in the final analysis and are reported in Supplemental Table S2. We validated five miRs (let-7g, let-7f, miR-26a, miR-638, and miR-107) by qRT-PCR using Taqman miR assays (Applied Biosystems) in 49 EAGLE samples normalized to Endogenous Control RNU6B that had sufficient tumor miR expression remaining after array analysis (17).
Epidemiologic and Quercetin-rich food data
At baseline, epidemiologic data were collected using both a computer-assisted personal interview and a self-administered questionnaire to address potential risk factors associated with lung cancer, including comprehensive data on smoking exposure and dietary intake specific to this Italian population (26). Dietary intake in the previous year was obtained from a self-administered 58 item food frequency questionnaire (FFQ) where frequency of consumption was designated using 11 possible response categories that ranged from ‘never’ to ‘2 or more times a day’. Quercetin-rich food items (apples, grapes, onions, artichoke/fennel/celery, beans/chick peas, plum, turnips, peppers, strawberries, tomatoes, and broccoli) in the FFQ were identified based on data published in the United States Department of Agriculture on food-specific quercetin content (>0.50mg/100g) (27). We created a summary measure of quercetin-rich foods by adding the seasonal frequency of intake reported for the individual food item (2).
Statistical analysis
Quercetin-rich food intake was divided into sex-specific tertiles based on the distribution of the controls from the EAGLE study (2). We further defined highest and lowest consumers of quercetin-rich foods to be those in the third and first tertile, respectively. We previously showed that miR levels differed by histology in this population (17) and miR expression might be associated with smoking status (28). To address potential residual confounding by smoking and differential effect by histology, all our analyses were performed for smoking-specific (former and current smokers) and lung cancer subtype (adenocarcinoma and squamous cell carcinoma). Although we examined the influence of quercetin-rich food intake on miR expression in never smokers, the results were unstable and not reported due to too few individuals (n=28).
Individual-miR-based analyses
We first compared the expression levels of 198 miRs between highest (T3) and lowest (T1) quercetin-rich food consumers using multivariate ANOVA tests. Models were adjusted for age (continuous), sex, body mass index (continuous), pack-years of smoking (continuous), consumption of non-quercetin-rich fruits and vegetables(continuous), red/processed meats (continuous), and lifetime alcohol consumption (continuous). In the larger EAGLE study, frequency of quercetin-rich intake was correlated with frequency of non-quercetin rich fruits and vegetables (r=0.64) and consumption of red/processed meat (r=0.07). Selection of other covariates was based on factors that have been associated with either miR expression or lung cancer risk. Individual food items comprised within the individual food groups are described in Supplemental Table S3.
To further address the issue of multiple comparisons we calculated a global p-value (pglobal). For this calculation, we randomly permuted the highest and lowest classes of quercetin intake 10,000× where the number of significant miRs from ANOVA testing was recorded (nP). The global p-value was then defined as one plus the number of times in which np was at least as large as the number of original significant miRs divided by 10,000.
There is a lack of data on quercetin-associated effects on miR expression in human tissues in the current literature, particularly at habitual consumable level. In order to minimize false negative findings at the initial analyses, we considered a permuted pglobal < 0.10 to be statistically significant.
Family-miR-based analyses
We grouped each miR into known biological functional families using MiRBase release 17.0. We assembled miRs into families based on unique ‘seed sequence’ (nucleotides 2–7 at the 5′ end) and identified 21 miR families (Supplemental Table S4). A piori, we restricted our family-miR-based analyses to 9 miR families with at least one miR identified at a p-value < 0.05. We used Functional Class Scoring (FCS) to compare the expression profile of each miR family between high-versus-low consumers of a quercetin-rich diet.
FCS computes p-values by assigning all miRs within a particular group (or family) an aggregate raw score (the arithmetic mean of the negative natural logarithm of each p-value obtained from miR analyses) (29). This raw score was then compared to the score of randomly derived groups of the same size through q repeated samplings (q=1,000). Each score was ordered in ascending order to build an empirically derived score distribution. The FCS p-value was determined as the fraction of randomly sampled groups having a higher score than the group score of interest. For these analyses, we defined statistical significance at a p-valueFCS <0.05. We further evaluated the statistical significance using the more conservative Bonferroni p-value (0.05/9= 0.006).
Permutations were performed in the statistical package R. All other analyses were performed using SAS, version 9.2.
Results
In the present study, lung cancer cases had a mean age at diagnosis of 65 years. Table 1 presents the distribution of selected characteristics by tertiles of quercetin intake, separately for adenocarcinoma and squamous cell carcinoma. Among ever smokers, highest (T3) consumers of quercetin-rich foods smoked less and were more likely to be AD cases compared with low consumers. Former smokers on average consumed more servings of quercetin-rich foods per day than current smokers (1.65±0.88 vs. 1.29±0.79). Compared with AD cases, SQ cases tended to be males, smoked more, and consumed more alcohol and meat across tertiles of quercetin intake. Never smokers (n=29) were all AD except for one individual.
Table 1.
Selected characteristics by sex-specific tertile (T1–T3) of quercetin-rich fooda intake in EAGLE, separately for histologic subtypes
| Subject characteristics | Adenocarcinoma
|
Squamous Cell Carcinoma
|
||||||
|---|---|---|---|---|---|---|---|---|
| T1 (n=57) | T2 (n=47) | T3 (n=40) | p-value | T1 (n=48) | T2 (n=42) | T3 (n=30) | p-value | |
| Quercetin-rich food intake b, median (IQR) | 0.79 (0.42) | 1.56 (0.46) | 2.45 (0.53) | 0.71 (0.37) | 1.47 (0.44) | 2.53 (0.67) | ||
| Age, mean (SD) | 62.58 ± 9.03 | 64.58 ± 8.21 | 65.57 ± 8.55 | 0.22c | 68.48 ± 7.09 | 69.02 ± 6.30 | 66.37± 8.62 | 0.29c |
| Male, n (%) | 38 (66.67) | 25 (53.19) | 20 (50.0) | 0.20d | 46 (95.83) | 42 (100) | 30 (100) | 0.22d |
| BMI, mean (SD) | 24.42 ± 3.76 | 25.59 ± 3.98 | 24.42 ± 3.17 | 0.21c | 25.42 ± 3.10 | 26.73 ± 3.71 | 27.74 ± 3.92 | 0.02c |
| Smoking status (%) | 0.01d | 0.08d | ||||||
| never | 4 (7.02) | 16 (34.04) | 7 (17.50) | 0 | 1 (2.38) | 0 | ||
| former | 22 (38.60) | 12 (25.53) | 22 (55.0) | 18 (37.50) | 20 (47.62) | 20 (66.67) | ||
| current | 31 (54.39) | 19 (40.43) | 11 (27.50) | 30 (62.50) | 21 (50.0) | 10 (33.33) | ||
| Pack-years, median (IQR) | 40.0 (24.0) | 34.5 (32.0) | 33.0 (28.0) | 0.63e | 54.0 (28.25) | 46.25 (21.70) | 42.50 (33.0) | 0.22e |
| Intake | ||||||||
| Vegetablesb,f median (IQR) | 1.01 (0.43) | 2.03 (0.79) | 2.46 (1.93) | <0.01e | 0.95 (0.51) | 1.68 (0.94) | 2.61 (1.83) | <0.01e |
| Fruitsb,g, median (IQR) | 0.96 (0.82) | 1.78 (1.28) | 2.80 (1.40) | <0.01e | 1.03 (0.89) | 1.55 (0.69) | 3.00 (1.37) | <0.01e |
| Meatsb,h, median (IQR) | 0.70 (0.68) | 1.07 (0.79) | 0.94 (0.98) | 0.01e | 1.07 (0.94) | 1.23 (1.27) | 1.33 (0.62) | 0.06e |
| Lifetime alcohol b, median (IQR) | 23.13 (25.17) | 14.79 (34.01) | 7.41 (19.58) | 0.01e | 30.69 (24.69) | 36.23 (20.30) | 31.18 (31.97) | 0.66e |
NOTE: Column percent totals may not sum to 100% due to rounding; Bolded p-values indicated statistical significance; T1–T3 = 1st tertile through 3rd tertile
IQR, interquartile range; SD, Standard Deviation.
Quercetin-rich foods: summary measure of apples, grapes, onions, artichoke/fennel/celery, beans, apricots, plums, turnips, peppers, strawberries, tomatoes, and broccoli.
Frequency (food groups, servings/day; alcohol, grams/day)
ANOVA test.
Chi-square test.
Non-parametric Kruskal-Wallis test.
Total vegetables intake: summary measure of tomatoes, peppers, carrots, salad, peas, beans/chickpeas, mushrooms, broccoli, turnips, savoy, black cabbage, onions, cooked spinach/Swiss
Total fruits intake: summary measure of apples, pears, bananas, kiwis, oranges/grapefruits, mandarins/clementines, grapes, peaches/clingstones, apricots, plums, strawberries, melons, and fruit cocktails.
Total meat intake: summary measure of cooked ham (prosciutto cotto), smoked ham (prosciutto crudo), cured ham (speck), salami, baloney (mortadella), wurstel, salted sliced beef, coppa, pancetta, and other types of processed meats.
Individual- miR-based expression
We identified 16 miRs that were differentially expressed for AD (4 miRs) and SQ (12 miRs) cases (p-values < 0.05) between high-versus-low consumers of quercetin-rich foods (Table 2). Likewise, 19 miRs were differentially expressed for former (12 miRs) and current (7 miRs) smokers (see Supplemental Table, S5).
Table 2.
MiRs that significantly (at P<0.05) differentiate highest (T3) versus lowest (T1) consumers of quercetin-rich food intake, separately by histology
| T1 mean ± SD | T3 mean ± SD | Fold Change* | P-value** | |
|---|---|---|---|---|
|
|
||||
| Adenocarcinoma (n=97) | ||||
| hsa-miR-502 | 0.085 ± 0.353 | 0.202 ± 0.350 | 1.124 | 0.017 |
| hsa-mir-564 | 0.565 ± 0.258 | 0.449 ± 0.273 | 0.890 | 0.030 |
| hsa-miR-124a | 0.232 ± 0.447 | 0.072 ± 0.423 | 0.852 | 0.044 |
| hsa-miR-125a | 0.625 ± 0.723 | 1.034 ± 0.715 | 1.505 | 0.045 |
| Squamous Cell Carcinoma (n=78) | ||||
| hsa-miR-510 | 0.283 ± 0.361 | 0.147 ± 0.295 | 0.872 | 0.003 |
| hsa-mir-605 | 2.118 ± 0.815 | 1.279 ± 0.765 | 0.432 | 0.004 |
| hsa-miR-155 | −5.113 ± 1.071 | −4.777 ± 0.784 | 1.399 | 0.012 |
| hsa-miR-373 | −0.005 ± 0.420 | −0.091 ± 0.330 | 0.917 | 0.014 |
| hsa-miR-453 | 0.597 ± 0.324 | 0.491 ± 0.211 | 0.899 | 0.017 |
| hsa-miR-502 | 0.318 ± 0.309 | 0.095 ± 0.223 | 0.801 | 0.017 |
| hsa-miR-18b | −2.621 ± 0.735 | −2.227 ± 0.724 | 1.483 | 0.020 |
| hsa-miR-183 | 1.160 ± 0.495 | 0.779 ± 0.470 | 0.683 | 0.022 |
| hsa-mir-573 | 0.267 ± 0.355 | 0.126 ± 0.406 | 0.869 | 0.024 |
| hsa-miR-524* | 0.074 ± 0.259 | −0.082 ± 0.341 | 0.855 | 0.036 |
| hsa-mir-612 | −0.171 ± 0.851 | −0.104 ± 0.719 | 1.069 | 0.042 |
| hsa-miR-363* | −0.076 ± 0.778 | 0.124 ± 0.703 | 1.222 | 0.046 |
Fold change is the ratio (T3/T1) of geometric means (>1.0 indicates upregulation and < 1.0 downregulation)
Coefficient P-value from ANOVA model adjusted for age, sex, BMI, smoking status, non-quercetin-rich fruits and vegetables, red/processed meat, alcohol, and cigarette packyears
Table 3 presents analyses examining the influence of quercetin-rich diet on miR expression within histologic subtypes for former and current smokers separately. Considering the four sub-groups, we identified overall 48 unique miRs that were differentially expressed between highest-vs-lowest quercetin-rich consumers (p-value < 0.05, Table 3).
Table 3.
Influence of quercetin-rich food intake (T3-vs-T1) on individual miR, stratified by histology and smoking status
| Adenocarcinoma | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Former smokers (n=44)
|
Current smokers (n=42)
|
||||||||
| miR name | T1 Mean SD | T3 Mean ± SD | Fold Change* | P-value** | miR name | T1 Mean ± SD | T3 Mean ± SD | Fold Change* | P-value** |
|
|
|
||||||||
| hsa-mir-641 | −0.048 ± 0.548 | −0.237 ± 0.879 | 0.828 | 0.003 | hsa-mir-580 | 0.492 ± 0.340 | 0.226 ± 0.286 | 0.767 | 0.003 |
| hsa-miR-29b | −1.110 ± 1.390 | −1.021 ± 1.158 | 1.092 | 0.003 | hsa-miR-215 | −0.665 ± 0.403 | −0.932 ± 0.456 | 0.766 | 0.004 |
| hsa-miR-146a | −4.714 ± 0.805 | −4.830 ± 1.334 | 0.890 | 0.006 | hsa-miR-194 | −0.726 ± 0.662 | −1.159 ± 0.883 | 0.648 | 0.011 |
| hsa-miR-500a | 0.646 ± 0.360 | 0.471 ± 0.539 | 0.839 | 0.008 | hsa-mir-598 | −0.119 ± 0.498 | −0.674 ± 0.538 | 0.574 | 0.016 |
| hsa-let-7e | −1.121 ± 0.808 | −0.882 ± 0.804 | 1.270 | 0.018 | hsa-miR-518a-2* | 0.077 ± 0.343 | 0.004 ± 0.228 | 0.929 | 0.020 |
| hsa-miR-134 | 0.404 ± 0.304 | 0.354 ± 0.322 | 0.952 | 0.020 | hsa-miR-503 | 0.147 ± 0.682 | −0.244 ± 0.451 | 0.677 | 0.037 |
| hsa-miR-26b | −1.624 ± 1.514 | −0.930 ± 1.157 | 2.003 | 0.021 | hsa-miR-146b | −4.682 ± 1.234 | −4.278 ± 1.388 | 1.497 | 0.043 |
| hsa-miR-302c* | 0.107 ± 0.333 | 0.244 ± 0.305 | 1.147 | 0.023 | hsa-miR-381 | 0.044 ± 0.432 | −0.127 ± 0.269 | 0.843 | 0.047 |
| hsa-miR-98 | −1.804 ± 1.199 | −1.798 ± 1.527 | 1.006 | 0.024 | |||||
| hsa-let-7c | −1.634 ± 1.559 | −1.265 ± 1.326 | 1.446 | 0.024 | |||||
| hsa-miR-27a | −1.351 ± 1.218 | −1.097 ± 1.002 | 1.290 | 0.025 | |||||
| hsa-let-7a | −2.044 ± 1.400 | −1.663 ± 1.288 | 1.464 | 0.026 | |||||
| hsa-let-7g | −2.283 ± 1.344 | −2.396 ± 1.469 | 0.893 | 0.026 | |||||
| hsa-let-7i | −2.153 ± 1.492 | −1.810 ± 1.300 | 1.409 | 0.028 | |||||
| hsa-let-7f | −2.512 ± 1.587 | −2.191 ± 1.433 | 1.377 | 0.030 | |||||
| hsa-miR-195 | −2.106 ± 1.343 | −2.141 ± 1.111 | 0.966 | 0.031 | |||||
| hsa-miR-16 | −2.852 ± 1.482 | −2.495 ± 1.081 | 1.429 | 0.032 | |||||
| hsa-miR-146b | −4.292 ± 1.012 | −4.254 ± 1.336 | 1.039 | 0.034 | |||||
| hsa-miR-26a | −0.943 ± 1.723 | −0.364 ± 1.552 | 1.783 | 0.034 | |||||
| hsa-miR-19b | −3.679 ± 1.048 | −3.947 ± 1.108 | 0.764 | 0.036 | |||||
| hsa-mir-564 | 0.556 ± 0.263 | 0.495 ± 0.220 | 0.941 | 0.037 | |||||
| hsa-miR-20a | −4.321 ± 1.456 | −4.262 ± 1.075 | 1.061 | 0.041 | |||||
| hsa-miR-106a | −3.747 ± 1.533 | −3.702 ± 0.988 | 1.047 | 0.044 | |||||
| hsa-miR-34a | −0.896 ± 0.743 | −0.779 ± 0.530 | 1.124 | 0.046 | |||||
| hsa-miR-92a | −3.729 ± 1.292 | −3.614 ± 1.051 | 1.121 | 0.048 | |||||
| Squamous cell carcinoma | |||||||||
| Former smokers (n=38) | Current smokers (n=40) | ||||||||
|
|
|
||||||||
| miR name | T1 Mean ± SD | T3 Mean ± SD | Fold Change* | P-value** | miR name | T1 Mean ± SD | T3 Mean ± SD | Fold Change* | P-value** |
|
|
|
||||||||
| hsa-miR-492 | −1.383 ± 0.540 | −1.262 ± 0.394 | 1.129 | 0.012 | hsa-miR-502 | 0.354 ± 0.222 | 0.108 ± 0.220 | 0.782 | 0.010 |
| hsa-miR-510 | 0.408 ± 0.374 | 0.166 ± 0.307 | 0.785 | 0.021 | hsa-mir-605 | 2.198 ± 0.861 | 1.105 ± 0.701 | 0.335 | 0.013 |
| hsa-miR-491 | −0.274 ± 0.798 | −0.121 ± 0.638 | 1.165 | 0.023 | hsa-miR-506 | 0.245 ± 0.222 | −0.195 ± 0.615 | 0.644 | 0.017 |
| hsa-mir-612 | −0.264 ± 1.072 | −0.109 ± 0.789 | 1.168 | 0.025 | hsa-miR-183 | 1.282 ± 0.518 | 0.357 ± 0.390 | 0.397 | 0.028 |
| hsa-miR-500a | 0.387 ± 0.278 | 0.291 ± 0.385 | 0.908 | 0.028 | hsa-miR-524* | 0.094 ± 0.294 | −0.170 ± 0.151 | 0.768 | 0.029 |
| hsa-mir-663 | −0.169 ± 0.551 | −0.117 ± 0.495 | 1.054 | 0.034 | |||||
| hsa-miR-503 | −0.069 ± 0.710 | −0.149 ± 0.417 | 0.923 | 0.034 | |||||
| hsa-miR-453 | 0.584 ± 0.324 | 0.482 ± 0.226 | 0.903 | 0.035 | |||||
| hsa-mir-654 | −1.239 ± 0.422 | −1.111 ± 0.366 | 1.137 | 0.041 | |||||
| hsa-mir-658 | 0.211 ± 0.793 | 0.123 ± 0.372 | 0.916 | 0.047 | |||||
Note: miRs are ordered by P-value within strata
Fold change is the ratio (T3/T1) of geometric means (>1.0 indicates upregulation and < 1.0 downregulation)
Coefficient P-value from ANOVA model adjusted for age, sex, BMI, non-quercetin-rich fruits and vegetables, red/processed meat, alchohol, and cigarette packyears.
Among former and current smokers with AD, 33 miRs were observed to be differentiated between highest and lowest quercetin-rich consumers (23 expected by chance; p-valueglobal = 0.047, Table 3). For SQ cases, we identified 15 miRs (p-valueglobal = 0.15, Table 3).
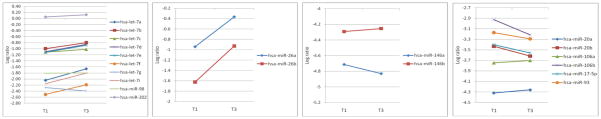
Quercetin-mediated miR expression profiles appeared to be more prevalent among former smokers with AD compared with current smokers. In this group, we identified 25 miRs with a p-value < 0.05 (22 expected by chance; p-valueglobal = 0.076; see Supplemental Figure 1 for heat map). The largest fold-change was observed for miR-26b, a proapoptotic miR (fold-change = 2.00; p-value = 0.020). Notably, among the identified significant miRs, all of the let-7 family members (let-7a, let-7b, let-7c, let-7d, let-7e, let-7f, let-7g, let-7i, and miR-98) were associated with a quercetin-rich diet. Moreover, the majority of let-7 family members were up-regulated with increasing frequency of quercetin intake (Figure 1).
Figure 1.
Mean expression levels for significant miR groups comparing the highest vs. lowest tertile of quercetin-rich food intake in former smokers with adenocarcinoma
In comparison to AD cases, far fewer miRs were identified at a p-value <0.05 among SQ cases. None of the SQ subgroups was statistically significant (p-valuesglobal > 0.10).
Family- miR-based expression
Table 4 presents the results examining the association of quercetin-rich foods with families of miRs in smoking-specific analyses for AD and SQ cases. A quercetin-rich diet appears to significantly differentiate miR expressions in former smokers with AD. Among this group, our data showed that the let-7 family was strongly differentiated by a quercetin-rich diet (p-valueFCS < 0.001, Table 4) followed by miR-146 (p-valueFCS = 0.002), miR-26 (p-valueFCS = 0.010), and miR-17 (p-valueFCS = 0.031). Both the let 7 and the miR-146 families remained significant after Bonferroni correction for multiple comparisons (pbonferonni=0.05/9 = 0.006). We observed no significant difference in miR expression for SQ cases and current smokers regardless of histology.
Table 4.
Influence of quercetin-rich food intake (T3-vs-T1) on family of functionala miR, stratified by histology and smoking status
| Family | miRNA members | Adenocarcinoma
|
Squamous Cell Carcinomas
|
||
|---|---|---|---|---|---|
| Function | Former | Current | Former | Current | |
|
|
|
||||
| P-value* | P-value* | P-value* | P-value* | ||
| Let-7 family | hsa-miR-let-7a | P <0.001 | P =0.426 | P =0.366 | P =0.988 |
| Tumor suppressor | hsa-miR-let-7b | ||||
| hsa-miR-let-7c | |||||
| hsa-miR-let-7d | |||||
| hsa-miR-let-7e | |||||
| hsa-miR-let-7f | |||||
| hsa-miR-let-7g | |||||
| hsa-miR-let-7i | |||||
| hsa-miR-98 | |||||
| hsa-miR-202 | |||||
| miR-146 family | hsa-miR-146a | P =0.002 | P =0.092 | P =0.753 | P =0.222 |
| Tumor growth and invasion | hsa-miR-146b | ||||
| miR-26 family | hsa-miR-26a | P =0.010 | P =0.623 | P =0.588 | P =0.664 |
| Apoptosis | hsa-miR-26b | ||||
| miR-17 family | hsa-miR-20a | P =0.031 | P =0.943 | P =0.766 | P =0.283 |
| Tumor progression | hsa-miR-20b | ||||
| hsa-miR-106a | |||||
| hsa-miR-106b | |||||
| hsa-miR-17-5p | |||||
| hsa-miR-93 | |||||
| miR-29 family | hsa-miR-29a | P =0.064 | P =0.373 | P =0.886 | P =0.137 |
| DNA methylation | hsa-miR-29b | ||||
| hsa-miR-29c | |||||
| miR-18 family | hsa-miR-18a | P =0.705 | P =0.220 | P =0.156 | P =0.392 |
| Tumor progression | hsa-miR-18b | ||||
| miR-34 family | hsa-miR-34a | P =0.142 | P =0.649 | P =0.275 | P =0.568 |
| Tumor suppressor | hsa-miR-34c | ||||
| miR-19 family | hsa-miR-19a | P =0.072 | P =0.991 | P =0.608 | P =0.103 |
| Tumor progression | hsa-miR-19b | ||||
| miR-15/16 family | hsa-miR-503 | P =0.286 | P =0.073 | P =0.307 | P =0.763 |
| Apoptosis | hsa-miR-15a | ||||
| hsa-miR-16 | |||||
| hsa-miR-195 | |||||
| hsa-miR-424 | |||||
NOTE: Only results of miR families that had at least one miR that were significant at p-value <0.05 from individual-based miR analyses (Table 2); Bolded p-values indicated results that remained significant after Bonferroni correction for multiple comparisons
Models adjusted for age, sex, BMI, non-quercetin-rich fruits and vegetables, red/processed meat, alcohol, and cigarette packyears.
Refer to Supplemental Table S5 for more detailed functions
P-value based on Functional Class Score as described in the Methods section
Figure 1 graphically depicts the directionality of quercetin-associated miR expression of let-7, miR-146, miR-26, and miR-17 families with a quercetin-rich diet for formers smokers with AD. In general, members of the let-7 family, miR-26 as well as miR-146b were up-regulated. In contrast, the expression of miR-146a was downregulated.
Discussion
We previously observed that higher consumption of quercetin-rich foods was associated with lower risk of lung cancer in a large population-based case-control study in Italy (2). The present study tests the hypothesis that a quercetin-rich diet modulates the expression of miRs in human lung tissues. In individual-miR-based analyses, we identified significant quercetin-mediated miR expression signatures for 48 unique miRs. These identified miRs have been shown to decrease tumor metastasis and invasion (miR-146a/b, -503, and -194), decrease cell proliferation (miR-125a, -155, let-7 family, -302c, -195, -26a, -503, and -215), increase apoptosis (miR-125a, -605, -26b, let-7g, -34a, -491, and -16), and target tumor suppressors (let-7 family, miR-125a, -183, -146a, -98, -19b, -106a, and -381). In family-miR-based analyses, we found that the large majority of members of the let 7 family was strongly up-regulated among former smokers with AD who consumed a higher intake of quercetin compared with low consumers (p-valueFCS < 0.001). We also observed similar family-based results for miR-146, miR-26, and miR-17 families (p-valueFCS < 0.05) in this group.
Due to its association with lung cancer (25) and possibly with polyphenols (20), we specifically focused on the let-7 class of miRs in relation to a quercetin-rich diet. In addition to being the most statistically significant result based on FCS in the family-based analyses, the let-7 family remained significant after Bonferroni correction at p-value < 0.006. Members of the let-7 family are known to function as tumor suppressors in lung carcinoma by repressing NSCLC cell proliferation (16, 30) and by negatively regulating the RAS oncogene (31). Among the let-7 miRs in the present study, let-7a, a known suppressor of k-RAS and c-Myc oncogenes (32), exhibited the largest fold change (fold-change = 1.46). Our data suggest a possible mechanism of quercetin-related tumor protection through the increased expression of these key tumor suppressors.
We also observed differential quercetin-mediated expression of the miR-17 family (miR-20a, -20b, -106a, -106b, -17, and -93) in former smokers with AD. MiR-17 family belongs to the oncogenic miR-17-92 cluster (33). Investigators have shown that the miR-17-92 cluster is frequently overexpressed in lung cancer (25, 34). Expression of miR-17 is associated with poorer prognosis and cellular proliferation (35) while miR-106b targets p21 and subsequently promotes cell cycle progression (36). Additionally, suppression of miR-20a induces apoptosis in lung cancer (37). In our study, the majority of the miRs (67%) in miR-17 family were downregulated in frequent consumers of quercetin-rich food among former smokers with AD.
Quercetin-rich food consumption also significantly differentiated miR-146 and miR-26 families in our study. Neither of these miR families has been extensively studied with respect to lung carcinogenesis; however, both families have been associated with tumor development. In lung alveolar epithelial cells, miR-146 was observed to negatively regulate pro-inflammatory chemokines (38). Additionally, miR-146a is one of two known miRs (miR-146a and -155) involved in inflammatory signaling pathways and has been observed to be up-regulated by quercetin in experimental study (21). We corroborated this up-regulation of miR-146a among higher consumers of quercetin in the present study. One study showed that miR-155, was downregulated with quercetin in murine cells (18). In lung carcinoma, miR-155 has often been seen to be up-regulated and to have prognostic impact (39). However, it has also been suggested to function as a tumor suppressor by repressing cell proliferation (40). In the present study, however, miR-155 was not significant in the individual-miR-based analyses by histology and smoking status.
Both miR-26a and miR-26b exhibited the greatest fold change in our individual miR-based analyses (miR-26a, FC=1.78) and (miR-26b, FC=2.00). Our data suggest that a quercetin-rich diet increases the expression of the miR-26 family, which has been shown to suppress cell proliferation in nasopharyngeal carcinoma through G1 phase arrest and repression of c-Myc (41) as well as to induce apoptosis in breast cancer cells (42). Proapoptotic characteristics of miR-26 make this particular group of miRs an important candidate for study in future research investigating quercetin-mediated miR-targets.
We previously identified a miR expression profile that strongly differentiated adenocarcinoma from squamous cell carcinoma with prognostic implications in EAGLE (17). Results from this present study showed consumption of quercetin-rich foods is associated with differential miR expression by histology. In general, our data suggest that quercetin-rich foods influenced miR expression in tumor tissues of former smokers with AD, but not for SQ and current smokers. The modest fold-change effect of dietary quercetin on miR expression might only be detectable in a milieu that is less saturated by smoking exposure, as in former smokers and in AD which is less associated with smoking than SQ (43). Furthermore, AD cases included in the present study on average smoked less intensely than SQ cases. The anti-carcinogenic capabilities associated with dietary quercetin may be weakened in SQ cases by competing tobacco-related carcinogens.
To our knowledge, the present study is the only investigation examining the association between dietary quercetin, at a habitual consumable level, and miR expression in lung tissues. In addition to having both dietary information and miR expression data, this study included several variables that allowed tight control for potential confounders. This richness of epidemiologic data coupled with epigenetic data from human tissues permitted an integrative approach—making it possible to explore underlying mechanisms that may explain the protective effect of quercetin and lung cancer risk seen in observational studies.
Despite its uniqueness, the study had a limited sample size, which reduces statistical precision, and explored a limited number of human miRs. Although the EAGLE questionnaire assessed food consumption a year prior to the study, we cannot exclude that the lung cancer diagnosis had influenced the patients’ responses. However, this potential recall bias should not have differentially affected the adenocarcinoma or squamous cell carcinoma cases, and could not explain the different association with quercetin by histology type. Secondly, the northern Italian population comprised of the EAGLE study consumed very high intake of meat products (44) and in contrast substantially lower intake of fruits and vegetables; therefore the FFQ captured a limited variety of food sources of quercetin as reflective of the consumption in this population. While our exposure measure of quercetin-rich diet is likely subject to measurement errors, the errors however would be nondifferential errors and would not alter our findings and conclusion. Moreover, we did not measured quercetin content directly; thus, we cannot rule out the contribution of other flavonoids or nutrients that are found in those foods.
In conclusion, we observed that a quercetin-rich diet is associated with differential expression of key miRs in lung tissue within smoking specific histology groups. Notably, expression of miRs in the let-7 family, a known tumor suppressor, was strongly associated with frequent quercetin-rich food intake in the present study. Our findings provide suggestive insights into a possible mechanism to explain the inverse association between quercetin-rich food consumption and lung cancer risk observed in epidemiological studies. Confirmation in a larger prospective study with both dietary quercetin information, miR expression from lung tissue, and clinical data is warranted.
Supplementary Material
Acknowledgments
We wish to express our gratitude to the participants of the Environment and Genetics in Lung Cancer Etiology study and the investigators who made the study possible (listed on the EAGLE website http://eagle.cancer.gov).
Footnotes
Disclosure of Potential Conflicts of Interests: None
Authors’ Contributions:
Conception and design: TKL, MTL
Development of methodology: TKL, SS, MTL
Acquisition of data: TKL, MTL, AP, FM, PAB
Analysis and interpretation of data: TKL, SS, MTL, YZ
Writing, review, and/or revision of the manuscript: TKL, SS, YZ, AP, PAB, NEC, MTL
Administrative, technical, or material support: FM, EW, YZ
Study supervision: TL, MTL
References
- 1.Murakami A, Ashida H, Terao J. Multitargeted cancer prevention by quercetin. Cancer Lett. 2008;269(2):315–25. doi: 10.1016/j.canlet.2008.03.046. [DOI] [PubMed] [Google Scholar]
- 2.Lam TK, Rotunno M, Lubin JH, Wacholder S, Consonni D, Pesatori AC, et al. Dietary quercetin, quercetin-gene interaction, metabolic gene expression in lung tissue and lung cancer risk. Carcinogenesis. 2010;31(4):634–42. doi: 10.1093/carcin/bgp334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cui Y, Morgenstern H, Greenland S, Tashkin DP, Mao JT, Cai L, et al. Dietary flavonoid intake and lung cancer-A population-based case-control study. Cancer. 2008;112(10):2241–8. doi: 10.1002/cncr.23398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirvonen T, Virtamo J, Korhonen P, Albanes D, Pietinen P. Flavonol and flavone intake and the risk of cancer in male smokers (Finland) Cancer Causes Control. 2001;12(9):789–96. doi: 10.1023/a:1012232008016. [DOI] [PubMed] [Google Scholar]
- 5.Knekt P, Kumpulainen J, Jarvinen R, Rissanen H, Heliovaara M, Reunanen A, et al. Flavonoid intake and risk of chronic diseases. Am J Clin Nutr. 2002;76(3):560–8. doi: 10.1093/ajcn/76.3.560. [DOI] [PubMed] [Google Scholar]
- 6.Murakami A, Ashida H, Terao J. Multitargeted cancer prevention by quercetin. Cancer Lett. 2008;269(2):315–25. doi: 10.1016/j.canlet.2008.03.046. [DOI] [PubMed] [Google Scholar]
- 7.Lotito SB, Zhang WJ, Yang CS, Crozier A, Frei B. Metabolic conversion of dietary flavonoids alters their anti-inflammatory and antioxidant properties. Free Radic Biol Med. 2011;51(2):454–63. doi: 10.1016/j.freeradbiomed.2011.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeh SL, Yeh CL, Chan ST, Chuang CH. Plasma rich in quercetin metabolites induces G2/M arrest by upregulating PPAR-γ expression in human A549 lung cancer cells. Planta Med. 2011;77(10):992–8. doi: 10.1055/s-0030-1250735. [DOI] [PubMed] [Google Scholar]
- 9.Khanduja KL, Gandhi RK, Pathania V, Syal N. Prevention of N-nitrosodiethylamine-induced lung tumorigenesis by ellagic acid and quercetin in mice. Food Chem Toxicol. 1999;37(4):313–8. doi: 10.1016/s0278-6915(99)00021-6. [DOI] [PubMed] [Google Scholar]
- 10.Spencer JP. Beyond antioxidants: the cellular and molecular interactions of flavonoids and how these underpin their actions on the brain. Proc Nutr Soc. 2010;69(2):244–60. doi: 10.1017/S0029665110000054. [DOI] [PubMed] [Google Scholar]
- 11.Parasramka MA, Ho E, Williams DE, Dashwood RH. MicroRNAs, diet, and cancer: new mechanistic insights on the epigenetic actions of phytochemicals. Mol Carcinog. 2012;51(3):213–30. doi: 10.1002/mc.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J, Xu X. Diet, epigenetic, and cancer prevention. Adv Genet. 2010;71:237–55. doi: 10.1016/B978-0-12-380864-6.00008-0. [DOI] [PubMed] [Google Scholar]
- 13.Milenkovic D, Deval C, Gouranton E, Landrier JF, Scalbert A, Morand C, et al. Modulation of miRNA Expression by Dietary Polyphenols in apoE Deficient Mice: A New Mechanism of the Action of Polyphenols. PLoS ONE. 2012;7(1):e29837. doi: 10.1371/journal.pone.0029837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lovat F, Valeri N, Croce CM. MicroRNAs in the pathogenesis of cancer. Semin Oncol. 2011;38(6):724–33. doi: 10.1053/j.seminoncol.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 15.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105(10):3903–8. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landi MT, Zhao Y, Rotunno M, Koshiol J, Liu H, Bergen AW, et al. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res. 2010;16(2):430–41. doi: 10.1158/1078-0432.CCR-09-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boesch-Saadatmandi C, Loboda A, Wagner AE, Stachurska A, Jozkowicz A, Dulak J, et al. Effect of quercetin and its metabolites isorhamnetin and quercetin-3-glucuronide on inflammatory gene expression: role of miR-155. J Nutr Biochem. 2011;22(3):293–9. doi: 10.1016/j.jnutbio.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 19.Arola-Arnal A, Blade C. Proanthocyanidins modulate microRNA expression in human HepG2 cells. PLoS ONE. 2011;6(10):e25982. doi: 10.1371/journal.pone.0025982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, VandenBoom TG, 2nd, Kong D, Wang Z, Ali S, Philip PA, et al. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 2009;69(16):6704–12. doi: 10.1158/0008-5472.CAN-09-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noratto GD, Kim Y, Talcott ST, Mertens-Talcott SU. Flavonol-rich fractions of yaupon holly leaves (Ilex vomitoria, Aquifoliaceae) induce microRNA-146a and have anti-inflammatory and chemopreventive effects in intestinal myofibroblast CCD-18Co cells. Fitoterapia. 2011;82(4):557–69. doi: 10.1016/j.fitote.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 22.Wen XY, Wu SY, Li ZQ, Liu ZQ, Zhang JJ, Wang GF, et al. Ellagitannin (BJA3121), an anti-proliferative natural polyphenol compound, can regulate the expression of MiRNAs in HepG2 cancer cells. Phytother Res. 2009;23(6):778–84. doi: 10.1002/ptr.2616. [DOI] [PubMed] [Google Scholar]
- 23.Tsang WP, Kwok TT. Epigallocatechin gallate up-regulation of miR-16 and induction of apoptosis in human cancer cells. J Nutr Biochem. 2010;21(2):140–6. doi: 10.1016/j.jnutbio.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 24.Ortholan C, Puissegur MP, Ilie M, Barbry P, Mari B, Hofman P. MicroRNAs and lung cancer: new oncogenes and tumor suppressors, new prognostic factors and potential therapeutic targets. Curr Med Chem. 2009;16(9):1047–61. doi: 10.2174/092986709787581833. [DOI] [PubMed] [Google Scholar]
- 25.Osada H, Takahashi T. let-7 and miR-17-92: small-sized major players in lung cancer development. Cancer Sci. 2011;102(1):9–17. doi: 10.1111/j.1349-7006.2010.01707.x. [DOI] [PubMed] [Google Scholar]
- 26.Landi MT, Consonni D, Rotunno M, Bergen AW, Goldstein AM, Lubin JH, et al. Environment And Genetics in Lung cancer Etiology (EAGLE) study: an integrative population-based case-control study of lung cancer. BMC Public Health. 2008;8:203. doi: 10.1186/1471-2458-8-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laboratory ND United States Department of Agriculture ARS, editor. Database for the Flavonoid Content of Selected Foods. Beltsville Human Nutrition Research Center, Nutrient Data Laboratory; Beltsville, MD: 2003. [Google Scholar]
- 28.Izzotti A, Calin GA, Arrigo P, Steele VE, Croce CM, De Flora S. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J. 2009;23(3):806–12. doi: 10.1096/fj.08-121384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pavlidis P, Lewis DP, Noble WS. Exploring gene expression data with class scores. Pac Symp Biocomput. 2002:474–85. [PubMed] [Google Scholar]
- 30.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18(10):505–16. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 31.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 32.He XY, Chen JX, Zhang Z, Li CL, Peng QL, Peng HM. The let-7a microRNA protects from growth of lung carcinoma by suppression of k-Ras and c-Myc in nude mice. J Cancer Res Clin Oncol. 2010;136(7):1023–8. doi: 10.1007/s00432-009-0747-5. [DOI] [PubMed] [Google Scholar]
- 33.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133(2):217–22. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Osada H, Takahashi T. let-7 and miR-17-92: small-sized major players in lung cancer development. Cancer Sci. 2011;102(1):9–17. doi: 10.1111/j.1349-7006.2010.01707.x. [DOI] [PubMed] [Google Scholar]
- 35.Yu J, Ohuchida K, Mizumoto K, Fujita H, Nakata K, Tanaka M. MicroRNA miR-17-5p is overexpressed in pancreatic cancer, associated with a poor prognosis, and involved in cancer cell proliferation and invasion. Cancer Biol Ther. 2010;10(8):748–57. doi: 10.4161/cbt.10.8.13083. [DOI] [PubMed] [Google Scholar]
- 36.Ivanovska I, Ball AS, Diaz RL, Magnus JF, Kibukawa M, Schelter JM, et al. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol Cell Biol. 2008;28(7):2167–74. doi: 10.1128/MCB.01977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsubara H, Takeuchi T, Nishikawa E, Yanagisawa K, Hayashita Y, Ebi H, et al. Apoptosis induction by antisense oligonucleotides against miR-17-5p and miR-20a in lung cancers overexpressing miR-17-92. Oncogene. 2007;26(41):6099–105. doi: 10.1038/sj.onc.1210425. [DOI] [PubMed] [Google Scholar]
- 38.Perry MM, Moschos SA, Williams AE, Shepherd NJ, Larner-Svensson HM, Lindsay MA. Rapid changes in microRNA-146a expression negatively regulate the IL-1beta-induced inflammatory response in human lung alveolar epithelial cells. J Immunol. 2008;180(8):5689–98. doi: 10.4049/jimmunol.180.8.5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raponi M, Dossey L, Jatkoe T, Wu X, Chen G, Fan H, et al. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 2009;69(14):5776–83. doi: 10.1158/0008-5472.CAN-09-0587. [DOI] [PubMed] [Google Scholar]
- 40.Dorsett Y, McBride KM, Jankovic M, Gazumyan A, Thai TH, Robbiani DF, et al. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28(5):630–8. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu J, He ML, Wang L, Chen Y, Liu X, Dong Q, et al. MiR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer Res. 2011;71(1):225–33. doi: 10.1158/0008-5472.CAN-10-1850. [DOI] [PubMed] [Google Scholar]
- 42.Liu XX, Li XJ, Zhang B, Liang YJ, Zhou CX, Cao DX, et al. MicroRNA-26b is underexpressed in human breast cancer and induces cell apoptosis by targeting SLC7A11. FEBS Lett. 2011;585(9):1363–7. doi: 10.1016/j.febslet.2011.04.018. [DOI] [PubMed] [Google Scholar]
- 43.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359(13):1367–80. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lam TK, Cross AJ, Consonni D, Randi G, Bagnardi V, Bertazzi PA, et al. Intakes of red meat, processed meat, and meat mutagens increase lung cancer risk. Cancer Res. 2009;69(3):932–9. doi: 10.1158/0008-5472.CAN-08-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.